Abstract
Background and purpose:
Certain nuclear receptors (NRs) such as the constitutive androstane receptor (CAR), pregnane X receptor (PXR) and farnesoid X receptor (FXR) mediate induction of some cytochrome P450 enzymes and ABC transporters but conflicting reports exist. The purpose of this study was to assess the reasons for these discrepancies and use a standardized approach to compare activators of NRs.
Experimental approach:
Dexamethasone, pregnenolone 16α-carbonitrile, rifampicin, phenobarbital and chenodeoxycholic acid were incubated with HepG2, Caco-2 and cryopreserved human hepatocytes prior to analysis of mRNA and protein for CYP2B6, CYP3A4, CYP3A5, ABCB1, ABCC1, ABCC2, PXR, CAR and FXR.
Key results:
Dexamethasone significantly up-regulated PXR, CYP3A4 and ABCB1 expression in HepG2 and Caco-2 cells. As a result, including dexamethasone as a media supplement masked the induction of these genes by pregnenolone 16α-carbonitrile, which may explain discrepancies between previous reports. In the absence of dexamethasone, significant activator-dependent induction was observed in all cell types. Significant correlations were observed between the fold increase in mRNA and in protein, which were, for most instances, logarithmic. Changes in mRNA expression were greater in cell lines than primary cells but for most transcripts correlations were observed between fold increases in HepG2 and hepatocytes.
Conclusions and implications:
Clearly, no in vitro system can imitate the physiology of a hepatocyte or intestinal cell within an intact organ in vivo, but these data explain some of the discrepancies reported between laboratories and have important implications for study design.
Keywords: PXR, CAR, FXR, P-glycoprotein, MDR1, cytochrome P450
Introduction
Cytochrome P450 (CYP) enzymes and ATP-binding cassette (ABC) transporters are predominantly expressed within the liver and intestine, and substantial substrate overlap is evident. Notably, overlap exists between CYP3A4 and ABCB1 and a functional interplay has been postulated, whereby the rate of CYP3A4-mediated metabolism is influenced by ABCB1 (Benet et al., 2004). It is also evident that the mechanisms that control gene expression are similar for some members of these two families of disposition genes.
Nuclear receptors (NRs), including the constitutive androstane receptor (CAR; NR1I3), pregnane X receptor (PXR; NR1I2) and farnesoid X receptor (FXR; NR1H4), regulate hepatic and intestinal enzymes and transporters in response to exogenous and endogenous activators. The best-studied orphan NR is PXR, which mediates induction of a wide variety of genes, including CYP2B6, CYP3A4, ABCB1 and ABCC2 (Bertilsson et al., 1998; Synold et al., 2001; Kast et al., 2002; Wang et al., 2003). Previous studies have illustrated species differences in activation with pregnenolone 16α-carbonitrile (PCN)-activating rodent but not human PXR and rifampicin (RIF)-activating human but not rodent PXR in CV-1 cells co-transfected with expression plasmids for PXR from different species and the (CYP3A1 DR3)2-tk-CAT reporter (Jones et al., 2000). However, other studies assessing genomic activation report that RIF increases CYP3A in rat hepatocytes and PCN increases CYP3A in human hepatocytes (Ogg et al., 1999; Swales et al., 2003). Data have also indicated that the human PXR/human glucocorticoid receptor augments activation of CYP3A4 reporters in HepG2 cells (El-Sankary et al., 2001). Clearly, controversy remains regarding species differences that may stem, at least in part, from differences in methodology.
Ligands such as methoxychlor and artemisinin have been shown to activate both PXR and CAR (Blizard et al., 2001; Burk et al., 2005b), indicating that some ligand overlap exists. CAR binds to response elements similar to those binding PXR, both mediating induction of CYP3A (Xie et al., 2000), CYP2B6 (Goodwin et al., 2001) and ABCB1 (Burk et al., 2005a) via the same everted repeat-6, direct repeat (DR)-4 and phenobarbital (PB)-responsive enhancer module, and an everted repeat-8 mediates induction of ABCC2 by PXR, CAR and FXR (Kast et al., 2002).
Due to its emerging role in the control of cholesterol, lipid and glucose metabolism, FXR has been proposed as a target for cardiovascular (Bishop-Bailey, 2004) and cholestatic liver disease (Willson et al., 2001). In addition to its role in the induction of ABCC2 by chenodeoxycholic acid (CDCA), CYP3A4 was recently shown to be upregulated by CDCA-activated-FXR binding to an everted repeat-8, inverted repeat-1 and DR-3 (Gnerre et al., 2004). FXR binds predominantly to inverted repeat-1 elements but has also been shown to downregulate apolipoprotein AI as a monomer, by binding to a single recognition motif (Claudel et al., 2002). Interestingly, electrophoretic mobility shift assays and reporter assays have illustrated that FXR is also able to bind and activate transcription via DR-4 and DR-5 motifs (Laffitte et al., 2000).
In vitro studies employ either transformed cell lines or primary cell cultures. In contrast to primary cells, transformed cells are readily available in substantial quantities, and data generated are more reproducible within an individual laboratory. However, key phenotypic differences have been reported between labs (Sambuy et al., 2005), and different culture media are often employed.
Molecular techniques such as mRNA and protein analyses, electrophoretic mobility shift assay and reporter assays are all used to study the mechanisms that underpin the regulation of gene expression by NRs (Faucette et al., 2006; Owen et al., 2006; Ripp et al., 2006). However, it must be noted that some of these methodologies have limitations. Electrophoretic mobility shift assays utilize synthetic oligonucleotides and artificial binding buffers and are often coupled with high concentrations of recombinant proteins. Electrophoretic mobility shift assays and response element-based reporter assays often focus on only part of the regulatory region and therefore regulation effected by distal sequences may be missed. Moreover, different investigators have utilized different reporter constructs (sometimes from different genes) to study PXR activation (Lamba et al., 2005; Faucette et al., 2006). This has resulted in an additional tier of complexity when interpreting data.
Clearly, there are disparate reports in the literature regarding the impact of NR activators on target genes. Such apparent discrepancies may be the result of variations in culture conditions, vehicles, methodology (sensitivity), cell source, passage number and so on. There is currently no comprehensive, systematic analysis of the effects of typical NR activators on multiple target genes conducted in the same cells and laboratory using the same methodology. The aim of this study was to apply consistent, sensitive and specific methodology to assess the effects of four, typical, well-established activators of NRs—PB, CDCA, PCN and RIF—on the expression of ABCB1, ABCC1, ABCC2, CYP2B6, CYP3A4, CYP3A5, PXR, CAR and FXR in HepG2, Caco-2 and cryopreserved human hepatocytes.
Materials and methods
Cell culture
HepG2 and Caco-2 cell lines were purchased from American Type Tissue Culture. HepG2 cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Sigma-Aldrich, Dorset, UK) supplemented with 10% fetal bovine serum (FBS; Bio-Whittaker, Berkshire, UK). Caco-2 cells were maintained in DMEM supplemented with 15% FBS. All cell lines were incubated at 37 °C and 5% CO2 and subcultured every 4–5 days. Basal levels of all transcripts were initially quantified from passages 82–101 and 72–91 in HepG2 and Caco-2 cells, respectively (data not shown). We observed decreases in a number of transcripts in both cell lines after 13 continuous passages in our laboratory, and therefore cells were not used after passage 96 for HepG2 or passage 86 for Caco-2.
Cryopreserved human hepatocytes were removed from liquid nitrogen storage (2 × 10 vials per donor; H1 and H2). The cells were thawed by gently shaking the vials in a 37 °C water bath (∼75–90 s). The pooled hepatocyte suspensions (10 ml per donor) were then transferred into two separate 50 ml centrifuge tubes on ice. Cold suspension medium (15 ml) was then slowly added to the suspensions at a rate of ∼1 ml per 10 s. The cells were then centrifuged at 50 g for 3 min and the resulting pellet was resuspended in 12 ml of DMEM supplemented with 10% FBS. Viability was then determined based on Trypan blue exclusion (Loretz et al., 1989; Li et al., 1999) and was found to be 57.4% for H1 and 65.5% for H2.
Assessment of protein binding
Equilibrium dialysis was used to determine protein binding of PB, CDCA, PCN and RIF within the culture supernatant. Briefly, Dianorm dialysis membranes (GmbH, Munich, Germany) with molecular weight cutoff (MWC) of 5000 were soaked for 1 h in DMEM (Sigma-Aldrich, UK). PB, PCN, CDCA and RIF were then individually added to DMEM containing either 10% (HepG2 media) or 15% (Caco-2 media) FBS to a final concentration of 10 μM. An aliquot (1 ml) was then dispensed into one side of the dialysis block divided by the pre-soaked membrane, the other side containing control (without additions) media. The dialysis block was then sealed and rotated in a water bath overnight at 37 °C. A 200 μl aliquot was subsequently removed from the control side of the dialysis block and placed into quench tubes containing 200 μl of ice-cold methanol. After centrifugation at 400 g for 20 min, the supernatant was transferred into 96-well plates and analysed using liquid chromotography/mass spectrometry/mass spectometry (LC-MS/MS) (AstraZeneca in-house methodology).
Treatment of cells
Cell lines were seeded at a density of 5 × 106 per well into the appropriate FBS-supplemented medium, in Nunclon Surface 6 well plates (Nunc A/S, Kamstrup, Denmark). For all analyses, cells were plated 24 h prior to addition of compounds and were therefore not permitted to differentiate. Initial experiments assessed the effect of dexamethasone (DEX) on the expression of CYP3A4, ABCB1 and PXR and on the inducibility of CYP3A4, ABCB1 and PXR by PCN in HepG2 and Caco-2 cells. Firstly, concentration–response experiments for DEX (Sigma, Sigma-Aldrich, Dorset, UK) were conducted at final concentrations of 0, 0.01, 0.1, 1.0, 10 and 100 μM. PCN (Sigma, UK) was then assessed at final concentrations of 0, 0.01, 0.1, 1.0, 10 and 100 μM with and without 500 nM DEX (here DEX was included as a media supplement and as such included in the media prior to addition of PCN). DMSO-treated controls were used for PCN and DEX (0.1% v/v for vehicles).
For subsequent experiments, DEX was not included in the culture media. Test compounds, PB, CDCA, PCN and RIF (all compounds from Sigma, UK) were added at final concentrations of 0, 0.01, 0.1, 1.0, 10 and 100 μM. DMSO-treated controls were used for PB, CDCA and PCN and methanol-treated controls were used for RIF (0.1% v/v for both vehicles). For all experiments in HepG2 and Caco-2, cells were incubated for 18 h at 37 °C and 5% CO2.
For primary hepatocytes, cells were seeded at a density of 1 × 106 per well into 10% FBS-supplemented medium, in Nunclon Surface 6-well plates (Nunc A/S, Denmark). The NR activators, PB, CDCA, PCN and RIF were added to a final concentration of 1.0 μM, and vehicle controls were as described above. Cells were then incubated at 37 °C and 5% CO2 and sampled at 0, 2, 4, 6 and 18 h.
Toxicity
Test compounds were assayed for toxicity in HepG2 and Caco-2 cells by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (Mosmann, 1983) at final concentrations of 0.01–100 μM. These assays were initially performed at 18 h to assess cell death at the point of analysis. Subsequently, toxicity was assessed after 5-day incubations, as toxic concentrations would not necessarily be evident after an18 h incubation.
Quantitative real-time PCR
For HepG2, Caco-2 and primary hepatocytes, total RNA was isolated and cDNA was constructed as described previously (Owen et al., 2004). For each transcript, real-time PCR assays were developed for quantification relative to β-actin (housekeeping gene). In each case, assays were optimized to limit formation of primer dimer to ensure no aberrant data as a result of non-specific intercalation of pico green (Molecular Probes, Paisley, UK). In addition, fragments were sequenced in order to confirm correct amplification, and, where possible, assays were validated against minor groove binder (MGB) probe-based methodology. Relative expression (ΔΔCT) of transcripts against the housekeeping gene β-actin was performed in an Opticon2 Fluorescence Detector (MJ Research, Bio-Rad, Hertfordshire, UK). Amplification was conducted in a reaction consisting of 2.5 μl 10 × Taqman Buffer II, 0.5 U Taq polymerase, 1.25 μM MgCl2 (Amplitaq Gold; Applied Biosystems, Warrington, UK), 1.25 μM dNTPs (Promega, Southampton, UK), 20 ng cDNA, 0.5 μl pico green (final concentration 1:5000) and 0.03 μM forward and reverse primers (0.3 μM forward and reverse primers for CYP2B6), and nuclease-free water was added to a final volume of 25 μl (Sigma-Aldrich, UK). All primer sequences are available on request.
For CYP2B6, CYP3A4 and ABCB1, pre-validated MGB probe-based methodology was used to cross-validate the pico-green assays. This was carried out by the ΔΔCT method using GAPDH as a housekeeping gene as described previously (Owen et al., 2004).
Immunoblotting
For CYP proteins, immunoblotting was conducted on crude protein homogenates, and for transporter proteins, crude membrane protein fractions were purified as described previously (Marshak, 1996). In both cases, protein concentration was determined using the bicinchoninic acid assay (Stoscheck, 1990), samples were normalized to 5 μg μl−1 and stored at −80 °C.
Western blotting of all proteins was conducted using NuPage 4–12% Bis-Tris Gels (Invitrogen, Paisley, UK). Blotting was conducted using nitrocellulose membranes and an iBlot Gel Transfer System (Invitrogen, UK). Membranes were blocked in 10% non-fat-dried milk overnight at 4 °C.
For CYP2B6, CYP3A4, CYP3A5 (1:1000), ABCC1, ABCC2 (1:2000), ABCB1 and β-actin (1:5000), membranes were incubated with ab22734 sheep anti-human CYP2B6 (Abcam, Cambridge, UK), AB1278 sheep anti-human CYP3A4 (Chemicon, Temecula, USA), AB1279 rabbit anti-human CYP3A5 (Chemicon, USA), ab3373 (M2 III-6) mouse anti-human MRP1 (Abcam, UK), ab24102 (MRPm5) mouse anti-human MRP2 (Abcam, UK), mdr(C-19) goat anti-human P-gp (Santa Cruz Biotechnology, CA, USA) and anti-β-actin (Sigma, UK) for 2 h at room temperature in 2% non-fat-dried milk. For CYP2B6, CYP3A4, CYP3A5 and β-actin, 0.01% T-TBS was used, and for others, 0.05% T-TBS was used.
Secondary antibodies were incubated for 1 h at room temperature and were as follows: STAR88P donkey anti-sheep HRP conjugated (Serotec, North Carolina, USA), ab6701 donkey anti-rabbit HRP conjugated (Abcam, UK) (1:10000), P0449 rabbit anti-goat HRP conjugated (DakoCytomation, Glostrup, Germany) and sheep anti-mouse HRP conjugated (Amersham Biosciences, UK) (1:15 000).
Visualization was performed using enhanced chemiluminescence technology (PerkinElmer, Massachusetts, USA) and quantification was achieved using Bio-Rad GS710 scanner and Bio-Rad Quantity One densitometric analysis software.
Data analysis and statistical procedures
All data are presented as the mean±s.d. of at least four separate experiments conducted in duplicate. A cautious approach was taken for data generated with ligand concentrations shown to be toxic at 5 days for HepG2 and Caco-2. Specifically, concentrations that produced toxicity after 5 days are illustrated in the figures as dashed lines and statistical analysis is only presented for data at which significant induction was observed below this threshold. This compromised the ability to generate robust EC50 and Emax estimates. Normality was assessed using a Shapiro–Wilk statistical test. Differences in mRNA and protein expression were assessed using a paired t-test. Logarithmic and/or linear regression were used to determine the relationship between change in mRNA and protein in cell lines. Finally, log-transformed average fold change at 1 μM for CDCA, PCN and PB in HepG2 and Caco-2 cells were linearly regressed against equivalent log-transformed average fold change observed in primary cells for each transcript. The equation of each line was then used to predict the fold induction for RIF in primary cells from that obtained in HepG2. The resulting predicted values for RIF were then regressed against the measured values in order to test the prediction.
Results
Toxicity
None of the compounds tested (up to 100 μM) were toxic in incubations up to 18 h as assessed by the MTT assay (as compared to vehicle controls). However, after 5 days, significant toxicity was detected at higher concentrations in HepG2 and Caco-2 cells and data generated at these concentrations were therefore interpreted with caution. For HepG2, PB was toxic at 10 μM (P<0.0001) but not until 100 μM for PCN (P<0.0001), CDCA (P=0.0028) and RIF (P=0.0002). For Caco-2, 10 μM was found to be toxic for PCN (P<0.0001) and CDCA (P=0.013) and 100 μM was found to be toxic for PB (P=0.0002) and RIF (P=0.0034).
Protein binding
For DMEM containing 10% FBS, PB, PCN, RIF and CDCA were found to be 98, 97, 98 and 82% unbound, respectively. In media containing 15% FBS, the corresponding free fractions were 97, 96, 98 and 80%, respectively, indicating low levels of binding to FBS and a similar free drug concentration was present for incubations with HepG2 and Caco-2 cells.
Pico-green assay validation
All pico-green assays used in this study were validated rigorously. Primer design, primer concentration, MgCl2 concentration, annealing temperatures and cycle number were first optimized so as to completely eliminate primer dimer formation by visualization after agarose gel electrophoresis (data not shown). All amplicons were then excised from the gel and sequenced with the forward and reverse primers used for the amplifications. Each amplicon was ensured to be specific for its target by a NCBI blast search (data not shown). For CYP2B6, CYP3A4 and ABCB1, pre-validated MGB probe-based methodology was available within our laboratory (Owen et al., 2004). Therefore, for these transcripts the pico-green assays were cross-validated using the samples obtained after incubation of Caco-2 cells with RIF. A significant positive correlation was observed between these assays (r2=0.99, P<0.0001 for ABCB1; r2=0.95, P<0.001 for CYP2B6; r2=0.89, P<0.005 for CYP3A4). Bland and Altman plots were also constructed to show the relationship between the pico-green-based assay and the MGB probe-based assay for the effect of RIF on ABCB1 in Caco-2 cells, and the assays were within 95% limits of agreement (data not shown). Finally, none of the compounds were shown to affect the β-actin C(t) values at any of the concentrations used, indicating that β-actin was an appropriate housekeeping gene for these studies (data not shown).
Effects of DEX on CYP3A4, ABCB1 and PXR
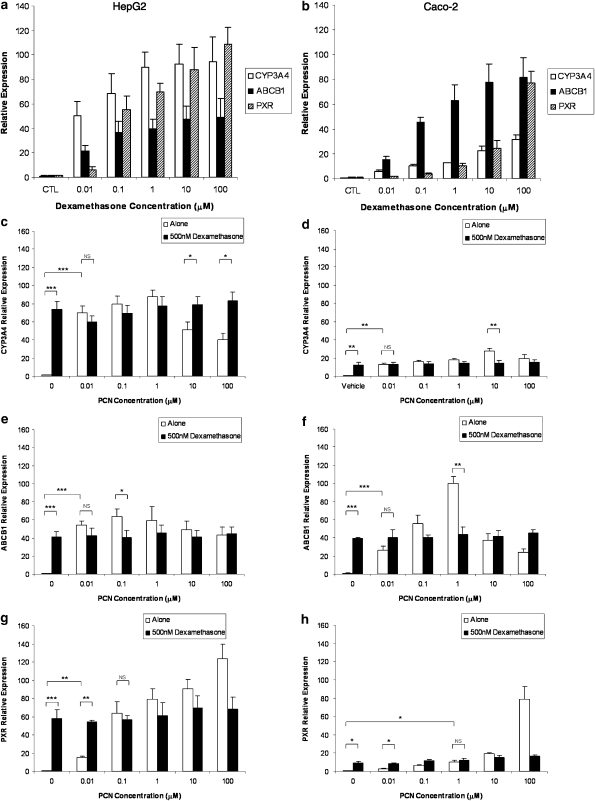
DEX potently induced CYP3A4, ABCB1 and PXR in HepG2 (Figure 1a) and Caco-2 (Figure 1b). Concentration-dependent induction was observed in each case with significant induction at 0.01 μM DEX for CYP3A4, ABCB1 and PXR in HepG2, 0.01 μM DEX for CYP3A4 and ABCB1 and 0.1 μM for PXR in Caco-2. Maximum induction of all transcripts was observed at 100 μM.
Figure 1.
Implications of using dexamethasone as a media supplement. (a, b) Impact of DEX (0–100 μM) on mRNA expression of CYP3A4, ABCB1 and PXR in HepG2 and Caco-2 cells. (c–h) Impact of DEX (500 nM) on inducibility of CYP3A4 (c, d), ABCB1 (e, f) and PXR (g, h) by PCN in HepG2 and Caco-2 cells. Data are the mean±s.d. of four experiments conducted in duplicate. For clarity, not all statistical analyses are given. For each transcript, a significant increase in expression was observed when cells were incubated with PCN alone, which was not seen if DEX was included in the culture media. *P<0.05; **P<0.01; ***P<0.001.
In HepG2 and Caco-2 cells, PCN significantly induced the mRNA of CYP3A4 (Figures 1c and d), ABCB1 (Figures 1e and f) and PXR (Figures 1g and h), as compared to the DEX-free control. However, when DEX was included in the culture media, no significant induction was observed for any of the transcripts, as compared to the DEX control (Figures 1c–h).
Induction of CYP2B6, CYP3A4 and CYP3A5 mRNA in HepG2, Caco-2 and primary hepatocytes
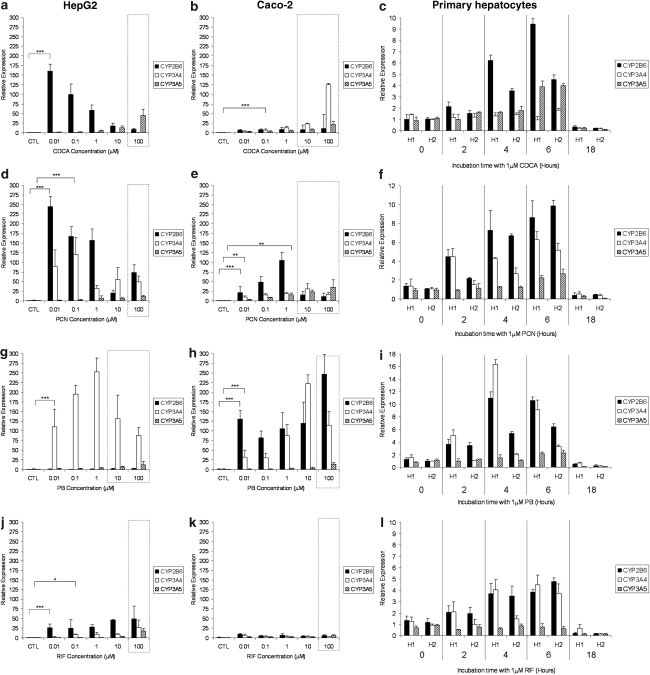
The impact of CDCA, PCN, PB and RIF on the expression of CYP2B6, CYP3A4 and CYP3A5 mRNA in HepG2, Caco-2 and primary hepatocytes is shown in Figure 2. For clarity, statistical analysis is only presented for the lowest concentration at which a significant induction was observed. CDCA significantly increased expression of CYP2B6 in HepG2 (Figure 2a), CYP3A4 in Caco-2 (Figure 2b) and CYP2B6 and CYP3A5 in primary cells (Figure 2c). For PCN, a significant increase in CYP2B6 and CYP3A4 was observed in all three cell types (Figures 2d–f) and a less marked induction of CYP3A5 was also observed in Caco-2 and primary cells (Figures 2e and f). PB elicited a significant induction of CYP3A4 in all three cell types (Figures 2g–i) as well as that of CYP2B6 in Caco-2 and primary cells (Figures 2h and i). PB did not affect CYP3A5 in any cell type. Finally, RIF significantly upregulated CYP2B6 and CYP3A4 in HepG2 and primary cells (Figures 2j and l) but not in Caco-2 cells (Figure 2k). No effects of RIF on CYP3A5 mRNA was observed.
Figure 2.
Impact of typical activators on CYP isoforms. Effect of CDCA (a–c), PCN (d–f), PB (g–i) and RIF (j–l) on CYP2B6, CYP3A4 and CYP3A5 mRNA expression in HepG2, Caco-2 and primary hepatocytes. Data are the mean±s.d. of four experiments conducted in duplicate. In cell lines, concentration dependency was investigated, whereas, in primary cells, time dependency was assessed in two cultures of hepatocytes (H1 and H2) at 1 μM of each compound. Dotted lines indicate concentrations at which toxicity was observed following 5-day incubations with drug. For clarity, statistical analyses are given only for the lowest concentration at which a significant difference was observed: *P<0.05; **P<0.01; ***P<0.001.
Induction of ABCB1, ABCC1 and ABCC2 mRNA in HepG2, Caco-2 and primary hepatocytes
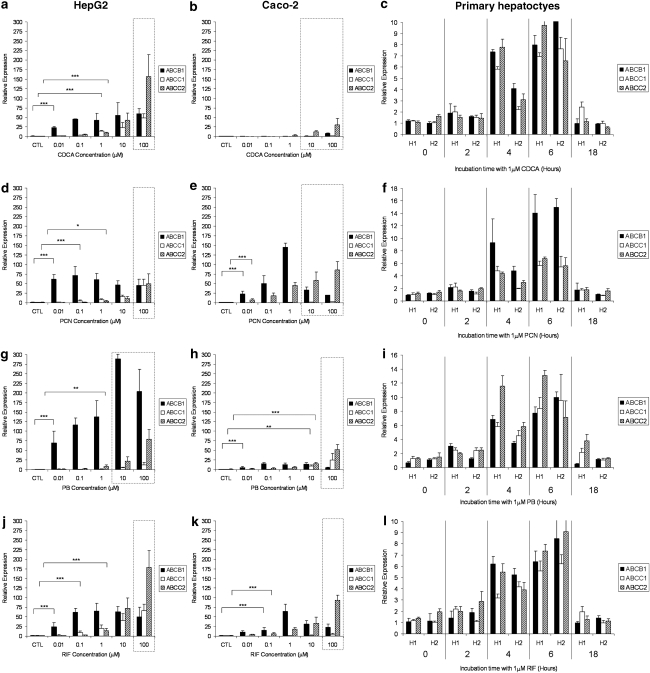
The effects of CDCA, PCN, PB and RIF on the expression of ABCB1, ABCC1 and ABCC2 mRNA in HepG2, Caco-2 and primary hepatocytes is shown in Figure 3. CDCA elicited significant upregulation of all three transporters in HepG2 (Figure 3a) and primary cells (Figure 3c) but not in Caco-2 cells (Figure 3b). PCN significantly increased ABCB1 and ABCC2 mRNA in all three cell types (Figures 3d and e) and ABCC1 mRNA in HepG2 and primary cells (Figures 3d and e). PB increased the expression of all three transporters in Caco-2 (Figure 3h) and primary cells (Figure 3i) but only that of ABCB1 and ABCC2 in HepG2 (Figure 3g). Conversely, RIF induced all three transporters in HepG2 (Figure 3j) and primary cells (Figure 3l) but only ABCB1 and ABCC2 in Caco-2 cells (Figure 3k).
Figure 3.
Impact of typical activators on transporters. Effect of CDCA (a–c), PCN (d–f), PB (g–i) and RIF (j–l) on ABCB1, ABCC1 and ABCC2 mRNA expression in HepG2, Caco-2 and primary hepatocytes. Data are the mean±s.d. of four experiments conducted in duplicate. In cell lines, concentration dependency was investigated, whereas, in primary cells, time dependency was assessed in two cultures of hepatocytes (H1 and H2) at 1 μM of each compound. Dotted lines indicate concentrations at which toxicity was observed following 5-day incubations with drug. For clarity, statistical analyses are given only for the lowest concentration at which a significant difference was observed: *P<0.05; **P<0.01; ***P<0.001.
Induction of CAR, FXR and PXR mRNA in HepG2, Caco-2 and primary hepatocytes
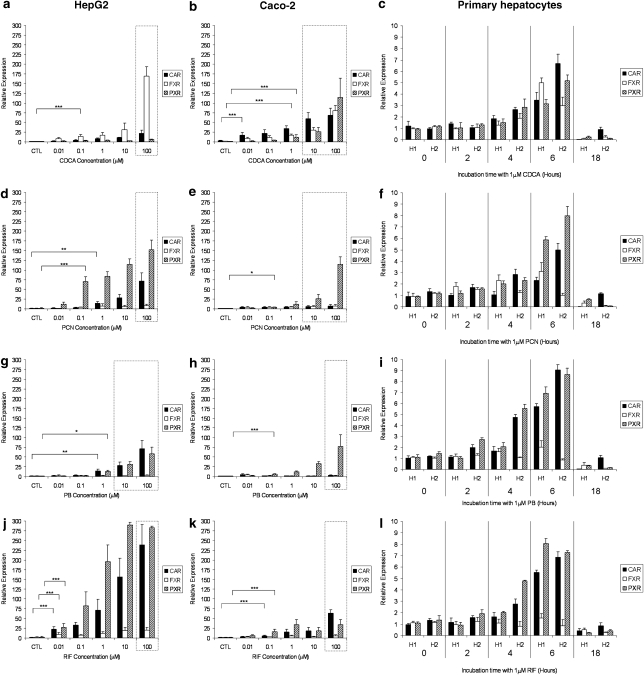
The effects of CDCA, PCN, PB and RIF on the expression of CAR, FXR and PXR mRNA in HepG2, Caco-2 and primary hepatocytes are shown in Figure 4. CDCA caused significant induction of CAR, FXR and PXR in Caco-2 (Figure 4b) and primary cells (Figure 4c) but only that of FXR in HepG2 (Figure 4a). PCN significantly upregulated mRNA for PXR and CAR in HepG2 (Figure 4d) and primary cells (Figure 4f) but only PXR in Caco-2 cells (Figure 4e). For PB, a significant induction of PXR was observed in all three cell types (Figures 4g–i), and CAR was also upregulated in HepG2 (Figure 4g) and primary cells (Figure 4i). RIF upregulated CAR and PXR but not FXR in all three cell types (Figures 4j–l).
Figure 4.
Impact of typical activators on nuclear receptors. Effect of CDCA (a–c), PCN (d–f), PB (g–i) and RIF (j–l) on CAR, FXR and PXR mRNA expression in HepG2, Caco-2 and primary hepatocytes. Data are the mean±s.d. of four experiments conducted in duplicate. In cell lines, concentration dependency was investigated, whereas, in primary cells, time dependency was assessed in two cultures of hepatocytes (H1 and H2) at 1 μM of each compound. Dotted lines indicate concentrations at which toxicity was observed following 5-day incubations with drug. For clarity, statistical analyses are given only for the lowest concentration at which a significant difference was observed: *P<0.05; **P<0.01; ***P<0.001.
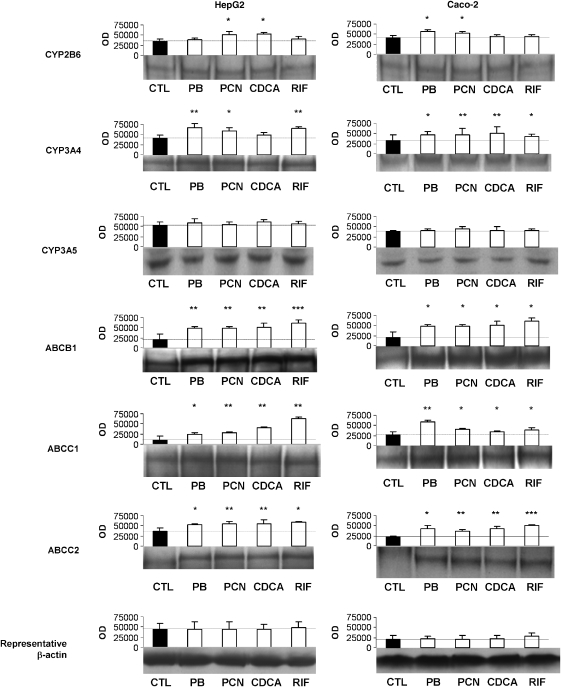
Induction of CYP2B6, CYP3A4, CYP3A5, ABCB1, ABCC1 and ABCC2 protein in HepG2 and Caco-2 cells
Figure 5 shows the effects of CDCA, PCN, PB and RIF on the expression of CYP2B6, CYP3A4, CYP3A5, ABCB1, ABCC1 and ABCC2 protein in HepG2 and Caco-2 cells. CDCA significantly upregulated CYP2B6, ABCB1, ABCC1 and ABCC2 protein in HepG2 and CYP3A4, ABCB1, ABCC1 and ABCC2 protein in Caco-2 cells. PCN upregulated all proteins except CYP3A5 in both HepG2 and Caco-2. PB significantly upregulated CYP2B6, CYP3A4, ABCB1, ABCC1 and ABCC2 protein in Caco-2 and CYP3A4, ABCB1, ABCC1 and ABCC2 protein in HepG2 cells. RIF elicited induction of CYP3A4, ABCB1, ABCC1 and ABCC2 in all three cell types.
Figure 5.
Effect of CDCA, PCN, PB and RIF on protein expression of CYP2B6, CYP3A4, CYP3A5, ABCB1, ABCC1 and ABCC2 in HepG2 and Caco-2 cells. Results for β-actin are also given to illustrate equal loading. A representative western blot as well as the mean (±s.d.) optical densitometric results from four experiments conducted in duplicate are shown. *P<0.05; **P<0.01; ***P<0.001.
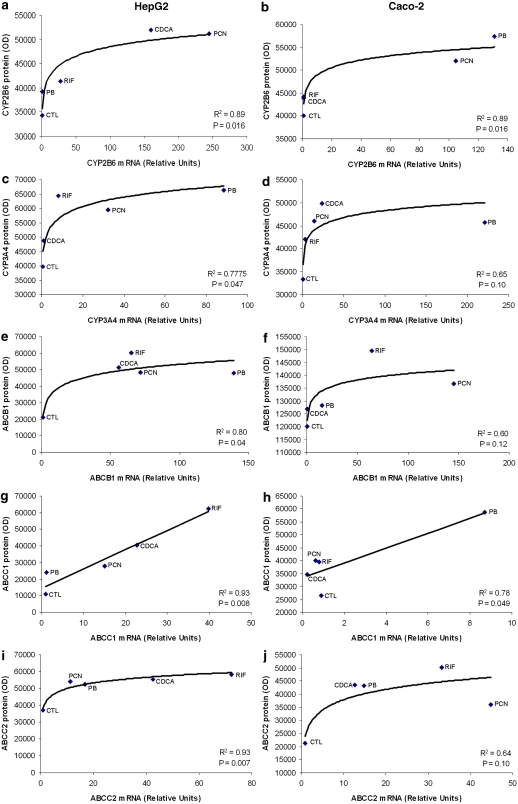
Relationship between mRNA and protein expression
In order to assess whether a relationship existed between induction of mRNA and protein, 1 μM mRNA data for each transcript (with the exception of CYP3A5, as no change in protein was observed with any compound) were plotted against the corresponding protein data (Figure 6). In HepG2, a significant logarithmic relationship was observed between mRNA and protein for CYP2B6 (Figure 6a), CYP3A4 (Figure 6c), ABCB1 (Figure 6e) and ABCC2 (Figure 6i). Conversely, the relationship between mRNA and protein was linear for ABCC1 (Figure 6g). Similar trends were observed in Caco-2 cells but were only statistically significant for CYP2B6 (Figure 6b) and ABCC1 (Figure 6h). For the latter, the correlation was driven predominantly by one data point (PB) and this was not statistically significant when this data point was removed (R2=0.07; P=0.7).
Figure 6.
Relationship between mRNA and protein expression. Correlation between mRNA and protein expression in HepG2 and Caco-2 cells for CYP2B6 (a, b), CYP3A4 (c, d), ABCB1 (e, f), ABCC1 (g, h) and ABCC2 (i, j). Data for CYP3A5 are not presented because no differences in the protein were observed. A best fit to the data was observed by logarithmic regression for all CYPs and transporters except ABCC1, which was best described by linear regression.
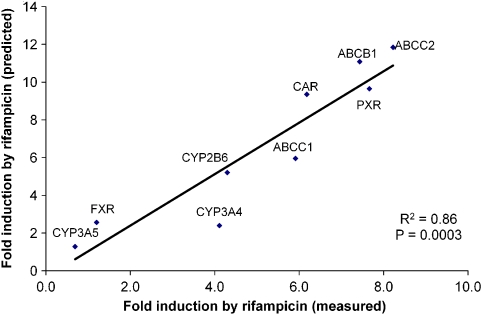
Relationship between fold change in primary cells and cell lines
Linear regression was used to assess the relationship between fold change of each transcript in HepG2 versus primary cells for CDCA, PCN and PB (Table 1). A significant linear relationship was observed for CYP3A4, ABCB1, ABCC2 and FXR. For all other transcripts (except ABCC1), a trend was observed (r2>0.41; 0.05 <P<0.15). The equations of these lines for each transcript were then used to assess whether the fold change in primary cells could be predicted from the HepG2 data for RIF. When the predicted values for RIF were regressed against the measured values, a significant linear correlation was observed (Figure 7).
Table 1.
Relationship between fold change observed in HepG2 and that observed in primary hepatocytes
| Transcript | r2 | P-value | Power (%) (for 5% significance) |
|---|---|---|---|
| CYP2B6 | 0.41 | 0.41 | 16.2 |
| CYP3A4 | 0.92 | 0.04 | 65.8 |
| CYP3A5 | 0.73 | 0.15 | 35.5 |
| ABCB1 | 0.91 | 0.04 | 63.4 |
| ABCC1 | 0.19 | 0.56 | 8.9 |
| ABCC2 | 0.98 | 0.01 | 87.2 |
| CAR | 0.85 | 0.08 | 51.1 |
| FXR | 0.96 | 0.02 | 80.5 |
| PXR | 0.75 | 0.13 | 37.8 |
The fold change observed for CDCAs, PCN and PB in HepG2 and an average of primary hepatocytes were log transformed and linearly regressed.
Figure 7.
Correlation between induction observed in primary cells (measured) with that predicted from HepG2 cells (predicted). For predicted values, the log-transformed average fold change from HepG2 for CDCA, PCN and PB was plotted against the log-transformed average fold change from primary cells for each transcript (see Table 1 for regression). The equations from the resultant lines were then used to calculate a prediction of the fold change in primary cells from the fold change in HepG2 for RIF.
Discussion and conclusions
Ease of culture of immortalized cells and the reproducibility of data obtained from them have resulted in their widespread use to study regulation of gene expression. Such studies have been pivotal in identifying key regulatory mechanisms. For example, HepG2 cells have been shown to lack expression of the co-chaperone, cytosolic CAR retention protein, resulting in nuclear accumulation of unliganded CAR (Kobayashi et al., 2003). However, many cell lines acquire mutations that result in dissociation from the phenotype in vivo (Castell et al., 2006), interindividual variability cannot be assessed, and there are likely to be significant inadequacies when extrapolating to the situation in vivo. Nonetheless, several cell lines still find widespread utility for evaluation of drug absorption and metabolism. Primary hepatocytes express typical hepatic functions, and quantitative similarities between in vitro and in vivo metabolism have been observed, making them an attractive alternative to immortalized cells (Vermeir et al., 2005).
HepG2 cells exist in at least two primary strains, and the level and activity of enzymes is influenced further by culture conditions and clonal selection in different laboratories (Rodriguez-Antona et al., 2002). Analysis of Caco-2 cells has shown that transporter expression resembles closely that of normal colon, although properties are also compromised at high passage (Calcagno et al., 2006). Another important difference between laboratories is the use of DEX as a media supplement. DEX prolongs cell viability (Bailly-Maitre et al., 2001) but is a known inducer of CYP3A4 (Chieli et al., 1994) and has been shown to modulate P-gp expression in rat hepatocytes (Fardel et al., 1993). DEX also upregulates PXR and CAR mRNA (Pascussi et al., 2000a, 2000b) and the order of DEX and PCN administration appears to be an important determinant of effects on CYP3A1 in rats (Hosoe et al., 2005). Data presented here show that DEX has a marked effect on the expression of CYP3A4, ABCB1 and PXR and its inclusion in media may therefore decrease the ability to detect induction. Similar observations have been reported in rat liver slices (Meredith et al., 2003). These data suggest that there is a maximal response to PCN. Therefore, the addition of DEX effectively increases the baseline but, as the maximum is fixed, a reduction in the potential effect is observed. Therefore, one hypothesis is that this competition allows the biological system to self-limit and prevent over-activation.
The mechanism by which this interaction between PCN and DEX occurs is currently being investigated. However, what is known is that sub-micromolar concentrations of PCN have previously been shown to activate rodent PXR (Shah et al., 2007) but antagonize the glucocorticoid receptor. In addition, activation of glucocorticoid receptors by DEX increases PXR expression and thus increases CYP3A1, whereas PCN activates PXR directly in rats (Hosoe et al., 2005). Clearly, the overlap in NR activation and function makes this area extremely complex, but, given that a considerable body of convincing structural and mechanistic data that support the concept that human PXR is not activated by PCN, one could hypothesize that this phenomenon is mediated via other NRs. It is important to note that the involvement of other NRs would also explain the surprising observations that the classical potent PXR ligand, RIF, produced one of the weaker activations in all systems. Nonetheless, because of these issues, DEX was not included in subsequent experiments, and this may explain some discrepancies with previous studies.
Consistent with some reports, similar inductions of CYP3A4 and ABCB1 by PCN and RIF were observed in HepG2, Caco-2 cells and primary hepatocytes (Faucette et al., 2006). Although studies with reporter assays have shown that PCN activates rodent but not human PXR, and the converse is true of RIF (Jones et al., 2000), PCN has been shown to increase human CYP3A4 (Ogg et al., 1999) and RIF increases the rodent orthologue (Swales et al., 2003). Clearly, this appears counter-intuitive, given the species differences in PXR activation. One possible explanation is that other mechanisms are involved in the observed induction. Indeed, PCN has been reported to activate the CYP3A4 promoter via the glucocorticoid receptor (Ogg et al., 1999). Administration of PCN to humans has also been shown to diminish the activity of some drugs, which is consistent with induction of metabolism in vivo (Szabo et al., 1975).
Some differences were observed between the profiles of induction in HepG2 versus Caco-2. Although it is tempting to invoke tissue-specific differences between hepatic and intestinal cells, it is unclear whether these differences are inherent between tissues or result from divergence during transformation or culture. Interestingly, the magnitude of induction was in the rank order of HepG2>Caco-2>primary hepatocytes, and as the rank order of transporter expression is primary hepatocytes>Caco-2>HepG2 and these compounds are substrates, it is tempting to speculate that this may be a consequence of lower intracellular inducer accumulation. This requires further experimentation and was beyond the scope of this study.
Data for PB and RIF are in agreement with other studies conducted in primary hepatocytes (Jigorel et al., 2006; Nishimura et al., 2006). Also, where overlap exists, the data agree with studies in HepG2, Caco-2 and primary hepatocytes (Schrenk et al., 2001). For Caco-2 cells, results with RIF conflict with previous studies that indicated no effect on ABCB1, ABCC1, ABCC2, CYP3A4 or PXR expression (Pfrunder et al., 2003; Collett et al., 2004). Induction of ABCB1, ABCC1, ABCC2 and PXR but not CYP3A4 was observed here. There are differences in methodology; Pfrunder et al. (2003) used media containing 10% FBS supplemented with gentamicin and up to 1% DMSO was used as a vehicle, whereas in this study 15% FBS and methanol were used, respectively. RIF has been shown to be stable for 3 months in methanol (Le Guellec et al., 1997) and DMSO (Karlson and Ulrich, 1969), and DMSO has been reported not to effect PXR and CYP3A4 expression below a concentration of 0.1% (Bowen et al., 2000). However, RIF reduces CYP3A4 and CYP3A5 mRNA levels when >0.5% DMSO is used (Nishimura et al., 2002).
Logarithmic correlations between changes in mRNA and protein in HepG2 and Caco-2 cells were observed. Lower changes in mRNA elicited linear increases in protein but there appeared to be a threshold beyond which no additional increase in protein was observed. Finally, although there are clearly important inadequacies of transformed cells that must be considered when studying mechanisms, an accurate prediction of the profile of induction in hepatocytes by RIF from HepG2 data was possible. This is exploratory and requires examination with a larger set of compounds, and its importance will ultimately be determined by the suitability of primary hepatocytes as surrogates for gene regulation in vivo.
For ABCB1, an increase in mRNA and protein was observed when cells were exposed to CDCA at non-toxic concentrations. However, in Caco-2 cells, an increase in protein was noted even though increases in mRNA were not noted until toxic concentrations (100 μM) were added. This is in agreement with a previous study that showed increased expression of ABCB1 and P-glycoprotein activity in Madin Darby canine kidney cells exposed to 100 μM CDCA (Kneuer et al., 2007). Nonetheless, the change in protein was comparable between HepG2 and Caco-2, despite marked differences between the two cell lines at the mRNA level. The reason for this requires further investigation, but one could speculate that there may be transcriptional, post-transcriptional and/or translational mechanisms involved in the upregulation of the protein. It is of interest that non-transcriptional mechanism has been reported to induce P-glycoprotein expression in other transformed cells (Yague et al., 2003).
It is important to recognize that the viability of the primary hepatocytes used here was <70%. This is in line with the majority of previous studies using cryopreserved human hepatocytes (Li et al., 1999; Shibata et al., 2002; Baccarani et al., 2005; Terry et al., 2005; Miyamoto et al., 2006). A number of investigators have examined the addition of media supplements and/or purification of cells through a Percoll gradient in order to improve the viability and therefore metabolic competency of these cells. As it was not our intention to investigate metabolism, we opted for a more simplistic approach so as not to add additional compounds that could potentially interfere with gene regulation themselves. In either case, it must be noted that a potential limitation to the use of primary hepatocytes is a selective loss of hepatocyte sub-types during isolation and/or cryopreservation (that is, diploid/tetraploid or mono-/bi-nuclear) with lower viabilities, in which the phenotype may be different.
Given the potential controversy of some of these findings, some samples were also analysed using MGB-probe assays. These results confirmed the data for RIF in HepG2 and validated the pico-green methodology (data not shown). Caco-2 cells are well known for exhibiting markedly different phenotypes according to passage number and culture conditions (Sambuy et al., 2005).
In these studies, we selected a range of concentrations of PB from 0.01 to 100 μM. However, many previous publications have reported studies with concentrations of PB above these concentrations (most commonly ⩾1000 μM). Furthermore, the vast majority of previous studies do not report concentration–response relationships, and it is not clear to us why such high concentrations were selected. Our data indicate that these previous concentrations (as high as 5000 μM in some studies) are associated with (a) toxicity (100 μM for HepG2 and 10 μM for Caco-2) and (b) a submaximal response for some genes (for example, for CYP3A4 and ABCB1, lower expression was observed at 10 and 100 μM than at lower concentrations). This issue may also explain some previous inconsistencies, as such high concentrations, which are unattainable in vivo, may compromise the ability to clearly define the phenotype.
As seen with PB, bell-shaped concentration–induction profiles were also observed in other cases, for example, induction by PCN of CYP3A4 and ABCB1 in HepG2 and Caco-2 cells. It is tempting to speculate, as others have done, that this may be due to toxicity. Indeed, in the case of PCN on ABCB1 in Caco-2 cells, this did correlate with the toxicity. However, in HepG2 cells, sub-maximal induction was observed at concentrations that were non-toxic. This has been observed in other studies of CYP3A4 with both expression (Ripp et al., 2006) and function (Reinach et al., 1999). In these cases, the underlying mechanisms are not understood. However, one could hypothesize that at higher concentrations, activation of other systems may result in repression or partial antagonism. In this regard, it is interesting to note that PCN has also been shown to activate liver-X-receptor response elements and liver-X-receptor reduces activation of the CYP3A4-XREM in response to PCN (Kocarek et al., 2002). Higher concentrations of PCN may therefore activate liver-X-receptor (and/or other factors) and elicit a negative effect on PXR-mediated induction of CYP3A4. These data indicate that using a single concentration may result in gross underestimation and may also explain some of the discrepancies between this and other studies, underscoring the importance of conducting full concentration–response experiments. Indeed, significant induction of these transcripts was seen for primary hepatocytes, which is in agreement with other studies (Phillips et al., 2005).
For RIF and PB, induction of both PXR and CAR was observed in all three cell types, and PB induced PXR and CAR in HepG2 and hepatocytes but not Caco-2. If PXR and CAR self-regulate, overlap may be expected, as both bind to similar response elements (Xie et al., 2000; Goodwin et al., 2001; Burk et al., 2005a). The mechanisms controlling PXR expression have not been studied in great detail, but some data are emerging (Aouabdi et al., 2006; Gibson et al., 2006). Recently, it was reported that PXR is controlled by PXR itself as well as PPARα (Aouabdi et al., 2006) and FXR (Jung et al., 2006). In support of this, CDCA increased expression of PXR in Caco-2 and primary hepatocytes in this study. Transfection of unliganded PXR and CAR was previously shown to downregulate expression of PXR (Aouabdi et al., 2006). Taken collectively with the data here, it appears that, although unliganded PXR and CAR downregulate expression, activation of these NRs may increase transcription. This is certainly an interesting subject for future study.
CDCA increased expression of FXR and CAR in HepG2 cells and all three NRs in both Caco-2 cells and primary hepatocytes. This may represent an additional avenue in the complex regulation of multiple NRs, but clearly the interpretation of these findings is dependent on the specificity of CDCA for FXR. The observation that CDCA elicits induction of ABCC2 and CYP3A4 is in agreement with other reports (Kast et al., 2002; Gnerre et al., 2004), but this is the first report that implicates CDCA in the regulation of ABCB1, ABCC1 and CYP2B6. However, other bile acids have been shown to regulate ABCC2 via non-FXR mediated pathways (Zollner et al., 2003), and so the relative role of FXR should be interpreted with caution. The clinical relevance of these observations will depend on the number of therapeutic compounds that activate FXR: there is significant interest in FXR as a therapeutic target for cardiovascular (Bishop-Bailey, 2004) and cholestatic liver diseases (Willson et al., 2001). As FXR modulators emerge, it will be interesting to characterize their effects on the expression of disposition genes.
The data presented herein provide a solid platform from which to examine the molecular mechanisms that underlie these observations.
Acknowledgments
This work was funded by AstraZeneca Charnwood. PM is sponsored by an AstraZeneca PhD Studentship.
Abbreviations
- CAR
constitutive androstane receptor
- CDCA
chenodeoxycholic acid
- CYP
cytochrome P450
- DEX
dexamethasone
- DMEM
Dulbecco's modified Eagle's medium
- DR
direct repeat
- FBS
fetal bovine serum
- FXR
farnesoid X receptor
- MGB
minor groove binder
- NR
nuclear receptor
- PB
phenobarbital
- PCN
pregnenolone 16α-carbonitrile
- PXR
pregnane X receptor
- RIF
rifampicin
- T-TBS
Tween–Tris-buffered saline
Conflict of interest
AO and DJB have received research funding from AstraZeneca, Boehringer Ingelheim, GlaxoSmithKline, Abbott laboratories, Pfizer, Tibotec and Roche Pharmaceuticals.
References
- Aouabdi S, Gibson G, Plant N. Transcriptional regulation of the PXR gene: identification and characterization of a functional peroxisome proliferator-activated receptor alpha binding site within the proximal promoter of PXR. Drug Metab Dispos. 2006;34:138–144. doi: 10.1124/dmd.105.006064. [DOI] [PubMed] [Google Scholar]
- Baccarani U, Sanna A, Cariani A, Sainz M, Adani GL, Lorenzin D, et al. Cryopreserved human hepatocytes from cell bank: in vitro function and clinical application. Transplant Proc. 2005;37:256–259. doi: 10.1016/j.transproceed.2004.12.230. [DOI] [PubMed] [Google Scholar]
- Bailly-Maitre B, de Sousa G, Boulukos K, Gugenheim J, Rahmani R. Dexamethasone inhibits spontaneous apoptosis in primary cultures of human and rat hepatocytes via Bcl-2 and Bcl-xL induction. Cell Death Differ. 2001;8:279–288. doi: 10.1038/sj.cdd.4400815. [DOI] [PubMed] [Google Scholar]
- Benet LZ, Cummins CL, Wu CY. Unmasking the dynamic interplay between efflux transporters and metabolic enzymes. Int J Pharm. 2004;277:3–9. doi: 10.1016/j.ijpharm.2002.12.002. [DOI] [PubMed] [Google Scholar]
- Bertilsson G, Heidrich J, Svensson K, Asman M, Jendeberg L, Sydow-Backman M, et al. Identification of a human nuclear receptor defines a new signaling pathway for CYP3A induction. Proc Natl Acad Sci USA. 1998;95:12208–12213. doi: 10.1073/pnas.95.21.12208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop-Bailey D. FXR as a novel therapeutic target for vascular disease. Drug News Perspect. 2004;17:499–504. doi: 10.1358/dnp.2004.17.8.863693. [DOI] [PubMed] [Google Scholar]
- Blizard D, Sueyoshi T, Negishi M, Dehal SS, Kupfer D. Mechanism of induction of cytochrome p450 enzymes by the proestrogenic endocrine disruptor pesticide-methoxychlor: interactions of methoxychlor metabolites with the constitutive androstane receptor system. Drug Metab Dispos. 2001;29:781–785. [PubMed] [Google Scholar]
- Bowen WP, Carey JE, Miah A, McMurray HF, Munday PW, James RS, et al. Measurement of cytochrome P450 gene induction in human hepatocytes using quantitative real-time reverse transcriptase-polymerase chain reaction. Drug Metab Dispos. 2000;28:781–788. [PubMed] [Google Scholar]
- Burk O, Arnold KA, Geick A, Tegude H, Eichelbaum M. A role for constitutive androstane receptor in the regulation of human intestinal MDR1 expression. Biol Chem. 2005a;386:503–513. doi: 10.1515/BC.2005.060. [DOI] [PubMed] [Google Scholar]
- Burk O, Arnold KA, Nussler AK, Schaeffeler E, Efimova E, Avery BA, et al. Antimalarial artemisinin drugs induce cytochrome P450 and MDR1 expression by activation of xenosensors pregnane X receptor and constitutive androstane receptor. Mol Pharmacol. 2005b;67:1954–1965. doi: 10.1124/mol.104.009019. [DOI] [PubMed] [Google Scholar]
- Calcagno AM, Ludwig JA, Fostel JM, Gottesman MM, Ambudkar SV. Comparison of drug transporter levels in normal colon, colon cancer, and Caco-2 cells: impact on drug disposition and discovery. Mol Pharm. 2006;3:87–93. doi: 10.1021/mp050090k. [DOI] [PubMed] [Google Scholar]
- Castell JV, Jover R, Martinez-Jimenez CP, Gomez-Lechon MJ. Hepatocyte cell lines: their use, scope and limitations in drug metabolism studies. Expert Opin Drug Metab Toxicol. 2006;2:183–212. doi: 10.1517/17425255.2.2.183. [DOI] [PubMed] [Google Scholar]
- Chieli E, Santoni-Rugiu E, Cervelli F, Sabbatini A, Petrini M, Romiti N, et al. Differential modulation of P-glycoprotein expression by dexamethasone and 3-methylcholanthrene in rat hepatocyte primary cultures. Carcinogenesis. 1994;15:335–341. doi: 10.1093/carcin/15.2.335. [DOI] [PubMed] [Google Scholar]
- Claudel T, Sturm E, Duez H, Torra IP, Sirvent A, Kosykh V, et al. Bile acid-activated nuclear receptor FXR suppresses apolipoprotein A-I transcription via a negative FXR response element. J Clin Invest. 2002;109:961–971. doi: 10.1172/JCI14505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collett A, Tanianis-Hughes J, Warhurst G. Rapid induction of P-glycoprotein expression by high permeability compounds in colonic cells in vitro: a possible source of transporter mediated drug interactions. Biochem Pharmacol. 2004;68:783–790. doi: 10.1016/j.bcp.2004.05.006. [DOI] [PubMed] [Google Scholar]
- El-Sankary W, Gibson GG, Ayrton A, Plant N. Use of a reporter gene assay to predict and rank the potency and efficacy of CYP3A4 inducers. Drug Metab Dispos. 2001;29:1499–1504. [PubMed] [Google Scholar]
- Fardel O, Lecureur V, Guillouzo A. Regulation by dexamethasone of P-glycoprotein expression in cultured rat hepatocytes. FEBS Lett. 1993;327:189–193. doi: 10.1016/0014-5793(93)80167-s. [DOI] [PubMed] [Google Scholar]
- Faucette SR, Sueyoshi T, Smith CM, Negishi M, Lecluyse EL, Wang H. Differential regulation of hepatic CYP2B6 and CYP3A4 genes by constitutive androstane receptor but not pregnane X receptor. J Pharmacol Exp Ther. 2006;317:1200–1209. doi: 10.1124/jpet.105.098160. [DOI] [PubMed] [Google Scholar]
- Gibson GG, Phillips A, Aouabdi S, Plant K, Plant N. Transcriptional regulation of the human pregnane-X receptor. Drug Metab Rev. 2006;38:31–49. doi: 10.1080/03602530600569810. [DOI] [PubMed] [Google Scholar]
- Gnerre C, Blattler S, Kaufmann MR, Looser R, Meyer UA. Regulation of CYP3A4 by the bile acid receptor FXR: evidence for functional binding sites in the CYP3A4 gene. Pharmacogenetics. 2004;14:635–645. doi: 10.1097/00008571-200410000-00001. [DOI] [PubMed] [Google Scholar]
- Goodwin B, Moore LB, Stoltz CM, McKee DD, Kliewer SA. Regulation of the human CYP2B6 gene by the nuclear pregnane X receptor. Mol Pharmacol. 2001;60:427–431. [PubMed] [Google Scholar]
- Hosoe T, Nakahama T, Inouye Y. Divergent modes of induction of rat hepatic and pulmonary CYP3A1 by dexamethasone and pregnenalone16alpha-carbonitrile. J Health Sci. 2005;51:75–79. [Google Scholar]
- Jigorel E, Le Vee M, Boursier-Neyret C, Parmentier Y, Fardel O. Differential regulation of sinusoidal and canalicular hepatic drug transporter expression by xenobiotics activating drug-sensing receptors in primary human hepatocytes. Drug Metab Dispos. 2006;34:1756–1763. doi: 10.1124/dmd.106.010033. [DOI] [PubMed] [Google Scholar]
- Jones SA, Moore LB, Shenk JL, Wisely GB, Hamilton GA, McKee DD, et al. The pregnane X receptor: a promiscuous xenobiotic receptor that has diverged during evolution. Mol Endocrinol. 2000;14:27–39. doi: 10.1210/mend.14.1.0409. [DOI] [PubMed] [Google Scholar]
- Jung D, Mangelsdorf DJ, Meyer UA. Pregnane X receptor is a target of farnesoid X receptor. J Biol Chem. 2006;281:19081–19091. doi: 10.1074/jbc.M600116200. [DOI] [PubMed] [Google Scholar]
- Karlson AG, Ulrich JA. Stability of rifampin in dimethylsulfoxide. Appl Microbiol. 1969;18:692–693. doi: 10.1128/am.18.4.692-693.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kast HR, Goodwin B, Tarr PT, Jones SA, Anisfeld AM, Stoltz CM, et al. Regulation of multidrug resistance-associated protein 2 (ABCC2) by the nuclear receptors pregnane X receptor, farnesoid X-activated receptor, and constitutive androstane receptor. J Biol Chem. 2002;277:2908–2915. doi: 10.1074/jbc.M109326200. [DOI] [PubMed] [Google Scholar]
- Kneuer C, Honscha W, Gabel G, Honscha KU. Adaptive response to increased bile acids: induction of MDR1 gene expression and P-glycoprotein activity in renal epithelial cells. Pflugers Arch. 2007;454:587–594. doi: 10.1007/s00424-007-0235-1. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Sueyoshi T, Inoue K, Moore R, Negishi M. Cytoplasmic accumulation of the nuclear receptor CAR by a tetratricopeptide repeat protein in HepG2 cells. Mol Pharmacol. 2003;64:1069–1075. doi: 10.1124/mol.64.5.1069. [DOI] [PubMed] [Google Scholar]
- Kocarek TA, Shenoy SD, Mercer-Haines NA, Runge-Morris M. Use of dominant negative nuclear receptors to study xenobiotic-inducible gene expression in primary cultured hepatocytes. J Pharmacol Toxicol Methods. 2002;47:177–187. doi: 10.1016/S1056-8719(03)00002-9. [DOI] [PubMed] [Google Scholar]
- Laffitte BA, Kast HR, Nguyen CM, Zavacki AM, Moore DD, Edwards PA. Identification of the DNA binding specificity and potential target genes for the farnesoid X-activated receptor. J Biol Chem. 2000;275:10638–10647. doi: 10.1074/jbc.275.14.10638. [DOI] [PubMed] [Google Scholar]
- Lamba J, Lamba V, Schuetz E. Genetic variants of PXR (NR1I2) and CAR (NR1I3) and their implications in drug metabolism and pharmacogenetics. Curr Drug Metab. 2005;6:369–383. doi: 10.2174/1389200054633880. [DOI] [PubMed] [Google Scholar]
- Le Guellec C, Gaudet ML, Lamanetre S, Breteau M. Stability of rifampin in plasma: consequences for therapeutic monitoring and pharmacokinetic studies. Ther Drug Monit. 1997;19:669–674. doi: 10.1097/00007691-199712000-00011. [DOI] [PubMed] [Google Scholar]
- Li AP, Lu C, Brent JA, Pham C, Fackett A, Ruegg CE, et al. Cryopreserved human hepatocytes: characterization of drug-metabolizing enzyme activities and applications in higher throughput screening assays for hepatotoxicity, metabolic stability, and drug–drug interaction potential. Chem Biol Interact. 1999;121:17–35. doi: 10.1016/s0009-2797(99)00088-5. [DOI] [PubMed] [Google Scholar]
- Loretz LJ, Li AP, Flye MW, Wilson AG. Optimization of cryopreservation procedures for rat and human hepatocytes. Xenobiotica. 1989;19:489–498. doi: 10.3109/00498258909042288. [DOI] [PubMed] [Google Scholar]
- Marshak D. Strategies for Protein Purification and Characterization: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press: Plainview, NY; 1996. [Google Scholar]
- Meredith C, Scott MP, Renwick AB, Price RJ, Lake BG. Studies on the induction of rat hepatic CYP1A, CYP2B, CYP3A and CYP4A subfamily form mRNAs in vivo and in vitro using precision-cut rat liver slices. Xenobiotica. 2003;33:511–527. doi: 10.1080/0049825031000085960. [DOI] [PubMed] [Google Scholar]
- Miyamoto Y, Suzuki S, Nomura K, Enosawa S. Improvement of hepatocyte viability after cryopreservation by supplementation of long-chain oligosaccharide in the freezing medium in rats and humans. Cell Transplant. 2006;15:911–919. doi: 10.3727/000000006783981404. [DOI] [PubMed] [Google Scholar]
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- Nishimura M, Koeda A, Suzuki E, Kawano Y, Nakayama M, Satoh T, et al. Regulation of mRNA expression of MDR1, MRP1, MRP2 and MRP3 by prototypical microsomal enzyme inducers in primary cultures of human and rat hepatocytes. Drug Metab Pharmacokinet. 2006;21:297–307. doi: 10.2133/dmpk.21.297. [DOI] [PubMed] [Google Scholar]
- Nishimura M, Yoshitsugu H, Naito S, Hiraoka I. Evaluation of gene induction of drug-metabolizing enzymes and transporters in primary culture of human hepatocytes using high-sensitivity real-time reverse transcription PCR. Yakugaku Zasshi. 2002;122:339–361. doi: 10.1248/yakushi.122.339. [DOI] [PubMed] [Google Scholar]
- Ogg MS, Williams JM, Tarbit M, Goldfarb PS, Gray TJ, Gibson GG. A reporter gene assay to assess the molecular mechanisms of xenobiotic-dependent induction of the human CYP3A4 gene in vitro. Xenobiotica. 1999;29:269–279. doi: 10.1080/004982599238669. [DOI] [PubMed] [Google Scholar]
- Owen A, Chandler B, Back DJ, Khoo SH. Expression of pregnane-X-receptor transcript in peripheral blood mononuclear cells and correlation with MDR1 mRNA. Antivir Ther. 2004;9:819–821. [PubMed] [Google Scholar]
- Owen A, Goldring C, Morgan P, Park BK, Pirmohamed M. Induction of P-glycoprotein in lymphocytes by carbamazepine and rifampicin: the role of nuclear hormone response elements. Br J Clin Pharmacol. 2006;62:237–242. doi: 10.1111/j.1365-2125.2006.02587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascussi JM, Drocourt L, Fabre JM, Maurel P, Vilarem MJ. Dexamethasone induces pregnane X receptor and retinoid X receptor-alpha expression in human hepatocytes: synergistic increase of CYP3A4 induction by pregnane X receptor activators. Mol Pharmacol. 2000a;58:361–372. doi: 10.1124/mol.58.2.361. [DOI] [PubMed] [Google Scholar]
- Pascussi JM, Gerbal-Chaloin S, Fabre JM, Maurel P, Vilarem MJ. Dexamethasone enhances constitutive androstane receptor expression in human hepatocytes: consequences on cytochrome P450 gene regulation. Mol Pharmacol. 2000b;58:1441–1450. doi: 10.1124/mol.58.6.1441. [DOI] [PubMed] [Google Scholar]
- Pfrunder A, Gutmann H, Beglinger C, Drewe J. Gene expression of CYP3A4, ABC-transporters (MDR1 and MRP1-MRP5) and hPXR in three different human colon carcinoma cell lines. J Pharm Pharmacol. 2003;55:59–66. doi: 10.1111/j.2042-7158.2003.tb02434.x. [DOI] [PubMed] [Google Scholar]
- Phillips A, Hood SR, Gibson GG, Plant NJ. Impact of transcription factor profile and chromatin conformation on human hepatocyte CYP3A gene expression. Drug Metab Dispos. 2005;33:233–242. doi: 10.1124/dmd.104.001461. [DOI] [PubMed] [Google Scholar]
- Reinach B, de Sousa G, Dostert P, Ings R, Gugenheim J, Rahmani R. Comparative effects of rifabutin and rifampicin on cytochromes P450 and UDP-glucuronosyl-transferases expression in fresh and cryopreserved human hepatocytes. Chem Biol Interact. 1999;121:37–48. doi: 10.1016/s0009-2797(99)00089-7. [DOI] [PubMed] [Google Scholar]
- Ripp SL, Mills JB, Fahmi OA, Trevena KA, Liras JL, Maurer TS, et al. Use of immortalized human hepatocytes to predict the magnitude of clinical drug–drug interactions caused by CYP3A4 induction. Drug Metab Dispos. 2006;34:1742–1748. doi: 10.1124/dmd.106.010132. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Antona C, Donato MT, Boobis A, Edwards RJ, Watts PS, Castell JV, et al. Cytochrome P450 expression in human hepatocytes and hepatoma cell lines: molecular mechanisms that determine lower expression in cultured cells. Xenobiotica. 2002;32:505–520. doi: 10.1080/00498250210128675. [DOI] [PubMed] [Google Scholar]
- Sambuy Y, De Angelis I, Ranaldi G, Scarino ML, Stammati A, Zucco F. The Caco-2 cell line as a model of the intestinal barrier: influence of cell and culture-related factors on Caco-2 cell functional characteristics. Cell Biol Toxicol. 2005;21:1–26. doi: 10.1007/s10565-005-0085-6. [DOI] [PubMed] [Google Scholar]
- Schrenk D, Baus PR, Ermel N, Klein C, Vorderstemann B, Kauffmann HM. Up-regulation of transporters of the MRP family by drugs and toxins. Toxicol Lett. 2001;120:51–57. doi: 10.1016/s0378-4274(01)00306-x. [DOI] [PubMed] [Google Scholar]
- Shah YM, Ma X, Morimura K, Kim I, Gonzalez FJ. Pregnane X receptor activation ameliorates DSS-induced inflammatory bowel disease via inhibition of NF-kappaB target gene expression. Am J Physiol Gastrointest Liver Physiol. 2007;292:1114–1122. doi: 10.1152/ajpgi.00528.2006. [DOI] [PubMed] [Google Scholar]
- Shibata Y, Takahashi H, Chiba M, Ishii Y. Prediction of hepatic clearance and availability by cryopreserved human hepatocytes: an application of serum incubation method. Drug Metab Dispos. 2002;30:892–896. doi: 10.1124/dmd.30.8.892. [DOI] [PubMed] [Google Scholar]
- Stoscheck C. Quantitation of protein. Methods in Enzymol. 1990;182:50–69. doi: 10.1016/0076-6879(90)82008-p. [DOI] [PubMed] [Google Scholar]
- Swales K, Plant N, Ayrton A, Hood S, Gibson G. Relative receptor expression is a determinant in xenobiotic-mediated CYP3A induction in rat and human cells. Xenobiotica. 2003;33:703–716. doi: 10.1080/0049825031000121626. [DOI] [PubMed] [Google Scholar]
- Synold TW, Dussault I, Forman BM. The orphan nuclear receptor SXR coordinately regulates drug metabolism and efflux. Nat Med. 2001;7:584–590. doi: 10.1038/87912. [DOI] [PubMed] [Google Scholar]
- Szabo S, Komblos S, Ignjatovic Z. Effect of pregnenolone-16alpha-carbonitrile (PCN) on drug response in man. J Pharm Pharmacol. 1975;27:113–118. doi: 10.1111/j.2042-7158.1975.tb09418.x. [DOI] [PubMed] [Google Scholar]
- Terry C, Dhawan A, Mitry RR, Lehec SC, Hughes RD. Preincubation of rat and human hepatocytes with cytoprotectants prior to cryopreservation can improve viability and function upon thawing. Liver Transpl. 2005;11:1533–1540. doi: 10.1002/lt.20503. [DOI] [PubMed] [Google Scholar]
- Vermeir M, Annaert P, Mamidi RN, Roymans D, Meuldermans W, Mannens G. Cell-based models to study hepatic drug metabolism and enzyme induction in humans. Expert Opin Drug Metab Toxicol. 2005;1:75–90. doi: 10.1517/17425255.1.1.75. [DOI] [PubMed] [Google Scholar]
- Wang H, Faucette S, Sueyoshi T, Moore R, Ferguson S, Negishi M, et al. A novel distal enhancer module regulated by pregnane X receptor/constitutive androstane receptor is essential for the maximal induction of CYP2B6 gene expression. J Biol Chem. 2003;278:14146–14152. doi: 10.1074/jbc.M212482200. [DOI] [PubMed] [Google Scholar]
- Willson TM, Jones SA, Moore JT, Kliewer SA. Chemical genomics: functional analysis of orphan nuclear receptors in the regulation of bile acid metabolism. Med Res Rev. 2001;21:513–522. doi: 10.1002/med.1023. [DOI] [PubMed] [Google Scholar]
- Xie W, Barwick JL, Simon CM, Pierce AM, Safe S, Blumberg B, et al. Reciprocal activation of xenobiotic response genes by nuclear receptors SXR/PXR and CAR. Genes Dev. 2000;14:3014–3023. doi: 10.1101/gad.846800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yague E, Armesilla AL, Harrison G, Elliott J, Sardini A, Higgins CF, et al. P-glycoprotein (MDR1) expression in leukemic cells is regulated at two distinct steps, mRNA stabilization and translational initiation. J Biol Chem. 2003;278:10344–10352. doi: 10.1074/jbc.M211093200. [DOI] [PubMed] [Google Scholar]
- Zollner G, Fickert P, Fuchsbichler A, Silbert D, Wagner M, Arbeiter S, et al. Role of nuclear bile acid receptor, FXR, in adaptive ABC transporter regulation by cholic and ursodeoxycholic acid in mouse liver, kidney and intestine. J Hepatol. 2003;39:480–488. doi: 10.1016/s0168-8278(03)00228-9. [DOI] [PubMed] [Google Scholar]