Abstract
The assembly and composition of human excision nuclease were investigated by electrophoretic mobility shift assay and DNase I footprinting. Individual repair factors or any combination of up to four repair factors failed to form DNA–protein complexes of high specificity and stability. A stable complex of high specificity can be detected only when XPA/RPA, transcription factor IIH, XPC⋅HHR23B, and XPG and ATP are present in the reaction mixture. The XPF⋅ERCC1 heterodimer changes the electrophoretic mobility of the DNA–protein complex formed with the other five repair factors, but it does not confer additional specificity. By using proteins with peptide tags or antibodies to the repair factors in electrophoretic mobility shift assays, it was found that XPA, replication protein A, transcription factor IIH, XPG, and XPF⋅excision repair cross-complementing 1 but not XPC⋅HHR23B were present in the penultimate and ultimate dual incision complexes. Thus, it appears that XPC⋅HHR23B is a molecular matchmaker that participates in the assembly of the excision nuclease but is not present in the ultimate dual incision complex. The excision nuclease makes an assymmetric DNase I footprint of ≈30 bp around the damage and increases the DNase I sensitivity of the DNA on both sides of the footprint.
Keywords: damage recognition, molecular matchmaker, xeroderma pigmentosum
Human excision nuclease is the enzymatic activity resulting from concerted action of 14–16 polypeptides in six repair factors (1); it makes dual incisions bracketing the lesion in the damaged strand and releases the damage in the form of 24–32 nt-long oligomers (2). Recently, the enzyme system has been reconstituted from highly purified subunits consisting of XPA, replication protein A (RPA), transcription factor IIH (TFIIH), XPC⋅HHR23B, XPG, and XPF⋅ERCC1 repair factors (3–5). By using substrates with single lesions at predetermined sites and either purified repair factors (6) or cell extracts from wild-type and mutant cell lines (7), it was found that the dual incision event is preceded by unwinding of DNA by 20–25 bp by the bidirectional helicase activity of the TFIIH subunit. The study with purified proteins identified three reaction intermediates, named preincision complexes 1, 2, and 3 (PIC1, 2, and 3) on the pathway to dual incisions (6). These studies have provided considerable insight into the reaction mechanism of human excision nuclease. However, two important issues regarding the mechanism have remained unresolved: (i) Is there a damage recognition factor as such, and, if there is, how does it recruit the other repair factors to the site of damage? (ii) Do all of the repair factors assemble together to form a complex capable of performing dual incisions or do some of the factors function as molecular matchmakers (8), that is, help in the assembly of the enzyme but are absent in the ultimate dual incision complex?
By using randomly damaged DNA as substrate and a variety of methods including filter binding and gel retardation assays for detecting DNA–protein complexes, it has been shown that RPA (9, 10), XPA (11, 12), the combination of XPA and RPA (13, 14), and XPC (15) bind with moderately higher affinity to damaged DNA compared with undamaged DNA. However, there are no reports on the interactions of these proteins with DNA fragments containing a single lesion. Hence, the designation of any of these proteins or combinations thereof as “the damage recognition factor” of human excinuclease must be considered provisional. Similarly, although it has been shown conclusively that all six factors are required for dual incisions (3, 4) and that five factors but not XPC⋅HHR23B are required for excision of a lesion adjacent to or within a mismatch “bubble” (6, 16), the composition of the reaction intermediate of the excision reaction has not been determined.
In this study, we have identified the minimum set of repair factors required for formation of a high affinity and specificity DNA–protein complex and physically have separated this complex from the free DNA and proteins in the reaction mixture by electrophoresis on nondenaturing polyacrylamide gels. Physical separation, in turn, has enabled us to probe the protein composition of the complex by “supershift” assays and the region of the substrate in contact with the proteins by DNase I footprinting. Our data show that XPA, RPA, TFIIH, XPC⋅HHR23B, and XPG are required for high specificity DNA–protein complex formation and that XPC⋅HHR23B is a molecular matchmaker that is not present in the ultimate dual incision complex that covers an ≈30-bp region of DNA around the lesion.
MATERIALS AND METHODS
Substrates.
The substrate used in most of our binding experiments was a 136-bp duplex containing a (6–4) photoproduct in the center. The sequence and preparation of this substrate have been described (17). The duplex contained 32P radiolabel either at the 5′ terminus of the damaged or complementary strand or at the 4th phosphodiester bond 5′ to the (6–4) photoproduct. The substrate with a 10-nt “bubble” 5′ to a cyclobutane thymine dimer, which is referred to as T<>T (5′-10), was prepared as described elsewhere (6).
Repair Factors.
Recombinant RPA (18), MBP-XPA (19), (His)6–XPA (4), XPC and XPC⋅HHR23B (15), XPF⋅ERCC1 (20), and XPG (21) were purified as described. The TFIIH was purified from HeLa cells by the method of Mu et al. (4) and contained eight subunits. In certain experiments, rat TFIIH of high purity (22) was substituted for the human TFIIH.
Antibodies.
Antibodies against the p70 and p34 subunits of RPA were obtained from Oncogene Science, and antibody against the p62 subunit of TFIIH was purchased from Santa Cruz Biotechnology. The polyclonal and monoclonal XPB antibodies were as described (23, 24). mAb against XPC was provided by Eva Lee (University of Texas, San Antonio, TX).
Electrophoretic Mobility Shift Assay.
DNA (3 fmol) and proteins at the indicated concentrations (XPA, 50 ng; RPA, 300 ng; TFIIH, 300 ng; XPC⋅HHR23B, 10 ng; XPG, 10 ng) were incubated in 25 μl of excision buffer containing 30 mM Hepes⋅KOH (pH 7.9), 50 mM KCl, 4 mM MgCl2, 0.2 mM EDTA, 0.1 mM DTT, and 2 mM ATP. Incubation was at 30°C for 30 min. Then, the sample was loaded directly onto a 0.25 × 20 × 40-cm polyacrylamide gel (3.5% polyacrylamide) in 0.5 × TBE (25 mM Tris⋅borate, pH 7.9/0.6 mM EDTA) containing 1 mM ATP and 10 mM MgCl2. Electrophoresis was carried out in 0.5 × TBE containing 1 mM ATP and 10 mM MgCl2 at 4°C and 150 V for 16–18 h. DNA–protein complexes were visualized by autoradiography. In reactions containing the XPF⋅ERCC1 complex, the DNA was first incubated with the other repair factors at 30°C for 30 min; then, the mixture was put on ice, XPF⋅ERCC1 was added to the desired amount (20 ng) and, after 10 min of incu- bation on ice, the sample was loaded onto the polyacrylamide gel.
DNase I Footprinting.
DNA plus protein mixtures were prepared as described above, and then CaCl2 was added to 4 mM followed by 0.002 units of DNaseI from Life Technologies (Grand Island, NY). The mixture was incubated at room temperature for 3 min and then loaded onto a 3.5% nondenaturing polyacrylamide gel to separate free DNA from DNA–protein complexes. The free and protein-bound DNA were located by autoradiography, the bands were excised, the DNAs were eluted from polyacrylamide matrix, and then the DNAs with equal amount of radioactivities were analyzed on 6% denaturing polyacrylamide gels along with Maxam-Gilbert G+A chemical sequence ladder.
RESULTS
Lack of High Specificity Damage Recognition by Individual Repair Factors.
Damage-specific binding has been reported for RPA (9, 10), XPA (11), and XPC⋅HHR23B (15). However, the discrimination between damaged and undamaged DNA reported in these studies was modest. Furthermore, these studies were conducted with randomly damaged DNAs and as a consequence had additional limitations vis-a-vis the affinity of a given protein for a given type of lesion and the ability of the repair protein to discriminate between damaged and undamaged DNA. Such data can be obtained most unambiguously by investigating the interaction of purified proteins with a pure substrate, that is, a substrate that contains a known lesion at a known site (25). The DNA–protein complexes that form under these conditions can be characterized qualitatively by DNase I footprinting and quantitatively by electrophoretic mobility shift assay (gel retardation). Repeated attempts to obtain conventional DNase I footprints of XPA, RPA, TFIIH, XPC⋅HHR23B, XPG, and XPF⋅ERCC1 or various pairwise combinations of these repair factors with various substrates failed to elicit specific DNA–protein complexes. Hence, we decided to search for specific DNA–protein complexes by the gel retardation assay.
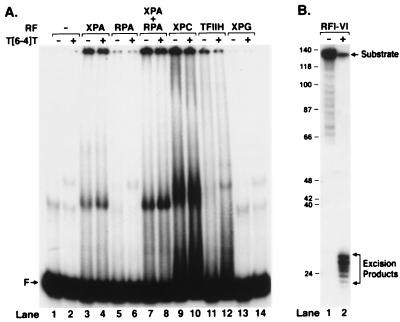
Fig. 1A shows the results of a gel retardation assay performed with five excision repair factors individually and with the XPA + RPA combination that has been reported to confer higher specificity than either factor alone (13, 14). The concentrations of the factors were chosen such that the combination of all six repair factors yielded high efficiency of excision (Fig. 1B). With a 136-mer containing a centrally located (6–4) photoproduct as a probe, XPA has a modestly higher (1.9-fold) affinity to damaged DNA than to undamaged DNA (Fig. 1, lanes 3 and 4); RPA does not have measurable affinity to either probe under our assay conditions (Fig. 1, lanes 5 and 6), but the presence of RPA in the XPA + DNA mixture increases the fraction of both nonspecific (Fig. 1, lane 7) and specific (Fig. 1, lane 8) DNA–protein complexes without affecting specificity. Of interest, the DNA–protein complexes that form with XPA + RPA have the same mobility as the complex formed with XPA alone (compare Fig. 1 lanes 3 and 4 with 7 and 8). However, because the XPA + RPA combination did not increase the specificity, the composition of the complex that forms with these two factors was not investigated any further. In agreement with a previous report (15), XPC⋅HHR23B also bound (6–4) photoproduct-containing DNA with a slightly higher affinity than undamaged DNA (Fig. 1, lanes 9 and 10). However, again, the discrimination between undamaged and damaged DNA was not sufficiently high to qualify the XPC⋅HHR23B protein as “the damage recognition subunit” of human excinuclease. Finally, neither TFIIH nor XPG bound to DNA under the conditions used in our experiment (lanes 11–14).
Figure 1.
Damage-specific binding and excision. (A) Binding of human excision repair factors to damaged DNA. Repair factors were incubated for 30 min with either a 136-bp control DNA or a duplex containing (6–4) photoproduct and were analyzed on a nondenaturing gel (lanes 1–14). (B) Excision reaction by repair factors used in binding assay. The (6–4) substrate was incubated for 2 h with the five repair factors (RFI–V: XPA, RPA, TFIIH, XPC, XPG) at the concentration used in binding experiment and XPF⋅ERCC1 (RFVI). Excision products were analyzed on 8% denaturing polyacrylamide gel. Quantitative analysis shows that 93% of the damage was excised.
Formation of a High Specificity DNA–Protein Complex with Human Excinuclease.
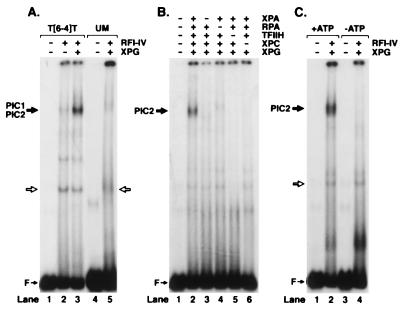
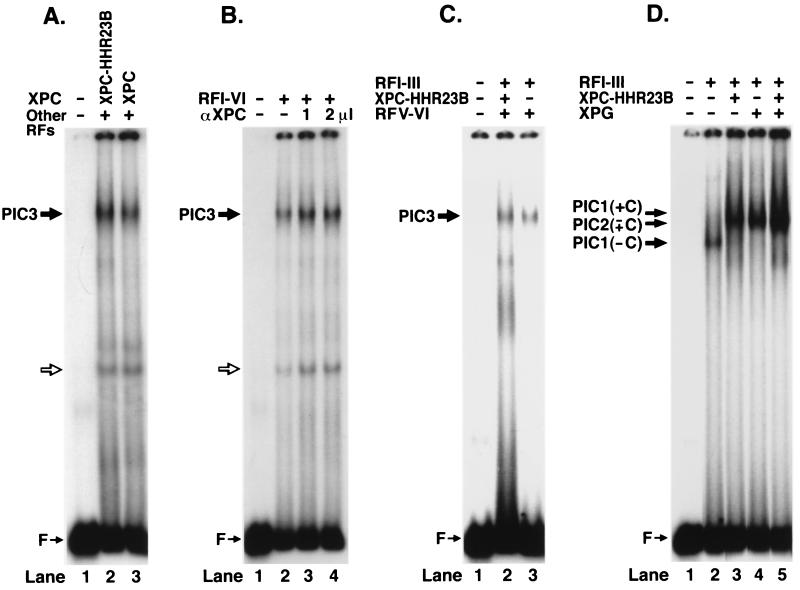
Experiments with all combinations of two and three repair factors failed to improve the specificity of binding in the gel retardation assay. In contrast, the four repair factors XPA, RPA, TFIIH, and XPC⋅HHR23B, known from permanganate footprinting experiments to unwind DNA around the lesion and hence to make a specific complex called PIC1 (6), did indeed form a slow-migrating DNA–protein band with the (6–4) photoproduct substrate (Fig. 2A, lane 2). However, the DNA within this complex was a minor fraction of input DNA indicating that the complex was rather unstable. Addition of XPG to the reaction mixture greatly increased the fraction of the DNA in the complex (Fig. 2, lane 3), again in agreement with the results of permanganate footprinting, which suggested that XPG greatly stabilizes the DNA–protein complex formed at the lesion site by XPA, RPA, TFIIH, and XPC⋅HHR23B proteins converting it to PIC2 (6). Important to note, no detectable complex is formed when unmodified DNA is used in gel retardation experiments (Fig. 2, lane 5) even in the presence of XPG. Thus, the damage specific complex we have identified with human excision nuclease (Fig. 2, lane 3) differs from all previously reported excision repair factor protein–DNA complexes in degree of its selectivity because the ratio of damage-specific to nonspecific binding with five repair factors is ≈50 (Fig. 2, lanes 3 and 5). Furthermore, the formation of this high specificity complex depends on all five repair factors; as can be seen from Fig. 2B, omission of any of the four factors, XPA, RPA, TFIIH, or XPC⋅HHR23B, completely abolishes specific complex formation. In comparison, XPG omission greatly destabilizes the complex without totally eliminating binding (compare Fig. 2A, lanes 2 and 3, with Fig. 2B, lanes 2–6). Finally, as expected, the formation of the damaged DNA–human excision nuclease complex has an absolute requirement for ATP (Fig. 2C, lanes 2 and 4), in agreement with the permanganate probing experiments that revealed specific unwinding of damaged DNA only in the presence of ATP (6).
Figure 2.
Detection of preincision complexes 1 and 2 by electrophoretic mobility shift assay. (A) Weak PIC1 forms with RFI-IV (XPA, RPA, TFIIH, and XPC) (lane 2), which is stabilized by XPG to form PIC2 (lane 3). No PIC2 can be detected with unmodified DNA (lane 5). (B) Formation of PIC2 requires all five repair factors. (C) PIC2 formation is ATP-dependent. Reaction mixtures were incubated in the presence or absence of ATP and separated on a 3.5% nondenaturing polyacrylamide gel as indicated. Open arrow shows the binding by a minor contaminant in the TFIIH factor.
Binding of XPF⋅ERCC1 to the Specific DNA–Protein Complex.
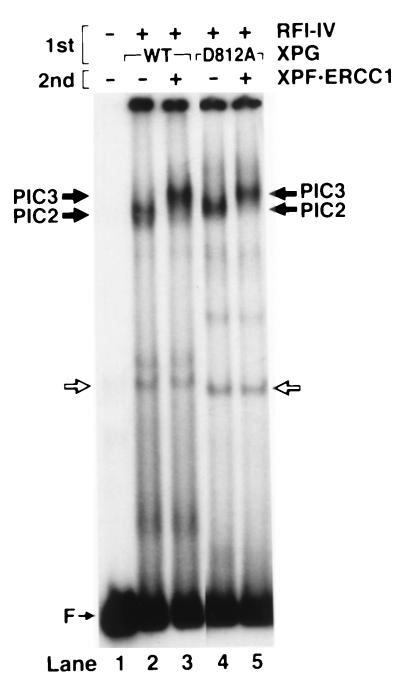
The data presented so far are consistent with the formation of a specific excision nuclease complex with five of the six repair factors that constitute the human excinuclease. If the damaged DNA–protein complex we detect is an intermediate on the pathway for dual incisions then it is expected to bind to the sixth repair factor, the XPF⋅ERCC1 heterodimer. This factor is the nuclease that makes the 5′ incision (21, 26), and kinetic experiments indicate that it enters the excinuclease complex last (4, 6). Hence, we decided to investigate the effect of XPF⋅ERCC1 on the electrophoretic mobility of the complex that forms with XPA, RPA, TFIIH, XPC⋅HHR23B, and XPG and that we refer to as PIC2.
Because incubation of substrate with the six repair factors leads to dual incisions and disassembly of the enzyme (4), we conducted these experiments under conditions not conducive to dual incisions. We used two experimental approaches for this purpose. In one, PIC2 was assembled at 30°C then put on ice, XPF⋅ERCC1 was added to the mixture, and after another 10-min incubation at this temperature the sample was loaded onto a gel. Fig. 3 (lanes 2 and 3) shows that XPF⋅ERCC1 does associate with PIC2 and causes further retardation (“supershift”) of the complex. In accordance with the nomenclature used previously (6), we refer to the complex that forms with all six repair factors PIC3. In the second experimental approach, we used an active site XPG mutant in the initial assembly reaction. Previously, we showed that association of XPF⋅ERCC1 with PIC2 was independent of the 3′ nuclease activity of XPG (6, 27). Indeed, as probed by electrophoretic mobility shift assay, the XPG active site mutant XPG-D812A forms PIC2 with the same efficiency as the wild-type XPG. Similarly, the PIC2 with the mutant XPG protein is supershifted by XPF⋅ERCC1 complex (Fig. 3, lanes 4 and 5). These data taken together with previous studies on the role of XPG in assembly (6, 27) are consistent with the notion that the damage-specific complex formed by the five repair factors (PIC2) is on the pathway to dual incision as is the complex (PIC3) that forms upon addition of the sixth repair factor, the XPF⋅ERCC1 heterodimer.
Figure 3.
Detection of preincision complex 3 by electrophoretic mobility shift assay. The substrate was incubated with RFI-IV (XPA, RPA, TFIIH, XPC) and either wild-type or mutant XPG at 30°C to form PIC2 (1st incubation), then XPF⋅ERCC1 was added on ice as indicated (second incubation), and the DNA–protein complexes were analyzed by electrophoretic mobility shift assay.
Subunit Composition of Human Excinuclease.
Previous studies on assembly of human excinuclease did not address the question of protein composition of preincision complexes. There are two possible scenarios. In one model, one or more of the repair factors act as molecular matchmakers, which in an ATP-dependent manner promote the formation of the dual incision complex but are not part of this complex (8). In the second scenario, the repair factors assemble in a preordained sequence and all are present in the ultimate, dual incision complex.
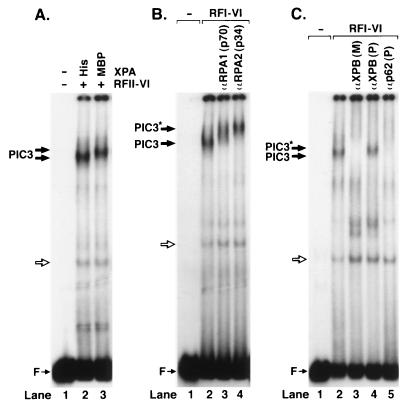
To differentiate between these two models, we probed the composition of PIC3 by using protein tags and antibodies to the repair factors in electrophoretic mobility supershift experiments. (i) We tested for the presence of XPA by comparing the migration of PIC3s formed with (His)6–XPA and MBP-XPA. Fig. 4A shows that PIC3 formed with the two forms of XPA have different mobilities, which proves that XPA is in the ultimate incision complex. (ii) Similarly, Fig. 4B shows that PIC3 is supershifted by antibodies against the p70 and p34 subunits of RPA, thus revealing that RPA also is in the incision complex. (iii) TFIIH also is in the complex because polyclonal antibodies against the XPB subunits supershift PIC3, and monoclonal and polyclonal antibodies against the XPB and p62 subunits disrupt PIC3 (Fig. 4B). (iv--v) Physical presence of XPG and XPF⋅ERCC1 is evident from the fact that incubating the complex at 30°C leads to dual incisions.
Figure 4.
Detection of repair factors in PIC3. (A) The PIC3 formed with MBP-XPA migrates slower than the complex formed with smaller (His)6–XPA. (B) Antibodies to p70 and p34 subunits of RPA supershift PIC3. (C) Polyclonal antibodies to the XPB subunit of TFIIH supershift (lane 4); monoclonal XPB and polyclonal p62 antibodies disrupt PIC3 (lanes 3 and 5). PIC3* indicates supershifted PIC3.
XPC as a Molecular Matchmaker.
Preliminary experiments suggested that XPC⋅HHR23B may not be present in PIC2 and PIC3. Because this was a rather unexpected result, we probed for XPC in these complexes by three different methods. First, we compared the electrophoretic mobilities of PIC3 with two forms of XPC. Under physiological conditions, nearly all of XPC is in a complex with HHR23B in the form of XPC⋅HHR23B heterodimer (28). However, it was found that XPC alone was as active as (15) or slightly less active (29) than the heterodimer in reconstituting human excinuclease. Thus, to find out whether XPC is present in PIC3, we used either the heterodimer or the XPC monomer in the reaction and compared the mobility of the two complexes. Fig. 5A shows that the PIC3s formed in the presence of these two forms of the XPC factor have the same mobility. However, the molecular mass of HHR23B is only 58 kDa (28), and hence it is conceivable that, even if XPC were part of PIC3, the effect of HHR23B on migration of a complex of ≈800–900 kDa would not be as pronounced as it is on the XPC⋅DNA complex alone (15) in our system, even though the effect of MBP (≈40 kDa) could be detected as shown in Fig. 4A. Hence, we tested for the presence of XPC in PIC3 by using anti-XPC antibodies in a supershift experiment. Fig. 5B shows that XPC antibodies do not alter the electrophoretic migration of PIC3 suggesting that XPC is absent from PIC3. This provisional conclusion was supported by experiments performed with T<>T (5′-10) substrate, which contains a 10-nt mismatch (“bubble”) 5′ to a cyclobutane thymine dimer. Previous work has shown that the T<>T can be excised from this substrate by human excinuclease reconstituted in the presence or absence of XPC⋅HHR23B (6). Hence, this substrate was used in electrophoretic mobility shift assay with excinuclease reconstituted with or without XPC⋅HHR23B. Fig. 5C shows that, although there is some quantitative difference in the level of PIC3 formed, the complexes formed under the two conditions co-migrate, again consistent with the notion that XPC is absent from PIC3. However, we were still concerned about whether the mass of XPC⋅HHR23B was of sufficient magnitude to change the mobility of PIC3 in our system. To address this point, we tested the effect of XPC⋅HHR23B and XPG (which is comparable in mass to XPC⋅HHR23B) on mobility of PIC1, which was formed without XPC. The result is shown in Fig. 5D; XPC⋅HHR23B (lane 3) and XPG (lane 4) supershift PIC1 formed without XPC⋅HHR23B (lane 2); however, the combination of the two does not cause further retardation (lane 5). This result is consistent with the notion that the entry of XPG to the preincision complex is coincident with departure of XPC⋅HHR23B from the complex. Thus, all of the data presented in Fig. 5 constitute strong evidence that XPC⋅HHR23B is a molecular matchmaker that does not participate in the dual incision step of excinuclease.
Figure 5.
Evidence for lack of XPC in PIC3. (A) PIC3s formed with either XPC⋅HHR23B or XPC have the same electrophoretic mobility. (B) Anti-XPC antibodies do not supershift PIC3. (C and D) Band mobility shift assays with T<>T(5′-10) “bubble” substrate. (C) PIC3s formed with (lane 2) and without (lane 3) XPC⋅HHR23B have the same electrophoretic mobility. (D) XPC⋅HHR23B (lane 3) and XPG (lane 4) cause supershift in PIC1 formed without XPC⋅HHR23B, but the combination of the two does not cause further retardation (lane 5).
Footprint of Human Excinuclease.
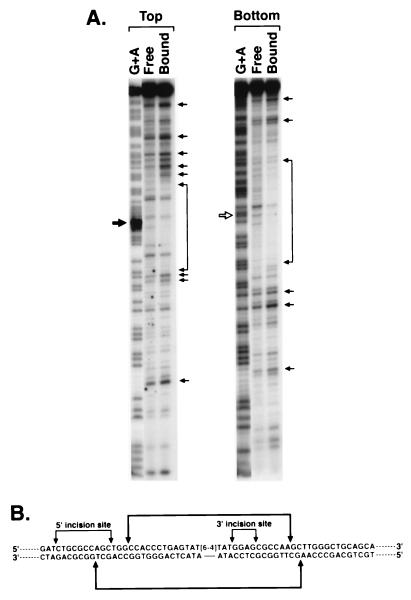
Previous work has shown that ≈50 bp on the 5′ side and 30 bp on the 3′ side of damage are required for optimal functioning of human excinuclease (30) and provides an insight into the approximate extent of DNA–protein interactions that would be expected in excinuclease–substrate complex. To better define the DNA–protein interactions, we performed DNase I footprinting experiments on PIC2, which is the first high specificity and high stability complex detectable with the human excinuclease. Even under optimum conditions for excision most of the DNA is unbound (see Figs. 1 and 2), so it is impossible to obtain footprint with the conventional DNase I footprinting method. Similarly, our repeated attempts to perform in situ footprinting on the retarded band in gel were unsuccessful presumably because during the manipulations necessary for this method PIC2 disassembles. Hence, we conducted footprinting subjecting the complex to DNase I before gel electrophoresis. The DNase I-treated reaction mixtures containing PIC2 were separated on nondenaturing polyacrylamide gels, and DNAs in free and bound fractions were eluted and analyzed on sequencing gels. Fig. 6 shows that the DNase I-protected region does not extend beyond 20 nt 5′ and 15 nt 3′ to the damage. Furthermore, it appears that the DNA outside of the protected region is largely hypersensitive to DNase I. It is possible that this hypersensitivity is caused by wrapping of DNA around the proteins in the PIC2 complex. Significantly, the DNA near the 5′ incision site is hypersensitive to DNase I, consistent with minor groove widening caused by kinking of DNA in this region.
Figure 6.
DNase I footprint of PIC2. The 136-bp substrate with 5′ label either in the strand containing the (6–4) photoproduct (top strand) or in the complementary strand (bottom strand) was incubated with the five repair factors (no XPF⋅ERCC1) and digested with DNase I, and then bound and unbound fractions were separated on nondenaturing polyacrylamide gels; DNA was eluted from the free and bound fractions and analyzed on 6% denaturing polyacrylamide gels. (A) Footprinting gels along with G+A sequence ladder. Solid and open arrows show the site of (6–4) photoproduct. Brackets and arrow indicate protected region and prominent hypersensitive sites. (B) Schematic illustration of footprint of human excinuclease indicating incision sites and protected region.
DISCUSSION
Excision repair in eukaryotes in general and in humans in particular has been investigated in considerable detail in recent years (1, 31, 32). These studies have led to reasonably detailed models for excision repair. However, two aspects of nucleotide excision repair in humans have remained ill-defined: damage recognition and the role of XPC⋅HHR23B.
Damage Recognition.
Substrates for human excinuclease cover the entire spectrum of damaged bases (2). Hence, specific chemical groups on DNA cannot be determinants for recognition. It appears that any abnormal DNA structure, but in particular adducts that destabilize the helix, is recognized by the enzyme system (17, 33, 34). Attempts to assign the damage recognition function to specific repair factors have had limited success, which is in part due to the fact that all six repair factors have affinity for DNA and in part due to the finding that those that have been reported to discriminate between damaged and undamaged DNA, XPA (11, 12), RPA (9, 10), and XPC⋅HHR23B (15) do not exhibit high enough specificity to account for the capacity of the human excinuclease system to find the rare lesions within ≈1010 bp of human diploid DNA. It has been reported that the combination of XPA and RPA improves specificity (13, 14). However, as can be seen in Fig. 1, the improvement in selectivity is modest and cannot account for the specificity of excision nuclease. Similarly, it has been reported that XPA helps recruit TFIIH to the damage site and thus promote a more stable complex (35, 36). However, permanganate footprinting has shown that the XPA + TFIIH combination alone does not lead to unwinding around the damage (6), and with the defined substrates used in this study, we failed to detect specific XPA⋅TFIIH⋅DNA complexes. Instead, the first high specificity complex we can detect with either the permanganate footprinting or gel retardation assay requires the XPA+RPA+ XPC⋅HHR23B+TFIIH combination, and the specificity and stability of the complex are further increased by XPG. Hence, until the development of higher resolution data capable of showing whether these four to five factors arrive at the damage site sequentially or simultaneously, it might be prudent to refrain from referring to any of these factors as the damage recognition subunit. Clearly, XPA and RPA contribute by their affinities to DNA, TFIIH by unwinding DNA around the lesion and facilitating the entry of XPC⋅HHR23B into the complex, and XPC⋅HHR23B and XPG contribute to specificity by binding to TFIIH and the “bubble” structure created by TFIIH and to RPA (21) within the complex.
XPC as a Molecular Matchmaker.
XP-C mutant cells and XPC protein exhibit some unique features. First, XP-C cells have ≈20% residual repair activity, all of which appears to be due to normal level of repair of the template strand of transcribed genes (37, 38). Second, in in vitro reconstitution experiments, it was found that XPC is not required for excision of an artificial lesion (cholesterol moiety substituted for a base) expected to cause substantial unwinding (4). Finally, when a T<>T is adjacent to a 10-nt bubble or within, a 20-nt bubble excision occurs independent of XPC⋅HHR23B (6, 16). Of all basal factors for excision repair, it thus appears that XPC⋅HHR23B is the only factor that is dispensable under certain circumstances, in particular, under conditions that favor DNA unwinding. It is possible that XPC⋅HHR23B helps to position correctly the repair factors within and around unwound DNA and in the process of doing so must dissociate from the substrate for formation of a productive preincision complex. Significantly, it appears that, during assembly, XPC⋅HHR23B and XPG cannot exist in the complex simultaneously and that the entry of XPG into the complex coincides with XPC⋅HHR23B leaving the complex. Whether XPC⋅HHR23B plays a direct role in recruiting XPG cannot be ascertained from our data. It is interesting that, depending on the purification procedure used, either XPC⋅HHR23B (23) or XPG (3), but not both, co-purify with TFIIH through several chromatographic steps. In addition, our data in conjunction with a previous report (6) [that showed that unwinding of DNA around the lesion to form PIC2 required TFIIH and XPC and could be inhibited by the nonhydrolyzable ATP analog ATPγS (6)] show that the formation of PIC2 and PIC3 promoted by XPC⋅HHR23B depends on the ATPase function of TFIIH (22).
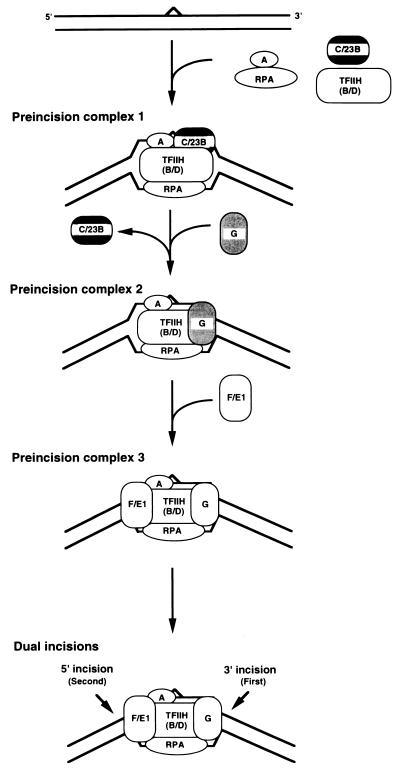
A molecular matchmaker is described as a protein that, in an ATP-dependent reaction, brings target DNA and an effector protein together, promotes a stable complex formation, and then dissociates from the complex to enable the effector molecule to engage in further protein–protein interactions or perform its catalytic function on target DNA (8). Thus, by all of these criteria, which originally were applied to the UvrA protein of Escherichia coli excinuclease (8, 39), XPC⋅HHR23B is a molecular matchmaker. Taking the molecular matchmaker function of XPC⋅HHR23B into account, we present the model shown in Fig. 7 as the current refinement of our understanding of the action mechanism of human excinuclease. This model differs from other models in two significant aspects. First, there is no “damage recognition” subunit as such. Several proteins acting together find the damage. Second, the entry of XPG into the complex coincides with the exit of XPC⋅HHR23B from the complex and hence XPC⋅HHR23B is absent from the dual incision complex.
Figure 7.
Model for reaction mechanism of human excinuclease. XPC⋅HHR23B is a molecular matchmaker that is not present in either the penultmate or the ultimate dual incision complex. A, B, and D, XPA, XPB, and XPD, respectively; C/23B, XPC⋅HHR23B, G, XPG; and F/E1, XPF⋅ERCC1 complex.
Acknowledgments
We thank Drs. T. Bessho, T. Matsunaga, D. Mu, J. T. Reardon, and C. P. Selby for providing the human excision repair factors, X. Zhao and T. Bessho for the site-specifically damaged oligonucleotide used in this study, and D. Mu and J. T. Reardon for useful discussions and critical comments on the manuscript. This work was supported by grant GM32833 from the National Institutes of Health.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: XP, xeroderma pigmentosum; RPA, replication protein A; TFIIH, transcription factor IIH; HHR23B, human homolog B of Rad23; ERCC, excision repair cross-complementing; PIC, preincision complex; RF, repair factor; MBP, maltose binding protein.
References
- 1.Sancar A. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 2.Huang J C, Svoboda D L, Reardon J T, Sancar A. Proc Natl Acad Sci USA. 1992;89:3664–3668. doi: 10.1073/pnas.89.8.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mu D, Park C H, Matsunaga T, Hsu D S, Reardon J T, Sancar A. J Biol Chem. 1995;270:2415–2418. doi: 10.1074/jbc.270.6.2415. [DOI] [PubMed] [Google Scholar]
- 4.Mu D, Hsu D S, Sancar A. J Biol Chem. 1996;271:8285–8294. doi: 10.1074/jbc.271.14.8285. [DOI] [PubMed] [Google Scholar]
- 5.Moggs J G, Yarema K J, Essigmann J M, Wood R D. J Biol Chem. 1996;271:7177–7186. doi: 10.1074/jbc.271.12.7177. [DOI] [PubMed] [Google Scholar]
- 6.Mu D, Wakasugi M, Hsu D S, Sancar A. J Biol Chem. 1997;272:28971–28979. doi: 10.1074/jbc.272.46.28971. [DOI] [PubMed] [Google Scholar]
- 7.Evans E, Fellows J, Coffer A, Wood R D. EMBO J. 1997;16:625–638. doi: 10.1093/emboj/16.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sancar A, Hearst J E. Science. 1993;259:1415–1420. doi: 10.1126/science.8451638. [DOI] [PubMed] [Google Scholar]
- 9.Clugston C K, McLauglin K, Kenny M K, Brown R. Cancer Res. 1992;52:6375–6379. [PubMed] [Google Scholar]
- 10.Burns J L, Guzder S N, Sung P, Prakash S, Prakash L. J Biol Chem. 1996;271:11607–11610. doi: 10.1074/jbc.271.20.11607. [DOI] [PubMed] [Google Scholar]
- 11.Jones C J, Wood R D. Biochemistry. 1993;32:12096–12104. doi: 10.1021/bi00096a021. [DOI] [PubMed] [Google Scholar]
- 12.Kuraoka I, Morita E H, Saijo M, Matsuda T, Morikawa K, Shirakawa M, Tanaka K. Mutation Res. 1996;362:87–95. doi: 10.1016/0921-8777(95)00038-0. [DOI] [PubMed] [Google Scholar]
- 13.He Z, Henricksen L A, Wold M S, Ingles C J. Nature (London) 1995;374:566–569. doi: 10.1038/374566a0. [DOI] [PubMed] [Google Scholar]
- 14.Li L, Lu X, Peterson C A, Legerski R J. Mol Cell Biol. 1995;15:5396–5402. doi: 10.1128/mcb.15.10.5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reardon J T, Mu D, Sancar A. J Biol Chem. 1996;271:19451–19456. doi: 10.1074/jbc.271.32.19451. [DOI] [PubMed] [Google Scholar]
- 16.Mu D, Sancar A. J Biol Chem. 1997;272:7570–7573. doi: 10.1074/jbc.272.12.7570. [DOI] [PubMed] [Google Scholar]
- 17.Mu D, Tursun M, Duckett D R, Drummond J T, Modrich P, Sancar A. Mol Cell Biol. 1997;17:760–769. doi: 10.1128/mcb.17.2.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henricksen L A, Umbricht C B, Wold M S. J Biol Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 19.Park C H, Sancar A. Nucleic Acids Res. 1993;21:5110–5116. doi: 10.1093/nar/21.22.5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bessho T, Sancar A, Thompson L A, Thelen M P. J Biol Chem. 1997;272:3833–3837. doi: 10.1074/jbc.272.6.3833. [DOI] [PubMed] [Google Scholar]
- 21.Matsunaga T, Park C H, Bessho T, Mu D, Sancar A. J Biol Chem. 1996;271:11047–11050. doi: 10.1074/jbc.271.19.11047. [DOI] [PubMed] [Google Scholar]
- 22.Dvir A, Conaway R C, Conaway J W. Proc Natl Acad Sci USA. 1997;94:9006–9010. doi: 10.1073/pnas.94.17.9006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drapkin R, Reardon J T, Ansari A, Huang J C, Zawel L, Ahn K, Sancar A, Reinberg D. Nature (London) 1994;368:769–772. doi: 10.1038/368769a0. [DOI] [PubMed] [Google Scholar]
- 24.Drapkin R, Roy G L, Cho H, Akoulitchev S, Reinberg D. Proc Natl Acad Sci USA. 1996;93:6488–6493. doi: 10.1073/pnas.93.13.6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reardon J T, Nichols A F, Keeney S, Smith C A, Taylor J-S, Linn S, Sancar A. J Biol Chem. 1993;268:21301–21308. [PubMed] [Google Scholar]
- 26.Matsunaga T, Mu D, Park C H, Reardon J T, Sancar A. J Biol Chem. 1995;270:20862–20869. doi: 10.1074/jbc.270.35.20862. [DOI] [PubMed] [Google Scholar]
- 27.Wakasugi M, Reardon J T, Sancar A. J Biol Chem. 1997;272:16030–16034. doi: 10.1074/jbc.272.25.16030. [DOI] [PubMed] [Google Scholar]
- 28.Masutani C, Sugasawa K, Yanagisawa J, Sonoyama T, Ui M, Enomoto T, Takio K, Tanaka K, van der Spek P J, Bootsma D, et al. EMBO J. 1994;13:1831–1843. doi: 10.1002/j.1460-2075.1994.tb06452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masutani C, Araki M, Sugasawa K, van der Spek P J, Yamada A, Uchida A, Maekawa T, Bootsma D, Hoeijmakers J H J, Hanaoka F. Mol Cell Biol. 1997;17:6915–6923. doi: 10.1128/mcb.17.12.6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang J C, Sancar A. J Biol Chem. 1994;269:19034–19040. [PubMed] [Google Scholar]
- 31.Guzder S N, Habraken Y, Sung P, Prakash L, Prakash S. J Biol Chem. 1995;270:12973–12976. doi: 10.1074/jbc.270.22.12973. [DOI] [PubMed] [Google Scholar]
- 32.Wood R D. Annu Rev Biochem. 1996;65:135–167. doi: 10.1146/annurev.bi.65.070196.001031. [DOI] [PubMed] [Google Scholar]
- 33.Gunz D, Hess M T, Naegeli H. J Biol Chem. 1996;271:25089–25098. doi: 10.1074/jbc.271.41.25089. [DOI] [PubMed] [Google Scholar]
- 34.Hess M T, Schwitter U, Petretta M, Giese B, Naegeli H. Proc Natl Acad Sci USA. 1997;94:6664–6669. doi: 10.1073/pnas.94.13.6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park C H, Mu D, Reardon J T, Sancar A. J Biol Chem. 1995;270:4896–4902. doi: 10.1074/jbc.270.9.4896. [DOI] [PubMed] [Google Scholar]
- 36.Nocentini S, Coin F, Saijo M, Tanaka K, Egly J M. J Biol Chem. 1997;272:22991–22994. doi: 10.1074/jbc.272.37.22991. [DOI] [PubMed] [Google Scholar]
- 37.Cleaver J E, Kraemer K H. In: The Metabolic Basis of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, editors. Vol. 2. New York: McGraw–Hill; 1989. pp. 2949–2971. [Google Scholar]
- 38.Venema J, van Hoffen A, Karcagi V, Natarajan A T, van Zeeland A A, Mullenders L H F. Mol Cell Biol. 1991;11:4128–4134. doi: 10.1128/mcb.11.8.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Orren D K, Selby C P, Hearst J E, Sancar A. J Biol Chem. 1992;267:780–788. [PubMed] [Google Scholar]