Abstract
Objective
To provide an updated overview of the methods used in genetic, transcriptomic and proteomic studies in Alzheimer’s disease and to demonstrate the importance of those methods for the improvement of the current diagnostic and therapeutic possibilities.
Data sources
Medline-based search of 234 peer-reviewed articles published between 1975 and 2006.
Data synthesis
Alzheimer’s disease is a genetically heterogeneous disorder. Rare mutations in the APP, presenilin 1, and presenilin 2 genes have shown the importance of the amyloid metabolism for its development. In addition, converging evidence from population-based genetic studies, gene expression studies and protein profile studies in the brain and in the cerebrospinal fluid suggest the existence of several pathogenetic pathways such as amyloid precursor protein processing, β-amyloid degradation, tau phosphorylation, proteolysis, protein misfolding, neuroinflammation, oxidative stress and lipid metabolism.
Conclusions
The development of high-throughput genotyping methods and of elaborated statistical analyses will contribute to the identification of genetic risk profiles related to the development and course of this devastating disease. The integration of knowledge derived from genetic, transcriptomic and proteomic studies will greatly advance our understanding of the causes of Alzheimer’s disease, it will improve our capability of establishing an early diagnosis, it will help defining disease subgroups and it will ultimately help to pave the road towards improved and tailored treatments.
I. Genetics of Alzheimer’s disease
Alzheimer’s disease (AD) is a neurodegenerative disorder, which preferentially affects individuals over 60 years of age with steadily increasing risk in older ages. The prevalence of AD in the general population increases from about 1% in persons younger than 65 years to about 40% in nonagenarians.1
Clinically, AD is characterized by progressive impairments in memory and other cognitive domains. With disease progression, non-cognitive symptoms such as delusions, agitation, changes in personality, and mood disturbances may also occur. Neuropathologically, AD is characterized by the presence of two histological hallmarks, neuritic plaques and neurofibrillary tangles. Aggregates of fibrillar β-amyloid peptide (Aβ) form the core of the neuritic plaques. The accumulation of Aβ (particularly the aggregated form of the protein containing 42 amino acids) has been strongly suggested to play a central pathophysiological role in the AD-related neurodegenerative cascade. The production of Aβ, which is derived from the amyloid precursor protein (APP), is under the control of the proteolytic activity of the alpha-, beta, and gamma-secretases. While the alpha-secretase cleavage site precludes the formation of Aβ, beta- and gamma-secretases generate amyloidogenic APP components.
Ia. Principles of AD genetics
AD is a multifactorial and genetically complex disorder. Several factors influence the risk for the development of AD and modify the age-at-onset and the course of the disease. These factors may be:
genetic (e.g. causative mutations, predisposing risk alleles, protective alleles)
sociodemographic (e.g. level of education, intelligence)
life style (e.g. aspects of nutrition, aerobic fitness, and mental exercise)
environment (e.g. head trauma)
clinical (e.g. comorbid medical conditions)
medications (e.g. non-steroidal anti-inflammatory drugs, statins)
Of these factors, genetic influences seem to be of major importance: twin studies suggest that about 74% of the risk for late-onset AD (i.e. onset after the 65th year) is genetic.2
Modes of inheritance
From a genetic point of view, AD may be subdivided into three forms according to the observed mode of inheritance within families:
autosomal-dominant familial AD
familial AD without clear mendelian inheritance (familial aggregation)
sporadic AD without familial aggregation
Only a minority of all AD cases may be fully explained by the presence of genetic factors (autosomal dominant AD). Most of these cases are caused by mutations in the genes encoding the amyloid precursor protein (APP), presenilin 1 (PSEN1), and presenilin 2 (PSEN2). Several studies demonstrated the existence of familial aggregation, such that relatives of AD patients show increased risk for developing dementia compared with relatives of healthy control subjects.3–12 The familial aggregation of AD may be due to shared genetic or, at least theoretically, environmental risk factors within families. Most cases are, however, sporadic, which is defined by the absence of evidence for familial aggregation.
Research strategies
There are generally two strategies for examining genetic risk factors of complex and common diseases: linkage studies and association studies.
In linkage studies, genetic markers are examined in families with multiple affected members. If a certain marker with known chromosomal localization is linked to the disease due to linkage disequilibrium with the disease-causing mutation, positional cloning and fine-mapping is performed to identify the disease-related gene. The strategy of linkage analysis and subsequent positional cloning has been very successful in monogenic disorders and in some rare and severe variants of complex diseases. However, the use of linkage mapping in complex traits may be problematic: the polygenic aetiology of these diseases reduces the possibility that a marker in linkage disequilibrium with a putative susceptibility locus will produce sufficient signal for statistical detection. Even if a signal is detected, subsequent positional cloning is impeded due to the considerable inaccuracy with respect to the correct localization of the target gene (the target gene may be located within a region of 40cM around the marker).13 Furthermore, practical problems, which are inherent to AD research (late onset of disease, lack of sufficient large informative families, uncertain diagnoses), may additionally reduce the feasibility of linkage studies.
Association studies follow a different strategy: Based on theoretical considerations and experimental findings, one or more genes involved in AD pathogenesis are chosen (candidate-genes). Polymorphisms of these genes, which ideally alter gene function, are expected to be associated with the risk for the development of disease. This hypothesis can be tested by assessment of the frequency of the genetic variation in a sample of AD patients and control subjects. Association mapping is an additional and hypothesis-free method of genetic association. It is used to detect chromosomal regions containing disease-susceptibility loci through the identification of marker loci at which some alleles are more frequent among cases than among controls. There are many advantages inherent to the genetic association approach to the study of AD:
The selection of candidate genes is plausible and based on empirical data about the molecular events contributing to AD histopathology: the focus may be upon biologically defined candidate genes, genes suggested by differential display experiments, or positional candidates from prior linkage studies.
In common diseases, the signal derived from association studies is expected to be greater than that derived from linkage studies. For example, numerous association studies identified the apolipoprotein E (APOE) e4 allele (APOE4) to be a risk factor for AD with an odds ratio of 3 to 4, and persons with two copies of this allele have an even higher odds ratio.14 Several linkage studies failed to detect this signal.
The genotyping and statistical methods used in association studies are easy to perform, thus enabling independent replication experiments.
As noted below, the development of high-density genotyping platforms with the ability to genotype >500,000 single nucleotide polymorphisms (SNPs) may, for the first time, provide sufficiently dense markers to permit an unbiased hypothesis-free association study of the entire human genome. It has been proposed that one needs at least 300,000 evenly distributed markers to conduct a whole genome study (assuming an adequate number of well characterized cases and controls are included).
However, there are two major issues, which must be considered when interpreting the results of association studies:
The validity of an association study depends critically upon a proper selection of patients and control subjects. While matching for age, gender, and educational level is an easy-to-achieve prerequisite, controlling for ethnicity (i.e. similar genetic background) may become a problem, especially in population-based association studies. Population admixture is difficult to control for and may lead to erroneous results. The development of such family-based association tests as the sibship disequilibrium test (TDT)15 and others aims at dealing with the problem of population stratification. In addition, methods such as the Structured Association analysis16 and Genomic Control17 may be used in case-control association studies to estimate the level of genetic heterogeneity and thereby to control for this possible bias.
The number of possible candidate genes, which can be examined in a case-control sample, is very high, thus many false positive results may be generated. Indeed, a considerable number of studies reported significant associations of genetic variants with AD, however, the number of at least partially replicated findings is limited.18 The statistical problem of type I errors (i.e., chance findings due to multiple comparisons) is particularly apparent in whole-genome association studies involving many thousands of comparisons. Clearly, the identification and control of the factors leading to inflation of type I error will be instrumental to the future genetic studies in AD.
Ib. Genetics of sporadic AD
Sporadic AD accounts for the majority of all AD cases. Genetic factors seem to influence the risk for the development of sporadic AD and case-control genetic association studies are broadly used for their assessment. Usually, the selection of candidate genes examined in association studies is hypothesis-driven and based upon pathophysiological criteria. In the case of sporadic AD, most candidate genes are involved in amyloid metabolism, in brain cholesterol homeostasis or are components of the AD-related immune reaction. APOE is the only hitherto well-established risk factor for sporadic AD. Research findings on all the other genes remain controversial.
Apolipoprotein E (APOE) polymorphisms
Located on chromosome 19, the APOE gene encodes the apolipoprotein E, which is known to play a central role in the regulation of the cholesterol and triglyceride metabolism19 and which has been more recently suggested to play a direct or indirect role in the development of AD pathology. There are three common APOE alleles, known as e2, e3, and e4. Each person can have any combination of these 3 alleles, resulting in the e2e2, e2e3, e3e3, e3e4, or e4e4 APOE genotypes. In comparison with the most common APOE e3e3 genotype, having an e2 allele is associated with a lower risk of AD and a slightly older median age at dementia onset. Even better established, each additional copy of the e4 allele in a person’s APOE genotype is associated with a higher risk of AD and a slightly younger median age at dementia onset.
The APOE e2, e3, and e4 alleles are distinguished from each other on the basis of two SNPs, resulting in two amino acid changes at positions 112 and 158. The APOE e2-allele is characterized by cysteine at positions 112 and 158, the APOE e3-allele by cysteine at position 112 and arginine at position 158, and the APOE e4-allele by arginine at both positions.
A significant association of the APOE e4-allele with AD was initially demonstrated in 1993.20;21 This finding was replicated in numerous studies and in now considered the best-established genetic association with AD. In Caucasian populations, APOE e4 heterozygous individuals have a threefold increased risk and homozygous persons at least an eightfold increased risk for developing AD by age 75 years compared to APOE e3 heterozygous individuals.14 The magnitude of the effect of the APOE e4-allele as a risk factor for AD is influenced by a person’s age and ethnicity. Although the APOE e4 effect is evident at all ages between 40 and 90 years, it becomes weaker after the age of 70 years. The highest odds ratios (ORs) are detected in the Japanese population (OR = 5.6 for APOE e4-allele heterozygous, OR = 33.1 for APOE e4-allele homozygous).14 The APOE e4 effect is attenuated in Hispanic populations (OR = 2.5 for APOE e4-allele homozygous), and, interestingly, in some African populations no association between APOE e4 and AD can be observed.22;23 These results together with the observation that the frequency of the APOE e4-allele in Pygmies, aborigines of Malaysia and Australia, Papuans, some Native Americans, and Lapps is significantly higher than in populations with long-established agricultural economy24 indicate that the association between AD and the APOE e4 allele can be explained in part by the interaction of this allele with contemporary environmental conditions.
Due to the high ORs, at least in Caucasian populations, the possibility of using the APOE genotype as a diagnostic tool has been considered.25;26 However, as stated by the American College of Medical Genetics/American Society of Human Genetics Working Group on APOE and Alzheimer disease,27 APOE genotyping is not currently recommended to predict a healthy person’s risk for AD, because the APOE genotype fails to predict with sufficient certainty whether or when a person will develop symptoms.28 However, if future treatments prove to be associated with a preferential response according to APOE genotype group, the possibility of disclosing genotype information to patients should be carefully evaluated. Indeed, randomized trials (i.e. the REVEAL study) are currently evaluating the benefits and risks of disclosing genotype information following a person’s informed consent.29;30
Genetic testing
In accordance with the American College of Medical Genetics/American Society of Human Genetics Working Group on APOE and Alzheimer disease,27 neither APOE nor any other susceptibility gene is currently recommended for diagnostic or prognostic genetic testing. However, when a person’s family history is compatible with a Mendelian mode of inheritance, which is the case for the known APP, PSEN1 and PSEN2 mutations, genetic testing may be highly informative and useful. On the other hand, the deterministic character of genetic testing in such incurable diseases as AD may have far-reaching consequences for the patients and their families. In such cases, genetic testing requires a highly sensible and professional approach. Paralleling the guidelines for genetic testing in Huntington's disease,31 genetic testing in AD must be accompanied by competent pre-and post-test counselling done by certified specialists.32
Additional risk genes for LOAD
Significant findings have been obtained for several additional genes, which may be involved in the AD-related pathophysiologic cascade, such as cholesterol 24S-hydroxylase (CYP46A1),33 cystatin-C (CST3),34 interleukin 1 (IL1),35;36 and interleukin 6 (IL6)37;38 and, very recently, apolipoprotein A1 (APOA1)39 and ubiquilin 1 (UBQLN1).40 Polymorphisms in these genes have been suggested as potential susceptibility factors for AD, which may also modify the onset of the disease. However, none of these findings has been consistently replicated thus far, which, among other factors, may be related to the current lack of systematic meta-analyses for each reported SNP in populations of different origin. By gathering data on published SNPs and association studies, the recently developed AlzGene database aims to provide an unbiased, publicly available and regularly updated collection of genetic association studies on AD (http://www.alzforum.org/res/com/gen/alzgene/). Although the integrated meta-analyses of this database are a very valuable tool, there is still no possibility to study gene-gene interactions, which undoubtedly exist in this polygenic disease.
New perspectives
Genetic clusters
Genetically complex phenotypes and diseases result from the interplay between genetic variants and environmental factors.41 Despite this fact, the vast majority of the genetic association studies has either evaluated one genetic variation at a time or has analyzed multiple variations independently. However, this approach ignores the polygenic nature of the studied phenotype, focuses on marginal gene effects, inflates the probability of type I error, and erroneously assumes that the impact of the variation under study is comparable to a major gene effects. Recent developments in high-throughput genotyping and computerized algorithms now facilitate multi-locus analyses for the localization of complex trait genes. As shown for schizophrenia42 and AD,43 multi-locus analyses acknowledge the polygenic nature of disease, provide sets of trait-associated markers and also control for multiple testing. Importantly, such analyses may provide individual genetic clusters with potentially important clinical implications for future diagnosis, prognosis, and personalized treatments.
Genome-wide association (GWA)
Candidate-gene association studies successfully identify genetic variations with significant impact on a polygenic trait. However, the success of such hypothesis-driven studies strongly depends upon pre-existing information, which limits their potential to identify novel genes and molecular pathways. Recent advances in the development of high-density genotyping platforms with the ability to genotype >500,000 of SNPs now allow for whole-genome association studies in polygenic phenotypes and diseases. The highest genomic resolution reported so far corresponds to 1 SNP per 26 kb (i.e. GeneChip® Human Mapping 100K Set). Although the current statistical approaches are not ideal for the analysis of the huge amount of data, the first genome-wide association studies already revealed highly reliable and novel genetic findings.44 Therefore, GWA is expected to substantially increase our knowledge on polygenic human traits.
AD endophenotypes
To further evaluate genetic and non-genetic modifiers of the risk for a neuropsychiatric disorder like AD, it may be possible to identify an “endophenotype,” i.e., a measurable feature that is more closely related to disease susceptibility than the clinical syndrome itself.45;46 For instance, patients with AD have characteristic and progressive declines in positron emission tomography (PET) measurements of the cerebral metabolic rate for glucose (CMRgl) in the posterior cingulate, parietotemporal, and frontal cortex. Studying 160 cognitively normal persons with two copies, one copy, and no copies of the APOE e4 allele, Reiman et al. recently showed a significant correlation between these three levels of genetic risk for AD and lower CMRgl in each of these brain regions. They have suggested how PET could be used as a quantitative, presymptomatic endophenotype to help evaluate individual and aggregate effects of putative genetic and nongenetic modifiers of AD risk.46 Brain imaging techniques like PET, and other biomarkers of AD susceptibility could complement retrospective case-control studies and prospective cohort studies in the evaluation of these putative modifiers of AD risk.
Ic. Genetics of autosomal-dominant AD
In some families, AD is inherited as a fully penetrant, autosomal dominant disease apparently resulting from a single gene defect (i.e., gene mutations by themselves sufficient to cause AD). These rare AD families have been of utmost importance for the identification of causative AD genes by using the methods of linkage analysis with subsequent positional cloning. This strategy has led to the identification of 3 hitherto known AD-related genes.
Amyloid precursor protein (APP)
In 1987, St. George-Hyslop et al.,47 located a genetic defect causing autosomal-dominant AD on the long arm of chromosome 21. The APP gene, which codes for the amyloid precursor protein, was found to map to this region.48 Interestingly, a mutation in exon 16 of APP was found to cause hereditary cerebral hemorrhage with amyloidosis of the Dutch type (HCHWA-D).49 HCHWA-D is associated with Aβ deposition in cerebral blood vessels with the consequence of recurrent cerebral hemorrhages. Moreover, amyloid plaques similar to those found in AD patients were described in the brain of patients with HCHWA-D. These observations strongly supported the notion that the likelihood of APP mutations also causing AD would be very high. In 1991, Goate et al. described the first missence mutation in exon 17 of APP co-segregating with familial AD.50 Subsequent studies identified additional APP mutations in families with presenile AD. Interestingly, all these mutations are located in exons 16 and 17 of the APP, which encode the Aβ region of APP (Table 1). By altering the proteolytic cleavage of the Aβ region, these mutations result in overproduction of the amyloidogenic, 42 amino acids-long Aβ (Aβ42). Despite extensive searching, no APP mutations away from the sites of proteolytic cleavage of the Aβ region have been discovered so far. Although APP mutations are sufficient to cause AD, their effect may be additionally modified by gene-gene interactions: In the majority of families bearing APP mutations, the APOE e4 allele results in an earlier age at onset.
Table 1.
Pathogenic APP gene mutations
The identification of APP mutations was instrumental for the understanding of the metabolic cascades leading to enhanced Aβ production and gave rise to the amyloid hypothesis of AD.51 However, only a small percentage of autosomal-dominant AD is caused by APP mutations. Linkage analysis excluding families with APP mutations led to the identification of a novel gene family, the presenilins.
Presenilin 1 (PSEN1)
A genetic locus involved in early-onset autosomal-dominant AD was identified on the long arm of chromosome 14 by Schellenberg et al. in 1992.52 Positional cloning and examination of various transcripts of this chromosomal region led to the discovery of the presenilin 1 (PSEN1) gene on 14q24.3, which contained five different missence mutations cosegregating with early-onset autosomal dominant AD.53 Since then, several PSEN1 mutations have been identified, often resulting in an aggressive, early form of the disorder between ages 35–65 years. Thus, PSEN1 mutations account for the majority of autosomal-dominant AD cases (Table 2). Most presenilin mutations are missence mutations scattered throughout the molecule. However, they tend to cluster within and in the vicinity of the transmembrane domains, which are important for the protein activity of the presenilins.
Table 2.
Pathogenic PSEN1 gene mutations
| Exon | Mutation | Citation |
|---|---|---|
| 4 | R35Q | 141 |
| 4 | A79V | 143 |
| 4 | V82L | 145 |
| 4 | Δ I83/M84 | 147 |
| 4 | L85P | 149 |
| 4 | V89L | 150 |
| 4 | C92S | 152 |
| 4 | V94M | 154 |
| 4 | V96F | 156 |
| 4 | F105L | 157 |
| 5 | L113P | 159 |
| 5 | IVS4 g.23024delG | 161 |
| 5 | Y115H | 145 |
| 5 | Y115C | 143 |
| 5 | T116N | 164 |
| 5 | T116I | 165 |
| 5 | P117L | 166 |
| 5 | P117S | 167 |
| 5 | P117R | 169 |
| 5 | E120K | 171 |
| 5 | E120D | 61 |
| 5 | E120D | 173 |
| 5 | K123E | 174 |
| 5 | N135D | 176 |
| 5 | M139V | 177 |
| 5 | M139T | 145 |
| 5 | M139I | 177 |
| 5 | M139K | 142 |
| 5 | I143F | 144 |
| 5 | I143T | 146 |
| 5 | M146L | 148 |
| 5 | M146L | 141 |
| 5 | M146V | 151 |
| 5 | M146I | 153 |
| 5 | M146I | 155 |
| 5 | M146I | 141 |
| 5 | T147I | 158 |
| 5 | L153V | 160 |
| 5 | Y154N | 162 |
| 5 | Y154C | 163 |
| 5 | g.25669_25670 insTTATAT | 141 |
| 6 | H163Y | 151 |
| 6 | H163R | 53 |
| 6 | W165C | 158 |
| 6 | L166R | 168 |
| 6 | L166P | 170 |
| 6 | ΔI167 | 163 |
| 6 | S169L | 172 |
| 6 | S169P | 57 |
| 6 | L171P | 175 |
| 6 | L173W | 158 |
| 6 | L174M | 178 |
| 6 | F177L | 141 |
| 6 | F177S | 141 |
| 6 | S178P | 141 |
| 6 | G183V | 180 |
| 7 | E184D | 182 |
| 7 | G206S | 141 |
| 7 | G206A | 141 |
| 7 | G206V | 185 |
| 7 | G209V | 173 |
| 7 | G209R | 186 |
| 7 | G209E | 141 |
| 7 | I213T | 156 |
| 7 | I213L | 141 |
| 7 | I213F | 169 |
| 7 | G217D | 191 |
| 7 | L219P | 193 |
| 7 | Q222R | 141 |
| 7 | Q222H | 179 |
| 7 | L226R | 196 |
| 7 | I229F | 163 |
| 7 | A231T | 145 |
| 7 | A231V | 143 |
| 7 | M233T | 198 |
| 7 | M233L | 200 |
| 7 | M233V | 202 |
| 7 | L235P | 145 |
| 7 | L235V | 163 |
| 7 | F237I | 204 |
| 7 | F237L | 163 |
| 7 | A246E | 53 |
| 7 | L250S | 171 |
| 7 | L250V | 207 |
| 7 | Y256S | 179 |
| 8 | A260V | 181 |
| 8 | V261F | 141 |
| 8 | L262P | 183 |
| 8 | C263R | 184 |
| 8 | C263F | 163 |
| 8 | P264L | 145 |
| 8 | P267S | 151 |
| 8 | P267L | 187 |
| 8 | R269G | 188 |
| 8 | R269H | 189 |
| 8 | L271V | 190 |
| 8 | V272A | 192 |
| 8 | E273A | 194 |
| 8 | T274R | 141 |
| 8 | R278K | 195 |
| 8 | R278I | 197 |
| 8 | R278T | 198 |
| 8 | E280A | 151 |
| 8 | E280G | 151 |
| 8 | L282R | 199 |
| 8 | L282V | 201 |
| 8 | P284L | 203 |
| 8 | A285V | 181 |
| 8 | L286V | 53 |
| Δ9 | 205 | |
| Δ9 | 198 | |
| Δ9 | 206 | |
| Δ9 | 203 | |
| 10 | InsR | 141 |
| 10 | T354I | 141 |
| 10 | R358Q | 141 |
| 10 | S365Y | 141 |
| 11 | R377M | 163 |
| 11 | G378E | 208 |
| 11 | G378V | 163 |
| 11 | G384A | 146 |
| 11 | S390I | 158 |
| 11 | L392V | 181 |
| 11 | L392P | 209 |
| 11 | G394V | 141 |
| 11 | N405S | 210 |
| 11 | A409T | 200 |
| 11 | C410Y | 145 |
| 12 | L418F | 141 |
| 12 | L424R | 211 |
| 12 | A426P | 173 |
| 12 | A431E | 141 |
| 12 | A431V | 212 |
| 12 | A434C | 213 |
| 12 | L435F | 141 |
| 12 | P436S | 214 |
| 12 | P436Q | 172 |
| 12 | I439V | 141 |
The APOE gene does not seem to interact with PSEN1 and does not further influence the onset age in PSEN1 families.54 However, other factors – probably of genetic origin – seem to interfere with PSEN1, since a considerable phenotypic variability (e.g. variable age of onset and variable clinical presentation) may be observed even within families carrying a specific mutation.55–61 Moreover, some PSEN1 mutations show incomplete penetrance, since not all mutation carriers will ultimately develop the disease.
After the identification of PSEN1 as a causative AD gene, the mechanism by which mutations of this gene caused the AD phenotype was an open matter and was not necessarily linked directly to Aβ production. Studies on fibroblast cell cultures of PSEN1 mutation carriers revealed a marked elevation of Aβ42 levels, suggesting that presenilin function is important for the regulation of APP processing.62 Modelling of PSEN1 mutations in cultured cells and in transgenic mice supported the notion that presenilin cleavage has a direct effect on APP procession and Aβ production. Presenilins are currently considered to be the catalytic subunits of the γ-secretase complex, which also contains three additional subunits (Nicastrin, Aph1, and Pen-2).63
Presenilin 2 (PSEN2)
In 1988, Bird et al. described a group of families with autosomal-dominant early-onset AD descending from a single German family that first immigrated to Russia and later to the United States (‘founder effect’).64 No APP or PSEN1 mutations were detected in these Volga-German kindreds. The search for proteins homologous to presenilin1 led to the cloning and characterization of the PSEN2 gene, which is located on chromosome 1q31-q42 and codes for a protein highly homologous to presenilin 1.65 Ten missence PSEN2 mutations have been described so far (Table 3). In analogy to PSEN1, the APOE gene does not seem to interact with PSEN2 and does not further influence the onset age in PSEN2 families. Similarly, incomplete penetrance and variable age of onset and clinical presentation are also characteristics of PSEN2 mutations.
Table 3.
Pathogenic PSEN2 gene mutations
Id. Conclusion
AD is a genetically heterogeneous disorder, in that several genes contribute to the disease risk. The identification of families with an autosomal-dominant mode of inheritance has been instrumental in the search for genetic defects and has led to the discovery of disease-causing mutations in the APP, PSEN1 and PSEN2 genes by linkage analysis and positional cloning. Most AD cases, however, do not follow a clear mendelian mode of inheritance and are considered polygenic diseases. The ε4 allele of the APOE genotype is the best-established genetic risk factor in these cases. However, it is obvious that several other genes, each exerting a minor effect, contribute to the overall genetically determined risk for the development of the disease. The recent advances in the characterization of the human genome, the identification of a large amount of polymorphic sites throughout human DNA and the development of high-throughput genotyping methods and elaborated statistical analyses will ultimately allow dechiphering these genetic susceptibility factors. It is possible that in the near future, genetic research will contribute to the estimation of individual disease risks and to the optimization of therapeutic strategies.
II. Transcriptional Profiling in AD
Genetic variability may be considered as the first source of phenotypic variability between individuals. However, the limited number of genes in the human genome (approximately 30000) suggests that a significant portion of interindividual differences is related to modifications at the transcriptional, translational, and post-translational level. The high-throughput analysis of gene transcripts (“transcriptomics”), which allows for the simultaneous assessment of thousands of mRNAs, is a valuable approach to identify genes that are differentially expressed in certain conditions.
Gene expression microarrays provide a powerful tool for understanding the alterations in cellular metabolism that occur during the course of AD and for developing targeted therapeutics to correct those alterations. The technology has, in the past several years, stabilized to the point where robust expression correlates of clinical traits can be ascertained, validated, and used to gain insight into the pathogenesis of a disorder. Numerous studies report specific gene dysregulation events associated with AD and have suggested important mechanisms whereby neuronal cells die in this disorder. More recent advances in single cell approaches to gene expression profiling allow the analysis of pathologically affected cell types specifically rather than of the heterogeneous brain sample as a whole and should allow more focused interrogation of disease mechanisms, thereby permitting development of therapeutics targeted specifically to pathologically affected cells.
There are two primary variations of expression arrays currently in use, cDNA and oligonucleotide microarrays. Oligonucleotide arrays provide the benefits of greater specificity since the probes used are of shorter sequence (~25–70 nucleotides) than those used for cDNA arrays (200-2000 nucleotides). cDNA arrays have greater sensitivity but cannot, for example, discriminate between splice variants because the size of DNA probes used often spans splice junctions. For each type of array, sequences of DNA that are homologous to different genes of interest are attached to a solid support at distinct locations. Each probe (or probe set for oligonucleotide arrays with built in redundancy such as the Affymetrx platform) on a slide corresponds to a single gene and each array can hold thousands of probes. One then hybridizes either labeled cRNA or cDNA from the cell type or tissue of interest to the microarray, rapidly generating an mRNA expression profile for thousands of genes.
Analysis of expression profiling data entails comparison of profiles from the state of interest to control profiles using statistical strategies which fall into two general categories: supervised and unsupervised approaches. Supervised analysis entails specifying the disease class before analysis proceeds. This approach allows generation of a list of genes whose expression differs significantly between the phenotypic classes of interest. Unsupervised approaches do not specify a phenotypic class a priori, rather allow similarity measures to be derived between expression data based strictly on the expression values, and phenotype is then superimposed on the arrays. Expression array analysis, while not completely straightforward, has matured to the point where the technology can be used as a robust screening tool and thus derive differences in gene expression which can then move forward into validation in independent sample sets at the protein level.
IIa. Whole tissue expression profiling in Alzheimer’s disease
Numerous expression microarray experiments to date have identified specific genes that are dysregulated under different AD-related conditions. The general conclusions from these studies are that there is a general downregulation of gene expression in the cortices of AD affected brains compared to unaffected individuals. Because each microarray experiment will identify over one hundred genes which are dysregulated between control and AD it is impossible to list all of the correlations that have been identified within the context of this review. Rather, we highlight several findings that have generalized significance to the pathological processes of AD. For example, alterations of the amyloid precursor protein, inflammatory pathways, and synaptic signaling components have been reported.66–68 All of these expression profiling results fit well with known pathological lesions of AD and provide interesting gene candidates that may underly these processes. Using cDNA microarrays, Ho et al. demonstrated the selective decrease in expression of synapsin-a splice variants I–III in the entorhinal, but not visual, cortex of post-mortem brains of early stage AD patients.66 Thirty-two genes were found to be differentially expressed ≥ 1.8-fold in samples from patients displaying moderate dementia compared to normal controls. Colangelo et al., using oligo-based microarrays on pooled hippocampal CA1 tissues from six individuals, demonstrated a generalized down-regulation in brain gene transcription, including decreases in RNA encoding transcription factors (ISGF, GATA-2, LYL-1 and GATA-4) as well as neurotrophic factors and synaptic signaling elements (synaptophysin, SLIT-2, NCP, CHAT, metallothionein III, and metal regulatory factor-1), which fits well with the observed synaptic dysfunction that occurs in AD.67 Expression increases in the βAPP gene were demonstrated along with increases in pro-apoptotic genes (DAXX, CDP5, and FAS) as well as pro-inflammatory genes (cPLA2, NF-κBp52/p100, DPP1, NFIL6, IL-1β, B94, HB15, COX-2, and CEX-1). Another group examined brain tissue from a single AD patient (hippocampal tissue containing neurofibrillary tangles vs. parietal cortex with no lesions) and showed a dramatic increase in the expression of the calcineurin Aβ gene.68 This group found a total of 310 differentially expressed genes, of which, 20 up- and 20 down-regulated were chosen for validation by in situ hybridization and RT-PCR. Calcineurin Aβ (protein phosphatase 2B, a serine/threonine phosphatase), a major calmodulin-binding protein in the brain was the most highly up-regulated gene identified. Its proposed mechanism of involvement in AD pathogenesis is its ability to dephosphorylate hyperphosphorylated tau protein.
More recent microarray experiments have focused on characterizing gene dysregulation in animal models of AD. In some cases, introduction of pathogenic mutations in the genes encoding APP, the presenilins, and tau into mice results in the development of AD pathologies, such as amyloid plaques and neurofibrillary tangles. Expression profiling has been used to identify the downstream effects of these pathogenic mutations in these transgenic animal models of AD.69–74 Specifically, analysis of PS-1 knockout mice revealed upregulation of inflammatory pathways, including complement component C1q.69 PS1 knockout and PS1 knock-in (DeltaE9 mutation) models of AD have also been directly compared.73 In this study, the PS1 knockout and mutant form of PS1 had opposite effects on the expression of specific genes, leading the authors to conclude the this particular pathogenic PS1 mutation may be a gain-of-function mutation. Analysis of APP/PS-1 (P264L) transgenic mice, which develop amyloid plaques, revealed upregulation of the immune response, carbohydrate metabolism, and proteolysis as well as downregulation of growth factor pathways (BDNF and IGF-1R).71 Each of these studies provides additional candidate genes that may mediate some of the known pathological hallmarks of AD, including the deposition of amyloid plaques, the aggregation of tau protein into neurofibrillary tangles, and the activation of glial cells in AD affected brain regions. The challenge now is to develop the appropriate functional assays and endpoints to determine the specific role that dysregulation of these genes plays in the development and progression of AD.
IIb. Single cell expression profiling in Alzheimer’s disease
All of the expression profiling experiments summarized above suffer from the major limitation of utilizing heterogeneous brain tissue samples, each of which are composed of at least half a dozen distinct neuronal and glial cell types and numerous neuronal subtypes. This adds significantly to the variability of these experiments and increases the likelihood that significant and pathological gene expression changes may be masked by the expression profiles of additional unaffected cell types. Such a masking would result if the cell types of interest are in the minority relative to the piece of whole brain used in the expression profiling experiment. Recent advances in microaspiration and microdissection techniques, in RNA amplification procedures, and in banking of high quality, clinically well characterized tissue now afford the opportunity to investigate the expression of specific pathologically affected cell types in clinically well-stratified patient cohorts to identify the full complement of gene expression changes that accompanies the pathological progression of Alzheimer’s disease.
To date, the application of single-cell expression profiling techniques to Alzheimer’s disease has been somewhat limited and includes an analysis of anterior nucleus basalis neurons from control and AD brain75 and characterization of expression changes associated with neurofibrillary tangle pathology in AD.76 These studies suggest important roles for the upregulation of neurotrophic signaling and downregulation of protein phosphatase activity in nucleus basalis neurons.75
Large scale efforts are currently underway to characterize the full complement of neuronal and glial gene expression changes that occur during the course of AD. This information will provide novel genes and mechanisms whereby neuronal cells are lost in AD. Clearly the ultimate goal of these expression studies is to identify the gene targets that can be used to develop novel and effective treatment strategies. Within our own groups, we have begun a comprehensive cataloging of neuronal malfunction relative to controls in a mild cognitive impairment patients as well as AD patients in six regions of the brain. We feel that looking at early stage disease will allow insights to be gleaned with respect to the early pathogenic cascades at work, and thus allow us to eventually design therapeutics aimed at the earliest cellular events leading to neuronal death. Initial findings from this effort highlight diverse molecular pathways associated with NFT formation in the entorhinal cortex during AD.77
IIc. The Future of Expression Profiling in Alzheimer’s Disease
The ultimate goal of the application of expression profiling technology to AD is to identify effective therapeutic targets, to assist in the development and testing of new drugs that hit those targets, and further, to identify which individuals are likely to respond to therapy. The realization of this goal depends, however, on the understanding of the experimental challenges and on the implementation of appropriate validation steps which complement expression studies. Indeed, different data mining methods, computational algorithms, and genes used for normalization are some of the numerous variables which influence an experiment’s outcome and reduce the comparability between studies.78;79 In addition, if the transcriptomics approach is limited only to cross-sectional comparisons, the identification of genes which represent rather a consequence, compensation or coincidence rather than the cause of the disease is likely. Therefore, appropriate corroborating procedures such as the analysis of specific cell types and the targeted manipulation of genetic pathways should be included in studies dealing with the expression pattern of several thousands of genes.
As advancement of single cell methods, coupled to ever-improving animal model outlets for functional validation of gene candidates proceeds, expression profiling in AD will move from an exploratory tool to interrogate disease mechanisms to an essential component in the development of safe and efficacious treatments. For example, a recent report uses expression profiling to track the changes associated with inhibition of CDK5, a gene thought to be important in the hyperphosphorylation of tau protein.80 Using this approach, the authors were able to show that the CDK5 inhibitor altered the expression of many genes involved in the pathological processes associated with Alzheimer’s disease and neuronal cell death, including numerous genes involved in synaptic transmission, neurite outgrowth, and neuronal survival.
Increasingly, as the application of microarrays to reveal disease mechanisms leads to the development of new drugs to slow or reverse those mechanisms, the focus of microarray technology will shift to analyzing therapeutic effectiveness of those drugs. The availability of a real-time monitoring system to track the molecular effects of therapy may be particularly useful for AD, where important molecular changes may precede clinical manifestation of disease by decades. Additionally, this approach may be increasingly useful in a healthcare environment where unforeseen drug side-effects could shelf any potentially useful drug at any time. Knowledge of the full molecular effects of a given drug may provide a harbinger for side-effects that would otherwise have passed current regulatory standards, thereby sparing the expense of withdrawing an otherwise useful pharmaceutical from the market. There has been little application of state-of-the-art expression profiling technologies to AD, but a multitude of studies are currently underway which should allow insight into the somatic changes in the brains of AD patients.
III. Proteomics in Alzheimer’s disease
Proteomics is a multidisciplinary, technology-driven science that is focused on the analysis of proteomes: i.e. the proteins of a biological system, their structures, interactions, post-translational modifications, and in particular the changes in their levels and their modifications, which are the result of specific diseases or are induced by various external factors, such as toxic agents. The goal of proteomics is to detect novel drug targets and diagnostic markers. Proteomics emerged during the last few years, and its development can be attributed to developments in mass spectrometry, computer and software sciences and to the enormous amount of information that became available from the sequencing of the genome of many organisms. Proteomics can detect gene products involved in various pathways which can not be detected in another way and its major difference from the previously existing protein analytical techniques is that proteomics does not analyze the proteins one by one, but in a possibly automated, large-scale mode.81
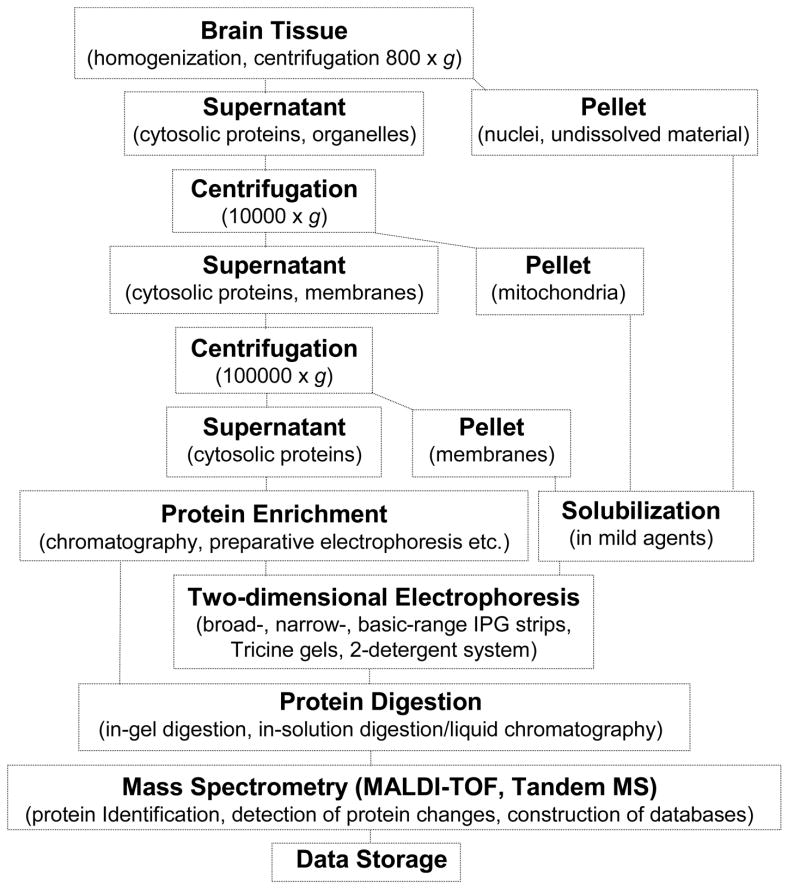
A proteomic analysis comprises two steps: (i) separation of the protein mixture in order to allow the efficient detection of the particular proteins included in the mixture and (ii) identification of the separated proteins by various analytical methods, mainly by mass spectrometry (MS). Separation is usually performed by two-dimensional (2-D) gel electrophoresis or liquid chromatography (LC) or more often by combination of the two approaches.82–84 Two-dimensional electrophoresis is the principal separation method in a proteomic analysis and has the advantage that it allows the simultaneous detection of thousands of proteins. It comprises two steps (dimensions), separation of the proteins on the basis of differences in their net charge by isoelectric focusing (IEF), and separation of the focused proteins on the basis of differences in their molecular masses by sodium dodecyl sulfate (SDS) polyacrylamide gels.85;86 When liquid chromatography is applied for protein separation, the intact proteins can be first fractionated by ion exchange and/or hydrophobic interaction chromatography or by a combination of various binding principles. In a next step, the proteins of the fractions collected are digested and the peptides are separated by reversed phase HPLC and analyzed by MS.
The most efficient and most widely used protein identification method in proteomics is the peptide mass fingerprint (PMF) approach which is mainly performed by matrix-assisted laser desorption/ionization time-of-flight mass spectroscopy (MALDI-TOF-MS).87 For MS analysis, protein spots are excised from 2-D gels, the proteins are in-gel digested, the digests together with the matrix are applied on the sample target and are measured in the mass spectrometer.88 In a high-throughput proteomic analysis, most of the operations are performed in automated mode and several thousands of protein spots can be analyzed by one person per day. The analysis of certain groups of proteins is more favorably performed by LC/MS, e.g. very acidic/basic, small proteins/peptides, and hydrophobic proteins. However compared to 2-D gels, isoforms, post-translational modifications, and processing products are very difficult to discern via LC/MS.89 Fig. 1 shows the workflow for the proteomic analysis of the brain, involving preparation of protein subfractions, protein separation by 2-D electrophoresis or LC and identification by mass spectrometry.
Figure 1.
Workflow for the proteomic analysis of the brain.
Proteomics is routinely used in clinical diagnosis today, and it has been applied in the investigation of infectious diseases, cancer, heart, and neurological disorders. The applications are summarized in several review articles.81;89–94 In the study of AD, proteomics has been mainly used for the analysis of human brain, of the brain of animals serving as models of AD and of cerebrospinal fluid (CSF) from patients with AD with the goal to collect information about gene products involved in the diseases i.e., alterations in protein levels and post-translational modifications.89 Proteomics has also been used in the investigation of Down syndrome (DS) and other neurodegenerative disorders. Almost all subjects with DS over 40 years show neuropathological and neurochemical abnormalities on post-mortem brain examinations that are indistinguishable from those seen in Alzheimer’s disease. Thus, results from the study of the DS brain may be useful in the AD studies.95–98
IIIa. Proteins with deranged levels and modifications in AD
Most neuroproteomics analyses involve 2-D electrophoresis and MALDI-TOF-MS steps, as this approach allows for a reliable quantification of changes in protein levels. Fig. 2 shows the proteins of total brain extract separated on a 2-D gel. The most abundant proteins detected in 2-D gels carrying total brain extract are structural proteins, like tubulin chains and actin, heat shock proteins, dihydropyrimidinase-related proteins-2 and -3, and house-keeping enzymes. The less abundant proteins are mainly hypothetical proteins, enzymes, as well as structural proteins. The relative levels of the low-abundance brain proteins vary in the range 0.005–0.01% and of the high-abundance about 3% in comparison with the levels of total proteins detected in the corresponding gel.89 In one study, the LC-MS/MS approach was used for the detection of proteins enriched in amyloid plaques in the AD brain.99
Figure 2.
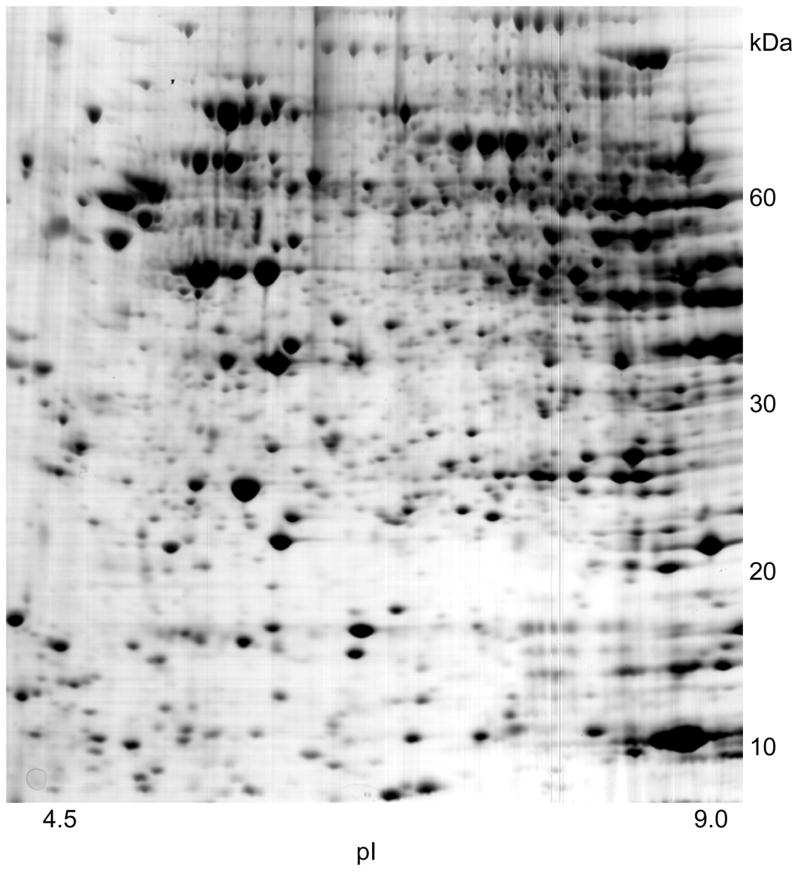
Proteins of total brain extract separated on a 2-D gel.
The levels of about 100 proteins were found to be changed in the AD brain and cerebrospinal fluid in comparison with the control brain and CSF, respectively. The observed changes in the protein levels usually vary in the order of 20–100% in comparison with the average levels of the control group. The largest changes were observed for the glial fibrillary acidic protein (GFAP), which distinguishes astrocytes from other glial cells during the development of the brain. For this protein, ten-fold or even higher levels were observed in individual AD brains, compared to the average levels of the control group.100 Fig. 3 shows an example of a protein, synaptosomal protein 25 kDa (snap25), which shows reduced levels in the AD brain in comparison with the control brain. The proteins detected by proteomics approaches to show changed levels or modifications in AD have various functions, mainly involved in conformational changes, neurotransmission, guidance, signal transduction, metabolism and detoxification. The CSF proteins with deranged levels are plasma proteins. The proteins detected by proteomics methods to have deranged levels in the AD brain and the CSF are listed in Table 4.
Figure 3.
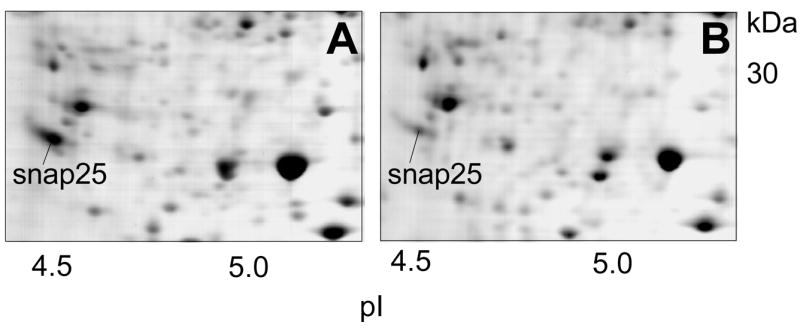
Example of a protein, in this case the synaptosomal protein 25 kDa (snap25), which shows reduced levels in the AD brain in comparison with the control brain.
A: controls brain, B: AD brain.
Table 4.
Altered proteins in AD
| Protein | Change | Reference |
|---|---|---|
| Heat Shock Proteins | ||
| Alpha crystallin B chain | I | 103;218 |
| Glucose regulated protein 75 kDa (GRP75) | D | 103;218 |
| Glucose regulated protein 75 kDa (GRP94) | I | 103;218 |
| Heat shock 60 kDa, mitochondrial | D | 219 |
| Heat shock 90kDa protein 1, beta | Enriched in amyloid plaques | 99 |
| Heat shock cognate 71 | Oxidation | 104 |
| Heat shock protein 70 RY | I | 103;218 |
| T-complex protein 1 (TCP-1) | D | 220 |
| Synaptosomal proteins | ||
| Beta-soluble N-ethylmaleimide-sensitive factor attachment protein (Beta SNAP) | D | 107 |
| Synaptosomal associated protein 25 kDa (Snap-25) | D | 100 |
| Synaptotagmin I | D | 107 |
| Guidance Proteins | ||
| Didyropyrimidinase-related protein 2 (DRP-2) | D | 108 |
| Signal Transduction Proteins | ||
| 14-3-3 beta/alpha | Enriched in amyloid plaques | 99 |
| 14-3-3 epsilon protein | I | 89;113 |
| 14-3-3 gamma protein | I | 89;113 |
| 14-3-3 zeta | Enriched in amyloid plaques | 99 |
| 2′-3′-Cyclic nucleotide 3-phosphodiesterase (CNPase) | D | 110 |
| Alpha-Endosulfine | D | 221 |
| ARPP-19 | D | 222 |
| cAMP-dependent protein kinase (PKA) | D | 222 |
| Fatty acid-binding protein, heart | D | 219 |
| Guanine nucleotide-binding protein beta subunit | D | 219 |
| Histamin-releasing factor | D | 223 |
| Nucleoside diphosphate kinase A (NDKA) | D | 111 |
| Stathmin | D | 112 |
| Antioxidant proteins | ||
| Alcohol dehydrogenase (ADH) | I | 117 |
| Antioxidant protein 2 (1-Cys peroxiredoxin) (1-Cys Prx) | I | 224 |
| Carbonyl reductase (CBR) | I | 117 |
| Peroxiredoxin I (Prx-I) | I | 118 |
| Peroxiredoxin II(Prx-II) | I | 118;119 |
| Peroxiredoxin III (Prx-III) | D | 119 |
| Peroxiredoxin VI | I | 119 |
| Superoxide dismutase | D | 116 |
| Oxidized Proteins | ||
| Alpha-enolase | Oxidation/Nitration | 104 |
| Beta-actin | Oxidation/D/Nitration | 122;123 |
| Creatine kinase BB | Oxidation/D | 105;225 |
| Didyropyrimidinase-related protein 2 | Oxidation/D | 104 |
| Gamma-Enolase | Oxidation/Nitration | 104 |
| Glutamate Transporter, EAAT2 | Oxidation/D | 123 |
| Glutamine synthase | Oxidation/D | 105 |
| Heat shock cognate 71 | Oxidation | 104 |
| L-Lactate dehydrogenase | Oxidation/Nitration | 104 |
| Neuropolypeptide h3 | Oxidation/Nitration | 104;123;226 |
| Triosephosphate Isomerase | Oxidation/Nitration | 122;226 |
| Ubiquitin carboxy-terminal hydrolase L-1 | Oxidation | 105 |
| Metabolism, Memory | ||
| ATP synthase alpha chain, mitochondrial | D | 219 |
| ATP synthase beta chain, mitochondrial | I | 219 |
| Beta-enolase | D | 219 |
| NADH-ubiquinone oxidoreductase 24 kDa subunit, mitochondrial [Precursor] | D | 227 |
| NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial [Precursor] | D | 227 |
| Phosphofructokinase, muscle ttype | Enriched in amyloid plaques | 99 |
| Phosphofructokinase, platelet type | Enriched in amyloid plaques | 99 |
| Ubiquinol-cytochrome-c reductase complex III core protein I | D | 228 |
| Voltage-dependent anion-selective channel protein 1 (VDAC1) | D | 229 |
| Voltage-dependent anion-selective channel protein 2 (VDAC2) | I | 229 |
| Apoptosis-related proteins | ||
| Bim/BOD (Bcl-2 interacting mediator of cell death/Bcl-2 related ovarian death gene) | I | 121 |
| DNA fragmentation factor 45 (DFF45) | D | 230 |
| p21 | I | 121 |
| Procaspase-3 | D | 230 |
| Procaspase-8 | D | 230 |
| Procaspase-9 | D | 230 |
| Receptor interacting protein (RIP)-like interacting CLARP kinase (RICK) | I | 230 |
| Zipper interacting protein kinase | I | 121 |
| Structural proteins | ||
| Collagen I, alpha-1, polypeptide | Enriched in amyloid plaques | 99 |
| Coronin, actin binding protein | Enriched in amyloid plaques | 99 |
| Drebrin | D | 231 |
| Fibrinogen, gamma | Enriched in amyloid plaques | 99 |
| NF-L | D | 232 |
| Tau | Enriched in amyloid plaques | 99 |
| Vimentin | Enriched in amyloid plaques | 99 |
| Proteolysis | ||
| Antitrypsin | Enriched in amyloid plaques | 99 |
| ATPase, Ca++ transporting plasma membrane protein 2 | Enriched in amyloid plaques | 99 |
| ATPase, H+ transporting, lysosomal V0 subunit A | Enriched in amyloid plaques | 99 |
| ATPase, H+ transporting, lysosomal V1 subunit B | Enriched in amyloid plaques | 99 |
| ATPase, H+ transporting, lysosomal V1 subunit D | Enriched in amyloid plaques | 99 |
| ATPase, H+ transporting, lysosomal V1 subunit E | Enriched in amyloid plaques | 99 |
| Cathepsin D | Enriched in amyloid plaques | 99 |
| Cystatin B | Enriched in amyloid plaques | 99 |
| Cystatin C | Enriched in amyloid plaques | 99 |
| Ubiquitin-activating enzyme E1 | Enriched in amyloid plaques | 99 |
| Vacuolar ATPase subunit H | Enriched in amyloid plaques | 99 |
| Glial associated proteins | ||
| Glial fibrillary acidic protein (GFAP) | I | 100 |
| Membrane trafficking proteins | ||
| Dynamin 1 | Enriched in amyloid plaques | 99 |
| Dynein, heavy polypeptide 1 | Enriched in amyloid plaques | 99 |
| Other | ||
| Amyloid beta-peptide | Enriched in amyloid plaques | 99 |
| CSF proteins | ||
| Alpha-1 antitrypsin | I | 233 |
| Alpha-1 beta glycoprotein | D | 233 |
| Apolipoprotein A1 | D | 233 |
| Apolipoprotein E | D | 233 |
| Apolipoprotein J | D | 233 |
| Beta-2 microglobulin | I | 234 |
| Cell cycle progression 8 protein | D | 233 |
| Kininogen | D | 233 |
| Proapolipoprotein | D | 234 |
| Retinol-binding protein | D | 233 |
| Transthyretin | I | 234 |
Heat shock proteins (HSP) are involved in conformational changes of the brain proteins, i.e. in protein folding and assembly into oligomeric structures.101;102 Several heat shock proteins showed deranged levels in the AD brain. Increased expression was observed for alpha crystallin B, glucose regulated protein 94 (GRP 94) and HSP70 RY, and decreased expression for heat shock cognate (HSC71), GRP 75, HSP60 and T-complex protein-1.103 The inconsistency in the direction of change of the heat shock proteins in AD may be explained by the different response of the individual heat shock proteins to stress conditions. Increased expression of heat shock proteins in AD may represent a defensive mechanism of response to amyloid fibril formation, as chaperone proteins are capable of preventing aggregation of other proteins and in particular inhibiting self-assembly of polyglutamine proteins into amyloid-like fibrils. Reduced levels of the HSP70 proteins, in particular of the HSC71, might be linked to their protective role in neuronal death associated with AD. HSC71 may be involved in the structural maintenance of the proteasome and conformational recognition of misfolded proteins by proteases.104;105 Thus, decreased activity of HSC71 which is attributed to either reduced levels103;106 or increased oxidative modification of the protein104 may be responsible for impaired protein clearance and deposition of amyloid-β peptide in the AD brain.
Synaptosomal proteins, soluble N-ethylmaleimide sensitive factor attachment proteins (SNAPs) forms α, β, γ, synaptososmal associated protein 25 (SNAP 25), synaptotagmin and vesicular proteins, are involved in synaptic transmission which is deranged in AD. Proteomics studies showed reduced expression for SNAP 25100 (Fig. 3), β-SNAP and synaptotagmin I107 in the AD brain and suggest impaired neurotransmission in AD.
Guidance is facilitated by proteins signaling the correct path to the growing axon. Dihydropyrimidinase-related protein 2 (DRP-2), also known as collapsin response mediator protein-2, is a cytosolic protein that shows a high homology to the rat turned on after division 64 kDa protein and the chicken collapsin response mediator protein-62, which are involved in path finding and migration of neurons. The functions of DRP-2 are not well known. It is possibly involved in neuronal repair and interacts with and modulates the activity of collapsin, a protein that elongates dendrites and directs them to adjacent neurons. Proteomics studies revealed deranged expression levels108 and oxidative modification104 of DRP-2 in the AD brain. The reduced levels of DRP-2 in the AD brain may be responsible for synaptic loss in AD as it can not respond to the formation of new synapses. Moreover, DRP-2 is immediately modified by oxidation and looses activity, which results in shortened dendrites that would cause decreased interneuronal communication and memory impairment.109
The levels of several signal transduction proteins have been found to be deranged in the AD brain. Thus, reduced levels were found for 2′, 3′-cyclic nucleotide-3′phosphodiesterase (CNPase), 110 nucleoside diphosphate kinase (NDK)-A111 and stathmin112 and increased expression for 14-3-3 gamma and epsilon proteins.113 CNPase is associated with oligodendroglia and myelination. NDKs are oligomeric enzymes, involved in the maintenance of the intracellular pool of deoxynucleotide triphophates and nucleotide triphosphates, playing regulatory roles in the activation of G-proteins.114 Stathmin, a major substrate of cyclin-dependent kinases and protein kinase A, is involved in multiple signal transduction pathways. Stathmin possibly plays a role in neurodegeneration and impaired signaling because of its negative correlation with neurofibrillary tangles, a hallmark of AD neuropathology, and interaction with tubulin to cause microtuble destabilization.112 14-3-3 proteins form homo- or heterodimers and are primarily, but not exclusively expressed in neurons and bind to and modulate the function of a wide variety of cellular proteins. They are involved in neuronal development, signal transduction, cell growth and cell death. They show increased levels in AD which may be rather linked to apoptosis than to other cellular activities, as they are highly enriched in areas of massive neuronal death caused by pathological processes.115 The increased levels of 14-3-3 proteins in AD may represent a protective response against apoptosis.
Oxidative stress, either detected as protein oxidation, lipid peroxidation, DNA oxidation and 3-nitrotyrosine formation, is considered to be a common mechanism for different age-related neurodegenerative pathologies. The levels of antioxidant proteins in the brain were studied using proteomics technologies. Superoxide dismuatase (CuZn, SOD1) catalyzes the dismutation of superoxide anion into hydrogen peroxide. The protein showed decreased levels in AD and the decrease may reflect cell loss in AD.116 The expression of two other proteins, carbonyl reductase and alcohol dehydrogenase, involved in oxidative stress, were found to be increased in AD.117 These two cytosolic enzymes catalyze reduction of carbonyls, which are toxic metabolic intermediates, increased in the AD brain104;105 and serve as markers for oxidative stress, suggesting a possible role of oxidation-related decrease in protein function in the process of neurodegeneration. The increased levels of these enzymes may represent an adaptive mechanism to detoxify the toxic intermediates. Peroxiredoxin I and II showed increased and peroxiredoxin III decreased levels in AD brain.118;119 Peroxiredoxins are a family of peroxidases that protect biomolecules by reducing hydrogen peroxide and alkyl hydroperoxides. There is evidence that over-expression of peroxiredoxins, in particular of peroxiredoxins I and II, in various cell lines abrogates the effects of agents that positively modulate apoptosis, including hydrogen peroxide.120 Enhanced apoptosis has been demonstrated in AD121 and up-regulation of these enzymes could represent a cellular response against apoptosis. In other neurodegenerative disorders, like DS and Pick’s disease, deranged levels for peroxiredoxins were observed as well.118;119
Oxidative stress is observed in the AD brain in regions where amyloid β-peptide 1-42 is present, but not in cerebellum where AD pathology is not observed.122 Several oxidized proteins were detected by immunoblots in the AD brain and identified by MALDI-TOF-MS (Table 4). One of the oxidized proteins is creatine kinase (BB isoform). Amyloid β-peptide inhibits creatine kinase and causes lipid peroxidation leading to decreased energy utilization, alterations in cytoskeletal proteins and increased excitotoxicity, all reported for AD. The other oxidized proteins are glutamine synthase, which is related to increased excitotoxicity, ubiquitin C-terminal hydrolase L-1, associated to aberrant proteasomal degradation of damaged or aggregated proteins, α-enolase, linked to altered energy production and dihydropyrimidinase-related protein 2, related to reduced growth cone elongation.122;123 The ubiquitin C-terminal hydrolases are suggested to hydrolyze small adducts of ubiquitin and generate free monomeric ubiquitins, thereby allowing recycling of ubiquitin, which is essential to the function of the proteolytic machinery. Thus, putative dysfunction of this oxidized deubiquitinating enzyme may be one way by which oxidative stress promotes ubiquitin-positive inclusion biogenesis, a pathological characteristic of the AD brain, which leads to neurodegeneration.
Using LC-MS/MS, Liao et al., identified 26 brain proteins enriched in amyloid plaques excised with laser capture microdissection from AD brains.99 The list includes cytoskeletal proteins (coronin, tau), membrane trafficking proteins (clathrin, dynamin 1, dynein), proteases (ATPases, cathepsin), signal transduction proteins (14-3-3 proteins), chaperons and others (Table 4).
IIIb. Limitations
The proteomic analysis of the brain has certain limitations which can be related to the sample and/or the analytical approach. In the analysis of the brain, many factors may be involved, such as differences among individuals, differences in age and sex, possibly other diseases, treatment with medicines, as well as technical, disease-unrelated factors, such as post-mortem time, improper treatment of the samples etc., all of which can affect a clear discrimination between healthy and diseased states of interest. The technical limitations involve inefficient detection of low-abundance gene products, of hydrophobic proteins (they do enter the IPG strips), acidic, basic, high- and low-molecular mass proteins. All these protein classes are underrepresented in 2-D gels.89;124 A combination of the proteomics methods with protein separation and enriching techniques and alternative methodology for detection will improve the detection of additional differences between AD and control brain. Such differences may be essential in the discovery of early disease markers and therapeutic approaches.
IV. Conclusions
AD is a genetically heterogeneous disorder. The recent advances in the detailed characterization of the human genome, the identification of millions of polymorphic sites in the human DNA and the development of high-throughput genotyping methods and elaborated statistical analyses will soon contribute to the identification of genetic risk profiles related to the development and course of this devastating disease. Additional, highly informative and relevant methods such as high-throughput gene and protein expression studies are now available. The intelligent integration of knowledge derived from genetic, transcriptomic and proteomic studies will greatly advance our understanding of the causes of AD, it will improve our capability of establishing an early diagnosis, it will help defining disease subgroups and it will ultimately help to pave the road towards improved and tailored treatments.
Acknowledgments
Financial support: Supported by grants from the Swiss National Science Foundation (PP00B-68859 to A.P.), the National Institute of Mental Health (R01MH057899 to E.M.R), the National Institute on Aging (P30AG19610 to E.M.R.), and the State of Arizona (D.A.S and E.M.R).
Reference List
- 1.Evans DA, Funkenstein HH, Albert MS, et al. Prevalence of Alzheimer’s disease in a community population of older persons. Higher than previously reported. JAMA. 1989;262:2551–2556. [PubMed] [Google Scholar]
- 2.Gatz M, Pedersen NL, Berg S, et al. Heritability for Alzheimer’s disease: the study of dementia in Swedish twins. J Gerontol A Biol Sci Med Sci. 1997;52:M117–M125. doi: 10.1093/gerona/52a.2.m117. [DOI] [PubMed] [Google Scholar]
- 3.Heston LL, Mastri AR, Anderson VE, et al. Dementia of the Alzheimer type. Clinical genetics, natural history, and associated conditions. Arch Gen Psychiatry. 1981;38:1085–1090. doi: 10.1001/archpsyc.1981.01780350019001. [DOI] [PubMed] [Google Scholar]
- 4.Heyman A, Wilkinson WE, Hurwitz BJ, et al. Alzheimer’s disease: genetic aspects and associated clinical disorders. Ann Neurol. 1983;14:507–515. doi: 10.1002/ana.410140503. [DOI] [PubMed] [Google Scholar]
- 5.Breitner JC, Folstein MF. Familial Alzheimer Dementia: a prevalent disorder with specific clinical features. Psychol Med. 1984;14:63–80. doi: 10.1017/s0033291700003081. [DOI] [PubMed] [Google Scholar]
- 6.Breitner JC, Folstein MF, Murphy EA. Familial aggregation in Alzheimer dementia--I A model for the age-dependent expression of an autosomal dominant gene. J Psychiatr Res. 1986;20:31–43. doi: 10.1016/0022-3956(86)90021-x. [DOI] [PubMed] [Google Scholar]
- 7.Huff FJ, Auerbach J, Chakravarti A, et al. Risk of dementia in relatives of patients with Alzheimer’s disease. Neurology. 1988;38:786–790. doi: 10.1212/wnl.38.5.786. [DOI] [PubMed] [Google Scholar]
- 8.Farrer LA, O’Sullivan DM, Cupples LA, et al. Assessment of genetic risk for Alzheimer’s disease among first-degree relatives. Ann Neurol. 1989;25:485–493. doi: 10.1002/ana.410250511. [DOI] [PubMed] [Google Scholar]
- 9.Korten AE, Jorm AF, Henderson AS, et al. Assessing the risk of Alzheimer’s disease in first-degree relatives of Alzheimer’s disease cases. Psychol Med. 1993;23:915–923. doi: 10.1017/s0033291700026386. [DOI] [PubMed] [Google Scholar]
- 10.Silverman JM, Raiford K, Edland S, et al. The Consortium to Establish a Registry for Alzheimer’s Disease (CERAD). Part VI Family history assessment: a multicenter study of first-degree relatives of Alzheimer’s disease probands and nondemented spouse controls. Neurology. 1994;44:1253–1259. doi: 10.1212/wnl.44.7.1253. [DOI] [PubMed] [Google Scholar]
- 11.Silverman JM, Li G, Zaccario ML, et al. Patterns of risk in first-degree relatives of patients with Alzheimer’s disease. Arch Gen Psychiatry. 1994;51:577–586. doi: 10.1001/archpsyc.1994.03950070069012. [DOI] [PubMed] [Google Scholar]
- 12.Heun R, Papassotiropoulos A, Jessen F, et al. A family study of Alzheimer disease and early- and late-onset depression in elderly patients. Arch Gen Psychiatry. 2001;58:190–196. doi: 10.1001/archpsyc.58.2.190. [DOI] [PubMed] [Google Scholar]
- 13.Roberts SB, MacLean CJ, Neale MC, et al. Replication of linkage studies of complex traits: an examination of variation in location estimates. Am J Hum Genet. 1999;65:876–884. doi: 10.1086/302528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farrer LA, Cupples LA, Haines JL, et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA. 1997;278:1349–1356. [PubMed] [Google Scholar]
- 15.Horvath S, Xu X, Laird NM. The family based association test method: strategies for studying general genotype--phenotype associations. Eur J Hum Genet. 2001;9:301–306. doi: 10.1038/sj.ejhg.5200625. [DOI] [PubMed] [Google Scholar]
- 16.Pritchard JK, Rosenberg NA. Use of unlinked genetic markers to detect population stratification in association studies. Am J Hum Genet. 1999;65:220–228. doi: 10.1086/302449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 18.Lohmueller KE, Pearce CL, Pike M, et al. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet. 2003;33:177–182. doi: 10.1038/ng1071. [DOI] [PubMed] [Google Scholar]
- 19.Breslow JL, Zannis VI, SanGiacomo TR, et al. Studies of familial type III hyperlipoproteinemia using as a genetic marker the apoE phenotype E2/2. J Lipid Res. 1982;23:1224–1235. [PubMed] [Google Scholar]
- 20.Poirier J, Davignon J, Bouthillier D, et al. Apolipoprotein E polymorphism and Alzheimer’s disease. Lancet. 1993;342:697–699. doi: 10.1016/0140-6736(93)91705-q. [DOI] [PubMed] [Google Scholar]
- 21.Saunders AM, Strittmatter WJ, Schmechel D, et al. Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer’s disease. Neurology. 1993;43:1467–1472. doi: 10.1212/wnl.43.8.1467. [DOI] [PubMed] [Google Scholar]
- 22.Osuntokun BO, Sahota A, Ogunniyi AO, et al. Lack of an association between apolipoprotein E epsilon 4 and Alzheimer’s disease in elderly Nigerians. Ann Neurol. 1995;38:463–465. doi: 10.1002/ana.410380319. [DOI] [PubMed] [Google Scholar]
- 23.Tang MX, Maestre G, Tsai WY, et al. Relative risk of Alzheimer disease and age-at-onset distributions, based on APOE genotypes among elderly African Americans, Caucasians, and Hispanics in New York City. Am J Hum Genet. 1996;58:574–584. [PMC free article] [PubMed] [Google Scholar]
- 24.Corbo RM, Scacchi R. Apolipoprotein E (APOE) allele distribution in the world. Is APOE*4 a ‘thrifty’ allele? Ann Hum Genet. 1999;63(Pt 4):301–310. doi: 10.1046/j.1469-1809.1999.6340301.x. [DOI] [PubMed] [Google Scholar]
- 25.Poirier J, Delisle MC, Quirion R, et al. Apolipoprotein E4 allele as a predictor of cholinergic deficits and treatment outcome in Alzheimer disease. Proc Natl Acad Sci U S A. 1995;92:12260–12264. doi: 10.1073/pnas.92.26.12260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roses AD. Apolipoprotein E and Alzheimer’s disease. A rapidly expanding field with medical and epidemiological consequences. Ann N Y Acad Sci. 1996;802:50–57. doi: 10.1111/j.1749-6632.1996.tb32598.x. [DOI] [PubMed] [Google Scholar]
- 27.American College of Medical Genetics/American Society of Human Genetics Working Group on ApoE and Alzheimer disease. Statement on use of apolipoprotein E testing for Alzheimer disease. JAMA. 1995;274:1627–1629. [PubMed] [Google Scholar]
- 28.McConnell LM, Sanders GD, Owens DK. Evaluation of genetic tests: APOE genotyping for the diagnosis of Alzheimer disease. Genet Test. 1999;3:47–53. doi: 10.1089/gte.1999.3.47. [DOI] [PubMed] [Google Scholar]
- 29.Cupples LA, Farrer LA, Sadovnick AD, et al. Estimating risk curves for first-degree relatives of patients with Alzheimer’s disease: the REVEAL study. Genet Med. 2004;6:192–196. doi: 10.1097/01.gim.0000132679.92238.58. [DOI] [PubMed] [Google Scholar]
- 30.LaRusse S, Roberts JS, Marteau TM, et al. Genetic susceptibility testing versus family history-based risk assessment: Impact on perceived risk of Alzheimer disease. Genet Med. 2005;7:48–53. doi: 10.1097/01.gim.0000151157.13716.6c. [DOI] [PubMed] [Google Scholar]
- 31.Guidelines for the molecular genetics predictive test in Huntington’s disease. International Huntington Association (IHA) and the World Federation of Neurology (WFN) Research Group on Huntington’s Chorea. Neurology. 1994;44:1533–1536. [PubMed] [Google Scholar]
- 32.Panegyres PK, Goldblatt J, Walpole I, et al. Genetic testing for Alzheimer’s disease. Med J Aust. 2000;172:339–343. doi: 10.5694/j.1326-5377.2000.tb123984.x. [DOI] [PubMed] [Google Scholar]
- 33.Papassotiropoulos A, Streffer JR, Tsolaki M, et al. Increased brain beta-amyloid load, phosphorylated tau, and risk of Alzheimer disease associated with an intronic CYP46 polymorphism. Arch Neurol. 2003;60:29–35. doi: 10.1001/archneur.60.1.29. [DOI] [PubMed] [Google Scholar]
- 34.Crawford FC, Freeman MJ, Schinka JA, et al. A polymorphism in the cystatin C gene is a novel risk factor for late-onset Alzheimer’s disease. Neurology. 2000;55:763–768. doi: 10.1212/wnl.55.6.763. [DOI] [PubMed] [Google Scholar]
- 35.Grimaldi LM, Casadei VM, Ferri C, et al. Association of early-onset Alzheimer’s disease with an interleukin-1alpha gene polymorphism. Ann Neurol. 2000;47:361–365. [PubMed] [Google Scholar]
- 36.Nicoll JA, Mrak RE, Graham DI, et al. Association of interleukin-1 gene polymorphisms with Alzheimer’s disease. Ann Neurol. 2000;47:365–368. [PMC free article] [PubMed] [Google Scholar]
- 37.Licastro F, Grimaldi LM, Bonafe M, et al. Interleukin-6 gene alleles affect the risk of Alzheimer’s disease and levels of the cytokine in blood and brain. Neurobiol Aging. 2003;24:921–926. doi: 10.1016/s0197-4580(03)00013-7. [DOI] [PubMed] [Google Scholar]
- 38.Papassotiropoulos A, Bagli M, Jessen F, et al. A genetic variation of the inflammatory cytokine interleukin-6 delays the initial onset and reduces the risk for sporadic Alzheimer’s disease. Ann Neurol. 1999;45:666–668. doi: 10.1002/1531-8249(199905)45:5<666::aid-ana18>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 39.Vollbach H, Heun R, Morris CM, et al. APOA1 polymorphism influences risk for early- onset nonfamiliar AD. Ann Neurol. 2005;58:436–441. doi: 10.1002/ana.20593. [DOI] [PubMed] [Google Scholar]
- 40.Bertram L, Hiltunen M, Parkinson M, et al. Family-based association between Alzheimer’s disease and variants in UBQLN1. N Engl J Med. 2005;352:884–894. doi: 10.1056/NEJMoa042765. [DOI] [PubMed] [Google Scholar]
- 41.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 42.Xu Q, Jia YB, Zhang BY, et al. Association study of an SNP combination pattern in the dopaminergic pathway in paranoid schizophrenia: a novel strategy for complex disorders. Mol Psychiatry. 2004;9:510–521. doi: 10.1038/sj.mp.4001472. [DOI] [PubMed] [Google Scholar]
- 43.Papassotiropoulos A, Wollmer MA, Tsolaki M, et al. A cluster of cholesterol-related genes confers susceptibility for Alzheimer’s disease. J Clin Psychiatry. 2005;66:940–947. [PubMed] [Google Scholar]
- 44.Klein RJ, Zeiss C, Chew EY, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 46.Reiman EM, Chen K, Alexander GE, et al. Correlations between apolipoprotein E epsilon4 gene dose and brain-imaging measurements of regional hypometabolism. Proc Natl Acad Sci U S A. 2005;102:8299–8302. doi: 10.1073/pnas.0500579102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.St George-Hyslop PH, Tanzi RE, Polinsky RJ, et al. The genetic defect causing familial Alzheimer’s disease maps on chromosome 21. Science. 1987;235:885–890. doi: 10.1126/science.2880399. [DOI] [PubMed] [Google Scholar]
- 48.Tanzi RE, Gusella JF, Watkins PC, et al. Amyloid beta protein gene: cDNA, mRNA distribution, and genetic linkage near the Alzheimer locus. Science. 1987;235:880–884. doi: 10.1126/science.2949367. [DOI] [PubMed] [Google Scholar]
- 49.Levy E, Carman MD, Fernandez-Madrid IJ, et al. Mutation of the Alzheimer’s disease amyloid gene in hereditary cerebral hemorrhage, Dutch type. Science. 1990;248:1124–1126. doi: 10.1126/science.2111584. [DOI] [PubMed] [Google Scholar]
- 50.Goate A, Chartier-Harlin MC, Mullan M, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature. 1991;349:704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- 51.Hardy JA, Higgins GA. Alzheimer’s disease: the amyloid cascade hypothesis. Science. 1992;256:184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- 52.Schellenberg GD, Bird TD, Wijsman EM, et al. Genetic linkage evidence for a familial Alzheimer’s disease locus on chromosome 14. Science. 1992;258:668–671. doi: 10.1126/science.1411576. [DOI] [PubMed] [Google Scholar]
- 53.Sherrington R, Rogaev EI, Liang Y, et al. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer’s disease. Nature. 1995;375:754–760. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- 54.Levy-Lahad E, Lahad A, Wijsman EM, et al. Apolipoprotein E genotypes and age of onset in early-onset familial Alzheimer’s disease. Ann Neurol. 1995;38:678–680. doi: 10.1002/ana.410380420. [DOI] [PubMed] [Google Scholar]
- 55.Axelman K, Basun H, Lannfelt L. Wide range of disease onset in a family with Alzheimer disease and a His163Tyr mutation in the presenilin-1 gene. Arch Neurol. 1998;55:698–702. doi: 10.1001/archneur.55.5.698. [DOI] [PubMed] [Google Scholar]
- 56.Campion D, Brice A, Hannequin D, et al. A large pedigree with early-onset Alzheimer’s disease: clinical, neuropathologic, and genetic characterization. Neurology. 1995;45:80–85. doi: 10.1212/wnl.45.1.80. [DOI] [PubMed] [Google Scholar]
- 57.Ezquerra M, Carnero C, Blesa R, et al. A presenilin 1 mutation (Ser169Pro) associated with early-onset AD and myoclonic seizures. Neurology. 1999;52:566–570. doi: 10.1212/wnl.52.3.566. [DOI] [PubMed] [Google Scholar]
- 58.Janssen JC, Hall M, Fox NC, et al. Alzheimer’s disease due to an intronic presenilin-1 (PSEN1 intron 4) mutation: A clinicopathological study. Brain. 2000;123(Pt 5):894–907. doi: 10.1093/brain/123.5.894. [DOI] [PubMed] [Google Scholar]
- 59.Kennedy AM, Newman SK, Frackowiak RS, et al. Chromosome 14 linked familial Alzheimer’s disease. A clinico-pathological study of a single pedigree. Brain. 1995;118(Pt 1):185–205. doi: 10.1093/brain/118.1.185. [DOI] [PubMed] [Google Scholar]
- 60.Lampe TH, Bird TD, Nochlin D, et al. Phenotype of chromosome 14-linked familial Alzheimer’s disease in a large kindred. Ann Neurol. 1994;36:368–378. doi: 10.1002/ana.410360308. [DOI] [PubMed] [Google Scholar]
- 61.Reznik-Wolf H, Treves TA, Davidson M, et al. A novel mutation of presenilin 1 in familial Alzheimer’s disease in Israel detected by denaturing gradient gel electrophoresis. Hum Genet. 1996;98:700–702. doi: 10.1007/s004390050288. [DOI] [PubMed] [Google Scholar]
- 62.Scheuner D, Eckman C, Jensen M, et al. Secreted amyloid beta-protein similar to that in the senile plaques of Alzheimer’s disease is increased in vivo by the presenilin 1 and 2 and APP mutations linked to familial Alzheimer’s disease. Nat Med. 1996;2:864–870. doi: 10.1038/nm0896-864. [DOI] [PubMed] [Google Scholar]
- 63.Marjaux E, Hartmann D, De SB. Presenilins in memory, Alzheimer’s disease, and therapy. Neuron. 2004;42:189–192. doi: 10.1016/s0896-6273(04)00218-1. [DOI] [PubMed] [Google Scholar]
- 64.Bird TD, Lampe TH, Nemens EJ, et al. Familial Alzheimer’s disease in American descendants of the Volga Germans: probable genetic founder effect. Ann Neurol. 1988;23:25–31. doi: 10.1002/ana.410230106. [DOI] [PubMed] [Google Scholar]
- 65.Levy-Lahad E, Wijsman EM, Nemens E, et al. A familial Alzheimer’s disease locus on chromosome 1. Science. 1995;269:970–973. doi: 10.1126/science.7638621. [DOI] [PubMed] [Google Scholar]
- 66.Ho L, Guo Y, Spielman L, et al. Altered expression of a-type but not b-type synapsin isoform in the brain of patients at high risk for Alzheimer’s disease assessed by DNA microarray technique. Neurosci Lett. 2001;298:191–194. doi: 10.1016/s0304-3940(00)01753-5. [DOI] [PubMed] [Google Scholar]
- 67.Colangelo V, Schurr J, Ball MJ, et al. Gene expression profiling of 12633 genes in Alzheimer hippocampal CA1: transcription and neurotrophic factor down-regulation and up-regulation of apoptotic and pro-inflammatory signaling. J Neurosci Res. 2002;70:462–473. doi: 10.1002/jnr.10351. [DOI] [PubMed] [Google Scholar]
- 68.Hata R, Masumura M, Akatsu H, et al. Up-regulation of calcineurin Abeta mRNA in the Alzheimer’s disease brain: assessment by cDNA microarray. Biochem Biophys Res Commun. 2001;284:310–316. doi: 10.1006/bbrc.2001.4968. [DOI] [PubMed] [Google Scholar]
- 69.Beglopoulos V, Sun X, Saura CA, et al. Reduced beta-amyloid production and increased inflammatory responses in presenilin conditional knock-out mice. J Biol Chem. 2004;279:46907–46914. doi: 10.1074/jbc.M409544200. [DOI] [PubMed] [Google Scholar]
- 70.Kim JR, Lee SR, Chung HJ, et al. Identification of amyloid beta-peptide responsive genes by cDNA microarray technology: involvement of RTP801 in amyloid beta-peptide toxicity. Exp Mol Med. 2003;35:403–411. doi: 10.1038/emm.2003.53. [DOI] [PubMed] [Google Scholar]
- 71.Wu ZL, Ciallella JR, Flood DG, et al. Comparative analysis of cortical gene expression in mouse models of Alzheimer’s disease. Neurobiol Aging. 2005 doi: 10.1016/j.neurobiolaging.2005.02.010. in press. [DOI] [PubMed] [Google Scholar]
- 72.Kong LN, Zuo PP, Mu L, et al. Gene expression profile of amyloid beta protein-injected mouse model for Alzheimer disease. Acta Pharmacol Sin. 2005;26:666–672. doi: 10.1111/j.1745-7254.2005.00129.x. [DOI] [PubMed] [Google Scholar]
- 73.Mirnics K, Korade Z, Arion D, et al. Presenilin-1-dependent transcriptome changes. J Neurosci. 2005;25:1571–1578. doi: 10.1523/JNEUROSCI.4145-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen F, Wollmer MA, Hoerndli F, et al. Role for glyoxalase I in Alzheimer’s disease. Proc Natl Acad Sci U S A. 2004;101:7687–7692. doi: 10.1073/pnas.0402338101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mufson EJ, Counts SE, Ginsberg SD. Gene expression profiles of cholinergic nucleus basalis neurons in Alzheimer’s disease. Neurochem Res. 2002;27:1035–1048. doi: 10.1023/a:1020952704398. [DOI] [PubMed] [Google Scholar]
- 76.Ginsberg SD, Hemby SE, Lee VM, et al. Expression profile of transcripts in Alzheimer’s disease tangle-bearing CA1 neurons. Ann Neurol. 2000;48:77–87. [PubMed] [Google Scholar]
- 77.Dunckley T, Beach TG, Ramsey KE, et al. Gene expression correlates of neurofibrillary tangles in Alzheimer’s disease. Neurobiol Aging. 2005 doi: 10.1016/j.neurobiolaging.2005.08.013. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.David DC, Hoerndli F, Gotz J. Functional Genomics meets neurodegenerative disorders Part I: transcriptomic and proteomic technology. Prog Neurobiol. 2005;76:153–168. doi: 10.1016/j.pneurobio.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 79.Hoerndli F, David DC, Gotz J. Functional Genomics meets neurodegenerative disorders. Part II: application and data integration. Prog Neurobiol. 2005;76:169–188. doi: 10.1016/j.pneurobio.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 80.Gillardon F, Steinlein P, Burger E, et al. Phosphoproteome and transcriptome analysis of the neuronal response to a CDK5 inhibitor. Proteomics. 2005;5:1299–1307. doi: 10.1002/pmic.200400992. [DOI] [PubMed] [Google Scholar]
- 81.Fountoulakis M. Proteomics: current technologies and applications in neurological disorders and toxicology. Amino Acids. 2001;21:363–381. doi: 10.1007/s007260170002. [DOI] [PubMed] [Google Scholar]
- 82.Fountoulakis M. Encyclopedia of Separation Science, II/Electrophoresis. London: Academic Press; 2000. Two-dimensional electrophoresis; pp. 1356–1363. [Google Scholar]
- 83.Fountoulakis M, Takacs B. Enrichment and proteomic analysis of low-abundance bacterial proteins. Methods Enzymol. 2002;358:288–306. doi: 10.1016/s0076-6879(02)58096-4. [DOI] [PubMed] [Google Scholar]
- 84.Righetti PG, Castagna A, Herbert B, et al. Prefractionation techniques in proteome analysis. Proteomics. 2003;3:1397–1407. doi: 10.1002/pmic.200300472. [DOI] [PubMed] [Google Scholar]
- 85.O’Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 86.Klose J. Protein mapping by combined isoelectric focusing and electrophoresis of mouse tissues. A novel approach to testing for induced point mutations in mammals. Humangenetik. 1975;26:231–243. doi: 10.1007/BF00281458. [DOI] [PubMed] [Google Scholar]
- 87.Lahm HW, Langen H. Mass spectrometry: a tool for the identification of proteins separated by gels. Electrophoresis. 2000;21:2105–2114. doi: 10.1002/1522-2683(20000601)21:11<2105::AID-ELPS2105>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 88.Jiang L, Lindpaintner K, Li HF, et al. Proteomic analysis of the cerebrospinal fluid of patients with schizophrenia. Amino Acids. 2003;25:49–57. doi: 10.1007/s00726-003-0356-6. [DOI] [PubMed] [Google Scholar]
- 89.Fountoulakis M. Application of proteomics technologies in the investigation of the brain. Mass Spectrom Rev. 2004;23:231–258. doi: 10.1002/mas.10075. [DOI] [PubMed] [Google Scholar]
- 90.Cash P. Proteomics in medical microbiology. Electrophoresis. 2000;21:1187–1201. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1187::AID-ELPS1187>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 91.Rohlff C. Proteomics in molecular medicine: applications in central nervous systems disorders. Electrophoresis. 2000;21:1227–1234. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1227::AID-ELPS1227>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 92.Cordwell SJ, Nouwens AS, Walsh BJ. Comparative proteomics of bacterial pathogens. Proteomics. 2001;1:461–472. doi: 10.1002/1615-9861(200104)1:4<461::AID-PROT461>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 93.Simpson RJ, Dorow DS. Cancer proteomics: from signaling networks to tumor markers. Trends Biotechnol. 2001;19:S40–S48. doi: 10.1016/S0167-7799(01)01801-7. [DOI] [PubMed] [Google Scholar]
- 94.Hanash S. Disease proteomics. Nature. 2003;422:226–232. doi: 10.1038/nature01514. [DOI] [PubMed] [Google Scholar]
- 95.Antonarakis SE. 10 years of Genomics, chromosome 21, and Down syndrome. Genomics. 1998;51:1–16. doi: 10.1006/geno.1998.5335. [DOI] [PubMed] [Google Scholar]
- 96.Cairns NJ. Neuropathology. J Neural Transm Suppl. 1999;57:61–74. doi: 10.1007/978-3-7091-6380-1_4. [DOI] [PubMed] [Google Scholar]
- 97.Engidawork E, Lubec G. Protein expression in Down syndrome brain. Amino Acids. 2001;21:331–361. doi: 10.1007/s007260170001. [DOI] [PubMed] [Google Scholar]
- 98.Cheon MS, Kim SH, Yaspo ML, et al. Protein levels of genes encoded on chromosome 21 in fetal Down syndrome brain: challenging the gene dosage effect hypothesis (Part I) Amino Acids. 2003;24:111–117. doi: 10.1007/s00726-002-0336-2. [DOI] [PubMed] [Google Scholar]
- 99.Liao L, Cheng D, Wang J, et al. Proteomic characterization of postmortem amyloid plaques isolated by laser capture microdissection. J Biol Chem. 2004;279:37061–37068. doi: 10.1074/jbc.M403672200. [DOI] [PubMed] [Google Scholar]
- 100.Greber S, Lubec G, Cairns N, et al. Decreased levels of synaptosomal associated protein 25 in the brain of patients with Down syndrome and Alzheimer’s disease. Electrophoresis. 1999;20:928–934. doi: 10.1002/(SICI)1522-2683(19990101)20:4/5<928::AID-ELPS928>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 101.Agashe VR, Hartl FU. Roles of molecular chaperones in cytoplasmic protein folding. Semin Cell Dev Biol. 2000;11:15–25. doi: 10.1006/scdb.1999.0347. [DOI] [PubMed] [Google Scholar]
- 102.Ellis RJ. Molecular chaperones ten years. Introduction Semin Cell Dev Biol. 2000;11:1–5. doi: 10.1006/scdb.1999.0345. [DOI] [PubMed] [Google Scholar]
- 103.Yoo BC, Kim SH, Cairns N, et al. Deranged expression of molecular chaperones in brains of patients with Alzheimer’s disease. Biochem Biophys Res Commun. 2001;280:249–258. doi: 10.1006/bbrc.2000.4109. [DOI] [PubMed] [Google Scholar]
- 104.Castegna A, Aksenov M, Thongboonkerd V, et al. Proteomic identification of oxidatively modified proteins in Alzheimer’s disease brain. Part II: dihydropyrimidinase-related protein 2, alpha-enolase and heat shock cognate 71. J Neurochem. 2002;82:1524–1532. doi: 10.1046/j.1471-4159.2002.01103.x. [DOI] [PubMed] [Google Scholar]
- 105.Castegna A, Aksenov M, Aksenova M, et al. Proteomic identification of oxidatively modified proteins in Alzheimer’s disease brain. Part I: creatine kinase BB, glutamine synthase, and ubiquitin carboxy-terminal hydrolase L-1. Free Radic Biol Med. 2002;33:562–571. doi: 10.1016/s0891-5849(02)00914-0. [DOI] [PubMed] [Google Scholar]
- 106.Yoo BC, Vlkolinsky R, Engidawork E, et al. Differential expression of molecular chaperones in brain of patients with Down syndrome. Electrophoresis. 2001;22:1233–1241. doi: 10.1002/1522-2683()22:6<1233::AID-ELPS1233>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 107.Yoo BC, Cairns N, Fountoulakis M, et al. Synaptosomal proteins, beta-soluble N-ethylmaleimide-sensitive factor attachment protein (beta-SNAP), gamma-SNAP and synaptotagmin I in brain of patients with Down syndrome and Alzheimer’s disease. Dement Geriatr Cogn Disord. 2001;12:219–225. doi: 10.1159/000051261. [DOI] [PubMed] [Google Scholar]
- 108.Lubec G, Nonaka M, Krapfenbauer K, et al. Expression of the dihydropyrimidinase related protein 2 (DRP-2) in Down syndrome and Alzheimer’s disease brain is downregulated at the mRNA and dysregulated at the protein level. J Neural Transm Suppl. 1999;57:161–177. doi: 10.1007/978-3-7091-6380-1_10. [DOI] [PubMed] [Google Scholar]
- 109.Butterfield DA, Boyd-Kimball D, Castegna A. Proteomics in Alzheimer’s disease: insights into potential mechanisms of neurodegeneration. J Neurochem. 2003;86:1313–1327. doi: 10.1046/j.1471-4159.2003.01948.x. [DOI] [PubMed] [Google Scholar]
- 110.Vlkolinsky R, Cairns N, Fountoulakis M, et al. Decreased brain levels of 2′,3′-cyclic nucleotide-3′-phosphodiesterase in Down syndrome and Alzheimer’s disease. Neurobiol Aging. 2001;22:547–553. doi: 10.1016/s0197-4580(01)00218-4. [DOI] [PubMed] [Google Scholar]
- 111.Kim SH, Fountoulakis M, Cairns NJ, et al. Human brain nucleoside diphosphate kinase activity is decreased in Alzheimer’s disease and Down syndrome. Biochem Biophys Res Commun. 2002;296:970–975. doi: 10.1016/s0006-291x(02)02035-1. [DOI] [PubMed] [Google Scholar]
- 112.Cheon MS, Fountoulakis M, Cairns NJ, et al. Decreased protein levels of stathmin in adult brains with Down syndrome and Alzheimer’s disease. J Neural Transm Suppl. 2001:281–288. doi: 10.1007/978-3-7091-6262-0_23. [DOI] [PubMed] [Google Scholar]
- 113.Fountoulakis M, Cairns N, Lubec G. Increased levels of 14-3-3 gamma and epsilon proteins in brain of patients with Alzheimer’s disease and Down syndrome. J Neural Transm Suppl. 1999;57:323–335. doi: 10.1007/978-3-7091-6380-1_23. [DOI] [PubMed] [Google Scholar]
- 114.Engidawork E, Lubec G. Molecular changes in fetal Down syndrome brain. J Neurochem. 2003;84:895–904. doi: 10.1046/j.1471-4159.2003.01614.x. [DOI] [PubMed] [Google Scholar]
- 115.Engidawork E, Lubec G. Protein expression in Down syndrome brain. Amino Acids. 2001;21:331–361. doi: 10.1007/s007260170001. [DOI] [PubMed] [Google Scholar]
- 116.Gulesserian T, Seidl R, Hardmeier R, et al. Superoxide dismutase SOD1, encoded on chromosome 21, but not SOD2 is overexpressed in brains of patients with Down syndrome. J Investig Med. 2001;49:41–46. doi: 10.2310/6650.2001.34089. [DOI] [PubMed] [Google Scholar]
- 117.Balcz B, Kirchner L, Cairns N, et al. Increased brain protein levels of carbonyl reductase and alcohol dehydrogenase in Down syndrome and Alzheimer’s disease. J Neural Transm Suppl. 2001:193–201. doi: 10.1007/978-3-7091-6262-0_15. [DOI] [PubMed] [Google Scholar]
- 118.Krapfenbauer K, Engidawork E, Cairns N, et al. Aberrant expression of peroxiredoxin subtypes in neurodegenerative disorders. Brain Res. 2003;967:152–160. doi: 10.1016/s0006-8993(02)04243-9. [DOI] [PubMed] [Google Scholar]
- 119.Kim SH, Fountoulakis M, Cairns N, et al. Protein levels of human peroxiredoxin subtypes in brains of patients with Alzheimer’s disease and Down syndrome. J Neural Transm Suppl. 2001:223–235. doi: 10.1007/978-3-7091-6262-0_18. [DOI] [PubMed] [Google Scholar]
- 120.Kim H, Lee TH, Park ES, et al. Role of peroxiredoxins in regulating intracellular hydrogen peroxide and hydrogen peroxide-induced apoptosis in thyroid cells. J Biol Chem. 2000;275:18266–18270. doi: 10.1074/jbc.275.24.18266. [DOI] [PubMed] [Google Scholar]
- 121.Engidawork E, Gulesserian T, Seidl R, et al. Expression of apoptosis related proteins in brains of patients with Alzheimer’s disease. Neurosci Lett. 2001;303:79–82. doi: 10.1016/s0304-3940(01)01618-4. [DOI] [PubMed] [Google Scholar]
- 122.Butterfield DA. Amyloid beta-peptide (1-42)-induced oxidative stress and neurotoxicity: implications for neurodegeneration in Alzheimer’s disease brain. A review. Free Radic Res. 2002;36:1307–1313. doi: 10.1080/1071576021000049890. [DOI] [PubMed] [Google Scholar]
- 123.Butterfield DA. Proteomics: a new approach to investigate oxidative stress in Alzheimer’s disease brain. Brain Res. 2004;1000:1–7. doi: 10.1016/j.brainres.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 124.Lubec G, Krapfenbauer K, Fountoulakis M. Proteomics in brain research: potentials and limitations. Prog Neurobiol. 2003;69:193–211. doi: 10.1016/s0301-0082(03)00036-4. [DOI] [PubMed] [Google Scholar]
- 125.Mullan M, Crawford F, Axelman K, et al. A pathogenic mutation for probable Alzheimer’s disease in the APP gene at the N-terminus of beta-amyloid. Nat Genet. 1992;1:345–347. doi: 10.1038/ng0892-345. [DOI] [PubMed] [Google Scholar]
- 126.Wakutani Y, Watanabe K, Adachi Y, et al. Novel amyloid precursor protein gene missense mutation (D678N) in probable familial Alzheimer’s disease. J Neurol Neurosurg Psychiatry. 2004;75:1039–1042. doi: 10.1136/jnnp.2003.010611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hendriks L, van Duijn CM, Cras P, et al. Presenile dementia and cerebral haemorrhage linked to a mutation at codon 692 of the beta-amyloid precursor protein gene. Nat Genet. 1992;1:218–221. doi: 10.1038/ng0692-218. [DOI] [PubMed] [Google Scholar]
- 128.Kamino K, Orr HT, Payami H, et al. Linkage and mutational analysis of familial Alzheimer disease kindreds for the APP gene region. Am J Hum Genet. 1992;51:998–1014. [PMC free article] [PubMed] [Google Scholar]
- 129.Grabowski TJ, Cho HS, Vonsattel JP, et al. Novel amyloid precursor protein mutation in an Iowa family with dementia and severe cerebral amyloid angiopathy. Ann Neurol. 2001;49:697–705. doi: 10.1002/ana.1009. [DOI] [PubMed] [Google Scholar]
- 130.Armstrong J, Boada M, Rey MJ, et al. Familial Alzheimer disease associated with A713T mutation in APP. Neurosci Lett. 2004;370:241–243. doi: 10.1016/j.neulet.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 131.Rossi G, Giaccone G, Maletta R, et al. A family with Alzheimer disease and strokes associated with A713T mutation of the APP gene. Neurology. 2004;63:910–912. doi: 10.1212/01.wnl.0000137048.80666.86. [DOI] [PubMed] [Google Scholar]
- 132.Pasalar P, Najmabadi H, Noorian AR, et al. An Iranian family with Alzheimer’s disease caused by a novel APP mutation (Thr714Ala) Neurology. 2002;58:1574–1575. doi: 10.1212/wnl.58.10.1574. [DOI] [PubMed] [Google Scholar]
- 133.Kumar-Singh S, De JC, Cruts M, et al. Nonfibrillar diffuse amyloid deposition due to a gamma(42)-secretase site mutation points to an essential role for N-truncated A beta(42) in Alzheimer’s disease. Hum Mol Genet. 2000;9:2589–2598. doi: 10.1093/hmg/9.18.2589. [DOI] [PubMed] [Google Scholar]
- 134.Ancolio K, Dumanchin C, Barelli H, et al. Unusual phenotypic alteration of beta amyloid precursor protein (betaAPP) maturation by a new Val-715 --> Met betaAPP-770 mutation responsible for probable early-onset Alzheimer’s disease. Proc Natl Acad Sci U S A. 1999;96:4119–4124. doi: 10.1073/pnas.96.7.4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.De JC, Esselens C, Kumar-Singh S, et al. Pathogenic APP mutations near the gamma-secretase cleavage site differentially affect Abeta secretion and APP C-terminal fragment stability. Hum Mol Genet. 2001;10:1665–1671. doi: 10.1093/hmg/10.16.1665. [DOI] [PubMed] [Google Scholar]
- 136.Eckman CB, Mehta ND, Crook R, et al. A new pathogenic mutation in the APP gene (I716V) increases the relative proportion of A beta 42(43) Hum Mol Genet. 1997;6:2087–2089. doi: 10.1093/hmg/6.12.2087. [DOI] [PubMed] [Google Scholar]
- 137.Murrell J, Farlow M, Ghetti B, et al. A mutation in the amyloid precursor protein associated with hereditary Alzheimer’s disease. Science. 1991;254:97–99. doi: 10.1126/science.1925564. [DOI] [PubMed] [Google Scholar]
- 138.Chartier-Harlin MC, Crawford F, Houlden H, et al. Early-onset Alzheimer’s disease caused by mutations at codon 717 of the beta-amyloid precursor protein gene. Nature. 1991;353:844–846. doi: 10.1038/353844a0. [DOI] [PubMed] [Google Scholar]
- 139.Murrell JR, Hake AM, Quaid KA, et al. Early-onset Alzheimer disease caused by a new mutation (V717L) in the amyloid precursor protein gene. Arch Neurol. 2000;57:885–887. doi: 10.1001/archneur.57.6.885. [DOI] [PubMed] [Google Scholar]
- 140.Kwok JB, Li QX, Hallupp M, et al. Novel Leu723Pro amyloid precursor protein mutation increases amyloid beta42(43) peptide levels and induces apoptosis. Ann Neurol. 2000;47:249–253. doi: 10.1002/1531-8249(200002)47:2<249::aid-ana18>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 141.Rogaeva EA, Fafel KC, Song YQ, et al. Screening for PS1 mutations in a referral-based series of AD cases: 21 novel mutations. Neurology. 2001;57:621–625. doi: 10.1212/wnl.57.4.621. [DOI] [PubMed] [Google Scholar]
- 142.Dumanchin C, Brice A, Campion D, et al. De novo presenilin 1 mutations are rare in clinically sporadic, early onset Alzheimer’s disease cases. French Alzheimer’s Disease Study Group. J Med Genet. 1998;35:672–673. doi: 10.1136/jmg.35.8.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cruts M, Van BC. Presenilin mutations in Alzheimer’s disease. Hum Mutat. 1998;11:183–190. doi: 10.1002/(SICI)1098-1004(1998)11:3<183::AID-HUMU1>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 144.Rossor MN, Fox NC, Beck J, et al. Incomplete penetrance of familial Alzheimer’s disease in a pedigree with a novel presenilin-1 gene mutation. Lancet. 1996;347:1560. doi: 10.1016/s0140-6736(96)90715-1. [DOI] [PubMed] [Google Scholar]
- 145.Campion D, Flaman JM, Brice A, et al. Mutations of the presenilin I gene in families with early-onset Alzheimer’s disease. Hum Mol Genet. 1995;4:2373–2377. doi: 10.1093/hmg/4.12.2373. [DOI] [PubMed] [Google Scholar]
- 146.Cruts M, Backhovens H, Wang SY, et al. Molecular genetic analysis of familial early-onset Alzheimer’s disease linked to chromosome 14q24.3. Hum Mol Genet. 1995;4:2363–2371. doi: 10.1093/hmg/4.12.2363. [DOI] [PubMed] [Google Scholar]
- 147.Houlden H, Baker M, McGowan E, et al. Variant Alzheimer’s disease with spastic paraparesis and cotton wool plaques is caused by PS-1 mutations that lead to exceptionally high amyloid-beta concentrations. Ann Neurol. 2000;48:806–808. [PubMed] [Google Scholar]
- 148.Sorbi S, Nacmias B, Forleo P, et al. Missense mutation of S182 gene in Italian families with early-onset Alzheimer’s disease. Lancet. 1995;346:439–440. doi: 10.1016/s0140-6736(95)92809-x. [DOI] [PubMed] [Google Scholar]
- 149.Ataka S, Tomiyama T, Takuma H, et al. A novel presenilin-1 mutation (Leu85Pro) in early-onset Alzheimer disease with spastic paraparesis. Arch Neurol. 2004;61:1773–1776. doi: 10.1001/archneur.61.11.1773. [DOI] [PubMed] [Google Scholar]
- 150.Queralt R, Ezquerra M, Lleo A, et al. A novel mutation (V89L) in the presenilin 1 gene in a family with early onset Alzheimer’s disease and marked behavioural disturbances. J Neurol Neurosurg Psychiatry. 2002;72:266–269. doi: 10.1136/jnnp.72.2.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Alzheimer’s Disease Collaborative Group. The structure of the presenilin 1 (S182) gene and identification of six novel mutations in early onset AD families. Nat Genet. 1995;11:219–222. doi: 10.1038/ng1095-219. [DOI] [PubMed] [Google Scholar]
- 152.Tedde A, Nacmias B, Ciantelli M, et al. Identification of new presenilin gene mutations in early-onset familial Alzheimer disease. Arch Neurol. 2003;60:1541–1544. doi: 10.1001/archneur.60.11.1541. [DOI] [PubMed] [Google Scholar]
- 153.Jorgensen P, Bus C, Pallisgaard N, et al. Familial Alzheimer’s disease co-segregates with a Met146I1e substitution in presenilin-1. Clin Genet. 1996;50:281–286. [PubMed] [Google Scholar]
- 154.Arango D, Cruts M, Torres O, et al. Systematic genetic study of Alzheimer disease in Latin America: mutation frequencies of the amyloid beta precursor protein and presenilin genes in Colombia. Am J Med Genet. 2001;103:138–143. doi: 10.1002/1096-8628(20011001)103:2<138::aid-ajmg1529>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 155.Gustafson L, Brun A, Englund E, et al. A 50-year perspective of a family with chromosome-14-linked Alzheimer’s disease. Hum Genet. 1998;102:253–257. doi: 10.1007/s004390050688. [DOI] [PubMed] [Google Scholar]
- 156.Kamino K, Sato S, Sakaki Y, et al. Three different mutations of presenilin 1 gene in early- onset Alzheimer’s disease families. Neurosci Lett. 1996;208:195–198. doi: 10.1016/0304-3940(96)12587-8. [DOI] [PubMed] [Google Scholar]
- 157.Finckh U, Muller-Thomsen T, Mann U, et al. High prevalence of pathogenic mutations in patients with early-onset dementia detected by sequence analyses of four different genes. Am J Hum Genet. 2000;66:110–117. doi: 10.1086/302702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Campion D, Dumanchin C, Hannequin D, et al. Early-onset autosomal dominant Alzheimer disease: prevalence, genetic heterogeneity, and mutation spectrum. Am J Hum Genet. 1999;65:664–670. doi: 10.1086/302553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Raux G, Gantier R, Thomas-Anterion C, et al. Dementia with prominent frontotemporal features associated with L113P presenilin 1 mutation. Neurology. 2000;55:1577–1578. doi: 10.1212/wnl.55.10.1577. [DOI] [PubMed] [Google Scholar]
- 160.Raux G, Gantier R, Martin C, et al. A novel presenilin 1 missense mutation (L153V) segregating with early-onset autosomal dominant Alzheimer’s disease. Hum Mutat. 2000;16:95. doi: 10.1002/1098-1004(200007)16:1<95::AID-HUMU28>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 161.Tysoe C, Whittaker J, Xuereb J, et al. A presenilin-1 truncating mutation is present in two cases with autopsy-confirmed early-onset Alzheimer disease. Am J Hum Genet. 1998;62:70–76. doi: 10.1086/301672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Hattori S, Sakuma K, Wakutani Y, et al. A novel presenilin 1 mutation (Y154N) in a patient with early onset Alzheimer’s disease with spastic paraparesis. Neurosci Lett. 2004;368:319–322. doi: 10.1016/j.neulet.2004.07.057. [DOI] [PubMed] [Google Scholar]
- 163.Janssen JC, Beck JA, Campbell TA, et al. Early onset familial Alzheimer’s disease: Mutation frequency in 31 families. Neurology. 2003;60:235–239. doi: 10.1212/01.wnl.0000042088.22694.e3. [DOI] [PubMed] [Google Scholar]
- 164.Romero I, Jorgensen P, Bolwig G, et al. A presenilin-1 Thr116Asn substitution in a family with early-onset Alzheimer’s disease. Neuroreport. 1999;10:2255–2260. doi: 10.1097/00001756-199908020-00006. [DOI] [PubMed] [Google Scholar]
- 165.La BV, Liguori M, Cittadella R, et al. A novel mutation (Thr116Ile) in the presenilin 1 gene in a patient with early-onset Alzheimer’s disease. Eur J Neurol. 2004;11:521–524. doi: 10.1111/j.1468-1331.2004.00828.x. [DOI] [PubMed] [Google Scholar]
- 166.Wisniewski T, Dowjat WK, Buxbaum JD, et al. A novel Polish presenilin-1 mutation (P117L) is associated with familial Alzheimer’s disease and leads to death as early as the age of 28 years. Neuroreport. 1998;9:217–221. doi: 10.1097/00001756-199801260-00008. [DOI] [PubMed] [Google Scholar]
- 167.Dowjat WK, Kuchna I, Wisniewski T, et al. A novel highly pathogenic Alzheimer presenilin-1 mutation in codon 117 (Pro117Ser): Comparison of clinical, neuropathological and cell culture phenotypes of Pro117Leu and Pro117Ser mutations. J Alzheimers Dis. 2004;6:31–43. doi: 10.3233/jad-2004-6105. [DOI] [PubMed] [Google Scholar]
- 168.Ezquerra M, Carnero C, Blesa R, et al. A novel presenilin 1 mutation (Leu166Arg) associated with early-onset Alzheimer disease. Arch Neurol. 2000;57:485–488. doi: 10.1001/archneur.57.4.485. [DOI] [PubMed] [Google Scholar]
- 169.Zekanowski C, Styczynska M, Peplonska B, et al. Mutations in presenilin 1, presenilin 2 and amyloid precursor protein genes in patients with early-onset Alzheimer’s disease in Poland. Exp Neurol. 2003;184:991–996. doi: 10.1016/S0014-4886(03)00384-4. [DOI] [PubMed] [Google Scholar]
- 170.Moehlmann T, Winkler E, Xia X, et al. Presenilin-1 mutations of leucine 166 equally affect the generation of the Notch and APP intracellular domains independent of their effect on Abeta 42 production. Proc Natl Acad Sci U S A. 2002;99:8025–8030. doi: 10.1073/pnas.112686799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hutton M, Busfield F, Wragg M, et al. Complete analysis of the presenilin 1 gene in early onset Alzheimer’s disease. Neuroreport. 1996;7:801–805. doi: 10.1097/00001756-199602290-00029. [DOI] [PubMed] [Google Scholar]
- 172.Taddei K, Kwok JB, Kril JJ, et al. Two novel presenilin-1 mutations (Ser169Leu and Pro436Gln) associated with very early onset Alzheimer’s disease. Neuroreport. 1998;9:3335–3339. doi: 10.1097/00001756-199810050-00034. [DOI] [PubMed] [Google Scholar]
- 173.Poorkaj P, Sharma V, Anderson L, et al. Missense mutations in the chromosome 14 familial Alzheimer’s disease presenilin 1 gene. Hum Mutat. 1998;11:216–221. doi: 10.1002/(SICI)1098-1004(1998)11:3<216::AID-HUMU6>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 174.Yasuda M, Maeda K, Hashimoto M, et al. A pedigree with a novel presenilin 1 mutation at a residue that is not conserved in presenilin 2. Arch Neurol. 1999;56:65–69. doi: 10.1001/archneur.56.1.65. [DOI] [PubMed] [Google Scholar]
- 175.Ramirez-Duenas MG, Rogaeva EA, Leal CA, et al. A novel Leu171Pro mutation in presenilin-1 gene in a Mexican family with early onset Alzheimer disease. Ann Genet. 1998;41:149–153. [PubMed] [Google Scholar]
- 176.Crook R, Ellis R, Shanks M, et al. Early-onset Alzheimer’s disease with a presenilin-1 mutation at the site corresponding to the Volga German presenilin-2 mutation. Ann Neurol. 1997;42:124–128. doi: 10.1002/ana.410420121. [DOI] [PubMed] [Google Scholar]
- 177.Boteva K, Vitek M, Mitsuda H, et al. Mutation analysis of presenillin 1 gene in Alzheimer’s disease. Lancet. 1996;347:130–131. doi: 10.1016/s0140-6736(96)90261-5. [DOI] [PubMed] [Google Scholar]
- 178.Bertoli Avella AM, Marcheco TB, Llibre Rodriguez JJ, et al. A novel presenilin 1 mutation (L174 M) in a large Cuban family with early onset Alzheimer disease. Neurogenetics. 2002;4:97–104. doi: 10.1007/s10048-002-0136-6. [DOI] [PubMed] [Google Scholar]
- 179.Miklossy J, Taddei K, Suva D, et al. Two novel presenilin-1 mutations (Y256S and Q222H) are associated with early-onset Alzheimer’s disease. Neurobiol Aging. 2003;24:655–662. doi: 10.1016/s0197-4580(02)00192-6. [DOI] [PubMed] [Google Scholar]
- 180.Dermaut B, Kumar-Singh S, Engelborghs S, et al. A novel presenilin 1 mutation associated with Pick’s disease but not beta-amyloid plaques. Ann Neurol. 2004;55:617–626. doi: 10.1002/ana.20083. [DOI] [PubMed] [Google Scholar]
- 181.Rogaev EI, Sherrington R, Rogaeva EA, et al. Familial Alzheimer’s disease in kindreds with missense mutations in a gene on chromosome 1 related to the Alzheimer’s disease type 3 gene. Nature. 1995;376:775–778. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- 182.Yasuda M, Maeda K, Ikejiri Y, et al. A novel missense mutation in the presenilin-1 gene in a familial Alzheimer’s disease pedigree with abundant amyloid angiopathy. Neurosci Lett. 1997;232:29–32. doi: 10.1016/s0304-3940(97)00569-7. [DOI] [PubMed] [Google Scholar]
- 183.Forsell C, Froelich S, Axelman K, et al. A novel pathogenic mutation (Leu262Phe) found in the presenilin 1 gene in early-onset Alzheimer’s disease. Neurosci Lett. 1997;234:3–6. doi: 10.1016/s0304-3940(97)00603-4. [DOI] [PubMed] [Google Scholar]
- 184.Wasco W, Pettingell WP, Jondro PD, et al. Familial Alzheimer’s chromosome 14 mutations. Nat Med. 1995;1:848. doi: 10.1038/nm0995-848a. [DOI] [PubMed] [Google Scholar]
- 185.Goldman JS, Reed B, Gearhart R, et al. Very early-onset familial Alzheimer’s disease: a novel presenilin 1 mutation. Int J Geriatr Psychiatry. 2002;17:649–651. doi: 10.1002/gps.657. [DOI] [PubMed] [Google Scholar]
- 186.Sugiyama N, Suzuki K, Matsumura T, et al. A novel missense mutation (G209R) in exon 8 of the presenilin 1 gene in a Japanese family with presenile familial Alzheimer’s disease. Mutation in brief no. 254. Online Hum Mutat. 1999;14:90. doi: 10.1002/(SICI)1098-1004(1999)14:1<90::AID-HUMU19>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 187.Kowalska A, Pruchnik-Wolinska D, Florczak J, et al. Presenilin 1 mutations in Polish families with early-onset Alzheimer’s disease. Folia Neuropathol. 2004;42:9–14. [PubMed] [Google Scholar]
- 188.Perez-Tur J, Croxton R, Wright K, et al. A further presenilin 1 mutation in the exon 8 cluster in familial Alzheimer’s disease. Neurodegeneration. 1996;5:207–212. doi: 10.1006/neur.1996.0028. [DOI] [PubMed] [Google Scholar]
- 189.Gomez-Isla T, Wasco W, Pettingell WP, et al. A novel presenilin-1 mutation: increased beta-amyloid and neurofibrillary changes. Ann Neurol. 1997;41:809–813. doi: 10.1002/ana.410410618. [DOI] [PubMed] [Google Scholar]
- 190.Kwok JB, Halliday GM, Brooks WS, et al. Presenilin-1 mutation L271V results in altered exon 8 splicing and Alzheimer’s disease with non-cored plaques and no neuritic dystrophy. J Biol Chem. 2003;278:6748–6754. doi: 10.1074/jbc.M211827200. [DOI] [PubMed] [Google Scholar]
- 191.Takao M, Ghetti B, Hayakawa I, et al. A novel mutation (G217D) in the Presenilin 1 gene ( PSEN1) in a Japanese family: presenile dementia and parkinsonism are associated with cotton wool plaques in the cortex and striatum. Acta Neuropathol (Berl) 2002;104:155–170. doi: 10.1007/s00401-002-0536-6. [DOI] [PubMed] [Google Scholar]
- 192.Jimenez-Escrig A, Rabano A, Guerrero C, et al. New V272A presenilin 1 mutation with very early onset subcortical dementia and parkinsonism. Eur J Neurol. 2004;11:663–669. doi: 10.1111/j.1468-1331.2004.00865.x. [DOI] [PubMed] [Google Scholar]
- 193.Smith MJ, Gardner RJ, Knight MA, et al. Early-onset Alzheimer’s disease caused by a novel mutation at codon 219 of the presenilin-1 gene. Neuroreport. 1999;10:503–507. doi: 10.1097/00001756-199902250-00011. [DOI] [PubMed] [Google Scholar]
- 194.Kamimura K, Tanahashi H, Yamanaka H, et al. Familial Alzheimer’s disease genes in Japanese. J Neurol Sci. 1998;160:76–81. doi: 10.1016/s0022-510x(98)00219-6. [DOI] [PubMed] [Google Scholar]
- 195.Assini A, Terreni L, Borghi R, et al. Pure spastic paraparesis associated with a novel presenilin 1 R278K mutation. Neurology. 2003;60:150. doi: 10.1212/01.wnl.0000040252.43269.83. [DOI] [PubMed] [Google Scholar]
- 196.Coleman P, Kurlan R, Crook R, et al. A new presenilin Alzheimer’s disease case confirms the helical alignment of pathogenic mutations in transmembrane domain 5. Neurosci Lett. 2004;364:139–140. doi: 10.1016/j.neulet.2004.04.030. [DOI] [PubMed] [Google Scholar]
- 197.Godbolt AK, Beck JA, Collinge J, et al. A presenilin 1 R278I mutation presenting with language impairment. Neurology. 2004;63:1702–1704. doi: 10.1212/01.wnl.0000143060.98164.1a. [DOI] [PubMed] [Google Scholar]
- 198.Kwok JB, Taddei K, Hallupp M, et al. Two novel (M233T and R278T) presenilin-1 mutations in early-onset Alzheimer’s disease pedigrees and preliminary evidence for association of presenilin-1 mutations with a novel phenotype. Neuroreport. 1997;8:1537–1542. doi: 10.1097/00001756-199704140-00043. [DOI] [PubMed] [Google Scholar]
- 199.Aldudo J, Bullido MJ, Arbizu T, et al. Identification of a novel mutation (Leu282Arg) of the human presenilin 1 gene in Alzheimer’s disease. Neurosci Lett. 1998;240:174–176. doi: 10.1016/s0304-3940(97)00950-6. [DOI] [PubMed] [Google Scholar]
- 200.Aldudo J, Bullido MJ, Valdivieso F. DGGE method for the mutational analysis of the coding and proximal promoter regions of the Alzheimer’s disease presenilin-1 gene: two novel mutations. Hum Mutat. 1999;14:433–439. doi: 10.1002/(SICI)1098-1004(199911)14:5<433::AID-HUMU10>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 201.Dermaut B, Kumar-Singh S, De JC, et al. Cerebral amyloid angiopathy is a pathogenic lesion in Alzheimer’s disease due to a novel presenilin 1 mutation. Brain. 2001;124:2383–2392. doi: 10.1093/brain/124.12.2383. [DOI] [PubMed] [Google Scholar]
- 202.Houlden H, Crook R, Dolan RJ, et al. A novel presenilin mutation (M233V) causing very early onset Alzheimer’s disease with Lewy bodies. Neurosci Lett. 2001;313:93–95. doi: 10.1016/s0304-3940(01)02254-6. [DOI] [PubMed] [Google Scholar]
- 203.Tabira T, Chui dH, Nakayama H, et al. Alzheimer’s disease with spastic paresis and cotton wool type plaques. J Neurosci Res. 2002;70:367–372. doi: 10.1002/jnr.10392. [DOI] [PubMed] [Google Scholar]
- 204.Sodeyama N, Iwata T, Ishikawa K, et al. Very early onset Alzheimer’s disease with spastic paraparesis associated with a novel presenilin 1 mutation (Phe237Ile) J Neurol Neurosurg Psychiatry. 2001;71:556–557. doi: 10.1136/jnnp.71.4.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Perez-Tur J, Froelich S, Prihar G, et al. A mutation in Alzheimer’s disease destroying a splice acceptor site in the presenilin-1 gene. Neuroreport. 1995;7:297–301. [PubMed] [Google Scholar]
- 206.Crook R, Verkkoniemi A, Perez-Tur J, et al. A variant of Alzheimer’s disease with spastic paraparesis and unusual plaques due to deletion of exon 9 of presenilin 1. Nat Med. 1998;4:452–455. doi: 10.1038/nm0498-452. [DOI] [PubMed] [Google Scholar]
- 207.Furuya H, Yasuda M, Terasawa KJ, et al. A novel mutation (L250V) in the presenilin 1 gene in a Japanese familial Alzheimer’s disease with myoclonus and generalized convulsion. J Neurol Sci. 2003;209:75–77. doi: 10.1016/s0022-510x(02)00466-5. [DOI] [PubMed] [Google Scholar]
- 208.Besancon R, Lorenzi A, Cruts M, et al. Missense mutation in exon 11 (Codon 378) of the presenilin-1 gene in a French family with early-onset Alzheimer’s disease and transmission study by mismatch enhanced allele specific amplification. Mutations in brief no. 141. Online. besancon@rockefeller1.univ.lyon1.fr. Hum Mutat. 1998;11:481. doi: 10.1002/(SICI)1098-1004(1998)11:6<481::AID-HUMU12>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 209.Tedde A, Forleo P, Nacmias B, et al. A presenilin-1 mutation (Leu392Pro) in a familial AD kindred with psychiatric symptoms at onset. Neurology. 2000;55:1590–1591. doi: 10.1212/wnl.55.10.1590. [DOI] [PubMed] [Google Scholar]
- 210.Yasuda M, Maeda S, Kawamata T, et al. Novel presenilin-1 mutation with widespread cortical amyloid deposition but limited cerebral amyloid angiopathy. J Neurol Neurosurg Psychiatry. 2000;68:220–223. doi: 10.1136/jnnp.68.2.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Kowalska A, Forsell C, Florczak J, et al. A Polish pedigree with Alzheimer’s disease determined by a novel mutation in exon 12 of the presenilin 1 gene: clinical and molecular characterization. Folia Neuropathol. 1999;37:57–61. [PubMed] [Google Scholar]
- 212.Matsushita S, Arai H, Okamura N, et al. Clinical and biomarker investigation of a patient with a novel presenilin-1 mutation (A431V) in the mild cognitive impairment stage of Alzheimer’s disease. Biol Psychiatry. 2002;52:907–910. doi: 10.1016/s0006-3223(02)01386-0. [DOI] [PubMed] [Google Scholar]
- 213.Devi G, Fotiou A, Jyrinji D, et al. Novel presenilin 1 mutations associated with early onset of dementia in a family with both early-onset and late-onset Alzheimer disease. Arch Neurol. 2000;57:1454–1457. doi: 10.1001/archneur.57.10.1454. [DOI] [PubMed] [Google Scholar]
- 214.Palmer MS, Beck JA, Campbell TA, et al. Pathogenic presenilin 1 mutations (P436S & I143F) in early-onset Alzheimer’s disease in the UK. Mutations in brief no. 223. Online Hum Mutat. 1999;13:256. doi: 10.1002/(sici)1098-1004(1999)13:3<256::aid-humu11>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 215.Cruts M, van Duijn CM, Backhovens H, et al. Estimation of the genetic contribution of presenilin-1 and -2 mutations in a population-based study of presenile Alzheimer disease. Hum Mol Genet. 1998;7:43–51. doi: 10.1093/hmg/7.1.43. [DOI] [PubMed] [Google Scholar]
- 216.Lao JI, Beyer K, Fernandez-Novoa L, et al. A novel mutation in the predicted TM2 domain of the presenilin 2 gene in a Spanish patient with late-onset Alzheimer’s disease. Neurogenetics. 1998;1:293–296. doi: 10.1007/s100480050044. [DOI] [PubMed] [Google Scholar]
- 217.Lleo A, Blesa R, Queralt R, et al. Frequency of mutations in the presenilin and amyloid precursor protein genes in early-onset Alzheimer disease in Spain. Arch Neurol. 2002;59:1759–1763. doi: 10.1001/archneur.59.11.1759. [DOI] [PubMed] [Google Scholar]
- 218.Yoo BC, Vlkolinsky R, Engidawork E, et al. Differential expression of molecular chaperones in brain of patients with Down syndrome. Electrophoresis. 2001;22:1233–1241. doi: 10.1002/1522-2683()22:6<1233::AID-ELPS1233>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 219.Tsuji T, Shiozaki A, Kohno R, et al. Proteomic profiling and neurodegeneration in Alzheimer’s disease. Neurochem Res. 2002;27:1245–1253. doi: 10.1023/a:1020941929414. [DOI] [PubMed] [Google Scholar]
- 220.Schuller E, Gulesserian T, Seidl R, et al. Brain t-complex polypeptide 1 (TCP- 1) related to its natural substrate beta1 tubulin is decreased in Alzheimer’s disease. Life Sci. 2001;69:263–270. doi: 10.1016/s0024-3205(01)01126-2. [DOI] [PubMed] [Google Scholar]
- 221.Kim SH, Lubec G. Brain alpha-endosulfine is manifold decreased in brains from patients with Alzheimer’s disease: a tentative marker and drug target? Neurosci Lett. 2001;310:77–80. doi: 10.1016/s0304-3940(01)02025-0. [DOI] [PubMed] [Google Scholar]
- 222.Kim SH, Nairn AC, Cairns N, et al. Decreased levels of ARPP-19 and PKA in brains of Down syndrome and Alzheimer’s disease. J Neural Transm Suppl. 2001:263–272. doi: 10.1007/978-3-7091-6262-0_21. [DOI] [PubMed] [Google Scholar]
- 223.Kim SH, Cairns N, Fountoulakisc M, et al. Decreased brain histamine-releasing factor protein in patients with Down syndrome and Alzheimer’s disease. Neurosci Lett. 2001;300:41–44. doi: 10.1016/s0304-3940(01)01545-2. [DOI] [PubMed] [Google Scholar]
- 224.Krapfenbauer K, Yoo BC, Fountoulakis M, et al. Expression patterns of antioxidant proteins in brains of patients with sporadic Creutzfeldt-Jacob disease. Electrophoresis. 2002;23:2541–2547. doi: 10.1002/1522-2683(200208)23:15<2541::AID-ELPS2541>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 225.Aksenov M, Aksenova M, Butterfield DA, et al. Oxidative modification of creatine kinase BB in Alzheimer’s disease brain. J Neurochem. 2000;74:2520–2527. doi: 10.1046/j.1471-4159.2000.0742520.x. [DOI] [PubMed] [Google Scholar]
- 226.Castegna A, Thongboonkerd V, Klein JB, et al. Proteomic identification of nitrated proteins in Alzheimer’s disease brain. J Neurochem. 2003;85:1394–1401. doi: 10.1046/j.1471-4159.2003.01786.x. [DOI] [PubMed] [Google Scholar]
- 227.Kim SH, Vlkolinsky R, Cairns N, et al. The reduction of NADH ubiquinone oxidoreductase 24- and 75-kDa subunits in brains of patients with Down syndrome and Alzheimer’s disease. Life Sci. 2001;68:2741–2750. doi: 10.1016/s0024-3205(01)01074-8. [DOI] [PubMed] [Google Scholar]
- 228.Kim SH, Vlkolinsky R, Cairns N, et al. Decreased levels of complex III core protein 1 and complex V beta chain in brains from patients with Alzheimer’s disease and Down syndrome. Cell Mol Life Sci. 2000;57:1810–1816. doi: 10.1007/PL00000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Yoo BC, Fountoulakis M, Cairns N, et al. Changes of voltage-dependent anion-selective channel proteins VDAC1 and VDAC2 brain levels in patients with Alzheimer’s disease and Down syndrome. Electrophoresis. 2001;22:172–179. doi: 10.1002/1522-2683(200101)22:1<172::AID-ELPS172>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 230.Engidawork E, Gulesserian T, Yoo BC, et al. Alteration of caspases and apoptosis-related proteins in brains of patients with Alzheimer’s disease. Biochem Biophys Res Commun. 2001;281:84–93. doi: 10.1006/bbrc.2001.4306. [DOI] [PubMed] [Google Scholar]
- 231.Shim KS, Lubec G. Drebrin, a dendritic spine protein, is manifold decreased in brains of patients with Alzheimer’s disease and Down syndrome. Neurosci Lett. 2002;324:209–212. doi: 10.1016/s0304-3940(02)00210-0. [DOI] [PubMed] [Google Scholar]
- 232.Bajo M, Yoo BC, Cairns N, et al. Neurofilament proteins NF-L, NF-M and NF-H in brain of patients with Down syndrome and Alzheimer’s disease. Amino Acids. 2001;21:293–301. doi: 10.1007/s007260170015. [DOI] [PubMed] [Google Scholar]
- 233.Puchades M, Hansson SF, Nilsson CL, et al. Proteomic studies of potential cerebrospinal fluid protein markers for Alzheimer’s disease. Brain Res Mol Brain Res. 2003;118:140–146. doi: 10.1016/j.molbrainres.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 234.Davidsson P, Westman-Brinkmalm A, Nilsson CL, et al. Proteome analysis of cerebrospinal fluid proteins in Alzheimer patients. Neuroreport. 2002;13:611–615. doi: 10.1097/00001756-200204160-00015. [DOI] [PubMed] [Google Scholar]