Abstract
In this article, we have characterized and compared gene expression profiles from laser capture microdissected neurons in six functionally and anatomically distinct regions from clinically and histopathologically normal aged human brains. These regions, which are also known to be differentially vulnerable to the histopathological and metabolic features of Alzheimer’s disease (AD), include the entorhinal cortex and hippocampus (limbic and paralimbic areas vulnerable to early neurofibrillary tangle pathology in AD), posterior cingulate cortex (a paralimbic area vulnerable to early metabolic abnormalities in AD), temporal and prefrontal cortex (unimodal and heteromodal sensory association areas vulnerable to early neuritic plaque pathology in AD), and primary visual cortex (a primary sensory area relatively spared in early AD). These neuronal profiles will provide valuable reference information for future studies of the brain, in normal aging, AD and other neurological and psychiatric disorders.
Keywords: Alzheimer’s disease, laser capture microdissection, Affymetrix microarrays, expression profiling, neuron, transcriptomics
A major goal of neurological research is to increase our understanding of the workings of the human brain. To fully comprehend the relationships that connect molecular mechanisms in the brain with their respective functional outcomes, we must understand more completely the processes that are normally enacted in the healthy brain. A powerful method that may be used to address this issue is gene expression profiling analysis, which allows the simultaneous monitoring of the steady-state expression of all human genes. In this study, we expression profiled pyramidal neurons specifically selected using laser capture microdissection from different areas of postmortem brains of neurologically healthy elderly individuals. The expression data generated from these analyses provide an invaluable reference database for a wide range of brain-focused research by establishing a baseline of normal pyramidal cell gene expression in the aged brain. This resource, along with other currently available references such as the Allen Brain Atlas (http://www.brainatlas.org), Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/), the mouse brain Gene Expression Map (BGEM) (http://www.stjudebgem.org/web/mainPage/mainPage.php), and GNF (Genomics Institute of the Novartis Research Foundation) Symatlas (http://symatlas.gnf.org/SymAtlas/) will play important roles in furthering current and future neurological research.
The brain regions-of-interest used in this study were originally selected based on previous research on Alzheimer’s disease (AD). These anatomically distinct regions, which include the entorhinal cortex [Brodmann’s areas (BA) 28 and 34], hippocampus, middle temporal gyrus (BA 21 and 37 and approximate BA 22), posterior cingulate cortex (BA 23 and 31), superior frontal gyrus (BA 10 and 11 and approximate BA 8), and primary visual cortex (BA 17), have been found to display metabolic and pathological differences in the brains of individuals afflicted with AD (2, 3, 5–9, 17, 18, 21, 25–27, 30–32, 44, 47, 50, 54, 58, 63). In particular, the posterior cingulate cortex has been found to show the heaviest abnormal measurements in positron emission tomography scans of cognitively normal late-middle-aged carriers of the APOE ε4 allele (56). Furthermore, functional brain imaging, structural brain imaging, and neuropathological studies suggest that the frontal lobes are preferentially affected by normal aging (14, 38, 60, 65). This study provides descriptive information about the genes that are differentially expressed in frontal regions, which are most strongly affected by normal aging compared with those that are most spared, such as the primary visual cortex.
Lastly, selection of these regions provides global coverage of functional zones of the human brain with representative regions of the limbic (hippocampus), paralimbic (entorhinal cortex and posterior cingulate cortex), heteromodal (superior frontal gyrus), unimodal (middle temporal gyrus), and primary sensory zones (primary visual cortex). By focusing on these areas, we will provide important reference information to help elucidate normal expression pathways in the brains of healthy, elderly individuals that may drive cognitive processes unique to each region and those that underlie the functional connectivity of these areas.
Currently there exists no similar comprehensive catalog of global neuronal expression signatures of disease-relevant brain regions obtained using tightly controlled patient sample sets. Well-known gene expression resources such as the Allen Brain Atlas and BGEM provide expression data on the mouse nervous system, whereas GEO and Symatlas are more global in encompassing publicly deposited expression data from different tissues in multiple model organisms. In this respect, this study will provide an important preliminary reference database focused purely on normal neuronal gene expression in the brain of elderly individuals and may be used as a resource for future research on deciphering the molecular underpinnings of region-specific functions of the human brain. In addition, establishing the normal gene expression patterns of elderly individuals will give us the foundation to locate dysregulated expression in disease-afflicted individuals. Understanding gene expression specific to normal neurons, and how it relates to gene expression in normal neurons in diseased brains, will serve as a springboard to help us find novel drug targets to develop improved therapies and interventions to common and devastating neurological diseases such as AD.
MATERIALS AND METHODS
Tissue collection
Brain samples were collected at the Sun Health Research Institute, an Alzheimer’s Disease Center, from individuals clinically classified as neurologically normal (10 males and 4 females) with a mean age of 79.8 ± 9.1 yr. All cases did not expire as a result of accident or suicide nor were kept alive heroically prior to death. Samples were collected (mean PMI of 2.5 h) from six brain regions that are either histopathologically or metabolically relevant to AD and aging; these include the entorhinal cortex (BA 28 and 34), superior frontal gyrus (BA 10 and 11 and approximate BA 8), hippocampus, primary visual cortex (BA 17), middle temporal gyrus (BA 21 and 37 and approximate BA 22), and the posterior cingulate cortex (BA 23 and 31). Following dissection, samples were frozen, sectioned (8 μm), and fixed on glass slides.
Brain sections were stained with 1% neutral red, and neurons were identified by their characteristic size, shape, and location within each region. In the entorhinal cortex, because layer II contains the pyramidal neurons, large stellate neurons were selected from layer II for this region. For the hippocampus, pyramidal neurons of CA1 were selected because this region is the most-affected and earliest affected hippocampal region with regards to tangle formation (CA3 was not evaluated because this region is affected in later stages of AD so that in normal subjects, changes aren’t expected to be seen in this area). For the other regions, layer III pyramidal neurons were selected. Approximately 1,000 normal pyramidal neurons were collected from cortical layer III per brain region per individual by laser capture microdissection with the Arcturus Autopix Automated Laser Capture Microdissection System (Mountain View, CA). Cells were collected onto CapSure Macro LCM Caps (Arcturus) and extracted according to the manufacturer’s protocol. Total RNA was isolated from the cell lysate using the PicoPure RNA Isolation Kit (Arcturus) with DNase I treatment using Qiagen’s RNase-free DNase Set (Valencia, CA).
Expression profiling
All total RNA isolated per brain section was double round amplified, cleaned, and biotin-labeled using Affymetrix’s GeneChip Two-Cycle Target Labeling kit (Santa Clara, CA) with a T7 promoter and Ambion’s MEGAscript T7 High Yield Transcription kit (Austin, TX) as per manufacturer’s protocol. Amplified and labeled cRNA was quantitated on a spectrophotometer and run on a 1% Tris-acetate-EDTA (TAE) gel to check for an evenly distributed range of transcript sizes. cRNA (20 μg) of was fragmented to ~35–200 bp by alkaline treatment (200 mM Tris-acetate, pH 8.2, 500 mM KOAc, 150 mM MgOAc) and run on a 1% TAE gel to verify fragmentation. Separate hybridization cocktails are made using 15 μg of fragmented cRNA from each sample as per Affymetrix’s protocol.
Microarray analysis
We separately hybridized 200 μl of each cocktail to an Affymetrix Human Genome U133 Plus 2.0 Array for 16 h at 45C in the Hybridization Oven 640. The Affymetrix Human Genome Arrays measure the expression of >47,000 transcripts and variants, including 38,500 characterized human genes. Arrays were washed on Affymetrix’s upgraded GeneChip Fluidics Station 450 using a primary streptavidin phycoerythrin (SAPE) stain, subsequent biotinylated antibody stain, and secondary SAPE stain. Arrays were scanned on Affymetrix’s GeneChip Scanner 3000 7G with Auto-Loader. Scanned images obtained by the Affymetrix GeneChip Operating Software (GCOS) v1.2 were used to extract raw signal intensity values per probe set on the array and calculate detection calls (absent, marginal, or present). Assignment of detection calls was based on probe-pair intensities for which one probe is a perfect match of the reference sequence and the other is a mismatch probe for which the 13th base (of the 25 oligonucleotide reference sequence) is changed. All raw chip data were scaled in GCOS to 150 to normalize signal intensities for interarray comparisons. Reports generated by GCOS were reviewed for quality control: at least 20% present calls, a maximum 3′/5′ GAPDH ratio of 30, and a scaling factor <10. Those arrays that failed to pass these standards were not included in further analyses.
Pyramidal cell quality control
To assess neuronal cell purity in the samples, expression of glial fibrillary acidic protein (GFAP), an astrocyte cell marker, was evaluated. Nine samples that had GFAP expression >2 SD from the mean were removed from statistical analyses; these samples included one hippocampal sample, two middle temporal gyrus samples, one posterior cingulate sample, three superior frontal gyrus samples, and two primary visual cortex samples. Removal of these samples from analysis left us with 75 total samples with ample coverage of each of the six brain regions of interest.
Statistical analysis
Raw signal intensity and detection call data obtained per array from GCOS were uploaded into Genespring v7.2 (Agilent Technologies, Palo Alto, CA) for analysis. MIAME-compliant microarray data files are located on the GEO site at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE5281 (project accession #GSE5281). Fold change and P value data for each of the six regions are available online at: http://www.tgen.org/neurogenomics/data. Posted lists show region-specific P values and fold changes, and normalized expression signals for genes that have at least seven present calls out of 75 total calls (no P value and fold change thresholds have been applied on these lists). Direct region-to-region comparisons were performed between all brain regions to analyze expression differences between every region. All genes were filtered for at least seven present calls out of all samples; genes that were not called present in at least ~10% of all samples were removed. Heteroscedatic (two-sample unequal variance), two-tailed t-tests were applied to each region-to-region pair-wise comparison to locate genes whose expression levels are statistically significant in differentiating expression between the two regions: for each region, genes that had either a maximum P value <0.05 or 0.01 were collected. From these regional lists, those genes that showed consistently high fold changes above a minimum threshold (between a 1.5- to a 4-fold increase or decrease) for separate regional comparisons were identified. In addition, genes that had all expression signals for each sample below a signal threshold of 100 were removed because dramatic fold changes under this threshold likely fall under background. P value and fold change thresholds were adjusted for each regional analysis to locate the genes that had the greatest statistical significance and the greatest fold changes relative to other regions. The least stringent thresholds were a maximum P value of 0.05- and a 1.5-fold change, whereas the most stringent thresholds were a maximum P value of 0.01- and a 4-fold change. Using this approach we identified sets of genes that were specifically up- or downregulated in each brain region. For example, the entorhinal cortex was separately compared with all five other regions to generate lists for each comparison. We then found the intersection of genes that fell within all five lists to find genes that were differentially expressed in the entorhinal cortex compared with all other brain regions tested. The most differentially expressed genes in each brain region were used to generate separate heat maps showing the distribution of differential gene expression. Heat maps for each brain region were created with Genespring v7.2.
To globally analyze the entire set of expression data by an unsupervised approach, we looked to find general expression themes that accounted for the greatest amount of change across all the regions-of-interest without placing emphasis on a single region. For this analysis, genes were first filtered for at least seven present calls out of a total of 75 detection calls to remove genes that were called absent ~90% of the time across all samples. Genes with at least a 0.95 correlation to zero variance in expression, or did not change greatly in expression, across all regions were removed. Statistical one-way ANOVA (parametric, assume variances unequal) on brain region was applied to this list to obtain genes that had high statistical significance in differentiating expression between regions; a threshold of a maximum P value of 0.05 was applied. To look at the overall general trends of expression over all regions, principal components analysis on genes was applied to this list with no additional rotation scheme in Genespring v7.2. This analysis, which is a multivariate mathematical technique, converts a multidimensional data set into a set with fewer dimensions by using a covariance matrix but also retains the variability in the data in doing so. Thus, principal components analysis (PCA) characterizes the major patterns of differential expression across all the regions that account for variability in the data. This analysis outputs six component signatures that best account for the variance of expression across all brain regions in addition to calculating the correlation of each gene of the input list to each individual component.
Lists of genes generated from Excel analyses were uploaded into MetaCore Pathway Analysis software (GeneGo, St. Joseph, MI) to identify relationships between the genes of interest and to uncover common processes and pathways that are up- and downregulated under normal conditions between the six regions. We would like to note that multiple testing corrections were not used since these methods are often overly stringent. Additionally, any candidate dys-regulated gene of interest still would need to be validated by secondary measures as we have done for a few candidates.
Immunohistochemistry
Fresh frozen sections from separate cases were used to validate expression profiling results at the protein level. The following protocol was first run to optimize staining at 1:100, 1:500, and 1:1,000 dilutions per antibody. After optimization, the protocol was run again at optimal antibody dilutions on triplicate sections.
Sections were thawed at room temperature and fixed in 100% acetone for 5 min. Slides were next equilibrated in TBS (Tris-buffered saline, pH 8.0; Sigma-Aldrich, St. Louis, MO) for 5 min, and endogenous peroxidases were blocked in 3% hydrogen peroxide (in TBS) for 10 min. Sections were then washed with TBST (Tris-buffered saline with Tween 20, pH 8.0; Sigma-Aldrich) and used to make slide pairs. All slides were blocked in 300 μl 2.5% horse serum from the ImmPRESS Anti-Mouse Ig and ImmPRESS Anti-Rabbit Ig kits (Vector Laboratories; Burlingame, CA) in TBST and incubated with three rinses of 100 μl of primary antibody (AbCam, Cambridge, MA) in normal horse serum (Jackson ImmunoResearch Laboratories, West Grove, PA) and TBST for a 1/2 h per rinse. Frizzled homolog 7 (Drosophila) (FZD7) polyclonal antibody was used at a 1:75 dilution, ITGA4 monoclonal antibody at a 1:25 dilution, DUSP4 polyclonal antibody at a 1:250 dilution, cAMP responsive element binding protein (CREB) monoclonal antibody at a 1:100 dilution, and nuclear transcription factor Y, beta (NFYB) polyclonal antibody at a 1:50 dilution. Sections were rinsed with 1 ml of TBST five times and incubated in 300 μl of secondary antibody from the ImmPRESS Anti-Mouse Ig and Anti-Rabbit Ig kits for 45 min. Slides were then rinsed again with 1 ml of TBST five times and incubated in 500 μl of substrate buffer treated with diaminobenzidine (DAB) chromogen and DAB enhancer (BioCare Medical, Concord, CA) for 5 min followed by a deionized water rinse, and 4 drops of DABSPARKLE treatment (BioCare Medical) for 1 min followed by a final deionized water rinse. Two drops of hematoxylin-2 (Richard-Allen Scientific, Kalamazoo, MI) were applied to each slide to counterstain, and slides were rinsed with and held in deionized water. Each slide was mounted and visualized under the AxioCam HRc Epifluorescence microscope (Carl Zeiss, Oberkochen, Germany) with AxioVision software at ×20.
RESULTS AND DISCUSSION
Establishing a reference data set for neuronal gene expression studies
In this study, we have established a valuable, high-quality gene expression reference data set describing neuronal gene expression in neurologically healthy individuals. The preliminary data set we present here is of an exploratory nature but will help us to gain insight into molecular mechanisms underlying neuronal function and region-specific brain function in healthy individuals. These data also serve as a baseline reference for studying causative mechanisms for a variety of neurological disorders, including AD.
Region-specific expression analysis
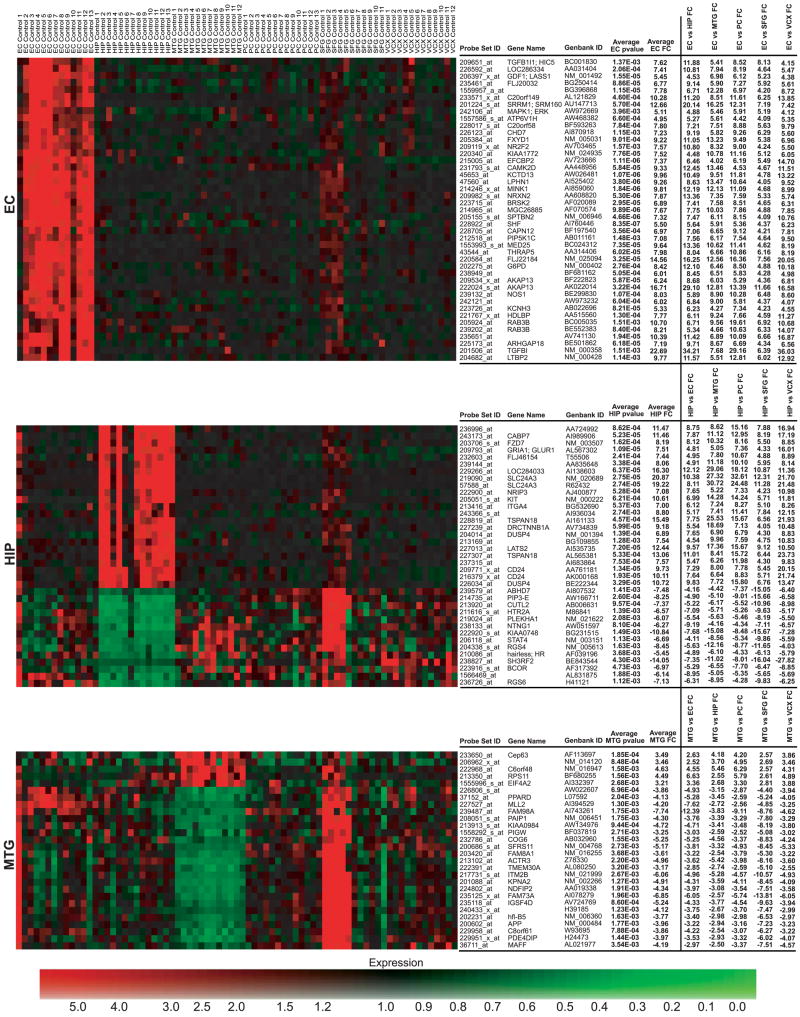
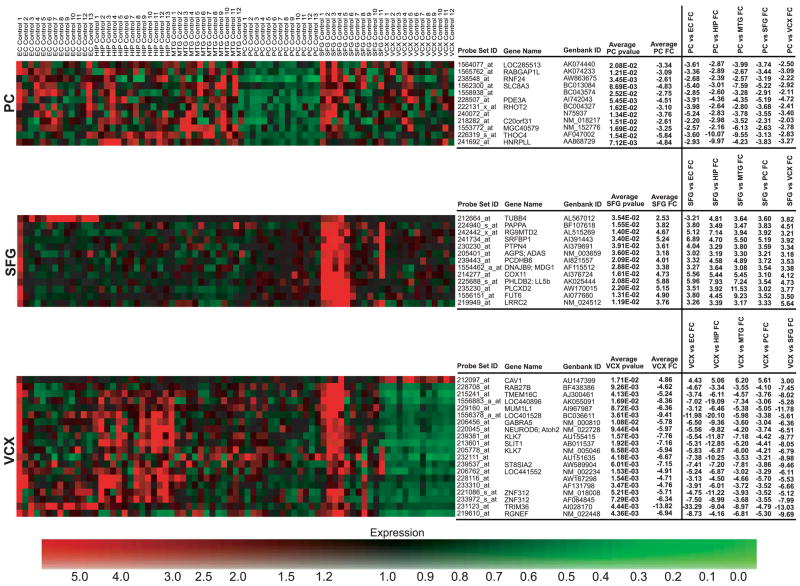
All neuronal populations were isolated and analyzed as described in MATERIALS AND METHODS. Additionally, all expression data are available online at http://www.tgen.org/neurogenomics/data. To identify region-specific gene expression differences in the brain, each of the six regions examined (hippocampus, entorhinal cortex, middle temporal gyrus, superior frontal gyrus, posterior cingulated cortex, and visual cortex) were separately compared with every other region to generate lists of genes unique to each area. Thresholds for P values and fold changes were adjusted for each region to find genes whose differential expression between regions had the largest fold changes with the highest statistical significance (see MATERIALS AND METHODS). Thresholds were accordingly adjusted to find approximately the top 50–100 genes whose expression is unique to the region-of-interest. The thresholds applied to the entorhinal cortex and hippocampus analyses were a maximum P value of 0.01 and a minimum fold change of 4. For the middle temporal gyrus analysis, a threshold of a maximum P value of 0.01 and a minimum fold change of 2.5 was applied. The posterior cingulate analysis had a threshold of a maximum P value of 0.05 and a minimum fold change of 2. Finally, the superior frontal gyrus and primary visual cortex analyses had thresholds of a maximum P value of 0.05 and a minimum fold change of 3. The entorhinal cortex and hippocampus had the highest number of significant genes specific to these regions with 43 and 37 genes, respectively. We found 28 genes for the middle temporal gyrus, 12 for the posterior cingulate cortex, 13 for the superior frontal gyrus, and 20 for the primary visual cortex. Top significant genes were used to generate heat maps that visualize differences in expression across all regions (Fig. 1). The profiles of these region-specific genes may serve as unique identifiers of the neurons in these respective regions. Genes for each region were input into GeneGo MetaCore software for network and pathway analysis.
Fig. 1.
heat maps of region-specific significant genes. Individual samples are listed across the top of the map with genes listed vertically along the right. Logged normalized expression levels were used to generate maps; colors range from red (high expression) to green (low expression) as indicated by the expression color bar. Significant genes found for each region were used to create separate heat maps to visualize dysregulation specific to the respective region. Fold changes for region comparisons for each set of region-specific genes are shown. Brain regions are abbreviated to the left of each map.
The entorhinal cortex (BA 28 and 34) is a component of the paralimbic zone of the cerebral cortex and is a major input into the hippocampal formation. As a part of the paralimbic zone, the entorhinal cortex receives major connections from heteromodal and unimodal areas and has massive extramural connections to the limbic zone (46). Although intermediary processing unique to the entorhinal cortex has yet to be fully elucidated, we have found statistically significant genes showing differential expression specific to this region. MetaCore pathway analysis of these genes identified known processes; one process that came through from this analysis is learning and/or memory, which is consistent with the involvement of the limbic association cortex in emotion, learning, and memory. Of the total 36 genes that fall within the learning and memory process of the MetaCore database and are linked to genes from the input list, eight statistically significant upregulated genes from the input list were identified as being associated with this process. These genes include DAB1 [disabled homolog 1 (Drosophila)], RELN (reelin), AKAP13 (A kinase anchor protein 13), APOE (apolipoprotein E), GRIK5 (glutamate receptor, ionotropic, kainate 5), GRINA (glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1), GSK3β (glycogen synthase kinase 3-β), and PRKCG (protein kinase C, γ). Although many details that define the pathways involved in learning and memory have yet to be entirely elucidated, these eight genes appear to play possible roles in these pathways. DAB1 and RELN1 coexpression have been found to be necessary for normal cortical development and normal mature brain functions (19). AKAP13 is part of a family of scaffolding proteins that bind PKA regulatory subunits; this binding regulates phosphorylation of proteins that have been found to play a role in synaptic plasticity and memory formation (15, 24, 59). APOE, which codes for the primary cholesterol transporter in the brain, may be involved with neuronal repair and plasticity (42) and also has allelic variants that are implicated in AD. Ionotropic glutamate receptors, such as GRIK5 and GRINA, are involved in synaptic plasticity, which underlies learning and memory processes (20). Interestingly, GSK3β is an enzyme that plays a role in the hyperphosphorylation of tau to lead to neurofibrillary tangle formation in AD brains (10, 33, 39) and here was found to be upregulated in the entorhinal cortex above all other regions analyzed in this study. Such upregulation in neurologically healthy individuals may suggest that neuroprotective functions are enacted in the neurologically healthy brain. Finally, PRKCG is activated by Ca2+ and diacylglycerol and has been found to play a vital role in long-term potentiation (1, 12, 29, 41, 43, 64), a mechanism involved with learning and memory.
Several other processes found through MetaCore analysis, cholinergic synaptic transmission, and phospholipase C activating pathway involve acetylcholine biosynthesis and metabolism; specifically, there is upregulated expression of ACM5 and ACM2, acetylcholine receptors. Because acetylcholine is a neurotransmitter that generally causes excitatory actions in the brain, increased expression of its receptors may suggest that such excitation may play a role in learning and memory functions specific to the entorhinal cortex. This finding may represent a compensatory upregulation as it is known that cortical cholinergic deafferentation occurs in normal aging and mesial temporal lobe structures are amongst the earliest affected regions (45, 52). Other top processes found for this region include nervous system development, synaptic transmission, neurotransmitter secretion, and signal transduction, all of which may be significant mechanisms in entorhinal cortex function. Additional genes showing differential expression in this region are shown in Fig. 1 and may represent a characteristic profile relevant to regional cognitive functions.
The hippocampus, a component of the allocortical formation that is physically located in the temporal lobe and functionally a part of the limbic zone, is also involved in learning and memory. As a part of the limbic zone, the hippocampus has major reciprocal connections with the hypothalamus and receives a large amount of input from the paralimbic zone (46). Statistically significant genes that have expression unique to the hippocampus may play a role in neural processing that underlies such connections. Among the significant genes unique to the hippocampus, we see an upregulation of expression of integral membrane and transmembrane proteins that serve as receptors for signal transmission into and out of the cell. These genes include ITGA4 (integrin, α4), ITGA7 (integrin, α7), and ITGA9 (integrin, α9), all of which code for the alpha chain of the heterodimeric integrin complex that consists of an alpha chain and beta chain. Additional genes coding for receptors showing increased expression include KIT (v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog), whose protein is a type 3 transmembrane receptor for mast cell growth factor, which uses tyrosine-protein kinase activity to transmit signals, and FZD7, which codes for the frizzled protein, a seven-transmembrane domain protein that acts as a receptor in the Wnt signaling pathway. Differential expression of genes coding for proteins involved in signal transduction regulation were also observed; these genes include RGS4 (regulator of G protein signaling 4), which was downregulated, and DUSP4 (dual specificity phosphatase 4), which was upregulated. RGS4 has been previously found to be expressed in the human brain but not in certain regions, such as the thalamus and basal ganglia (22) and, based on our findings, also has decreased expression in the hippocampus. RGS4, by modulating G protein activity, seems to regulate dopamine receptors (22), or more specifically, possibly D5 dopamine receptors (of the D1 receptor family), the one type of dopamine receptor highly found in the hippocampus (68). DUSP4, also known as MKP-2 (MAP kinase phosphatase 2), has been found to regulate mitogenic signal transduction by inactivating kinases such as ERK1, ERK2, and JNK, through dephosphorylation of threonine and tyrosine residues (48).
Another gene showing statistically significant decreased expression is RPH3A, which codes for rabphilin 3A homolog (mouse). This protein is a member of the RAB3A regulation pathway and is involved in docking and fusion of synaptic vesicles (16) and may play a role in neurotransmitter release through membrane flow regulation. Interestingly, rabphilin-3 has been known to be highly expressed in the murine brain (49, 62).
Over all, such regulated expression of factors involved in neurotransmitter transport and signaling may possibly be a hallmark of processing significant to the limbic zone, although additional limbic areas would need to be studied to determine any generalities of these findings.
The middle temporal gyrus (BA 21 and 37 and approximate BA 22) is found in the temporal lobe between the superior and inferior gyri and has functions involved in memory and emotions, perception, and localization of sounds, in addition to language functions that are primarily localized to the left side of the brain. This gyrus is a part of the modality-specific, or unimodal, zone of the association isocortex and in some parts falls in the heteromodal zone as well. As a component of these zones, this brain region receives a great deal of sensory information from up to only a few synapses away in auditory and visual association pathways and outputs to heteromodal and paralimbic areas (46). Since primary and secondary sensory synapses involved in information processing originate in the unimodal zone, this area of the brain is highly developed (46). Genes falling through from specific analysis of this region may be correlated to such unique processing in the middle temporal gyrus. Differentially expressed genes in this region were primarily downregulated relative to the five other regions. The majority of these downregulated genes code for enzymes and factors involved in regulation of lipid and carbohydrate metabolism. These genes include PCCB, which codes for propionyl coenzyme A carboxylase (beta polypeptide), ATF2, which codes for activating transcription factor 2, SCP2, which codes for the sterol carrier protein 2, and PRKAG1, which codes for the gamma 1 noncatalytic subunit of AMP-activated protein kinase (AMPK). Activating transcription factor-2 forms either a homodimer or heterodimer with c-jun to stimulate cAMP-responsive element (CRE)-dependent transcription and may also act as a histone acetyltransferase for transcription activation (28). Sterol carrier protein 2 is thought to be an intracellular lipid transfer protein, and so its downregulation may indicate decreased transport of lipids within neurons. AMPK, which is normally activated under conditions of cellular metabolic stress, phosphorylates and inactivates acetyl-CoA carboxylase and beta-hydroxy beta-methylglutaryl-CoA reductase, enzymes that help regulate fatty acid and cholesterol biosynthesis. These results suggest a possible overall “slowing down” of neuronal biosynthetic and metabolic processes specific to the medial temporal gyrus. Although it is, however, unclear as to how this regulation is involved with regional functions, these results may provide some clues about the possible requirement for a coordinated decrease in certain cellular processes in unimodal areas. Though the majority of significant genes showed decreased expression, we also saw an increase in expression of eIF4A2, which codes for a subunit of eIF4A, a factor required for binding of mRNA to 40S ribosomal subunits.
The posterior cingulate cortex (BA 23 and 31) is situated in the parietal lobe adjacent to the splenium of the corpus callosum and, like the entorhinal cortex, is a part of the paralimbic zone, which has functions involved with emotion formation and processing, learning, and memory. Like the entorhinal cortex, the posterior cingulate is involved in intermediary processing with its receipt of information from heteromodal and unimodal areas and its subsequent output to the hippocampus and other limbic areas of the brain (46). This region has also been found to be a component of the “default system” that is normally active during the baseline state of the human brain as it monitors sensory information (55). In this region there was an overall upregulation of DPYS, which codes for dihydropyrimidinase, an enzyme involved in the formation of uracil and beta-alanine metabolism. The posterior cingulate also demonstrated an overall decrease in expression of statistically significant genes. There was a downregulation of GABBR2, which codes for GAB (γ-aminobutyric acid) B receptor 2. B-type receptors for GABA have been found to inhibit neuronal activity through G protein-coupled second-messenger systems (23). There was also a downregulation of expression of members of the MAPK cascade and interleukin signaling pathway, including the mitogen-activated protein kinase kinase kinase 14 (MAP3K14), the alpha isoform of regulatory subunit B for protein phosphatase 3 (PPP3R1, also known as calcineurin B, type I), and member 1A of the tumor necrosis factor receptor superfamily (TNFRSF1A). In the interleukin signaling pathway, decreased expression of MAPKKK14 and TNFRSF1A ultimately leads to decreased activation of the IKK (IκB kinase) complex, which is needed to phosphorylate IκB proteins to mark them for the ubiquitination pathway (35). Since IκB proteins inhibit NF-κB, if they are not targeted for destruction, NF-κB will resultingly have decreased activation and may have downstream effects in inflammation, immunity, cell proliferation, and apoptosis, all of which may play a role in the normal aging process (67). Calcineurin B normally complexes with calcineurin A to form the calcineurin complex, which functions as a Ca2+/calmodulin-regulated protein phosphatase (36). Decreased expression of calcineurin B may lead to decreased formation of the entire calcineurin complex, which may possibly lead to dysregulation of Ca2+ homeostasis. There is also a downregulation of neurotransmitter receptors, including GABBR2 (γ-aminobutyric acid B receptor, 2) and GNRHR (gonadotropin-releasing hormone receptor), both of which are participants in the G-coupled receptor systems and likely play a role in synaptic plasticity (40). This overall downregulation of expression of genes whose products are involved with transcription, signal transduction, and metabolism may characterize posterior cingulate cortex neurons from other brain neurons and may play a role in the region’s maintenance of unique functions with integrating incoming information necessary for cognitive processes such as learning, memory, and emotions.
The superior frontal gyrus (BA 10 and 11 and approximate BA 8) is located centrally at the apex of the frontal lobe and has been found to be involved with working memory, reasoning, planning, attention, and some aspects of language. This gyrus is a member of the heteromodal, or high-order, zone of the brain and also has some overlap with unimodal areas. Under the heteromodal functional subtype, the superior frontal gyrus has major incoming connections from unimodal areas and major outgoing connections to the paralimbic zone (46). The role of this region in sensory experience may be linked to controlled gene expression found solely in this region. Over all there was an upregulation of genes involved in metabolism, spindle assembly and chromosomal separation, and cell cycle progression. NFYB, KPNB1 [karyopherin (importin) beta 1], ACLY (ATP citrate lyase), and CREB1 are all upregulated in the superior frontal gyrus relative to the other five regions and are all involved in lipid biosynthesis and metabolism. These proteins activate SREB2 and SREBP1, which further activate LDLR, the major receptor that transports low-density lipoprotein (LDL) into cells. Phosphorylation of CREB has also been implicated in inducing expression of immediate early genes (IEGs), which play a role in neural plasticity (11); increased expression of CREB consequently increases its availability for phosphorylation and activation of IEG transcription.
Expression of genes involved in cellular metabolism was also increased in superior frontal gyrus. These genes include GCLM (glutamate-cysteine ligase, modifier subunit), GCLC (glutamate-cysteine ligase, catalytic subunit), CCND1 (cyclin D1), and MAPK8 (mitogen-activated protein kinase 8). Upregulation of proteins participating in metabolic pathways suggests that such mechanisms may be involved with processing specific to the superior frontal gyrus. Increased expression of proteins involved in spindle assembly and chromosomal separation, including DCTN4 (dynactin 4), ANAPC1 (anaphase promoting complex subunit 1), and ANAPC7 (anaphase promoting complex subunit 7) may indicate increased cell cycle progression and cell proliferation. Dynactin 4 helps provide a contact between microtubules and the chromosome kinetochore by recruiting dynein to the kinetochore (61). Previous research has also shown that dynactin is specifically expressed in human neurons and may play a role in slow-anterograde microtubule transport along axons (66). Anaphase promoting complex subunits 1 and 7 help make up the multimeric anaphase promoting complex (APC), which is a ubiquitin ligase that targets cell cycle proteins for degradation. Thus, the APC can both promote and inhibit cell cycle progression by degrading protein inhibitors, cyclins and cyclin-dependent kinases (69).
To further evaluate the differentially expressed genes specific to the superior frontal gyrus, this list of genes was compared with findings from a previous study that also used microarray analysis to evaluate the frontal lobes of young and aged individuals (40). Overlap of genes differentially expressed in frontal vs. nonfrontal regions of the aged brain (which could reflect aging or some other specialized regional function) with genes found to be differentially expressed in older vs. younger brains may provide clues about the molecular substrates of normal aging. Although no specific genes fell in both lists, likely due to the fact that the frontal lobe project analyzed entire tissue sections as opposed to this study’s neuron-specific analysis of the superior frontal gyrus, there was significant overlap in the ontologies of the identified genes. These ontologies include synaptic transmission, G protein signaling, vesicular transport, protein turnover, and transcription. Identification of such an overlap may help us to find new targets at which to aim new age-slowing treatments and provides information that might be used to develop new imaging or nonimaging biomarkers of aging to study the pathological aging process.
The primary visual cortex (BA 17), which is located within the calcarine fissure of the occipital lobe, is involved in the initial stages of visual processing and is, thus, a part of the primary sensory cortex which acts as the entrance through which information from the environment enters the brain. This region, also known as striate cortex, contains highly organized neurons whose detailed connectivity plays a role in processing information on color, length, spatial frequency, movement, and orientation (46). Information collected and processed in the primary visual cortex is ultimately output to unimodal areas for downstream intermediary processing (46). Genes whose regulated expression is unique to this region may provide clues about the underlying processes specific to this area’s functional role.
In contrast to the upregulation of CREB1 seen in the superior frontal gyrus, the primary visual cortex had decreased expression of a gene involved in the CREB pathway. CAMK2D (calcium/calmodulin-dependent protein kinase II delta), which is significantly upregulated in the entorhinal cortex and downregulated in the primary visual cortex, codes for the calcium/calmodulin-dependent protein kinase II delta. CAMK2D is one of four subunits that make up CaMK II, a serine/threonine protein kinase that phosphorylates CREB. Because CREB phosphorylation leads to the activation of IEG expression, decreased expression of the delta subunit indicates there may be a resulting inhibition of IEG expression. Previous studies have shown that IEGs are likely involved with neural plasticity and long-term memories (34). Consequently, decreased expression of IEGs suggests that their presence and action may not be needed for the initial stages of visual processing.
There is also a downregulation of GABRA5 (GABA receptor, α5) in the primary visual cortex compared with other regions. GABA is the major inhibitory neurotransmitter in the mammalian brain and causes membrane hyperpolarization and subsequent reduced neuronal activity (23). In this region there was also uniquely decreased expression of TMEM16C (transmembrane protein 16C) and KCNA5 (potassium voltage-gated channel, shaker-related subfamily, member 5). Transmembrane proteins and potassium channels are involved in modulation of processes including neurotransmitter release and neuronal excitability; such regulation of expression of these factors may have a role with processing visual information in this region. One particular gene that shown upregulation was CAV1, which codes for caveolin 1, a scaffolding protein located in caveolar membranes that binds cholesterol and which directly interacts with G protein α-subunits to create an inhibitory effect on G-protein activity (51). Such inhibition parallels the downregulated expression of other gene products involved in numerous cellular processes and may help to gain insight into understanding the visual processing specific to this region.
Global expression analysis
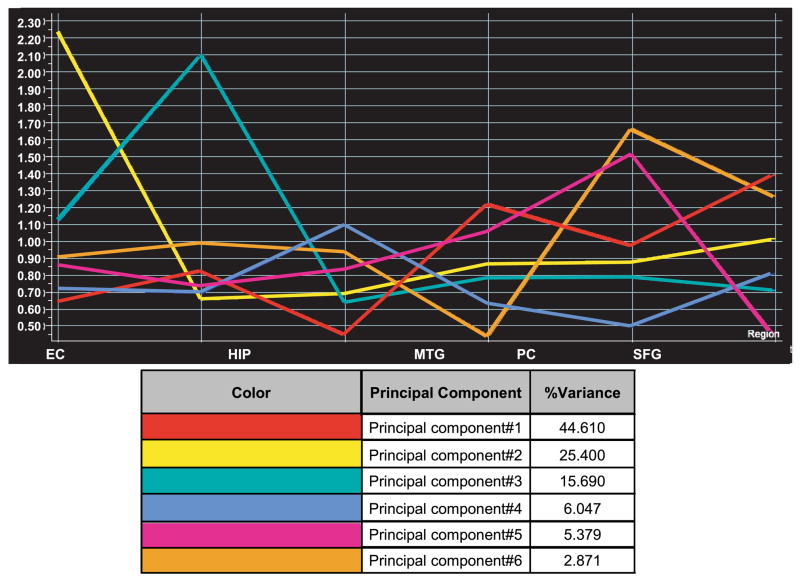
To gain insight into the general expression trends across all brain regions of interest, we globally analyzed all 75 samples using Genespring v7.2 to perform PCA on genes. This analysis found patterns for which each gene has a correlation value and indicated that one defining expression pattern accounts for 46.03% of all variance in gene expression, whereas the five other patterns accounted for a smaller amount of expression variation. This pattern, which illustrates the greatest amount of variance, is referred to as principal component 1 and demonstrates that the greatest overall differences in gene expression correlated to increased expression in the posterior cingulate, superior frontal gyrus, and primary visual cortex and less expression in the entorhinal cortex, hippocampus, and middle temporal gyrus as shown in Fig. 2 (principal component 1 is represented by the red graph line). It is also important to note here that PCA does not measure the significance of each component, so that the ordering of the components is purely based on the extent of expression variance. There were 102 genes with expression profiles having at least a 0.98 correlation with the principal component 1, while 19 genes had at least a 0.99 correlation (Table 1). Correlation values range from −1, representing a complete opposite theme, and 1, representing an expression pattern identical to the principal component. These genes may define the normal theme of differential gene expression with the greatest variance that occurs across the six brain regions in healthy individuals and may help to characterize the primary sensory, unimodal, heteromodal, paralimbic, and limbic functional zones that are represented by these six brain regions. The rest of the components detected by PCA are shown in Fig. 2 and represent separate patterns that account for a smaller amount of variation across all brain regions.
Fig. 2.
Principal components analysis. Six principal component eigenvectors were generated from principal components analysis of all 75 samples; the first component represents the gene expression signature that accounts for 46.03% of the total variance in all gene expression across all brain regions. The y-axis represents the logged normalized intensity value, and the x-axis designates the brain region. EC, entorhinal cortex; HIP, hippocampus; MTG, middle temporal gyrus; PC, posterior cingulate; SFG, superior frontal gyrus; VCX, primary visual cortex.
Table 1.
Principal component 1
| Affymetrix ID | GenBank Accession | Common | PCA 1 Correlation |
|---|---|---|---|
| 1568627_at | BC032531 | KIAA1387 | 0.990 |
| 201013_s_at | AA902652 | PAICS | 0.990 |
| 202258_s_at | U50532 | PFAAP5; CG005; 92M18.3 | 0.990 |
| 202850_at | NM_002858 | ABCD3; ABC43; PMP70; PXMP1 | 0.991 |
| 212470_at | AB011088 | KIAA0516; SPAG9; HSS; JLP | 0.991 |
| 219064_at | NM_030569 | ITIH5; pp14776; MGC10848 | 0.992 |
| 219976_at | NM_015888 | HOOK1; HK1 | 0.994 |
| 222111_at | AU145293 | KIAA1164 | 0.992 |
| 223535_at | AL136592 | DKFZp761I172; NUDT12 | 0.992 |
| 223766_at | AF130105 | MRIP2 | 0.993 |
| 227383_at | AW340595 | 0.992 | |
| 229956_at | AI659426 | 0.991 | |
| 235089_at | AI122770 | FBXL20 | 0.993 |
| 236126_at | AI188710 | 0.992 | |
| 236165_at | AA904502 | MSL3L1 | 0.993 |
| 238604_at | AA768884 | 0.994 | |
| 238919_at | R49295 | PCDH9 | 0.995 |
| 241721_at | AW515022 | 0.990 | |
| 244778_x_at | N63691 | M11S1 | 0.992 |
Genes showing at least a 0.99 correlation in expression with the principal component (PCA) 1 eigenvector from global principal component analysis of all samples.
The 102 genes that showed a 0.98 correlation to principal component 1 were used for pathway analysis with GeneGo MetaCore software. Among the top maps found include calcium signaling and the CREB pathway, both of which have been previously implicated in age-related changes in neuronal expression (11). Specifically, the three genes that were identified in the calcium signaling pathway include SLC8A1 [solute carrier family 8 (sodium/calcium exchanger), member 1], PPP2CA (protein phosphatase 2, catalytic subunit, α isoform), and CALM2 (calmodulin 2), while the three identified in the CREB pathway also include PPP2CA and CALM2 in addition to PRKAR1A (protein kinase, cAMP-dependent, regulatory, type I, α). SLC8A1, which is activated by protein phosphatase 2A, is a membrane transport protein that transports Ca2+ out of cells and Na+ into cells during relaxation to prevent intracellular calcium buildup. Calmodulin uses intracellular Ca2+ to control enzymes such as protein kinases and phosphatases. In regards to the CREB pathway, previous studies have shown that CREB phosphorylation is involved in activating expression of IEGs that ultimately affect neural synaptic plasticity in ageing animals and long term memory (13, 34, 53). The products of all three correlative genes in the CREB pathway work upstream of CREB to regulate its phosphorylation; these results suggest that modulation of CREB phosphorylation may play a role in neuronal aging processes in a healthy brain.
Other maps and processes that contain genes that correlate to principal component 1 include regulation of G1/S transition, cytoplasmic/mitochondrial transport of proapoptotic proteins, and the interferon-γ signaling pathway, which plays a role in cell cycle and translation regulation. Such processes may represent basal neuronal activity that is normally increased in the posterior cingulate, superior frontal gyrus, and primary visual cortex and normally decreased in the entorhinal cortex, hippocampus, and middle temporal gyrus (as defined by principal component 1). It is also interesting to note that principal component 1 demonstrates a very small amount of expression variation between the entorhinal cortex and hippocampus, regions that have been found to have similar functions in learning and memory. On a functional level, these two regions have intense bidirectional connections and may be broadly called the hippocampo-entorhinal cortex (46).
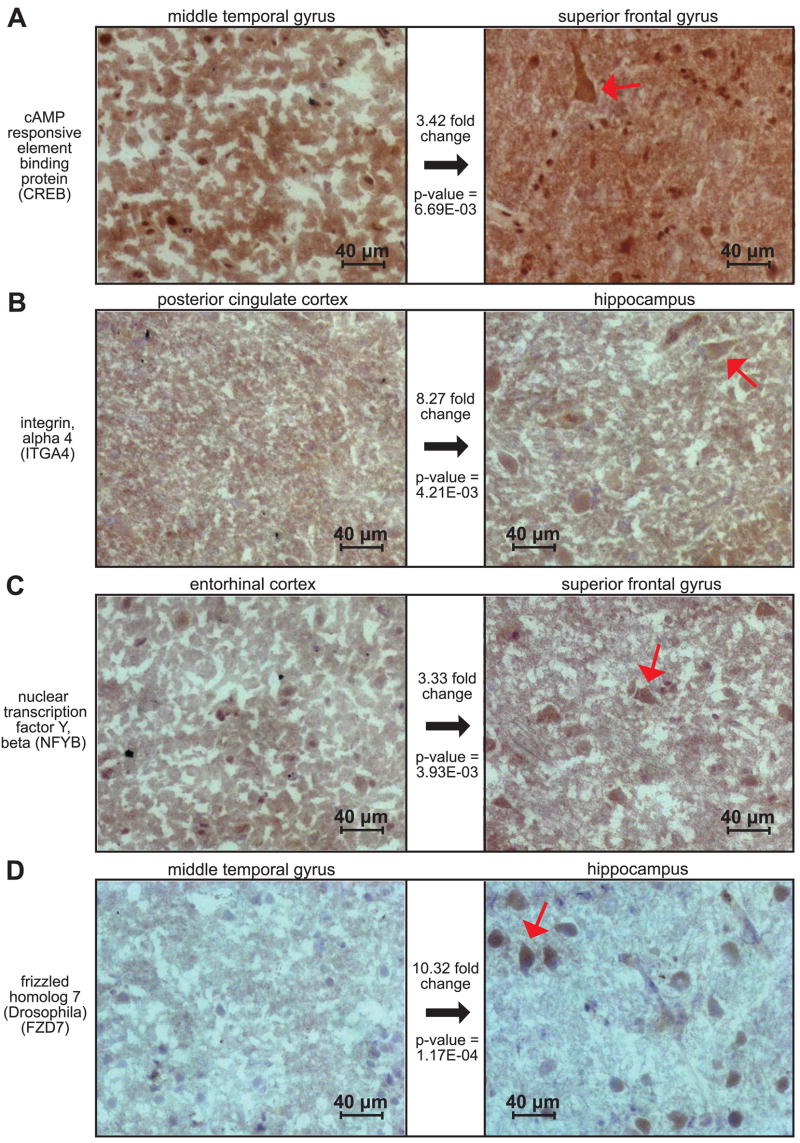
Immunohistochemistry validation of selected upregulated genes
We have performed immunohistochemistry on additional fresh frozen brain sections to validate genes, at the protein level, selected from statistical analyses (Fig. 3). In expression analyses, CREB was upregulated uniquely in the superior frontal gyrus compared with all other regions. Over all, CREB had an average 2.82 increased fold change in the superior frontal gyrus compared with all five other brain regions with an average P value of 1.61E-02. Staining of CREB in Fig. 3A (right, red arrow) shows clear immunperoxidase staining of layer III pyramidal neurons in the superior frontal gyrus compared with an absence of neuronal staining (Fig. 3A, left) in the middle temporal gyrus, the region that demonstrated the lowest expression of CREB. A direct comparison between the superior frontal and middle temporal gyri with respect to CREB expression showed a 3.42-fold change increase in the superior frontal gyrus with a P value of 6.69E-03.
Fig. 3.
Immunohistochemistry confirms region specific upregulation of multiple gene candidates. To validate expression profiling results at the protein level, we selected multiple genes showing significant upregulated expression in several regions and performed immunohistochemistry on additional fresh frozen sections. Because of the overall increase in expression of significant genes in the hippocampus and superior frontal gyrus, these areas were our focus for validation. The rightmost panels of A, B, C, and D show the region for which the respective protein was found to have the highest gene expression. The leftmost panels show the region that was found have the lowest levels of expression compared with the respective right panel for the gene and protein of interest. Gene expression fold changes and corresponding P values are shown between the 2 panels for each protein. Preferential staining of layer III pyramidal cells, as pointed out by the red arrows in the rightmost panels, is consistent with the gene expression results.
Integrin, alpha 4 (ITGA4) was found to have significantly upregulated expression in the hippocampus, with an average increased fold change of 6.99 over all other regions with a mean P value of 5.37E-03. Results from a one-to-one regional comparison between the hippocampus and posterior cingulate cortex showed an 8.27-fold change increase in the hippocampus with a P value of 4.21E-03. In Fig. 3B (right), our immunostaining confirms increased expression with the staining of layer III pyramidal cells, contrasting with Fig. 3B (left), which demonstrates an overall lack of neuronal staining in the posterior cingulate cortex.
NFYB also showed uniquely significant expression in the superior frontal gyrus. Compared with all other regions, NFYB had an average increased fold change of 2.73 with a P value of 1.24E-02. The entorhinal cortex showed the greatest decrease in expression of NFYB; specifically compared with the entorhinal cortex, NFYB had an increased 3.33-fold change in the superior frontal gyrus with a P value of 3.93E-03. These results correlate with the staining seen in Fig. 3C, in which layer III neurons are visibly stained in the right panel (superior frontal gyrus), and little if any staining is seen in the same layer in the entorhinal cortex (left panel).
Lastly, FZD7 displayed statistically significant expression in the hippocampus with an average fold increase of 8.19 with a P value of 1.62E-04. FZD7 showed the lowest expression in the middle temporal gyrus so that, directly compared with this region, FZD7 had an increase in fold change of 10.32 in the hippocampus with a P value of 1.17E-04. Again, these values are consistent with immunohistochemistry results in Fig. 3D. In the right panel, we see darkened neuronal staining in layer III of the hippocampus and an absence of neuronal staining in layer III of the medial temporal gyrus in the left panel.
Agonal conditions
Previous research has shown that agonal conditions of cases from which postmortem brain tissue is collected for study may influence gene expression analyses (37). This research found that individuals who experienced prolonged agonal states had a lower brain pH and that this was correlated with expression variation. Although speed of death of the case individuals analyzed in the study presented here is not classified, none of the individuals expired as a result of accident or suicide nor were maintained heroically, e.g., using a ventilator. Analysis of cerebrospinal fluid (CSF) pH of cases collected with the same methodology at the Sun Health Research Institute showed a narrow pH range with an approximate range of 6.4–7.2 (data not published). CSF pH consistency thus suggests that less gene expression variation would be expected. Furthermore, any presence of expression variability as a result of agonal state would result in the identification of fewer significant expression changes because of higher inherent variability of expression.
Summary
This study characterized and compared gene expression profiles from laser capture microdissected neurons in six different brain regions of the normal aged human brain. Previous microarray studies have focused on identifying differential expression in entire tissue sections of the cerebral cortex of healthy elderly and AD-affected individuals (57) and hippocampal CA1 and CA3 regions of AD-affected individuals (4). In this respect, this study provides a more focused analysis by evaluating the expression of individually selected normal neurons in six specific brain regions and helps to establish a baseline for future neuro-related studies as those mentioned. In particular, this study’s analysis has shown that there are significant expression changes that occur in the normal aged human brain and that these differences may play a role in age-related cognitive deficiencies. We have generated and laid out a descriptive reference data set that can be used as a foundation for future studies of normal neuronal function and for comparisons to numerous neurological disorders, including AD.
Acknowledgments
We thank the National Institute on Aging’s Alzheimer’s Disease Centers program and the National Alzheimer’s Coordinating Center for help in obtaining samples for analysis.
GRANTS
This project was funded by National Institute on Aging Grants K01AG-024079 to T. Dunckley, 1-RO1-AG-023193 to D. A. Stephan, P30 AG-19610 to E. M. Reiman, P50 AG-05681 to J. C. Morris, P01 AG-03991 to J. C. Morris, AG-05128 for the Duke University Alzheimer’s Disease Research Center, the National Alzheimer’s Coordinating Center Grant U01AG016976; by the Arizona Alzheimer’s Research Center (to E. M. Reiman) under a collaborative agreement from the National Institute on Aging; and by the State of Arizona to the Arizona Parkinson’s Disease Center (Arizona Biomedical Research Commission contract 0011).
Footnotes
Article published online before print. See web site for date of publication (http://physiolgenomics.physiology.org).
References
- 1.Abeliovich A, Chen C, Goda Y, Silva AJ, Stevens CF, Tonegawa S. Modified hippocampal long-term potentiation in PKC gamma-mutant mice. Cell. 1993;75:1253–1262. doi: 10.1016/0092-8674(93)90613-u. [DOI] [PubMed] [Google Scholar]
- 2.Angelie E, Bonmartin A, Boudraa A, Gonnaud PM, Mallet JJ, Sappey-Marinier D. Regional differences and metabolic changes in normal aging of the human brain: proton MR spectroscopic imaging study. AJNR Am J Neuroradiol. 2001;22:119–127. [PMC free article] [PubMed] [Google Scholar]
- 3.Beach TG, Walker R, McGeer EG. Patterns of gliosis in Alzheimer’s disease and aging cerebrum. Glia. 1989;2:420–436. doi: 10.1002/glia.440020605. [DOI] [PubMed] [Google Scholar]
- 4.Blalock EM, Geddes JW, Chen KC, Porter NM, Markesbery WR, Landfield PW. Incipient Alzheimer’s disease: microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc Natl Acad Sci USA. 2004;101:2173–2178. doi: 10.1073/pnas.0308512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bliss TV, Collingridge GL. A synaptic model of memory: long-term potentiation in the hippocampus. Nature. 1993;361:31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- 6.Bobinski M, de Leon MJ, Convit A, De Santi S, Wegiel J, Tarshish CY, Saint Louis LA, Wisniewski HM. MRI of entorhinal cortex in mild Alzheimer’s disease. Lancet. 1999;353:38–40. doi: 10.1016/s0140-6736(05)74869-8. [DOI] [PubMed] [Google Scholar]
- 7.Bouras C, Hof PR, Giannakopoulos P, Michel JP, Morrison JH. Regional distribution of neurofibrillary tangles and senile plaques in the cerebral cortex of elderly patients: a quantitative evaluation of a one-year autopsy population from a geriatric hospital. Cereb Cortex. 1994;4:138–150. doi: 10.1093/cercor/4.2.138. [DOI] [PubMed] [Google Scholar]
- 8.Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol (Berl) 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 9.Braak H, Braak E. The human entorhinal cortex: normal morphology and lamina-specific pathology in various diseases. Neurosci Res. 1992;15:6–31. doi: 10.1016/0168-0102(92)90014-4. [DOI] [PubMed] [Google Scholar]
- 10.Buée L, Bussiere T, Buee-Scherrer V, Delacourte A, Hof PR. Tau protein isoforms, phosphorylation and role in neurodegenerative disorders. Brain Res Rev. 2000;33:95–130. doi: 10.1016/s0165-0173(00)00019-9. [DOI] [PubMed] [Google Scholar]
- 11.Burke SN, Barnes CA. Neural plasticity in the ageing brain. Nat Rev. 2006;7:30–40. doi: 10.1038/nrn1809. [DOI] [PubMed] [Google Scholar]
- 12.Carroll RC, Zukin RS. NMDA-receptor trafficking and targeting: implications for synaptic transmission and plasticity. Trends Neurosci. 2002;25:571–577. doi: 10.1016/s0166-2236(02)02272-5. [DOI] [PubMed] [Google Scholar]
- 13.Clayton DF. The genomic action potential. Neurobiol Learn Mem. 2000;74:185–216. doi: 10.1006/nlme.2000.3967. [DOI] [PubMed] [Google Scholar]
- 14.Coffey CE, Wilkinson WE, Parashos IA. Quantitative cerebral anatomy of the aging human brain: a cross-sectional study using magnetic resonance imaging. Neurology. 1992;42:527–536. doi: 10.1212/wnl.42.3.527. [DOI] [PubMed] [Google Scholar]
- 15.Colledge M, Dean RA, Scott GK, Langeberg LK, Huganir RL, Scott JD. Targeting of PKA to glutamate receptors through a MAGUK-AKAP complex. Neuron. 2000;27:107–119. doi: 10.1016/s0896-6273(00)00013-1. [DOI] [PubMed] [Google Scholar]
- 16.Dalfo E, Barrachina M, Rosa JL, Ambrosio S, Ferrer I. Abnormal alpha-synuclein interactions with rab3a and rabphilin in diffuse Lewy body disease. Neurobiol Dis. 2004;16:92–97. doi: 10.1016/j.nbd.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 17.Davies L, Wolska B, Hilbich C, Multhaup G, Martins R, Simms G, Beyreuther K, Masters CL. A4 amyloid protein deposition and the diagnosis of Alzheimer’s disease: prevalence in aged brains determined by immunocytochemistry compared with conventional neuropathologic techniques. Neurology. 1988;38:1688–1693. doi: 10.1212/wnl.38.11.1688. [DOI] [PubMed] [Google Scholar]
- 18.De Leon MJ, George AE, Stylopoulos LA, Smith G, Miller DC. Early marker for Alzheimer’s disease: the atrophic hippocampus. Lancet. 1989;2:672–673. doi: 10.1016/s0140-6736(89)90911-2. [DOI] [PubMed] [Google Scholar]
- 19.Dequchi K, Inoue K, Avila WE, Lopez-Terrada D, Antalffy BA, Quattrocchi CC, Sheldon M, Mikoshiba K, D’Arcangelo G, Armstrong DL. Reelin and disabled-1 expression in developing and mature human cortical neurons. J Neuropathol Exp Neurol. 2003;62:676–684. doi: 10.1093/jnen/62.6.676. [DOI] [PubMed] [Google Scholar]
- 20.Dingledine R, Borges K, Bowie D, Traynelis SF. The glutamate receptor ion channels. Pharmacol Rev. 1999;51:7–61. [PubMed] [Google Scholar]
- 21.Du AT, Schuff N, Zhu XP, Jagust WJ, Miller BL, Reed BR, Kramer JH, Mungas D, Yaffe K, Chui HC, Weiner MW. Atrophy rates of entorhinal cortex in AD and normal aging. Neurology. 2003;60:481–486. doi: 10.1212/01.wnl.0000044400.11317.ec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Erdely HA, Lahti RA, Lopez MB, Myers CS, Roberts RC, Tamminga CA, Vogel MW. Regional expression of RGS4 mRNA in human brain. Eur J Neurosci. 2004;19:3125–3128. doi: 10.1111/j.0953-816X.2004.03364.x. [DOI] [PubMed] [Google Scholar]
- 23.Farrant M. Amino acids: inhibitory. In: Webster RA, editor. Neurotransmitters, Drugs, and Brain Function. Chichester, UK: John Wiley & Sons; 2001. pp. 225–245. [Google Scholar]
- 24.Feliciello A, Li Y, Avvedimento EV, Gottesman ME, Rubin CS. A-kinase anchor protein 75 increases the rate and magnitude of cAMP signaling to the nucleus. Curr Biol. 1997;7:1011–1014. doi: 10.1016/s0960-9822(06)00424-6. [DOI] [PubMed] [Google Scholar]
- 25.Fox NC, Warrington EK, Stevens JM, Rossor MN. Atrophy of the hippocampal formation in early familial Alzheimer’s disease. A longitudinal MRI study of at-risk members of a family with an amyloid precursor protein 717Val-Glymutation. Ann NY Acad Sci. 1996;777:226–232. doi: 10.1111/j.1749-6632.1996.tb34423.x. [DOI] [PubMed] [Google Scholar]
- 26.Frackowiak RSJ, Friston KJ, Frith CD, Dolan RJ, Mazziotta JC. Human Brain Function. San Diego, CA: Academic; 1997. [Google Scholar]
- 27.Frisoni GB, Laakso MP, Beltramello A, Geroldi C, Bianchetti A, Soininen H, Trabucchi M. Hippocampal and entorhinal cortex atrophy in frontotemporal dementia and Alzheimer’s disease. Neurology. 1999;52:91–100. doi: 10.1212/wnl.52.1.91. [DOI] [PubMed] [Google Scholar]
- 28.Galvin JE, Ginsberg SD. Expression profiling in the ageing brain: a perspective. Ageing Res Rev. 2005;4:529–547. doi: 10.1016/j.arr.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 29.Hu GY, Hvalby O, Walaas SI, Albert KA, Skjeflo P, Anderson P, Greengard P. Protein kinase C injection into hippocampal pyramidal cells elicits features of long term potentiation. Nature. 1987;328:426–429. doi: 10.1038/328426a0. [DOI] [PubMed] [Google Scholar]
- 30.Hyman BT, Van Hoesen GW, Damasio AR, Barnes CL. Alzheimer’s disease: cell-specific pathology isolates the hippocampal formation. Science. 1984;225:1168–1170. doi: 10.1126/science.6474172. [DOI] [PubMed] [Google Scholar]
- 31.Ibanez V, Pietrini P, Alexander GE, Furey ML, Teichberg D, Rajapakse JC, Rapoport SI, Schapiro MB, Horwitz B. Regional glucose abnormalities are not the result of atrophyin Alzheimer’s Disease. Neurology. 1998;50:1585–1593. doi: 10.1212/wnl.50.6.1585. [DOI] [PubMed] [Google Scholar]
- 32.Jack CR, Petersen RC, Xu Y, O’Brien PC, Smith GE, Ivnik RJ, Tangalos EG, Kokmen E. Rate of medial temporal lobe atrophy in typical aging and Alzheimer’s disease. Neurology. 1998;51:993–999. doi: 10.1212/wnl.51.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnston GV, Hartigan JA. Tau protein in normal and Alzheimer’s disease brain: an update. J Alzheimers Dis. 1999;1:329–351. doi: 10.3233/jad-1999-14-512. [DOI] [PubMed] [Google Scholar]
- 34.Jones MW, Errington ML, French PJ, Fine A, Bliss TV, Garel S, Charnay P, Bozon B, Laroche S, Davis S. A requirement for the immediate early gene Zif268 in the expression of late LTP and long-term memories. Nature Neurosci. 2001;4:289–296. doi: 10.1038/85138. [DOI] [PubMed] [Google Scholar]
- 35.Katsoulidis E, Li Y, Mears H, Platanias LC. The p38 mitogen-activated protein kinase pathway in interferon signal transduction. J Interferon Cytokine Res. 2005;25:749–756. doi: 10.1089/jir.2005.25.749. [DOI] [PubMed] [Google Scholar]
- 36.Klee CB, Draetta GF, Hubbard MJ. Calcineurin. Adv Enzymol Relat Areas Mol Biol. 1988;61:149–200. doi: 10.1002/9780470123072.ch4. [DOI] [PubMed] [Google Scholar]
- 37.Li JZ, Vawter MP, Walsh DM, Tomita H, Evans SJ, Choudary PV, Lopez JF, Avelar A, Shokoohi V, Chung T, Mesarwi O, Jones EG, Watson SJ, Akil H, Bunney WE, Jr, Myers RM. Systematic changes in gene expression in postmortem human brains associated with tissue pH and terminal medical conditions. Hum Mol Genet. 2004;13:609–616. doi: 10.1093/hmg/ddh065. [DOI] [PubMed] [Google Scholar]
- 38.Loessner A, Alavi A, Lewandrowski KU, Mozley D, Souder E, Gur RE. Regional cerebral function determined by FDG-PET in healthy volunteers: normal patterns and changes with age. J Nucl Med. 1995;36:1141–1149. [PubMed] [Google Scholar]
- 39.Lovestone S, Hartley CL, Pearce J, Anderton BH. Phosphorylation of tau by glycogen synthase kinase-3β in intact mammalian cells: the effects on the organization and stability of microtubules. Neuroscience. 1996;73:1145–1157. doi: 10.1016/0306-4522(96)00126-1. [DOI] [PubMed] [Google Scholar]
- 40.Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan J, Yankner BA. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- 41.MacDonald JF, Kotecha SA, Lu WY, Jackson MF. Convergence of PKC-dependent kinase signal cascade on NMDA receptors. Curr Drug Targets. 2001;2:299–312. doi: 10.2174/1389450013348452. [DOI] [PubMed] [Google Scholar]
- 42.Mahley RW, Rall SC., Jr Apolipoprotein E: far more than a lipid transport protein. Annu Rev Genomics Hum Genet. 2000;1:507–537. doi: 10.1146/annurev.genom.1.1.507. [DOI] [PubMed] [Google Scholar]
- 43.Malenka RC, Nicoll RA. Long-term potentiation-a decade of progress? Science. 1999;285:1870–1874. doi: 10.1126/science.285.5435.1870. [DOI] [PubMed] [Google Scholar]
- 44.Martin JH. Neuroanatomy: Text and Atlas. 3. New York: McGraw-Hill; 2003. [Google Scholar]
- 45.McGeer PL, McGeer EG, Suzuki J, Dolman CE, Nagai T. Aging, Alzheimer’s disease, and the cholinergic system of the basal forebrain. Neurology. 1984;34:741–745. doi: 10.1212/wnl.34.6.741. [DOI] [PubMed] [Google Scholar]
- 46.Mesulam MM. Principles of Behavioral and Cognitive Neurology. Cary, NC: Oxford University Press; 2000. Behavioral neuroanatomy; pp. 1–120. [Google Scholar]
- 47.Mielke R, Herholz K, Grond M. Clinical deterioration in probable Alzheimer’s disease correlates with progressive metabolic impairment of association areas. Dementia. 1994;5:36–41. doi: 10.1159/000106692. [DOI] [PubMed] [Google Scholar]
- 48.Misra-Press A, Rim CS, Yao H, Roberson MS, Stork PJ. A novel mitogen-activated protein kinase phosphatase. Structure, expression, and regulation. J Biol Chem. 1995;70:14587–14596. doi: 10.1074/jbc.270.24.14587. [DOI] [PubMed] [Google Scholar]
- 49.Mizoguchi A, Yano Y, Hamaguchi H, Yanagida H, Ide C, Zahraoui A, Shirataki H, Sasaki T, Takai Y. Localization of Rabphilin-3A on the synaptic vesicle. Biochem Biophys Res Commun. 1994;202:1235–1243. doi: 10.1006/bbrc.1994.2063. [DOI] [PubMed] [Google Scholar]
- 50.Morris JC, Price AL. Pathologic correlates of nondemented aging, mild cognitive impairment, and early-stage Alzheimer’s disease. J Mol Neurosci. 2001;17:101–118. doi: 10.1385/jmn:17:2:101. [DOI] [PubMed] [Google Scholar]
- 51.Okamoto T, Schlegel A, Scherer PE, Lisanti MP. Caveolins, a family of scaffolding proteins for organizing ’preassembled signaling complexes’ at the plasma membrane. J Biol Chem. 1998;273:5419–5422. doi: 10.1074/jbc.273.10.5419. [DOI] [PubMed] [Google Scholar]
- 52.Perry EK, Johnson M, Kerwin JM, Piggott MA, Court JA, Shaw PJ, Ince PG, Brown A, Perry RH. Convergent cholinergic activities in aging and Alzheimer’s disease. Neurobiol Aging. 1992;13:393–400. doi: 10.1016/0197-4580(92)90113-c. [DOI] [PubMed] [Google Scholar]
- 53.Platenik J, Kuramoto N, Yoneda Y. Molecular mechanisms associated with long-term consolidation of the NMDA signals. Life Sci. 2000;67:335–364. doi: 10.1016/s0024-3205(00)00632-9. [DOI] [PubMed] [Google Scholar]
- 54.Price JL, Davis PB, Morris JC, White DL. The distribution of tangles, plaques and related immunohistochemical markers in healthy aging and Alzheimer’s disease. Neurobiol Aging. 1991;12:295–312. doi: 10.1016/0197-4580(91)90006-6. [DOI] [PubMed] [Google Scholar]
- 55.Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL. A default mode of brain function. Proc Natl Acad Sci USA. 2001;98:676–682. doi: 10.1073/pnas.98.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reiman EM, Chen K, Alexander GE, Caselli RJ, Bandy D, Osborne D, Saunders AM, Hardy J. Correlations between apolipoprotein E epsilon4 gene dose and brain-imaging measurements of regional hypermetabolism. Proc Natl Acad Sci USA. 2005;102:8299–8302. doi: 10.1073/pnas.0500579102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ricciarelli R, d’Abramo C, Massone S, Marinari U, Pronzato M, Tabaton M. Microarray analysis in Alzheimer’s disease and normal aging. IUBMB Life. 2004;56:349–354. doi: 10.1080/15216540412331286002. [DOI] [PubMed] [Google Scholar]
- 58.Rogers J, Morrison JH. Quantitative morphology and regional and laminar distributions of senile plaques in Alzheimer’s disease. J Neurosci. 1985;5:2801–2808. doi: 10.1523/JNEUROSCI.05-10-02801.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rosenmund C, Carr DW, Bergeson SE, Nilaver G, Scott JD, Westbrook GL. Anchoring of protein kinase A is required for modulation of AMPA/kainate receptors on hippocampal neurons. Nature. 1994;368:853–856. doi: 10.1038/368853a0. [DOI] [PubMed] [Google Scholar]
- 60.Salmon E, Maquet P, Sadzot B, Degueldre C, Lemaire C, Franck G. Decrease of frontal metabolism demonstrated by positron emission tomography in a population of healthy elderly volunteers. Acta Neurol Belg. 1991;91:288–295. [PubMed] [Google Scholar]
- 61.Schroer TA. Dynactin. Annu Rev Cell Dev Biol. 2004;20:759–779. doi: 10.1146/annurev.cellbio.20.012103.094623. [DOI] [PubMed] [Google Scholar]
- 62.Shirataki H, Kaibuchi K, Sakoda T, Kishida S, Yamaguchi T, Wada K, Miyazaki M, Takai Y. Rabphilin-3A, a putative target protein for smg p25A/rab3A p25 small GTP-binding protein related to synaptotagmin. Mol Cell Biol. 1993;13:2061–2068. doi: 10.1128/mcb.13.4.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Small GW, Ercoli LM, Silverman DH, Huang SC, Komo S, Bookheimer SY, Lavretsky H, Miller K, Siddarth P, Rasgon NL, Mazziotta JC, Saxena S, Wu HM, Mega MS, Cummings JL, Saunders AM, Pericak-Vance MA, Roses AD, Barrio JR, Phelps ME. Cerebral metabolic and cognitive decline in persons at genetic risk for Alzheimer’s disease. Proc Natl Acad Sci USA. 2000;97:6037–6042. doi: 10.1073/pnas.090106797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Soderling TR, Derkach VA. Post-synaptic protein phosphorylation and LTP. Trends Neurosci. 2000;23:75–80. doi: 10.1016/s0166-2236(99)01490-3. [DOI] [PubMed] [Google Scholar]
- 65.Terry RD, DeTeresa R, Hansen LA. Neocortical cell counts in normal human adult aging. Ann Neurol. 1987;21:530–539. doi: 10.1002/ana.410210603. [DOI] [PubMed] [Google Scholar]
- 66.Tokito MK, Howland DS, Lee VM, Holzbaur EL. Functionally distinct isoforms of dynactin are expressed in human neurons. Mol Biol Cell. 1996;7:1167–1180. doi: 10.1091/mbc.7.8.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Viatour P, Merville MP, Bours V, Chariot A. Phosphorylation of NF-kappaB and IkappaB proteins: implications in cancer and inflammation. Trends Biochem Sci. 2005;30:43–52. doi: 10.1016/j.tibs.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 68.Webster RA. Dopamine (DA) In: Webster RA, editor. Neurotransmitters, Drugs, and Brain Function. Chichester, UK: John Wiley & Sons; 2001. pp. 147–148. [Google Scholar]
- 69.Zachariae W, Nasmyth K. Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev. 1999;13:2039–2058. doi: 10.1101/gad.13.16.2039. [DOI] [PubMed] [Google Scholar]