Abstract
Human serum albumin (HSA) nanoparticles represent a promising tool for targeted drug delivery to tumor cells. The coupling of the antibody trastuzumab to nanoparticles uses the capability of human epidermal growth factor receptor 2 (HER2)-positive cells to incorporate agents linked to HER2. In our present study, we developed targeted nanoparticles loaded with antisense oligonucleotides (ASOs) against polo-like kinase 1 (Plk1). We evaluated the receptor-mediated uptake into HER2-positive and -negative breast cancer and murine cell lines. We performed quantitative real-time PCR and Western blot analyses to monitor the impact on Plk1 expression in HER2-positive breast cancer cells. Antibody-conjugated nanoparticles showed a specific targeting to HER2-overexpressing cells with cellular uptake by receptor-mediated endocytosis and a release into HER2-positive BT-474 cells. We observed a significant reduction of Plk1 mRNA and protein expression and increased activation of Caspase 3/7. Thus, this is the first report about ASO-loaded HSA nanoparticles, where an impact on gene expression could be observed. The data provide the basis for the further development of carrier systems for Plk1-specific ASOs to reduce off-target effects evoked by systemically administered ASOs and to achieve a better penetration into primary and metastatic target cells. Treatment of tumors using trastuzumab-conjugated ASO-loaded HSA nanoparticles could be a promising approach to reach this goal.
Introduction
Antibodies are well-established tools to target drugs or colloidal carriers to specific cell types [1–3]. This targeted delivery reduces possible side effects and off-target effects. In addition, increasing knowledge about the genetic control of cellular proliferation provides the basis for specific therapeutic strategies to combat proliferative disorders such as cancer. Key regulators for mitosis in mammalian cells are the polo-like kinases (Plks), which represent highly conserved serine/threonine kinases [4]. Polo-like kinase 1 (Plk1) activity is elevated in all cancer cells analyzed to date [5]. The importance of Plk1 for the aggressiveness of a tumor and for predicting outcomes in cancer patients results from its contribution to transformation and from overriding the checkpoint control of the cell cycle [6–8]. The inhibition of Plk1 with antibodies, antisense oligonucleotides (ASOs), small interfering RNA (siRNA), or dominant-negative mutants leads to mitotic catastrophe, increased apoptosis, and tumor inhibition [9–15]. Antisense oligonucleotides can, when targeted to key elements of proliferation-relevant signal transduction pathways, prevent the development of specific human cancers. Several phosphorothioate ASOs are currently being evaluated in patients suffering from different types of cancer [16], suggesting ASOs as valuable agents for therapeutic approaches. Although the power of ASOs remains unrivaled, the dose-limiting side effects and toxicities of phosphorothioate ASOs in their relevant doses [16,17] or inadequate penetration into the target tumor tissue emphasize the necessity of further improvements in ASO chemistry and delivery systems [18], especially of targeted delivery through specific cell surface receptors [19–21].
Trastuzumab (Herceptin) is a humanized IgG1 monoclonal antibody directed against the extracellular domain of the human epidermal growth factor receptor 2 (HER2), which offers an excellent strategy for drug targeting to cancer cells because HER2 is an easily accessible cell surface receptor overexpressed on the primary tumor as well as on metastatic sites [22]. Biodegradable human serum albumin (HSA)-based nanoparticles represent a nontoxic colloidal carrier system with high drug loading capacity [23], which have been extensively studied in clinical settings with the cremophor-free formulation of paclitaxel (Abraxane, ABI-007) [24,25]. Drugs can be incorporated within the particle matrix, adsorbed on the particle surface, or bound by covalent linkage [26,27]. By this, the drug is protected against degradation, and controlled release is enabled [28].
The aim of the present study was to develop ASO-loaded HSA nanoparticles as cell type-specific drug delivery system with receptor-mediated uptake, release into the cytoplasm, and a functional readout concerning Plk1 expression. We observed for the first time a receptor-mediated cellular uptake of ASO-loaded trastuzumabmodified HSA nanoparticles with an inhibitory effect on Plk1 expression. Hence, this drug delivery system could provide the basis for a targeted therapy of HER2-positive cancers with Plk1-specific ASOs.
Materials and Methods
Reagents and Chemicals
Human serum albumin (fraction V) and glutaraldehyde, 8% aqueous solution, were obtained from Sigma (Steinheim, Germany). 2-Iminothiolane (Traut's reagent), and D-Salt Dextran Desalting columns were purchased from Pierce (Rockford, IL). The succinimidyl ester of methoxy poly(ethylene glycol)propionic acid with an average molecular weight of 5.0 kDa (mPEG5000-SPA) and the cross-linker poly(ethylene glycol)-α-maleimide-ϖ-NHS ester with an average molecular weight of 5.0 kDa (NHS-PEG5000-Mal) were purchased from Nektar Therapeutics (Huntsville, AL). Monoclonal anti-human Plk1 (#SC-17783), polyclonal rabbit Plk2 (#SC-25421) polyclonal goat Plk4 antibody (#SC-49101), polyclonal rabbit p38 antibody (#SC-535), and goat anti-mouse (#SC-2031), goat anti-rabbit (#SC-2030), and donkey anti-goat (#SC-2033) secondary antibodies were from Santa Cruz Biotechnology, Inc. (Heidelberg, Germany); monoclonal antibody against HER2 (#OP15L) was from Oncogene Research Products (Calbiochem, Nottingham, UK); and monoclonal antibody against β-actin (#A-5316) was from Sigma-Aldrich (Taufkirchen, Germany). Trastuzumab (Herceptin) was obtained from Roche (Mannheim, Germany). All reagents were of analytical grade and were used as received.
The phosphorothioate antisense oligonucleotides were P12 (targeted against Plk1: acc agt ccg gag ggg agg gc) and HSV (targeted against the Herpes Simplex Virus: gcg gag gtc cat gtc gta cgc) as described before [10,29]. For cellular distribution studies a Cy5-labeled phosphorothioate oligonucleotide (cta cga tct act ggc tcc at) was purchased from MWG-Biotech GmbH (Ebersberg, Germany).
Cell Culture
The breast cancer cell lines MCF-7 and BT-474 and the murine fibroblast cells NIH-3T3 and murine myoblast cells C2C12 were obtained from DSMZ (Braunschweig, Germany), and SK-BR-3 cells were from ATCC (LGC Promochem, Wesel, Germany). All cell lines were cultured according to the supplier's instructions without antibiotics. Fetal calf serum was purchased from PAA Laboratories (Cölbe, Germany). McCoy's medium was from BioWhittaker (Apen, Germany), RPMI 1640, sodium pyruvate, phosphate-buffered saline (PBS), glutamine, and trypsin were from Invitrogen (Karlsruhe, Germany), and bovine insulin was from Calbiochem (Schwalbach, Germany).
Preparation of ASO-Loaded HSA Nanoparticles
Human serum albumin (20.0 mg) was dissolved in 500 µl of purified water, and the pH was adjusted to 5.5. An amount of 300.0 µg of the respective ASO (P12 and HSV) was added, and the mixture was stirred for 15 minutes. To form nanoparticles, 2.7 ml of 96% ethanol was added at a rate of 1 ml/min with a tubing pump (IPN; Ismatec, Glattbrugg, Switzerland) under constant stirring (550 rpm) at room temperature as established in earlier publications [30,31]. Particles were stabilized by cross-linking with 0.03% glutaraldehyde solution (11.8 µl from the 8% stock solution) for at least 12 hours under constant stirring at room temperature. Particles were purified by centrifugation (16,100g for 8 minutes) and redispersion of the pellet in phosphate buffer, pH 8.0.
The coupling reaction of trastuzumab with the ASO-loaded nanoparticles was performed as described [31]. Nanoparticles (10 mg) were activated with NHS-PEG5000-Mal (8.8 mg). Trastuzumab was thiolated with 50-fold molar excess of 2-iminothiolane. For coupling reactions, the nanoparticle suspension was incubated with thiolated trastuzumab for at least 12 hours. Samples were purified as described earlier. Particles with PEG-modified surface instead of trastuzumab coupling were prepared as described previously [31].
Unloaded particles were prepared as described [31] at a pH of 7.5. Modifications were performed according to the ASO-loaded particles.
Particle Characterization
The amount of ASO bound to the nanoparticles was calculated as the difference between the total amount of the initial ASO added and the amount of ASO determined in the supernatants obtained during the purification steps. The ASO content was determined by a strong ion-exchange HPLC assay as described [30], using a 4x 250-mm column (DNApac PA100; Dionex, Idstein, Germany) and an HPLC system (Hitachi; Merck, Darmstadt, Germany). The amount of trastuzumab bound to the particle surface was analyzed by size exclusion chromatography as described previously [31].
Particle diameter and polydispersity were measured by photon correlation spectroscopy, and zeta potential was determined by microelectrophoresis using Zetasizer 3000 HSa (Malvern Instruments, Malvern, UK). Before measurement, the samples were diluted with purified water. Particle content was determined by gravimetry.
Storage Stability
Trastuzumab-modified particles loaded with P12 were prepared and analyzed as described earlier. Without any additional agents, the particle samples were stored in purified water at 4°C for a period of 6 weeks. Once a week, particle diameter, polydispersity, and zeta potential were measured. Additionally, an aliquot of the particle suspension was centrifuged, and the supernatant was analyzed for trastuzumab, HSA, and P12 using the chromatographic methods described earlier.
Treatment of Breast Cancer Cells with ASO-Loaded Trastuzumab-Modified Nanoparticles
To investigate the cell-specific binding, uptake and release efficiency of trastuzumab-modified compared to PEGylated HSA nanoparticles and to analyze the inhibitory effect of the incorporated ASOs on Plk1 expression, cells were seeded in 12-well plates, 75-cm2 cell culture flasks or on slide flasks, respectively, and were grown to 40% to 50% confluence. Cells were treated with trastuzumab-modified and with PEGylated nanoparticles in a concentration of 100 µg/ml in cell culture medium at 37°C and 5% CO2 as described [30]. Additionally, to confirm the specificity of particle binding to HER2-overexpressing cells, in the case of SK-BR-3 and BT-474 cells, experiments were performed with and without preincubating with 2.5 µg/ml trastuzumab for 30 minutes at 37°C.
After nanoparticle incubation for 30 minutes, 60 minutes, 3 hours, 5 hours, and 24 hours cells were harvested for fluorescence-activated cell sorting (FACScan) analysis, after 48 hours for reverse transcription-polymerase chain reaction (RT-PCR), after 24 and 48 hours for quantitative real-time PCR, after 24, 48, and 72 hours for Western blot analysis, and after 72 hours for apoptosis analyses. For the RT-PCR, quantitative real-time PCR, Western blot, and apoptosis analyses, nanoparticle suspensions were removed from the cells after an incubation time of 60 minutes, and fresh medium was added.
FACScan Analysis of ASO-Loaded HSA Nanoparticles
FACScan analysis to detect the autofluorescence of the particles within the cells was performed using a Becton Dickinson FACScan apparatus (Heidelberg, Germany) as described [31]. In brief, cells were harvested, washed with PBS, resuspended in cold PBS (pH 7.4), and ethanol was added. After 30 minutes of incubation on ice, cells were centrifuged and resuspended in PBS. For each experiment, 10,000 cells were analyzed in triplicate. Subsequent analysis was performed with CELLQuest software (Becton Dickinson).
Confocal Laser Scanning Microscopy Analysis of Nanoparticle Uptake in BT-474 Cells
For the analysis of the cellular uptake by confocal laser scanning microscopy, the nanoparticles were prepared with Cy5-labeled ASO instead of P12. After incubating with trastuzumab-modified or PEGylated nanoparticles, cells were washed with PBS for 15 minutes. Membrane staining was carried out using Alexa594 Concanavalin A (Molecular Probes, Göttingen, Germany; 500 µl, 0.0005%) for 2 minutes. Cells were washed with PBS, fixed with -20°C cold methanol for 15 minutes, and washed with PBS. Then the slides were embedded with Vectashield (Vector, Burlingame, CA).
RNA Preparation and RT-PCR Analysis of Plk1 and GAPDH Expression
Total RNA from cultured cells was isolated using RNeasy Mini Kits according to the manufacturer's protocol 48 hours after incubating with the nanoparticle formulations (Qiagen, Hilden, Germany). For RT-PCR, the QIAGEN OneStep RT-PCR Kit was used according to the supplier's instructions (Qiagen). Two micrograms of total RNA were added to the PCR mix containing Plk1-specific or GAPDH-specific primers, respectively, and the RTreactions were carried out as described [29]. Resulting DNA fragments were separated on 1% agarose gels and visualized by ethidium bromide staining.
Quantitative Real-Time PCR Analysis
After isolation of total RNA using RNeasy Mini Kits (Qiagen) according to the manufacturer's protocol 24 and 48 hours after incubating with the nanoparticle formulations, the mRNA was transcribed into cDNA using the High-Capacity cDNA Reverse Transcription Kits (Applied Biosystems, Darmstadt, Germany). Thereafter, 50 ng of cDNA was subjected to quantitative real-time PCR analyses targeting Plk1 and GAPDH using the FastStart Universal Probe Master ROX and Universal ProbeLibrary #30 (Plk1) and #60 (GAPDH) (Roche). Primer sequences were chosen as suggested by Roche: Plk1 forward 5′-cacagtgtcaatgcctccaa, Plk1 reverse 5′-ttgctgacccagaagatgg, GAPDH forward 5′-agccacatcgctcagacac, GAPDH reverse 5′-gcccaatacgaccaaatcc. Analysis was performed using the StepOne Real-Time PCR System and the StepOne v2.0 software (Applied Biosystems). Relative gene expression values were determined by the ΔΔCT method using the StepOne v2.0 software (Applied Biosystems). Data are presented as the fold difference in Plk1 expression normalized to the housekeeping gene GAPDH as endogenous reference, and relative to the untreated control cells.
Western Blot Analysis
For Western blot analysis, breast cancer and murine cells were lysed after 24, 48, and 72 hours after incubating with the nanoparticle formulations, and protein concentration was determined as previously described [9,29]. An amount of 50 µg of total protein was separated on 10% Bis-Tris-polyacrylamide gels and then transferred (30 V, 1 hour) to Immobilon-P membranes (Millipore, Bedford, MA) according to the Invitrogen protocol. Membranes were incubated for 1 hour in 5% powdered nonfat milk in PBS with monoclonal antibodies against HER2 (1:250), Plk1 (1:100), Plk2 (1:200), Plk4 (1:200), p38 (1:20,000), and β-actin (1:200,000) and for 30 minutes in 5% nonfat dry milk with goat anti-mouse, goat anti-rabbit, or donkey anti-goat serum, respectively (1:2000), and visualized as described [9].
In Western blot and RT-PCR experiments, Plk1 expression levels were routinely normalized to levels of GAPDH, p38, or β-actin expression, respectively. The resulting GAPDH-, p38-, or β-actin-normalized Plk1, Plk2, or Plk4 levels, respectively, are presented relative to GAPDH-, p38-, or β-actin-normalized levels in untreated cells. Polo-like kinase 1, Plk2, Plk4, HER2, GAPDH, p38, and β-actin expressions were quantified with a gel documentation system (Model 1D 3.5; Kodak, Stuttgart, Germany) as described previously [9].
Apoptosis Assay
We performed Caspase-Glo assays to detect activation of Caspase 3/7 using the Caspase-Glo 3/7 Assay System (Promega, Mannheim, Germany). In brief, cells were analyzed 72 hours after incubating with the nanoparticle formulations. We used an amount of 10 or 20 µg of total protein, respectively, to detect the activation of Caspase 3/7. Cell lysates were mixed with the Caspase-Glo substrate, incubated for approximately 30 minutes, and thereafter analyzed using a multilabel counter (Victor 1420; Perkin Elmer Wallac, Freiburg, Germany). These assay systems provide substrates that are processed by activated caspases to aminoluciferin which, itself, is a substrate for luciferase. The emitted light can be measured at 562 nm referred to as relative luminescence units (RLUs). RLU levels of nanoparticletreated cells were presented relative to RLU levels of untreated control cells.
Statistical Methods
Two-way analysis of variance (GraphPad Prism; GraphPad Software, Inc., San Diego, CA) was performed as described [9].
Results
HER2 Expression of Breast Cancer and Murine Cell Lines
In a first set of cell culture experiments, we analyzed the expression of endogenous HER2 protein levels in different breast cancer and murine cells by Western blot analysis. We included two murine cell lines in our studies to evaluate a potential nanoparticle uptake in murine cells for future cancer xenograft experiments.
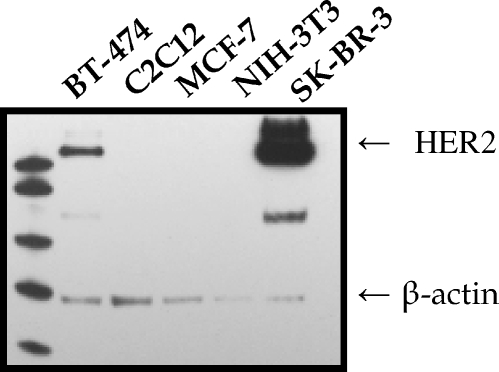
Membranes were incubated with antibodies against HER2 and β-actin. While in the case of MCF-7, NIH-3T3 and C2C12 cells, the level of HER2 was below the limit of detection, strong expression was observed in BT-474 and especially in SK-BR-3 cells, which is in agreement with previous observations [31,32] (Figure 1). This confirms suitable conditions for the analysis of trastuzumab-conjugated nanoparticles.
Figure 1.
Expression of HER2 in breast cancer and murine cell lines. A Western blot analysis was performed using anti-HER2 antibodies. To control for variability of loading, membranes were reprobed with antibodies against β-actin. The expression of HER2 in BT-474, C2C12, MCF-7, NIH-3T3, and SK-BR-3 cells is depicted in alphabetical order.
Preparation and Characterization of ASO-Loaded HSA Nanoparticles
HSA nanoparticles loaded with ASO were prepared using a desolvation technique. Before desolvation, the HSA solution was incubated with ASOs allowing the ASO to adsorb to the HSA molecules. During desolvation, the ASO precipitates with the HSA molecules, forming nanoparticles. After stabilization and purification, the particle surface was modified with the cross-linker NHS-PEG5000-Mal. The succinimidyl group reacts with the amino groups of the particle surface, and the maleimide group reacts with the thiol groups introduced in trastuzumab, forming a thioether linkage (Figure 2). The PEG modification prevents unspecific binding of plasma components to the particle surface and uptake in cells of the reticuloendothelial system [33]. The coupling of trastuzumab enables a site-specific targeting of the nanoparticles to HER2-overexpressing cancer cells. As control, nanoparticles without antibody coupling were prepared using a monofunctional PEG derivative.
Figure 2.
Schematic representation of the antibody surface modification of the ASO-loaded nanoparticles. Trastuzumab was covalently attached to the particle surface by a PEG-based heterobifunctional cross-linker.
The ASO-loaded nanoparticles were 267 to 278 nm in diameter, and particles without ASO loading were 192 to 196 nm in diameter (Table 1). All preparations showed a monodisperse diameter distribution, the surface charge was between -30 and -40 mV. The particles contained approximately 20 µg/mg ASO. No differences in loading could be observed between P12 and HSV. Trastuzumab was attached quantitatively to the nanoparticle surface, leading to 17 µg of trastuzumab/mg nanoparticle.
Table 1.
Physico-chemical Characteristics of Trastuzumab-Modified and PEGylated HSA Nanoparticles Loaded With P12- or HSV-ASO or Unloaded (n = 15).
| Preparation | P12 + Trastuzumab | P12 + mPEG | HSV + Trastuzumab | HSV + mPEG | Unloaded + Trastuzumab | Unloaded + mPEG | |
| Particle diameter | (nm) | 277.9 ± 42.6 | 267.2 ± 39.6 | 273.5 ± 35.3 | 271.5 ± 34.8 | 196.0 ± 12.7 | 191.6 ± 12.2 |
| Polydispersity | 0.060 ± 0.038 | 0.026 ± 0.020 | 0.035 ± 0.014 | 0.030 ± 0.015 | 0.049 ± 0.059 | 0.028 ± 0.020 | |
| Zeta potential | (mV) | -36.5 ± 4.6 | -38.4 ± 7.1 | -37.7 ± 8.8 | -39.8 ± 7.0 | -35.5 ± 10.4 | -35.9 ± 7.5 |
| Particle content | (mg/ml) | 15.3 ± 2.6 | 14.0 ± 1.4 | 14.7 ± 1.8 | 14.2 ± 1.8 | 17.9 ± 2.8 | 17.2 ± 2.3 |
| Antibody binding efficiency | (%) | 100 | 0 | 100 | 0 | 100 | 0 |
| (µg/mg) | 17.59 ± 3.87 | 0 | 17.92 ± 3.89 | 0 | 15.14 ± 3.74 | 0 | |
| ASO loading | (µg/mg) | 19.37 ± 2.89 | 20.63 ± 1.98 | 20.56 ± 2.35 | 21.21 ± 2.21 | 0 | 0 |
Values are presented as mean ± SD.
Storage Stability
For stability investigations, the physico-chemical properties of the P12-loaded trastuzumab-modified particles were monitored for a period of 6 weeks. Particle diameter, polydispersity, and zeta potential did not change significantly (Table 2). Furthermore, the particle suspensions were centrifuged, and the supernatants were analyzed for unbound trastuzumab and dissolved HSA by size exclusion chromatography. Over time, neither trastuzumab nor HSA was detectable. The analyses of the supernatants by ion exchange chromatography revealed a drug leakage of P12 of up to 6% of the total drug loading. Therefore, the particle system was stable over the observed time span and can be stored after preparation.
Table 2.
Storage Stability of P12-Loaded Trastuzumab-Modified Nanoparticles.
| Parameter | Week 0 | Week 1 | Week 2 | Week 3 | Week 4 | Week 5 | Week 6 | |
| Diameter | (nm) | 263.8 ± 0.3 | 260.7 ± 3.8 | 259.6 ± 4.3 | 258.6 ± 3.5 | 265.7 ± 1.4 | 263.7 ± 3.5 | 266.6 ± 3.8 |
| Polydispersity | 0.092 ± 0.091 | 0.030 ± 0.030 | 0.025 ± 0.020 | 0.027 ± 0.025 | 0.022 ± 0.012 | 0.020 ± 0.012 | 0.067 ± 0.071 | |
| Zeta potential | (mV) | -43.1 ± 5.2 | -32.0 ± 2.9 | -36.9 ± 5.9 | -31.5 ± 7.6 | -37.9 ± 6.5 | -36.1 ± 0.4 | -36.5 ± 2.7 |
| Trastuzumab | (%) | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| HSA | (%) | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| P12 | (%) | 3.5 ± 1.0 | 2.5 ± 0.4 | 3.2 ± 1.2 | 4.6 ± 1.2 | 5.3 ± 1.1 | 5.6 ± 1.5 | 6.3 ± 1.5 |
Physico-chemical properties were measured every week. The particle suspension was centrifuged and trastuzumab. HSA and P12 were determined in the supernatants. The time point Week 0 describes the loss during preparation (n = 3).
Values are presented as mean ± SD.
Specific Cellular Uptake of Trastuzumab-Modified Nanoparticles
To analyze whether the conjugation of trastuzumab is an effective method for specific targeting of HER2-positive breast cancer cells, we investigated the time-dependent uptake of trastuzumab-modified versus PEGylated ASO-loaded nanoparticles in HER2-positive SK-BR-3 and BT-474 cells and HER2-negative MCF-7 cells (Figure 3) by FACScan analysis. The uptake of unloaded HSA nanoparticles has been intensively examined in a previous study [31].
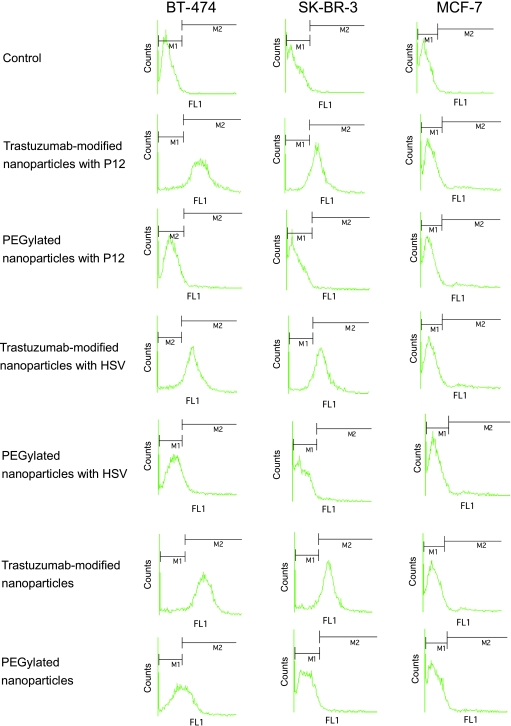
Figure 3.
Uptake of trastuzumab-modified and PEGylated nanoparticles with and without ASOs into HER2-positive and -negative breast cancer cell lines. FACScan analysis of BT-474 (left panels), SK-BR-3 (middle panels), and MCF-7 cells (right panels) after 60 minutes of incubating with the nanoparticles. M1 defines the cells without fluorescence (control), and M2 defines cells that show green fluorescence, indicating the uptake of nanoparticles.
In a first experiment, we analyzed the uptake in BT-474 cells (Figure 3, left panels). After 60 minutes of incubation, we observed a significant enrichment of trastuzumab-modified nanoparticles loaded with P12 in 60.09% of BT-474 cells compared to 1.91% in the case of P12-loaded PEGylated nanoparticles. For trastuzumab-modified HSV-loaded nanoparticles, the uptake was 65.23% compared to 5.30% for PEGylated nanoparticles. In SK-BR-3 cells, the uptake followed a similar kinetics (Figure 3, middle panels): P12-loaded trastuzumab-modified HSA nanoparticles showed receptor-mediated uptake with enrichment of 65.36% compared to 3.17% for PEGylated P12-loaded nanoparticles after 60 minutes of incubation. HSV-loaded trastuzumab-modified nanoparticles showed enrichment in 60.69% of SK-BR-3 cells compared to 4.45% for PEGylated nanoparticles. For both HER2-positive cell lines, fluorescence after 60 minutes of incubating with PEGylated nanoparticles was comparable to untreated control cells, indicating almost no affinity of the PEGylated nanoparticles to breast cancer cells. HER2-independent uptake or uptake mediated by recycled HER2 receptors of P12- and HSV-loaded nanoparticles increased over the time of 24 hours (Figure 4, A and B). Only the P12-loaded PEGylated nanoparticles showed slight HER2-independent uptake over the period of 24 hours.
Figure 4.
Uptake of trastuzumab-modified and PEGylated nanoparticles with and without ASOs into HER2-positive and -negative breast cancer cell lines. Graphical summary of time-dependent uptake of trastuzumab-modified and PEGylated nanoparticles with and without ASOs into (A) BT-474, (B) SK-BR-3, and (C) MCF-7 cells (n = 3, mean ± SD).
HER2-negative MCF-7 cells showed no significant differences in the uptake of trastuzumab-modified and PEGylated nanoparticles after 30 minutes to 24 hours of incubation (Figures 3, right panels, and 4C). Uptake increased in a time-dependent, HER2-independent manner for all nanoparticle formulations. For the P12- and HSV-loaded nanoparticles, the difference between trastuzumab-modified and PEGylated nanoparticles was even stronger than that for the unloaded particles, which might be due to the slightly smaller diameter of the unloaded particles compared to ASO-loaded particles.
In addition, we analyzed the effect of a preincubation of HER2-positive cells (BT-474 and SK-BR-3) with trastuzumab on the uptake of ASO-loaded and -unloaded trastuzumab-conjugated nanoparticles. Therefore, we incubated the cells with 2.5 µg/ml trastuzumab for 30 minutes at 37°C before the addition of the nanoparticle formulations. We observed that, after this preincubation, cell binding sites could be effectively blocked, and after 30 and 60 minutes, no significant difference in cellular accumulation between trastuzumab-modified and PEGylated nanoparticles loaded with P12 or HSV, respectively, was detectable (Figure 4, A and B). After incubation periods of 3, 5, and 24 hours, HER2-independent uptake or uptake mediated by recycled HER2 receptors increased up to levels comparable with the specific antibody-mediated uptake of trastuzumabmodified nanoparticles.
Cellular Uptake of Trastuzumab-Modified Nanoparticles into Normal Murine Cells
Next we analyzed whether the trastuzumab-modified or PEGylated unloaded or ASO-loaded HSA nanoparticles could be enriched in murine cells. We used NIH-3T3 and C2C12 cells and incubated these cell lines with the nanoparticle preparations as described earlier for the breast cancer cells. We observed only a slight time-dependent, HER2-independent uptake of the different nanoparticle preparations: in NIH-3T3 cells, uptake levels were below 20% after 30 minutes to 24 hours of incubation (data not shown), and in C2C12, uptake levels were below 20% up to 5 hours of incubation and showed a slight increase up to 40% after 24 hours of incubation (data not shown).
Uptake and Release of ASO-Loaded Trastuzumab-Modified Nanoparticles
We investigated the time course of the release of trastuzumab-modified nanoparticles with a Cy5-labeled ASO in HER2-positive BT-474 cells compared to PEGylated nanoparticles containing the same ASO. For this purpose, we performed confocal laser scanning microscopy studies, where we observed a time-dependent release of the Cy5-labeled ASO over a period of 1 to 24 hours (Figure 5). After 60 minutes, the released ASO could only be detected for the trastuzumab-modified nanoparticles and not for the PEGylated nanoparticles, underlining the receptor-mediated uptake.
Figure 5.
Time-dependent release of trastuzumab-modified and PEGylated nanoparticles with a Cy5-labeled ASO into BT-474 cells: (upper panels) PEGylated nanoparticles; (lower panels) trastuzumab-modified nanoparticles. Red: membrane staining with Alexa594 Concanavalin A; Blue: Cy5-labeled ASO.
Downregulation of Plk1 mRNA and Protein Expression
To investigate whether the trastuzumab-modified ASO-loaded nanoparticles show a biologic activity, we performed RT-PCR, quantitative real-time PCR, and Western blot analyses to determine the Plk1 mRNA and protein expression.
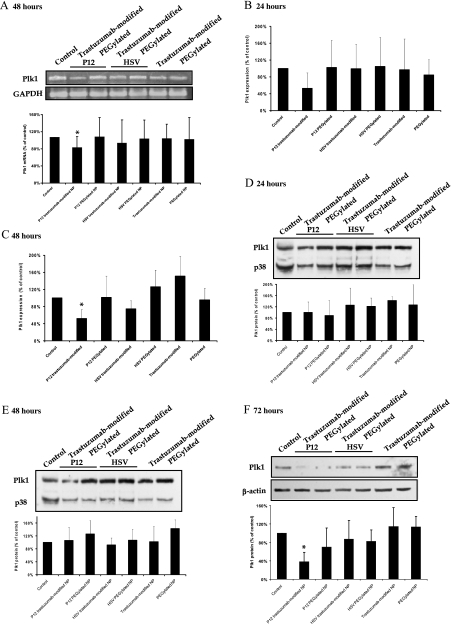
First, we incubated HER2-positive BT-474 cells with trastuzumabmodified and PEGylated nanoparticles loaded with P12 and HSV and with unloaded nanoparticles. Using RT-PCR, we observed a reduction of Plk1 mRNA 48 hours after incubating with P12-loaded trastuzumab-modified nanoparticles to levels of 77% (P < .05; Figure 6A) compared to control cells. Incubation with all other nanoparticle formulations did not alter Plk1 mRNA levels significantly.
Figure 6.
Plk1 mRNA and protein expression in BT-474 cells after incubating with nanoparticles. (A) An RT-PCR analysis was performed 48 hours after incubating with nanoparticle formulations using primers specific for Plk1 and for GAPDH for standardization. Representative gels show Plk1 cDNA (top) and GAPDH cDNA for standardization (bottom). Percentage of Plk1 cDNA expression is given as percentage of GAPDH-standardized Plk1 cDNA expression in control cells (n = 8, mean ± SD). A quantitative real-time PCR analysis was performed (B) 24 and (C) 48 hours after incubating with the nanoparticle formulations using Plk1- and GAPDH-specific primers. Graphical summary of gene expression values of treated cells standardized to control cells are shown (n = 3–5, mean ± SD). (D–F) Western blot analyses were performed using anti-Plk1 antibodies 24, 48, and 72 hours after incubating with the nanoparticle formulations. To control for variability of loading, membranes were reprobed with antibodies against p38 (24 and 48 hours) or β-actin (72 hours). Representative Western blots show Plk1 (top) and β-actin or p38 protein (bottom) for standardization. Percentage of Plk1 protein expression is given as percentage of β-actin- or p38-standardized Plk1 levels in control cells (n = 3, mean ± SD).
To investigate this effect in more detail, we did quantitative real-time PCR analyses 24 and 48 hours after incubating with the nanoparticle formulations. The analysis of Plk1 gene expression standardized to GAPDH expression revealed a nonsignificant reduction of Plk1 mRNA expression 24 hours after incubating with P12-loaded trastuzumab-modified nanoparticles compared to controls (P = .15; Figure 6B). When we compared Plk1 mRNA expression 48 hours after incubating with P12-loaded trastuzumab-modified nanoparticles with Plk1 mRNA expression in untreated control cells, the reduction was statistically significant (P < .01; Figure 6C).
To analyze whether this reduction in Plk1 mRNA was accompanied by a reduction of Plk1 protein, we performed Western blot analyses in BT-474 cells 24, 48, and 72 hours after incubating with the various nanoparticle formulations. Therefore, we were able to additionally investigate the time dependency of the Plk1 protein reduction. We did not observe a reduction of Plk1 protein after 24 or 48 hours, respectively. Polo-like kinase 1 levels were comparable for all nanoparticle formulations (Figure 6D, 24 hours; Figure 6E, 48 hours; P > .05 for all formulations compared to untreated control cells). After 72 hours, we observed a significant reduction of Plk1 protein expression after incubating with P12-loaded trastuzumab-modified nanoparticles to levels of 38% compared to control cells (P < .05; Figure 6F). Polo-like kinase 1 protein levels were not reduced significantly after incubating with PEGylated P12-loaded nanoparticles, with HSV-loaded, and with unloaded nanoparticles compared to controls (P > .05 for all treatments compared to controls).
Treatment of BT-474 Cells with Plk1-Specific ASO-Loaded Nanoparticles Does Not Influence Plk2 and Plk4 Protein Expression
To analyze whether incubation of BT-474 cells with P12-loaded trastuzumab-modified nanoparticles does not only influence Plk1 expression but also the expression of other members of the Plk family, we did Western blot analyses targeting Plk2 and Plk4, respectively, 72 hours after incubating with the nanoparticle formulations. We did not observe significant changes in Plk2 and Plk4 protein expression standardized to p38 expression after incubating BT-474 cells with P12-loaded trastuzumab-modified nanoparticles compared to untreated control cells and compared to the other nanoparticle formulations (Figure 7, A and B; P > .05 for all nanoparticle formulations compared to untreated control cells).
Figure 7.
Plk2 and Plk4 expression and activation of Caspases in BT-474 cells 72 hours after incubating with nanoparticles. (A) A Western blot analysis was performed using anti-Plk2 antibodies. To control for variability of loading, membranes were reprobed with antibodies against p38. Representative Western blots show Plk2 (top) and p38 protein (bottom) for standardization. Percentage of Plk2 protein expression is given as percentage of p38-standardized Plk2 levels in control cells (n = 3, mean ± SD). (B) A Western blot analysis was performed using anti-Plk4 antibodies. To control for variability of loading, membranes were reprobed with antibodies against p38. Representative Western blots show Plk4 (top) and p38 protein (bottom) for standardization. Percentage of Plk4 protein expression is given as percentage of p38-standardized Plk4 levels in control cells (n = 3, mean ± SD). (C) Graphical summary of Caspase 3/7 activation in BT-474 cells 72 hours after incubating with nanoparticles. Percentage of luminescence is given as percentage of luminescence levels in control cells (n = 3, mean ± SD).
Induction of Apoptosis
Because Plk1 inhibition has been shown to be associated with apoptosis induction in earlier studies [9,12,29,34], we performed apoptosis assays to evaluate the potential activation of Caspase 3/7 in BT-474 cells after incubating with P12-loaded trastuzumab-modified nanoparticles compared to control cells, to P12-loaded PEGylated nanoparticles, and to HSV-loaded nanoparticles. Caspase 3/7 was induced to levels of 165% (P < .01) using 10 µg of total protein after treating with trastuzumab-modified nanoparticles and to 136% (P < .05) with PEGylated nanoparticles compared to untreated controls (Figure 7C). The control ASO targeting HSV did not significantly induce Caspase 3/7 compared to control cells (trastuzumab-modified: 115%, P = .21; PEGylated: 115%, P = .22). Using 20 µg of total protein, Caspase 3/7 was induced to levels of 151% (P < .01) with trastuzumab-modified P12-loaded nanoparticles and to levels of 128% (P < .05) with PEGylated P12-loaded nanoparticles compared to controls (data not shown).
Discussion
In the present study, we developed trastuzumab-modified nanoparticles for an HER2-specific targeting of breast cancer cells with Plk1-specific ASOs. Such biodegradable nanoparticles are well-established systems for the delivery of drugs to cancer cells [35]. One option of drug delivery is the passive targeting due to the enhanced permeability and retention effect appearing in solid tumors, because once injected into the blood stream, nanoparticles accumulate in the tumors [36,37]. Nevertheless, active targeting is a highly promising strategy, because cancer cells often overexpress receptors for various ligands compared to benign tissues [38]. These receptors provide a promising option for the active and specific targeting of colloidal drug carriers, such as liposomes or nanoparticles. Antibodies or other ligands can be used to modify the surface of the nanoparticles to achieve better delivery efficiency and tumor-specific targeting [39,40]. Typically, the antibody has to be modified to achieve reactive groups for the linkage to the nanoparticle surface. Our previous study showed the successful receptor-mediated uptake of trastuzumab-modified HSA nanoparticles into different HER2-positive breast cancer cells in culture [31]. The HER2 receptor is overexpressed in approximately 20% to 30% of all breast cancer cases, and there are more than 200,000 newly diagnosed breast cancer cases within 1 year in the US and 55,000 in Germany [41], demonstrating the great prominence for advancement of treatment, especially for HER2-positive patients.
ASOs are one potential therapeutic option for the treatment of various human cancers, and several phosphorothioate ASOs are currently being evaluated in patients [16], suggesting ASOs as valuable agents for therapeutic approaches. Despite the promising antitumor activity in vitro and in vivo, many ASOs have shown side effects and toxicities in their relevant doses [16,17]. In addition, antisense therapy is hindered because of poor stability in physiological fluids and inadequate penetration into the target tumor tissue [37]. These facts emphasize the necessity to develop smart targeted delivery systems for ASOs [18–21]. In the present study, we developed trastuzumabmodified and PEGylated HSA nanoparticles loaded with ASOs targeting Plk1 and a control ASO. Polo-like kinase 1 is a promising target for cancer therapy because of its overexpression in diverse cancer types and because of its correlation to poor prognosis in cancer patients [5]. Polo-like kinase 1 overexpression in normal cells leads to transformation of cells [6], and conversely, inhibition of Plk1 induces cell cycle arrest, apoptosis, and mitotic catastrophe, indicating a reliable approach to inhibit cancer cell proliferation and tumor growth. Because of these facts, Plk1 is an attractive cancer target for many pharmaceutical companies [42–46]. Hence, it is very important to find delivery strategies for nucleic acid-based therapeutics, for example, targeting Plk1 to minimize unwanted side effects [47].
In our experiments, we achieved a specific uptake of HSA nanoparticles in HER2-positive breast cancer cells, such as SK-BR-3 and BT-474, whereas in HER2-negative MCF-7, NIH-3T3, and C2C12 cells, no significant difference between the uptake of trastuzumab-modified and PEGylated nanoparticles was observed. The cell-specific uptake was time-dependent in the breast cancer cells with longer incubation times, leading to increased HER2-independent interactions between nanoparticles and cells and to uptake mediated by recycled HER2 receptors. The specific ASO-loaded nanoparticle uptake was blocked by the preincubation with the free antibody trastuzumab as shown before for the unloaded nanoparticles [31]. Here, after 30 and 60 minutes of incubation, uptake was comparable to PEGylated nanoparticles, and the increase observed after 3 to 24 hours could be due to recycled HER2 receptors or could be mediated by HER2-independent uptake. Next, we analyzed whether this receptor-mediated uptake is followed by a release of the ASO into the cells. We observed strong Cy5 fluorescence after 60 minutes of incubating with trastuzumab-modified nanoparticles and almost no fluorescence for PEGylated nanoparticles. In addition, an HER2-independent increase in Cy5 fluorescence was observed over a period of 1 to 24 hours parallel to the HER2-independent uptake of the nanoparticles. These data indicate that the ASOs can be released within the cell.
In addition, we analyzed whether the HSA nanoparticles show uptake in HER2-negative murine cells. In NIH-3T3 and C2C12 cells, no enrichment of the nanoparticles, even no HER2-independent, time-dependent uptake was detectable. We observed an uptake of only approximately 20% in NIH-3T3 and 40% in C2C12 cells after 24 hours of incubation. Therefore, we can conclude that this system is suitable for in vivo studies in nude mice.
However, the most interesting aspect is the question whether the ASOs are intact and are able to exert biologic activity against their target gene after being released into the cytoplasm of the cancer cells. To address this question, we performed RT-PCR, quantitative realtime PCR, and Western blot analyses against Plk1 after incubating the cells with the different nanoparticle formulations. We made the observation that Plk1 mRNA and protein were statistically significantly reduced after treatment of HER2-positive BT-474 cells with trastuzumab-modified P12-loaded HSA nanoparticles compared to control cells. This reduction of Plk1 expression was time-dependent over a period of 24 to 48 hours for Plk1 mRNA and over a period of 24 to 72 hours for Plk1 protein. According to earlier studies [30], we achieved a concentration of ASOs in the cells, which corresponds to approximately 100 nM for a normal transfection, which is in concordance with our studies using pure phosphorothioate ASOs targeting Plk1 [10,29]. The extent of Plk1 mRNA reduction (to levels of 77% in RT-PCR analyses and to 52% in quantitative real-time PCR analyses compared to controls) and protein reduction (to levels of 38% compared to controls) was comparable with our latest observations after transfection of ASOs using the oligofectamine protocol [29], so we can assume that the ASOs are released in an intact form that can interfere with Plk1 mRNA. We did not see a reduction of Plk1 protein earlier than 72 hours after incubating with the nanoparticles, confirming the data about the release of ASOs from nanoparticles 24 hours after incubation [30] and in accordance to the half-life time of the Plk1 protein of approximately 9 hours. Thus, a significant reduction of Plk1 protein can be seen, for example, 48 hours after transfection with pure ASOs or siRNA targeting Plk1 [9,10], but not earlier than 72 hours after incubating with nanoparticles.
To substantiate the specificity of the Plk1 reduction, we analyzed the effect of nanoparticles loaded with Plk1-specific ASO P12 and with HSV ASO on the expression of two other family members of the Plk family, Plk2 and Plk4. Seventy-two hours after incubating with the different nanoparticles, the Plk2 and Plk4 protein expressions did not change significantly comparing P12-loaded trastuzumab-modified nanoparticles with HSV and untreated control cells. These observations confirm the specificity of the Plk1 reduction by Plk1-specific ASOs.
As a consequence of the reduction of Plk1 expression 72 hours after incubating with the P12-loaded trastuzumab-modified nanoparticles, we observed an activation of Caspase 3/7. This observation of elevated apoptosis is in concordance with our earlier studies about Plk1 inhibition using ASOs or siRNA [9,10,29,34]. It can be concluded that the P12-loaded trastuzumab-modified nanoparticles moderately activated the apoptotic pathway, resulting in an activation of the Caspases 3/7.
Taken together, the ASO-loaded nanoparticles have several advantages: simple and fast preparation, storage stability, efficient specific trastuzumab-mediated uptake in HER2-positive breast cancer cells, and minimal HER2-independent uptake in HER2-negative cells. Compared to the efforts from various other groups for the development of specific targeting strategies to deliver ASOs or other nucleic acid-based agents into tumor cells [37,48–50], we developed a very simple, safe, and cheap nanoparticle system based on HSA, which has already achieved FDA approval as paclitaxel formulation [24,25]. In conclusion, we could show for the first time a receptor-mediated targeting of tumor cells followed by a significant reduction of Plk1 expression in vitro and apoptosis after treatment of breast cancer cells with ASO-loaded trastuzumab-modified HSA nanoparticles. This provides the basis for an efficient targeted delivery of drugs into tumor cells and the prevention of side effects induced by unwanted uptake of drugs into benign tissues.
Acknowledgments
The authors thank Monika Raab for her helpful discussions concerning the Caspase assays.
Abbreviations
- ASO
antisense oligonucleotides
- HSA
human serum albumin
- Plk1
polo-like kinase 1
Footnotes
This work was supported by the Wilhelm-Sander-Stiftung (2001.007.2), the Held and Hecker-Stiftung, the Deutsche Forschungsgemeinschaft (SP 1092/1-1), the Messer-Stiftung, the Schleussner-Stiftung, and the Else Kröner-Fresenius-Stiftung.
References
- 1.Brannon-Peppas L, Blanchette JO. Nanoparticle and targeted systems for cancer therapy. Adv Drug Deliv Rev. 2004;56:1649–1659. doi: 10.1016/j.addr.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 2.Schrama D, Reisfeld RA, Becker JC. Antibody targeted drugs as cancer therapeutics. Nat Rev Drug Discov. 2006;5:147–159. doi: 10.1038/nrd1957. [DOI] [PubMed] [Google Scholar]
- 3.Kirpotin DB, Drummond DC, Shao Y, Shalaby MR, Hong K, Nielsen UB, Marks JD, Benz CC, Park JW. Antibody targeting of long-circulating lipidic nanoparticles does not increase tumor localization but does increase internalization in animal models. Cancer Res. 2006;66:6732–6740. doi: 10.1158/0008-5472.CAN-05-4199. [DOI] [PubMed] [Google Scholar]
- 4.Barr FA, Sillje HH, Nigg EA. Polo-like kinases and the orchestration of cell division. Nat Rev Mol Cell Biol. 2004;5:429–440. doi: 10.1038/nrm1401. [DOI] [PubMed] [Google Scholar]
- 5.Strebhardt K, Ullrich A. Targeting polo-like kinase 1 for cancer therapy. Nat Rev Cancer. 2006;6:321–330. doi: 10.1038/nrc1841. [DOI] [PubMed] [Google Scholar]
- 6.Smith MR, Wilson ML, Hamanaka R, Chase D, Kung H, Longo DL, Ferris DK. Malignant transformation of mammalian cells initiated by constitutive expression of the polo-like kinase. Biochem Biophys Res Commun. 1997;234:397–405. doi: 10.1006/bbrc.1997.6633. [DOI] [PubMed] [Google Scholar]
- 7.Ando K, Ozaki T, Yamamoto H, Furuya K, Hosoda M, Hayashi S, Fukuzawa M, Nakagawara A. Polo-like kinase 1 (Plk1) inhibits p53 function by physical interaction and phosphorylation. J Biol Chem. 2004;279:25549–25561. doi: 10.1074/jbc.M314182200. [DOI] [PubMed] [Google Scholar]
- 8.Smits VA, Klompmaker R, Arnaud L, Rijksen G, Nigg EA, Medema RH. Polo-like kinase-1 is a target of the DNA damage checkpoint. Nat Cell Biol. 2000;2:672–676. doi: 10.1038/35023629. [DOI] [PubMed] [Google Scholar]
- 9.Spankuch-Schmitt B, Bereiter-Hahn J, Kaufmann M, Strebhardt K. Effect of RNA silencing of polo-like kinase-1 (PLK1) on apoptosis and spindle formation in human cancer cells. J Natl Cancer Inst. 2002;94:1863–1877. doi: 10.1093/jnci/94.24.1863. [DOI] [PubMed] [Google Scholar]
- 10.Spankuch-Schmitt B, Wolf G, Solbach C, Loibl S, Knecht R, Stegmuller M, von Minckwitz G, Kaufmann M, Strebhardt K. Downregulation of human polo-like kinase activity by antisense oligonucleotides induces growth inhibition in cancer cells. Oncogene. 2002;21:3162–3171. doi: 10.1038/sj.onc.1205412. [DOI] [PubMed] [Google Scholar]
- 11.Spankuch B, Matthess Y, Knecht R, Zimmer B, Kaufmann M, Strebhardt K. Cancer inhibition in nude mice after systemic application of U6 promoter-driven short hairpin RNAs against PLK1. J Natl Cancer Inst. 2004;96:862–872. doi: 10.1093/jnci/djh146. [DOI] [PubMed] [Google Scholar]
- 12.Liu X, Erikson RL. Polo-like kinase (Plk) 1 depletion induces apoptosis in cancer cells. Proc Natl Acad Sci USA. 2003;100:5789–5794. doi: 10.1073/pnas.1031523100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lane HA, Nigg EA. Antibody microinjection reveals an essential role for human polo-like kinase 1 (Plk1) in the functional maturation of mitotic centrosomes. J Cell Biol. 1996;135:1701–1713. doi: 10.1083/jcb.135.6.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cogswell JP, Brown CE, Bisi JE, Neill SD. Dominant-negative polo-like kinase 1 induces mitotic catastrophe independent of cdc25C function. Cell Growth Differ. 2000;11:615–623. [PubMed] [Google Scholar]
- 15.Liu X, Lei M, Erikson RL. Normal cells, but not cancer cells, survive severe Plk1 depletion. Mol Cell Biol. 2006;26:2093–2108. doi: 10.1128/MCB.26.6.2093-2108.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gleave ME, Monia BP. Antisense therapy for cancer. Nat Rev Cancer. 2005;5:468–479. doi: 10.1038/nrc1631. [DOI] [PubMed] [Google Scholar]
- 17.Jason TL, Koropatnick J, Berg RW. Toxicology of antisense therapeutics. Toxicol Appl Pharmacol. 2004;201:66–83. doi: 10.1016/j.taap.2004.04.017. [DOI] [PubMed] [Google Scholar]
- 18.Zhang C, Pei J, Kumar D, Sakabe I, Boudreau HE, Gokhale PC, Kasid UN. Antisense oligonucleotides: target validation and development of systemically delivered therapeutic nanoparticles. Methods Mol Biol. 2007;361:163–185. doi: 10.1385/1-59745-208-4:163. [DOI] [PubMed] [Google Scholar]
- 19.Li D, Yu H, Huang H, Shen F, Wu X, Li J, Wang J, Cao X, Wang Q, Tang G. FGF receptor-mediated gene delivery using ligands coupled to polyethylenimine. J Biomater Appl. 2007;22:163–180. doi: 10.1177/0885328206074503. [DOI] [PubMed] [Google Scholar]
- 20.Cirstoiu-Hapca A, Bossy-Nobs L, Buchegger F, Gurny R, Delie F. Differential tumor cell targeting of anti-HER2 (Herceptin®) and anti-CD20 (Mabthera®) coupled nanoparticles. Int J Pharm. 2007;331:190–196. doi: 10.1016/j.ijpharm.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 21.Hayes ME, Drummond DC, Hong K, Zheng WW, Khorosheva VA, Cohen JA, Noble CO, VI, Park JW, Marks JD, Benz CC, et al. Increased target specificity of anti-HER2 genospheres by modification of surface charge and degree of PEGylation. Mol Pharm. 2006;3:726–736. doi: 10.1021/mp060040v. [DOI] [PubMed] [Google Scholar]
- 22.Nahta R, Esteva FJ. HER-2-targeted therapy: lessons learned and future directions. Clin Cancer Res. 2003;9:5078–5084. [PubMed] [Google Scholar]
- 23.Wartlick H, Michaelis K, Balthasar S, Strebhardt K, Kreuter J, Langer K. Highly specific HER2-mediated cellular uptake of antibody-modified nanoparticles in tumour cells. J Drug Target. 2004;12:461–471. doi: 10.1080/10611860400010697. [DOI] [PubMed] [Google Scholar]
- 24.Stinchcombe TE, Socinski MA, Walko CM, O'neil BH, Collichio FA, Ivanova A, Mu H, Hawkins MJ, Goldberg RM, Lindley C, et al. Phase I and pharmacokinetic trial of carboplatin and albumin-bound paclitaxel, ABI-007 (Abraxane®) on three treatment schedules in patients with solid tumors. Cancer Chemother Pharmacol. 2007;60:759–766. doi: 10.1007/s00280-007-0423-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sparreboom A, Scripture CD, Trieu V, Williams PJ, De T, Yang A, Beals B, Figg WD, Hawkins M, Desai N. Comparative preclinical and clinical pharmacokinetics of a cremophor-free, nanoparticle albumin-bound paclitaxel (ABI-007) and paclitaxel formulated in cremophor (Taxol) Clin Cancer Res. 2005;11:4136–4143. doi: 10.1158/1078-0432.CCR-04-2291. [DOI] [PubMed] [Google Scholar]
- 26.Langer K. Peptide nanoparticles. In: Kumar C, editor. Biological and Pharmaceutical Nanomaterials Weinheim. Germany: Wiley-VCH; 2006. pp. 145–184. [Google Scholar]
- 27.Moghimi SM, Hunter AC, Murray JC. Nanomedicine: current status and future prospects. FASEB J. 2005;19:311–330. doi: 10.1096/fj.04-2747rev. [DOI] [PubMed] [Google Scholar]
- 28.Arnedo A, Espuelas S, Irache JM. Albumin nanoparticles as carriers for a phosphodiester oligonucleotide. Int J Pharm. 2002;244:59–72. doi: 10.1016/s0378-5173(02)00300-9. [DOI] [PubMed] [Google Scholar]
- 29.Spankuch B, Heim S, Kurunci-Csacsko E, Lindenau C, Yuan J, Kaufmann M, Strebhardt K. Down-regulation of polo-like kinase 1 elevates drug sensitivity of breast cancer cells in vitro and in vivo. Cancer Res. 2006;66:5836–5846. doi: 10.1158/0008-5472.CAN-06-0343. [DOI] [PubMed] [Google Scholar]
- 30.Wartlick H, Spankuch-Schmitt B, Strebhardt K, Kreuter J, Langer K. Tumour cell delivery of antisense oligonucleotides by human serum albumin nanoparticles. J Control Release. 2004;96:483–495. doi: 10.1016/j.jconrel.2004.01.029. [DOI] [PubMed] [Google Scholar]
- 31.Steinhauser I, Spankuch B, Strebhardt K, Langer K. Trastuzumab-modified nanoparticles: optimisation of preparation and uptake in cancer cells. Biomaterials. 2006;27:4975–4983. doi: 10.1016/j.biomaterials.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 32.Chen X, Yeung TK, Wang Z. Enhanced drug resistance in cells coexpressing ErbB2 with EGF receptor or ErbB3. Biochem Biophys Res Commun. 2000;277:757–763. doi: 10.1006/bbrc.2000.3731. [DOI] [PubMed] [Google Scholar]
- 33.Lin W, Garnett MC, Schacht E, Davis SS, Illum L. Preparation and in vitro characterization of HSA-mPEG nanoparticles. Int J Pharm. 1999;189:161–170. doi: 10.1016/s0378-5173(99)00253-7. [DOI] [PubMed] [Google Scholar]
- 34.Spankuch B, Kurunci-Csacsko E, Kaufmann M, Strebhardt K. Rational combinations of siRNAs targeting Plk1 with breast cancer drugs. Oncogene. 2007;26:5793–5807. doi: 10.1038/sj.onc.1210355. [DOI] [PubMed] [Google Scholar]
- 35.Vasir JK, Labhasetwar V. Targeted drug delivery in cancer therapy. Technol Cancer Res Treat. 2005;4:363–374. doi: 10.1177/153303460500400405. [DOI] [PubMed] [Google Scholar]
- 36.Maeda H, Wu J, Sawa T, Matsumura Y, Hori K. Tumor vascular permeability and the EPR effect in macromolecular therapeutics: a review. J Control Release. 2000;65:271–284. doi: 10.1016/s0168-3659(99)00248-5. [DOI] [PubMed] [Google Scholar]
- 37.Li SD, Huang L. Targeted delivery of antisense oligodeoxynucleotide and small interference RNA into lung cancer cells. Mol Pharm. 2006;3:579–588. doi: 10.1021/mp060039w. [DOI] [PubMed] [Google Scholar]
- 38.Allen TM. Ligand-targeted therapeutics in anticancer therapy. Nat Rev Cancer. 2002;2:750–763. doi: 10.1038/nrc903. [DOI] [PubMed] [Google Scholar]
- 39.Schiffelers RM, Ansari A, Xu J, Zhou Q, Tang Q, Storm G, Molema G, Lu PY, Scaria PV, Woodle MC. Cancer siRNA therapy by tumor selective delivery with ligand-targeted sterically stabilized nanoparticle. Nucleic Acids Res. 2004;32:e149. doi: 10.1093/nar/gnh140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang L, Hou S, Mao S, Wei D, Song X, Lu Y. Uptake of folate-conjugated albumin nanoparticles to the SKOV3 cells. Int J Pharm. 2004;287:155–162. doi: 10.1016/j.ijpharm.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 41.Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, Thun MJ. Cancer statistics, 2006. CA Cancer J Clin. 2006;56:106–130. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 42.Lenart P, Petronczki M, Steegmaier M, Di FB, Lipp JJ, Hoffmann M, Rettig WJ, Kraut N, Peters JM. The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr Biol. 2007;17:304–315. doi: 10.1016/j.cub.2006.12.046. [DOI] [PubMed] [Google Scholar]
- 43.Steegmaier M, Hoffmann M, Baum A, Lenart P, Petronczki M, Krssak M, Gurtler U, Garin-Chesa P, Lieb S, Quant J, et al. BI 2536, a potent and selective inhibitor of polo-like kinase 1, inhibits tumor growth in vivo. Curr Biol. 2007;17:316–322. doi: 10.1016/j.cub.2006.12.037. [DOI] [PubMed] [Google Scholar]
- 44.Lansing TJ, McConnell RT, Duckett DR, Spehar GM, Knick VB, Hassler DF, Noro N, Furuta M, Emmitte KA, Gilmer TM, et al. In vitro biological activity of a novel small-molecule inhibitor of polo-like kinase 1. Mol Cancer Ther. 2007;6:450–459. doi: 10.1158/1535-7163.MCT-06-0543. [DOI] [PubMed] [Google Scholar]
- 45.Schmidt M, Hofmann HP, Sanders K, Sczakiel G, Beckers TL, Gekeler V. Molecular alterations after polo-like kinase 1 mRNA suppression versus pharmacologic inhibition in cancer cells. Mol Cancer Ther. 2006;5:809–817. doi: 10.1158/1535-7163.MCT-05-0455. [DOI] [PubMed] [Google Scholar]
- 46.McInnes C, Mazumdar A, Mezna M, Meades C, Midgley C, Scaerou F, Carpenter L, Mackenzie M, Taylor P, Walkinshaw M, et al. Inhibitors of polo-like kinase reveal roles in spindle-pole maintenance. Nat Chem Biol. 2006;2:608–617. doi: 10.1038/nchembio825. [DOI] [PubMed] [Google Scholar]
- 47.McInnes C, Mezna M, Fischer PM. Progress in the discovery of polo-like kinase inhibitors. Curr Top Med Chem. 2005;5:181–197. doi: 10.2174/1568026053507660. [DOI] [PubMed] [Google Scholar]
- 48.Chiu SJ, Marcucci G, Lee RJ. Efficient delivery of an antisense oligodeoxyribonucleotide formulated in folate receptor-targeted liposomes. Anti-cancer Res. 2006;26:1049–1056. [PubMed] [Google Scholar]
- 49.Hogrefe RI, Lebedev AV, Zon G, Pirollo KF, Rait A, Zhou Q, Yu W, Chang EH. Chemically modified short interfering hybrids (siHYBRIDS): nanoimmunoliposome delivery in vitro and in vivo for RNAi of HER-2. Nucleosides Nucleotides Nucleic Acids. 2006;25:889–907. doi: 10.1080/15257770600793885. [DOI] [PubMed] [Google Scholar]
- 50.Jeong JH, Kim SH, Kim SW, Park TG. In vivo tumor targeting of ODN-PEG-folic acid/PEI polyelectrolyte complex micelles. J Biomater Sci Polym Ed. 2005;16:1409–1419. doi: 10.1163/156856205774472335. [DOI] [PubMed] [Google Scholar]