Abstract
Upper urinary tract transitional cell carcinoma (UUT-TCC) is quite an uncommon disease, and its prognosis differs among individuals irrespective of tumor stage. DNA repair gene polymorphisms are reported to result in the modulation of the repair capacity and might influence the prognosis of UUT-TCC. We examined the associations between functional polymorphisms in five DNA repair genes, and the prognosis of UUT-TCC in 103 UUT-TCC patients. Variant alleles in xeroderma pigmentosum complementation group C, more than three total variant alleles in all DNA repair genes studied and more than two total variant alleles in three nucleotide excision repair genes were independently associated with improved overall and disease-specific survival of UUT-TCC patients in multivariate analysis (P = .0063 and P = .0005 for xeroderma pigmentosum complementation group C, P = .016 and P = .0016 for all genes, and P = .0053 and P = .018 for nucleotide excision repair genes, respectively). These results suggest that some DNA repair gene polymorphisms may preoperatively be valuable as prognostic factors for UUT-TCC beyond tumor stage and grade, helping to provide optimal treatment strategies for individual patients.
Introduction
Upper urinary tract transitional cell carcinoma (UUT-TCC), which arises from the renal pelvis or ureter, is quite an uncommon disease, accounting for only 5% of all cases of urothelial carcinoma [1]. Although the standard treatment for localized UUT-TCC is surgical resection, local recurrence or distant metastasis sometimes occurs during the follow-up after surgery, and the survival rate at 5 years is less than 50% for patients with T2–T3 tumors [1]. Therefore, some patients with localized UUT-TCC need combined-modality therapy, chemotherapy, and/or radiotherapy and nephroureterectomy [2,3]. Meanwhile, systemic chemotherapy is offered as a viable therapeutic option for patients with metastatic disease at the time of initial diagnosis. The survival rate at 5 years is normally less than 10% for patients with T4 or N+/M+ tumors [1]; however, some of these patients have a response to therapy and a relatively long-term survival [4]. It is therefore acknowledged that the prognosis of UUT-TCC differs among individuals irrespective of tumor stage. Thus, although tumor stage and grade have been considered the main prognostic factors for UUT-TCC [1,5], more accurate prognostic markers may help to provide optimal treatment strategies for individual patients.
The complex system of DNA repair enzymes plays a vital role in protecting the genome from the consequences of exogenous and endogenous mutagenic exposure [6]. There are at least four known pathways of DNA repair, namely, base excision repair, nucleotide excision repair (NER), double-strand break (including homologous recombination and nonhomologous end-joining) repair, and mismatch repair, each of which operates on a specific type of damaged DNA and each of which involves numerous molecules [7]. Any decreased repair capacity of these enzymes causes alterations to the genome and subsequent cancer development [6]. There are several common polymorphisms in genes encoding DNA repair enzymes, and some of these polymorphisms are reported to result in subtle structural alterations in the repair enzyme and modulation of its repair capacity. The wild-type and variant genotypes of xeroderma pigmentosum complementation groups C (XPC), D (XPD), and G (XPG) and X-ray repair cross-complementing groups 1 (XRCC1) and 3 (XRCC3) have been shown to be associated with different levels of DNA repair activity using various assays [8–13]. In this manner, these polymorphisms may contribute to interindividual cancer susceptibility in the general population. The associations between DNA repair gene polymorphisms and bladder cancer risk or prognosis have been demonstrated in several reports [7,14–20]. However, a study on the association of these polymorphisms with UUT-TCC is lacking.
Risk factors for UUT-TCC include cigarette smoking and exposure to certain industrial dyes or solvents [21], and some of the DNA damage that may occur as a result of these factors would be repaired by the DNA repair enzymes. The accumulation of multiple genetic changes is considered to cause the development and progression of UUT-TCC, such as bladder TCC [22–24]. We previously reported associations between DNA repair gene polymorphisms and tumor stage, grade, p53 alteration, or prognosis in patients with bladder cancer [17,18,25]. These polymorphisms, as host genetic factors, may also influence the progression and prognosis of UUT-TCC due to the accumulation of multiple genetic changes caused by the modulated DNA repair capacity. In the current study, we investigated the associations between polymorphisms in the DNA repair genes, XPC (Lys939Gln, A/C), XPD (Lys751Gln, A/C), and XPG (Asp1104His, G/C) (involved in NER), XRCC1 (Arg399Gln, G/A) (involved in base excision repair), and XRCC3 (Thr241Met, C/T) (involved in double-strand break repair), and clinicopathologic parameters and survival in patients with UUT-TCC to determine the value of these polymorphisms as prognostic markers. Germline genetic polymorphisms are thought to be suitable markers for UUT-TCC patients, because polymorphisms are available without operation. The polymorphisms studied in this report were generally selected according to prior data on functional effects or reports of association with malignancies, to increase the likelihood of positive findings [7,12,26], and we have consecutively investigated the effects of these polymorphisms on cancer biology [16–18,25,27]. In addition, we also examined the association of these polymorphisms with susceptibility to UUT-TCC in the present report, using the case-control study. To our knowledge, there have been few case-control studies on genetic polymorphisms, limited to UUT-TCC, to date.
Materials and Methods
Patients and Control Subjects
The study group comprised 103 patients (69 men and 34 women; median age, 70 years; age range, 44 to 93 years) with histopathologically confirmed UUT-TCC at Yamaguchi University Hospital, Ube, Japan, between August 1990 and June 2006. As control subjects, 215 healthy volunteers were chosen from the same geographical area (148 men and 67 women; median age, 67 years; age range, 29 to 87 years), as previously reported [27]. This study was approved by the institutional ethical committee at the Graduate School of Medicine, Yamaguchi University, and written informed consent was obtained from each patient and control. The characteristics of the UUT-TCC patients and control subjects, all of whom were native Japanese, are shown in Table 1. Regarding control subjects, only age and gender were recorded as personal data. The tumor staging system was based on the American Joint Commission on Cancer staging system [28], and the tumors were histopathologically graded according to the World Health Organization's classification. Of the 103 patients with UUT-TCC, 97 underwent an operation that was normally nephroureterectomy with removal of the bladder cuff. Platinum-based chemotherapy, radiotherapy, or both were performed in 24, 4, or 16 patients, respectively. Follow-up information was available for 100 of 103 patients with UUT-TCC. Of these, at a median follow-up of 36 months (mean, 44.5 months; range, 2 to 167 months), 39 and 24 suffered death from any cause and disease-specific death during the follow-up, respectively.
Table 1.
Clinicopathologic Characteristics of UUT-TCC Patients and Control Subjects.
| Cases, n (%) | Controls, n (%) | P | ||
| Age, years | Median (range) | 70 (44–93) | 67 (29–87) | |
| < 70 | 48 (46.6) | 122 (56.7) | ||
| ≥70 | 55 (53.4) | 93 (43.3) | .090* | |
| Gender | Male | 69 (67.0) | 148 (68.8) | |
| Female | 34 (33.0) | 67 (31.2) | .74* | |
| Smoking history | No | 41 (39.8) | ||
| Yes | 54 (52.4) | |||
| Unknown | 8 (7.8) | |||
| Tumor location | Renal pelvis | 40 (38.8) | ||
| Ureter | 54 (52.4) | |||
| Both | 9 (8.7) | |||
| Tumor stage | Tis | 9 (8.7) | ||
| Ta | 5 (4.9) | |||
| T1 | 27 (26.2) | |||
| T2 | 18 (17.5) | |||
| T3 | 34 (33.0) | |||
| T4 | 5 (4.9) | |||
| TX | 5 (4.9) | |||
| Lymph node metastasis | Negative | 83 (80.6) | ||
| Positive | 10 (9.7) | |||
| Unknown | 10 (9.7) | |||
| Distant metastasis | Negative | 89 (86.4) | ||
| Positive | 5 (4.9) | |||
| Unknown | 9 (8.7) | |||
| Tumor grade | G1 | 3 (2.9) | ||
| G2 | 48 (46.6) | |||
| G3 | 47 (45.6) | |||
| GX | 5 (4.9) | |||
| Histopathology | Pure TCC | 98 (95.1) | ||
| TCC with SCC | 5 (4.9) | |||
| Operation | Yes | 97 (94.2) | ||
| No | 6 (5.8) | |||
| Chemotherapy/radiotherapy | None | 55 (53.4) | ||
| Chemotherapy | 24 (23.3) | |||
| Radiotherapy | 4 (3.9) | |||
| Both | 16 (15.5) | |||
| Unknown | 4 (3.9) |
UUT-TCC indicates upper urinary tract transitional cell carcinoma; SCC, squamous cell carcinoma.
Difference between cases and controls (chi-square analysis).
DNA Extraction and Genotyping
Peripheral blood samples were collected from each patient at initial diagnosis and from each control subject, and lymphocyte DNA was extracted using the QIAamp DNA Mini Kit (VWR International, West Chester, PA). Polymorphisms in XPC, XPD, XPG, XRCC1, and XRCC3 genes were genotyped using polymerase chain reaction (PCR)-restriction fragment length polymorphism. This method, including the primer sequences, PCR conditions, and restriction endonucleases for each polymorphism, has been described previously [12,16,18]. Briefly, the DNA fragments were amplified from 10 ng of DNA in 10 µl of PCR reactions containing 1.5 mM MgCl2, 0.2 mM each deoxyribonucleotide triphosphate, 0.3 U of AmpliTaq Gold DNA polymerase (Applied Biosystems, Foster City, CA), and 0.3 µM each primer. The PCR products were digested with the appropriate restriction endonucleases that recognized and cut either wild-type or variant sequences at 37°C for at least 3 hours. The digested PCR products were electrophoresed on 2% agarose gels and were stained with ethidium bromide for visualization under ultraviolet light. To control the restriction digestion of the PCR products, genotyping assays were randomly repeated and results were checked for concordance. About 10% of the amplified fragments were also randomly checked by direct DNA sequencing and comparison of the PCR-restriction fragment length polymorphism and sequencing results showed 100% concordance in all experiments. Variant alleles were defined as Gln for XPC, Gln for XPD, His for XPG, Gln for XRCC1, and Met for XRCC3, according to previous reports [12,16].
Statistical Analysis
Differences in the genotype and allele frequencies of the DNA repair genes between the UUT-TCC cases and the control subjects, and associations between the genotypes and the clinicopathologic characteristics at the time of initial diagnosis of the UUT-TCC patients, were assessed for statistical significance using the chi-square analysis or two-sided Fisher's exact test; odds ratios (ORs) with 95% confidence intervals (CIs) were calculated. P values less than .005 were considered to indicate statistical significance, because multiple comparisons were conducted [29]. Overall and disease-specific survival of the UUT-TCC patients were calculated from the day of initial diagnosis until the last follow-up or death from any cause and from UUT-TCC, respectively; patients who were alive at the last follow-up were censored at that time. The associations between categorical variables, including DNA repair genotypes, and survival were estimated by calculating the hazard ratios (HRs) with 95% CIs using Cox proportional hazard regression models. In addition, overall and disease-specific survival was analyzed by plotting Kaplan-Meier curves, and the survival probability distributions were compared using the log-rank test. Because the prognosis of cancer patients likely involves multistep, multigenic pathways, it is unlikely that any one single genetic polymorphism would have a dramatic effect on clinical outcome, and it is important to undertake a pathway-based analysis of multiple polymorphic genes [19]. Thus, we also analyzed the numbers of total variant alleles in all DNA repair genes studied and in NER genes (XPC, XPD, and XPG). P values less than .05 were considered to indicate statistical significance in these tests and variables with P < .05 in univariate analysis were also assessed for their relationship with survival by multivariate analysis. Data were processed using JMP software (SAS Institute Inc., Cary, NC).
Results
Genotype and Allele Frequencies of the DNA Repair Genes in UUT-TCC Cases and Control Subjects
The genotype frequencies of the XPC, XPD, XPG, XRCC1, and XRCC3 genes, in both the 103 UUT-TCC cases and the 215 control subjects, were found to be in Hardy-Weinberg equilibrium. The genotype and allele frequencies in cases and controls are shown in Table 2. The Lys/Gln + Gln/Gln genotypes of the XPD gene were less frequent in cases than in controls (OR: 0.26, 95% CI: 0.061–0.77, P = .032; OR adjusted for age and gender: 0.29, 95% CI: 0.067–0.89, P = .053; two-sided Fisher's exact test). The Gln alleles of the XPD gene were also less frequent in cases than in controls (OR: 0.24, 95% CI: 0.072–0.80, P = .013; two-sided Fisher's exact test). However, these differences were not statistically significant when considering multiple comparisons (see Materials and Methods section). No significant differences in genotype or allele frequencies, between cases and controls, were observed for the other polymorphisms in the XPC, XPG, XRCC1, or XRCC3 genes.
Table 2.
Genotype and Allele Frequencies of DNA Repair Genes in UUT-TCC Cases and Controls.
| Gene (polymorphism) | Genotype or allele | Cases, n (%) | Controls, n (%) | Crude | Adjusted* | |||
| OR (95% CI) | p† | OR (95% CI) | p† | |||||
| XPC (Lys939Gln, A/C) | Lys/Lys | 30 (31.9) | 70 (32.6) | Ref. | Ref. | |||
| Lys/Gln | 46 (48.9) | 114 (53.0) | 0.94 (0.55–1.6) | .83 | 1.0 (0.57–1.8) | .97 | ||
| Gln/Gln | 18 (19.1) | 31 (14.4) | 1.4 (0.65–2.8) | .41 | 1.4 (0.66–2.9) | .38 | ||
| Lys/Gln + Gln/Gln | 64 (68.1) | 145 (67.4) | 1.0 (0.62–1.7) | .91 | 1.1 (0.64–1.9) | .75 | ||
| Lys/Lys + Lys/Gln | 76 (80.9) | 184 (85.6) | Ref. | Ref. | ||||
| Gln/Gln | 18 (19.1) | 31 (14.4) | 1.4 (0.73–2.6) | .30 | 1.4 (0.71–2.7) | .33 | ||
| Lys allele | 106 (56.4) | 254 (59.1) | Ref. | |||||
| Gln allele | 82 (43.6) | 176 (40.9) | 1.1 (0.79–1.6) | .53 | ||||
| XPD (Lys751Gln, A/C) | Lys/Lys | 96 (97.0) | 192 (89.3) | Ref. | Ref. | |||
| Lys/Gln | 3 (3.0) | 20 (9.3) | 0.30 (0.070–0.90) | .057 | 0.33 (0.074–1.0) | .082 | ||
| Gln/Gln | 0 (0.0) | 3 (1.4) | Not calculated | .83 | Not calculated | .84 | ||
| Lys/Gln + Gln/Gln | 3 (3.0) | 23 (10.7) | 0.26 (0.061–0.77) | .032 | 0.29 (0.067–0.89) | .053 | ||
| Lys/Lys + Lys/Gln | 99 (100.0) | 212 (98.6) | Ref. | Ref. | ||||
| Gln/Gln | 0 (0.0) | 3 (1.4) | Not calculated | .83 | Not calculated | .84 | ||
| Lys allele | 195 (98.5) | 404 (94.0) | Ref. | |||||
| Gln allele | 3 (1.5) | 26 (6.0) | 0.24 (0.072–0.80) | .013 | ||||
| XPG (Asp1104His, G/C) | Asp/Asp | 19 (19.6) | 51 (23.7) | Ref. | Ref. | |||
| Asp/His | 43 (44.3) | 99 (46.0) | 1.2 (0.62–2.2) | .64 | 1.1 (0.56–2.2) | .80 | ||
| His/His | 35 (36.1) | 65 (30.2) | 1.4 (0.75–2.9) | .28 | 1.2 (0.60–2.4) | .61 | ||
| Asp/His + His/His | 78 (80.4) | 164 (76.3) | 1.3 (0.72–2.4) | .42 | 1.1 (0.61–2.1) | .74 | ||
| Asp/Asp + Asp/His | 62 (63.9) | 150 (69.8) | Ref. | Ref. | ||||
| His/His | 35 (36.1) | 65 (30.2) | 1.3 (0.78–2.2) | .31 | 1.2 (0.74–2.1) | .41 | ||
| Asp allele | 81 (41.8) | 201 (46.7) | Ref. | |||||
| His allele | 113 (58.2) | 229 (53.3) | 1.2 (0.87–1.7) | .25 | ||||
| XRCC1 (Arg399Gln, G/A) | Arg/Arg | 52 (52.5) | 120 (55.8) | Ref. | Ref. | |||
| Arg/Gln | 39 (39.4) | 85 (39.5) | 1.1 (0.64–1.7) | .82 | 1.0 (0.60–1.7) | > 0.99 | ||
| Gln/Gln | 8 (8.1) | 10 (4.7) | 1.8 (0.67–4.9) | .22 | 2.0 (0.72–5.6) | .17 | ||
| Arg/Gln + Gln/Gln | 47 (47.5) | 95 (44.2) | 1.1 (0.71–1.8) | .59 | 1.1 (0.67–1.8) | .70 | ||
| Arg/Arg + Arg/Gln | 91 (91.9) | 205 (95.3) | Ref. | Ref. | ||||
| Gln/Gln | 8 (8.1) | 10 (4.7) | 1.8 (0.67–4.7) | .23 | 2.0 (0.73–5.4) | .17 | ||
| Arg allele | 143 (72.2) | 325 (75.6) | Ref. | |||||
| Gln allele | 55 (27.8) | 105 (24.4) | 1.2 (0.81–1.7) | .37 | ||||
| XRCC3 (Thr241Met, C/T) | Thr/Thr | 82 (84.5) | 173 (82.0) | Ref. | Ref. | |||
| Thr/Met | 15 (15.5) | 33 (15.6) | 0.96 (0.48–1.8) | .90 | 1.0 (0.50–2.0) | .98 | ||
| Met/Met | 0 (0.0) | 5 (2.4) | Not calculated | .85 | Not calculated | .85 | ||
| Thr/Met + Met/Met | 15 (15.5) | 38 (18.0) | 0.83 (0.42–1.6) | .58 | 0.89 (0.45–1.7) | .74 | ||
| Thr/Thr + Thr/Met | 97 (100.0) | 206 (97.6) | Ref. | Ref. | ||||
| Met/Met | 0 (0.0) | 5 (2.4) | Not calculated | .85 | Not calculated | .85 | ||
| Thr allele | 179 (92.3) | 379 (89.8) | Ref. | |||||
| Met allele | 15 (7.7) | 43 (10.2) | 0.74 (0.40–1.4) | .33 | ||||
UUT-TCC indicates upper urinary tract transitional cell carcinoma; OR, odds ratio; CI, confidence interval; Ref., Reference.
Adjusted for age and gender.
Chi-square analysis or two-sided Fisher's exact test.
Association Between DNA Repair Genotypes and Age, Gender, Tumor Stage, and Grade in UUT-TCC Patients
The associations between DNA repair genotypes and age, gender, tumor stage, and grade, in UUT-TCC patients, are presented in Table 3. His/His genotypes of the XPG gene were more frequent than Asp/Asp (OR: 4.5, 95% CI: 1.1–18, P = .038) or Asp/Asp + Asp/His (OR: 2.4, 95% CI: 1.0–5.8, P = .045) genotypes in female patients with UUT-TCC. In addition, Gln/Gln genotypes of the XPC gene were less frequent than Lys/Lys (OR: 0.23, 95% CI: 0.066–0.83, P = .032) or Lys/Lys + Lys/Gln (OR: 0.30, 95% CI: 0.10–0.91, P = .028) genotypes in patients with T2/T3/T4 tumors. However, these associations were not statistically significant. There were no significant associations between DNA repair genotypes and patient age or tumor grade.
Table 3.
DNA Repair Genotypes and Age, Gender, Tumor Stage, and Grade in UUT-TCC Patients.
| Gene (polymorphism) | Genotype | Age (years), n | OR (95% CI) | p* | Gender, n | OR (95% CI) | p* | Tumor stage, n | OR (95% CI) | p* | Tumor grade, n | OR (95% CI) | p* | ||||
| < 70 | ≥ 70 | Men | Women | Tis/Ta/T1 | T2/T3/T4 | G1/G2 | G3 | ||||||||||
| XPC | Lys/Lys | 11 | 19 | Ref. | 18 | 12 | Ref. | 9 | 21 | Ref. | 15 | 14 | Ref. | ||||
| (Lys939Gln, A/C) | Lys/Gln | 21 | 25 | 0.69 (0.27–1.8) | .44 | 34 | 12 | 0.53 (0.20–1.4) | .20 | 17 | 26 | 0.66 (0.24–1.8) | .40 | 22 | 22 | 1.1 (0.42–2.7) | .89 |
| Gln/Gln | 10 | 8 | 0.46 (0.14–1.5) | .20 | 11 | 7 | 0.96 (0.29–3.2) | .94 | 11 | 6 | 0.23 (0.066–0.83) | .032 | 11 | 6 | 0.58 (0.17–2.0) | .39 | |
| Lys/Gln + Gln/Gln | 31 | 33 | 0.62 (0.25–1.5) | .28 | 45 | 19 | 0.63 (0.26–1.6) | .32 | 28 | 32 | 0.49 (0.19–1.2) | .13 | 33 | 28 | 0.91 (0.38–2.2) | .83 | |
| Lys/Lys + Lys/Gln | 32 | 44 | Ref. | 52 | 24 | Ref. | 26 | 47 | Ref. | 37 | 36 | Ref. | |||||
| Gln/Gln | 10 | 8 | 0.58 (0.21–1.6) | .30 | 11 | 7 | 1.4 (0.48–4.0) | .55 | 11 | 6 | 0.30 (0.10–0.91) | .028 | 11 | 6 | 0.56 (0.19–1.7) | .30 | |
| XPD | Lys/Lys | 45 | 51 | Ref. | 65 | 31 | Ref. | 37 | 54 | Ref. | 48 | 43 | Ref. | ||||
| (Lys751Gln, A/C)† | Lys/Gln + Gln/Gln | 0 | 3 | Not calculated | .43 | 2 | 1 | 1.0 (0.092–12) | > .99 | 2 | 1 | 0.34 (0.030–3.9) | .65 | 1 | 2 | 2.2 (0.20–26) | .65 |
| XPG | Asp/Asp | 11 | 8 | Ref. | 16 | 3 | Ref. | 9 | 8 | Ref. | 8 | 9 | Ref. | ||||
| (Asp1104His, G/C) | Asp/His | 15 | 28 | 2.6 (0.85–7.8) | .090 | 30 | 13 | 2.3 (0.57–9.3) | .48 | 17 | 25 | 1.7 (0.53–5.1) | .40 | 22 | 19 | 0.77 (0.25–2.4) | .77 |
| His/His | 17 | 18 | 1.5 (0.47–4.5) | .51 | 19 | 16 | 4.5 (1.1–18) | .038 | 13 | 21 | 1.8 (0.56–5.9) | .38 | 19 | 16 | 0.75 (0.23–2.4) | .77 | |
| Asp/His + His/His | 32 | 46 | 2.0 (0.72–5.5) | .18 | 49 | 29 | 3.2 (0.85–12) | .15 | 30 | 46 | 1.7 (0.60–5.0) | .42 | 41 | 35 | 0.76 (0.26–2.2) | .79 | |
| Asp/Asp + Asp/His | 26 | 36 | Ref. | 46 | 16 | Ref. | 26 | 33 | Ref. | 30 | 28 | Ref. | |||||
| His/His | 17 | 18 | 0.77 (0.33–1.8) | .53 | 19 | 16 | 2.4 (1.0–5.8) | .045 | 13 | 21 | 1.3 (0.54–3.0) | .58 | 19 | 16 | 0.90 (0.39–2.1) | .81 | |
| XRCC1 | Arg/Arg | 23 | 29 | Ref. | 35 | 17 | Ref. | 24 | 27 | Ref. | 27 | 23 | Ref. | ||||
| (Arg399Gln, G/A)† | Arg/Gln + Gln/Gln | 22 | 25 | 0.90 (0.41–2.0) | .80 | 31 | 16 | 1.1 (0.46–2.5) | .89 | 14 | 29 | 1.8 (0.79–4.3) | .15 | 22 | 22 | 1.2 (0.52–2.6) | .70 |
| XRCC3 | Thr/Thr | 37 | 45 | Ref. | 52 | 30 | Ref. | 29 | 49 | Ref. | 38 | 40 | Ref. | ||||
| (Thr241Met, C/T)† | Thr/Met + Met/Met | 8 | 7 | 0.72 (0.24–2.2) | .59 | 12 | 3 | 0.43 (0.11–1.7) | .37 | 8 | 6 | 0.44 (0.14–1.4) | .33 | 9 | 5 | 0.53 (0.16–1.7) | .39 |
UUT-TCC indicates upper urinary tract transitional cell carcinoma; OR, odds ratio; CI, confidence interval; Ref., Reference.
Chi-square analysis or two-sided Fisher's exact test.
For those polymorphisms with few homozygous variant alleles, only the combined results of the heterozygous and homozygous variant alleles were shown.
Univariate Analysis of DNA Repair Genotypes and Overall Survival of UUT-TCC Patients
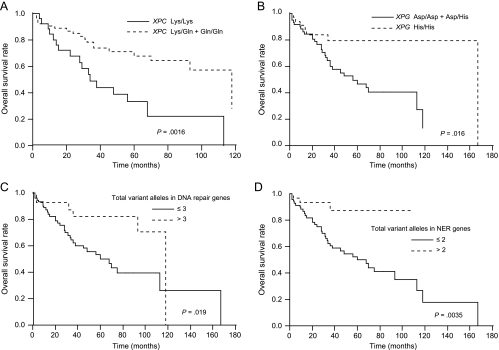
A univariate Cox regression analysis for predicting the overall survival of UUT-TCC patients is presented in Table 4. XPC and XPG genotypes, and the numbers of total variant alleles in all DNA repair genes studied and in NER genes, were associated with the probability of overall survival in UUT-TCC patients. Patients with Gln/Gln or Lys/Gln + Gln/Gln genotypes of the XPC gene had significantly improved overall survival compared with those with Lys/Lys genotypes (HR: 0.48, 95% CI: 0.25–0.80, P = .0041 for Gln/Gln; HR: 0.60, 95% CI: 0.43–0.84, P = .0032 for Lys/Gln + Gln/Gln). In addition, patients with more than two variant alleles in NER genes had significantly improved overall survival compared with those with two or less variant alleles (HR: 0.45, 95% CI: 0.22–0.76, P = .0011). Overall survival was plotted for the Lys/Gln + Gln/Gln genotypes of the XPC gene, His/His genotypes of the XPG gene, more than three variant alleles in all DNA repair genes and more than two variant alleles in NER genes, compared with the remaining groups, using Kaplan-Meier survival curves (Figure 1, A–D, respectively). These genotypes were associated with improved overall survival (P = .0016 for the XPC gene, P = .016 for the XPG gene, P = .019 for the total variant alleles in all DNA repair genes, and P = .0035 for the total variant alleles in NER genes; log-rank test). Clinicopathologic variables were also assessed for their relationship with the overall survival of UUT-TCC patients by univariate analysis (Table 4). Tumor stage (T2/T3/T4), distant metastasis and grade (G3) were associated with an unfavorable outcome.
Table 4.
Univariate and Multivariate Cox Regression Analyses for Predicting Overall Survival of UUT-TCC Patients.
| Variable (n) | Deaths, n | Univariate Analysis | Multivariate Model One | Multivariate Model Two | Multivariate Model Three | ||||
| HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | ||
| Age | |||||||||
| < 70 (46) | 17 | Ref. | |||||||
| ≥ 70 (54) | 22 | 1.3 (0.92–1.8) | .15 | ||||||
| Gender | |||||||||
| Men (68) | 28 | Ref. | |||||||
| Women (32) | 11 | 0.98 (0.68–1.4) | .93 | ||||||
| Tumor location | |||||||||
| Renal pelvis (40) | 15 | Ref. | |||||||
| Ureter + Both (60) | 24 | 0.95 (0.69–1.3) | .77 | ||||||
| Tumor stage | |||||||||
| Tis/Ta/T1 (39) | 12 | Ref. | Ref. | Ref. | Ref. | ||||
| T2/T3/T4 (56) | 25 | 1.6 (1.1–2.4) | .0061 | 1.4 (0.93–2.1) | .12 | 1.3 (0.90–1.9) | .17 | 1.4 (0.94–2.0) | .11 |
| Lymph node metastasis | |||||||||
| Negative (80) | 30 | Ref. | |||||||
| Positive (10) | 5 | 1.5 (0.87–2.3) | .13 | ||||||
| Distant metastasis | |||||||||
| Negative (86) | 32 | Ref. | Ref. | Ref. | Ref. | ||||
| Positive (5) | 4 | 2.3 (1.2–3.8) | .012 | 2.2 (1.1–4.0) | .022 | 2.1 (1.1–3.6) | .026 | 1.8 (0.95–3.1) | .067 |
| Tumor grade | |||||||||
| G1/G2 (48) | 13 | Ref. | Ref. | Ref. | Ref. | ||||
| G3 (47) | 23 | 1.5 (1.1–2.2) | .015 | 1.5 (1.0–2.2) | .048 | 1.3 (0.94–1.9) | .11 | 1.3 (0.88–1.9) | .21 |
| Histopathology | |||||||||
| Pure TCC (95) | 36 | Ref. | |||||||
| TCC with SCC (5) | 3 | 1.2 (0.59–2.0) | .55 | ||||||
| Operation | |||||||||
| Yes (94) | 37 | Ref. | |||||||
| No (6) | 2 | 1.2 (0.50–2.3) | .58 | ||||||
| Chemotherapy/Radiotherapy | |||||||||
| None (52) | 18 | Ref. | |||||||
| Chemotherapy (24) | 9 | 0.91 (0.59–1.3) | .64 | ||||||
| Radiotherapy (4) | 2 | 1.1 (0.45–2.2) | .73 | ||||||
| Both (16) | 7 | 1.2 (0.72–1.8) | .53 | ||||||
| XPC (Lys939Gln, A/C) | |||||||||
| Lys/Lys (27) | 17 | Ref. | Ref. | ||||||
| Lys/Gln (46) | 15 | 0.65 (0.45–0.93) | .019 | ||||||
| Gln/Gln (18) | 4 | 0.48 (0.25–0.80) | .0041 | ||||||
| Lys/Gln + Gln/Gln (64) | 19 | 0.60 (0.43–0.84) | .0032 | 0.59 (0.40–0.86) | .0063 | ||||
| Lys/Gln + Gln/Gln (64) | 19 | 0.60 (0.43–0.84) | .0032 | 0.59 (0.40–0.86) | .0063 | ||||
| Lys/Lys + Lys/Gln (73) | 32 | Ref. | |||||||
| Gln/Gln (18) | 4 | 0.61 (0.33–0.97) | .035 | ||||||
| XPD (Lys751Gln, A/C)* | |||||||||
| Lys/Lys (93) | 35 | Ref. | |||||||
| Lys/Gln + Gln/Gln (3) | 1 | 0.64 (0.15–1.5) | .36 | ||||||
| XPG (Asp1104His, G/C) | |||||||||
| Asp/Asp (19) | 7 | Ref. | |||||||
| Asp/His (41) | 23 | 1.3 (0.86–2.0) | .22 | ||||||
| His/His (34) | 7 | 0.70 (0.40–1.2) | .20 | ||||||
| Asp/His + His/His (75) | 30 | 1.0 (0.71–1.7) | .82 | ||||||
| Asp/Asp + Asp/His (60) | 30 | Ref. | Ref. | ||||||
| His/His (34) | 7 | 0.60 (0.37–0.89) | .010 | 0.79 (0.48–1.2) | .30 | ||||
| XRCC1 (Arg399Gln, G/A)* | |||||||||
| Arg/Arg (50) | 20 | Ref. | |||||||
| Arg/Gln + Gln/Gln (46) | 17 | 0.91 (0.65–1.3) | .59 | ||||||
| XRCC3 (Thr241Met, C/T)* | |||||||||
| Thr/Thr (79) | 31 | Ref. | |||||||
| Thr/Met + Met/Met (15) | 4 | 0.68 (0.37–1.1) | .12 | ||||||
| Total variant alleles in DNA repair genes | |||||||||
| ≤ 3 (73) | 33 | Ref. | Ref. | ||||||
| > 3 (27) | 6 | 0.60 (0.37–0.91) | .013 | 0.61 (0.37–0.92) | .016 | ||||
| Total variant alleles in NER genes† | |||||||||
| ≤ 2 (70) | 36 | Ref. | Ref. | ||||||
| > 2 (29) | 3 | 0.45 (0.22–0.76) | .0011 | 0.49 (0.24–0.83) | .0053 | ||||
Bold font indicates statistical significance.
UUT-TCC indicates upper urinary tract transitional cell carcinoma; HR, hazard ratio; CI, confidence interval; SCC, squamous cell carcinoma; NER, nucleotide excision repair; Ref., Reference.
For those polymorphisms with few homozygous variant alleles, only the combined results of the heterozygous and homozygous variant alleles were shown.
NER genes included XPC, XPD, and XPG.
Figure 1.
Kaplan-Meier overall survival curves for patients with UUT-TCC by DNA repair genotypes (log-rank tests). (A) Lys/Gln + Gln/Gln versus Lys/Lys of XPC; P = .0016. (B) His/His versus Asp/Asp + Asp/His of XPG; P = .016. (C) Total variant alleles in all DNA repair genes studied: > 3 versus ≤ 3; P = .019. (D) Total variant alleles in NER genes (XPC, XPD, and XPG): > 2 versus ≤ 2; P = .0035.
Multivariate Analysis of DNA Repair Genotypes and Overall Survival of UUT-TCC Patients
Using multivariate analysis (model one) including all factors with P < .05 in univariate analysis (tumor stage, distant metastasis, grade, and XPC and XPG genotypes), XPC genotypes (HR: 0.59, 95% CI: 0.40–0.86, P = .0063), as well as distant metastasis and grade, were independently associated with overall survival (Table 4). When the number of variant alleles in all DNA repair genes was used instead of XPC and XPG genotypes (model two), the number of variant alleles (HR: 0.61, 95% CI: 0.37–0.92, P = .016), as well as distant metastasis, was independently associated with overall survival. When the number of variant alleles in NER genes was used instead of XPC and XPG genotypes (model three), patients with more than two variant alleles were independently associated with a favorable clinical outcome (HR: 0.49, 95% CI: 0.24–0.83, P = .0053).
Univariate Analysis of DNA Repair Genotypes and Disease-Specific Survival of UUT-TCC Patients
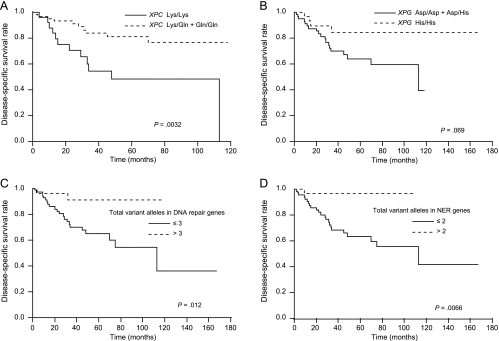
A univariate Cox regression analysis for predicting the disease-specific survival of UUT-TCC patients is presented in Table 5. XPC genotypes, and the numbers of total variant alleles in all DNA repair genes and in NER genes, were significantly associated with the probability of disease-specific survival in UUT-TCC patients. Patients with Gln/Gln genotypes of the XPC gene had significantly improved disease-specific survival compared with those with Lys/Lys genotypes (HR: 0.32, 95% CI: 0.076–0.74, P = .0039). In addition, patients with more than three variant alleles in all DNA repair genes had significantly improved disease-specific survival compared with those with three or less variant alleles (HR: 0.43, 95% CI: 0.17–0.80, P = .0048), and patients with more than two variant alleles in NER genes also had significantly improved disease-specific survival compared with those with two or less variant alleles (HR: 0.32, 95% CI: 0.076–0.70, P = .0015). Disease-specific survival was plotted for the Lys/Gln + Gln/Gln genotypes of the XPC gene, His/His genotypes of the XPG gene, more than three variant alleles in all DNA repair genes and more than two variant alleles in NER genes, compared with the remaining groups, using Kaplan-Meier survival curves (Figure 2, A–D, respectively). These genotypes were associated with improved disease-specific survival, except for XPG gene (P = .0032 for the XPC gene, P = .069 for the XPG gene, P = .012 for the total variant alleles in all DNA repair genes, and P = .0066 for the total variant alleles in NER genes; log-rank test). Clinicopathologic variables were also assessed for their relationship with the disease-specific survival of UUT-TCC patients by univariate analysis (Table 5). Tumor stage (T2/T3/T4), lymph node metastasis, distant metastasis, and grade (G3) were associated with an unfavorable outcome.
Table 5.
Univariate and Multivariate Cox Regression Analyses for Predicting Disease-Specific Survival of UUT-TCC Patients.
| Variable (n) | Deaths, n | Univariate Analysis | Multivariate Model One | Multivariate Model Two | Multivariate Model Three | ||||
| HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | ||
| Age | |||||||||
| < 70 (46) | 13 | Ref. | |||||||
| ≥ 70 (54) | 11 | 0.97 (0.64–1.5) | .87 | ||||||
| Gender | |||||||||
| Men (68) | 18 | Ref. | |||||||
| Women (32) | 6 | 0.87 (0.52–1.3) | .54 | ||||||
| Tumor location | |||||||||
| Renal pelvis (40) | 8 | Ref. | |||||||
| Ureter + both (60) | 16 | 1.0 (0.67–1.6) | .93 | ||||||
| Tumor stage | |||||||||
| Tis/Ta/T1 (39) | 4 | Ref. | Ref. | Ref. | Ref. | ||||
| T2/T3/T4 (56) | 18 | 2.3 (1.4–4.2) | .0008 | 1.8 (0.98–4.1) | .060 | 1.7 (0.98–3.5) | .058 | 1.8 (1.0–3.5) | .033 |
| Lymph node metastasis | |||||||||
| Negative (80) | 16 | Ref. | Ref. | Ref. | Ref. | ||||
| Positive (10) | 5 | 2.0 (1.2–3.3) | .016 | 0.66 (0.20–2.6) | .52 | 0.76 (0.34–1.5) | .46 | 0.71 (0.30–1.5) | .38 |
| Distant metastasis | |||||||||
| Negative (86) | 18 | Ref. | Ref. | Ref. | Ref. | ||||
| Positive (5) | 4 | 3.0 (1.6–5.3) | .0022 | 6.6 (1.7–22) | .0069 | 3.4 (1.5–7.4) | .0034 | 2.9 (1.3–6.5) | .012 |
| Tumor grade | |||||||||
| G1/G2 (48) | 6 | Ref. | Ref. | Ref. | Ref. | ||||
| G3 (47) | 15 | 1.7 (1.1–2.9) | .018 | 1.5 (0.86–3.0) | .16 | 1.5 (0.88–2.6) | .14 | 1.4 (0.82–2.5) | .22 |
| Histopathology | |||||||||
| Pure TCC (95) | 21 | Ref. | |||||||
| TCC with SCC (5) | 3 | 1.6 (0.76–2.7) | .20 | ||||||
| Operation | |||||||||
| Yes (94) | 22 | Ref. | |||||||
| No (6) | 2 | 1.6 (0.63–2.9) | .28 | ||||||
| Chemotherapy/Radiotherapy | |||||||||
| None (52) | 8 | Ref. | |||||||
| Chemotherapy (24) | 6 | 1.1 (0.63–1.9) | .71 | ||||||
| Radiotherapy (4) | 1 | 1.2 (0.28–2.9) | .74 | ||||||
| Both (16) | 7 | 1.7 (0.98–2.8) | .058 | ||||||
| XPC (Lys939Gln, A/C) | |||||||||
| Lys/Lys (27) | 12 | Ref. | Ref. | ||||||
| Lys/Gln (46) | 9 | 0.63 (0.40–0.98) | .038 | ||||||
| Gln/Gln (18) | 1 | 0.32 (0.076–0.74) | .0039 | ||||||
| Lys/Gln + Gln/Gln (64) | 10 | 0.55 (0.35–0.84) | .0058 | 0.37 (0.19–0.65) | .0005 | ||||
| Lys/Lys + Lys/Gln (73) | 21 | Ref. | |||||||
| Gln/Gln (18) | 1 | 0.38 (0.090–0.84) | .012 | ||||||
| XPD (Lys751Gln, A/C)* | |||||||||
| Lys/Lys (93) | 22 | Ref. | |||||||
| Lys/Gln + Gln/Gln (3) | 0 | Not calculated | .13 | ||||||
| XPG (Asp1104His, G/C) | |||||||||
| Asp/Asp (19) | 4 | Ref. | |||||||
| Asp/His (41) | 15 | 1.3 (0.79–2.5) | .31 | ||||||
| His/His (34) | 4 | 0.74 (0.36–1.5) | .41 | ||||||
| Asp/His + His/His (75) | 19 | 1.1 (0.67–2.0) | .73 | ||||||
| Asp/Asp + Asp/His (60) | 19 | Ref. | |||||||
| His/His (34) | 4 | 0.62 (0.33–1.0) | .054 | ||||||
| XRCC1 (Arg399Gln, G/A)* | |||||||||
| Arg/Arg (50) | 14 | Ref. | |||||||
| Arg/Gln + Gln/Gln (46) | 9 | 0.81 (0.52–1.2) | .32 | ||||||
| XRCC3 (Thr241Met, C/T)* | Thr/Thr (79) | 20 | Ref. | ||||||
| Thr/Met + Met/Met (15) | 2 | 0.61 (0.24–1.1) | .13 | ||||||
| Total variant alleles in DNA repair genes | |||||||||
| ≤ 3 (73) | 22 | Ref. | Ref. | ||||||
| > 3 (27) | 2 | 0.43 (0.17–0.80) | .0048 | 0.31 (0.073–0.70) | .0016 | ||||
| Total variant alleles in NER genes† | |||||||||
| ≤ 2 (70) | 23 | Ref. | Ref. | ||||||
| > 2 (29) | 1 | 0.32 (0.076–0.70) | .0015 | 0.39 (0.090–0.87) | .018 | ||||
Bold font indicates statistical significance.
UUT-TCC indicates upper urinary tract transitional cell carcinoma; HR, hazard ratio; CI, confidence interval; SCC, squamous cell carcinoma; NER, nucleotide excision repair; Ref., Reference.
For those polymorphisms with few homozygous variant alleles, only the combined results of the heterozygous and homozygous variant alleles were shown.
NER genes included XPC, XPD, and XPG.
Figure 2.
Kaplan-Meier disease-specific survival curves for patients with UUT-TCC by DNA repair genotypes (log-rank tests). (A) Lys/Gln + Gln/Gln versus Lys/Lys of XPC; P = .0032. (B) His/His versus Asp/Asp + Asp/His of XPG; P = .069. (C) Total variant alleles in all DNA repair genes studied: > 3 versus ≤ 3; P = .012. (D) Total variant alleles in NER genes (XPC, XPD, and XPG): > 2 versus ≤ 2; P = .0066.
Multivariate Analysis of DNA Repair Genotypes and Disease-Specific Survival of UUT-TCC Patients
Using multivariate analysis (model one) including all factors with P < .05 in univariate analysis (tumor stage, lymph node metastasis, distant metastasis, grade, and XPC genotypes), XPC genotypes (HR: 0.37, 95% CI: 0.19–0.65, P = .0005), as well as distant metastasis, were independently associated with disease-specific survival (Table 5). The XPC genotypes were the most powerful prognostic markers, followed by distant metastasis (HR: 6.6, 95% CI: 1.7–22, P = .0069). When the number of variant alleles in all DNA repair genes was used instead of XPC genotypes (model two), the number of variant alleles (HR: 0.31, 95% CI: 0.073–0.70, P = .0016), as well as distant metastasis, was independently associated with disease-specific survival. When the number of variant alleles in NER genes was used instead of XPC genotypes (model three), the number of variant alleles (HR: 0.39, 95% CI: 0.090–0.87, P = .018), as well as tumor stage and distant metastasis, was independently associated with disease-specific survival.
Discussion
In the present study, we found that some DNA repair genotypes were independently associated with the overall or disease-specific survival of UUT-TCC patients beyond tumor stage and grade. Recently, there have been some reports that have assessed molecular markers for their independent prognostic potential in patients with UUT-TCC [5,30–32]. However, most of these series include 80 cases or less, and the power may be limited in the analyses. In addition, almost all of the reported molecular markers need tumor tissue samples obtained by operation and complex techniques [5,30–32], unlike the evaluation of germline genetic polymorphisms, which requires little more than a peripheral blood sample and standard PCRs, making these tests potentially useful in the clinical setting [33]. In UUT-TCC, it is difficult to obtain enough tumor tissue samples for the molecular analyses on biopsy. DNA repair genotypes may therefore be preoperatively valuable as prognostic factors for UUT-TCC.
Recently, Brown et al. [34] emphasized the importance of neo-adjuvant chemotherapy as a first-line strategy for localized UUT-TCC patients with a high risk of recurrence or metastasis, to improve the outcome; however, they also mentioned the absence of a useful marker for risk assessment. Patients with UUT-TCC often sustain a significant loss of renal reserve as a result of nephroureterectomy, frequently in the setting of advanced age or chronic renal insufficiency. This factor essentially precludes the ability to deliver effective doses of cytotoxic chemotherapy after surgery [4]. The DNA repair genotypes could therefore be helpful in preoperative patient selection for neoadjuvant chemotherapy for UUT-TCC. Predicting the prognosis of UUT-TCC based on DNA repair genotypes might help to stratify the patients by their risk of progression and to provide personalized treatments and follow-up strategies.
In this study, some DNA repair gene polymorphisms potentially influenced the malignant phenotypes of UUT-TCC. This result may be due to the modulated DNA repair capacity caused by the DNA repair gene polymorphisms. Various assays have been developed to quantify DNA repair activity for DNA repair gene polymorphisms, including the Comet assay and the host cell reactivation assay [9,33]. Despite some inconsistencies in the literature, it seems likely that the wild-type and variant genotypes are associated with different levels of DNA repair activity [8–12]. Our patients with variant alleles in the XPC gene, those with more than three total variant alleles in all DNA repair genes studied and those with more than two total variant alleles in NER genes had significantly improved survival. The wild-type genotypes conferring suboptimal DNA repair in the tumor might lead to the accumulation of multiple genetic changes and to biologically more aggressive UUT-TCC. It has been shown that the wild-type genotypes of the XPC, XPD, XPG, and XRCC1 genes are associated with decreased DNA repair capacity [8,10–12], and these results are in accord with our findings.
On the contrary, another study showed that genetic polymorphisms in NER pathway could explain only a small amount of the variability in DNA repair capacity using an assay measuring the removal of in vitro-induced benzo[a]pyrene diolepoxide-DNA adducts in lymphoblastoid cells in sisters discordant for breast cancer from the New York Registry [35]. Although the accurate cause for the discrepancy in the findings between this and our studies is uncertain, it might be due to the differences in ethnic backgrounds, methods for DNA repair capacity assay, or cancer types.
DNA repair gene polymorphisms potentially influenced malignant phenotypes of UUT-TCC in our present study, although they were germline variations not somatic mutations in tumor cells. Loktionov [36] demonstrated that both cancer initiation risk and later neoplastic events (tumor growth, invasion, metastatic spread, response to therapeutic interventions, and finally, survival) are strongly affected by factors that are predetermined by the individual's genetic background. Hunter [37] also stated that recent evidence implies a significant role of germline polymorphisms in cancer progression. Germline polymorphisms may play an important role in cancer progression because cancer development and progression strongly depend on altered interactions between malignant cells and their normal neighbors [27].
Because the population of our UUT-TCC patients had heterogeneity with multiple stages and therapies, we included these factors as categorical variables in multivariate analyses for survival using Cox proportional hazard regression models. In these analyses, some DNA repair genotypes were found to be independently associated with survival beyond tumor stage and grade, whereas therapeutic modalities did not significantly affect the survival. If our findings that the modulated DNA repair capacity caused by the DNA repair genotypes might have a significant impact on the survival of UUT-TCC patients are confirmed in functional and larger studies, modification of the DNA repair enzyme level or DNA repair pathway signaling might be a novel therapeutic target for UUT-TCC.
In the case-control study, no significant associations were observed between DNA repair genotypes and individual risk for UUT-TCC. We have therefore concluded this study to be negative, with respect to UUT-TCC susceptibility. However, the sample size was limited to detect these associations, and larger studies are needed, including the analysis of smoking status both in cases and controls, such as some other types of cancer [14,15,38,39].
Concerning bladder cancer, there have been several reports on the significant associations between DNA repair genotypes and risk or progression [7,14–20,38]. Sanyal et al. [20] showed that bladder cancer patients that were simultaneous carriers of variant alleles at the XPD (Lys751Gln) and XPC (Lys939Gln) polymorphisms were at lower risk of death than other patients (P = .001). This result is partly compatible with our findings in UUT-TCC patients. Cigarette smoking and exposure to certain industrial dyes or solvents are associated with an increased risk of UUT-TCC, similar to bladder TCC [21]. However, phenacetin-containing analgesics and Balkan nephropathy are stronger risk factors for UUT-TCC than for bladder TCC [40,41]. In addition, UUT-TCC is associated with hereditary nonpolyposis colorectal cancer (HNPCC), because UUT-TCC occurs at a higher rate in patients with HNPCC [42,43]. Microsatellite instability, which is a well-established feature of HNPCC [44], appeared to be at a high level in patients with UUT-TCC, whereas it is a rare event in those with bladder TCC [43,45,46]. Thus, there are a few, but substantial, differences in the etiology and genetic alterations between UUT and bladder TCCs. Comparing the associations between DNA repair polymorphisms and cancer risk and prognosis in patients with UUT-TCC, with those in patients with bladder TCC, might provide a better understanding of the mechanism underlying the development and progression of UUT-TCC.
In conclusion, our study provides evidence that some DNA repair genotypes are independently associated with survival and may preoperatively be valuable as prognostic factors for UUT-TCC beyond tumor stage and grade, possibly due to altered DNA repair capacity caused by these polymorphisms. However, our results with a limited sample size allow only preliminary conclusions, and larger studies are needed to confirm the associations between DNA repair gene polymorphisms and the clinical outcomes of patients with UUT-TCC.
Abbreviations
- HNPCC
hereditary nonpolyposis colorectal cancer
- NER
nucleotide excision repair
- UUT-TCC
upper urinary tract transitional cell carcinoma
- XPC
xeroderma pigmentosum complementation group C
- XPD
xeroderma pigmentosum complementation group D
- XPG
xeroderma pigmentosum complementation group G
- XRCC1
X-ray repair cross-complementing group 1
- XRCC3
X-ray repair cross-complementing group 3
Footnotes
This work was supported by a Grant-in-Aid for Scientific Research (C) (19591853) from the Japan Society for the Promotion of Science.
References
- 1.Hall MC, Womack S, Sagalowsky AI, Carmody T, Erickstad MD, Roehrborn CG. Prognostic factors, recurrence, and survival in transitional cell carcinoma of the upper urinary tract: a 30-year experience in 252 patients. Urology. 1998;52:594–601. doi: 10.1016/s0090-4295(98)00295-7. [DOI] [PubMed] [Google Scholar]
- 2.Bamias A, Deliveliotis C, Fountzilas G, Gika D, Anagnostopoulos A, Zorzou MP, Kastritis E, Constantinides C, Kosmidis P, Dimopoulos MA. Adjuvant chemotherapy with paclitaxel and carboplatin in patients with advanced carcinoma of the upper urinary tract: a study by the Hellenic Cooperative Oncology Group. J Clin Oncol. 2004;22:2150–2154. doi: 10.1200/JCO.2004.09.043. [DOI] [PubMed] [Google Scholar]
- 3.Czito B, Zietman A, Kaufman D, Skowronski U, Shipley W. Adjuvant radiotherapy with and without concurrent chemotherapy for locally advanced transitional cell carcinoma of the renal pelvis and ureter. J Urol. 2004;172:1271–1275. doi: 10.1097/01.ju.0000137910.38441.8a. [DOI] [PubMed] [Google Scholar]
- 4.Lerner SE, Blute ML, Richardson RL, Zincke H. Platinum-based chemotherapy for advanced transitional cell carcinoma of the upper urinary tract. Mayo Clin Proc. 1996;71:945–950. [PubMed] [Google Scholar]
- 5.Roupret M, Fromont G, Azzouzi AR, Catto JW, Vallancien G, Hamdy FC, Cussenot O. Microsatellite instability as predictor of survival in patients with invasive upper urinary tract transitional cell carcinoma. Urology. 2005;65:1233–1237. doi: 10.1016/j.urology.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 6.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 7.Goode EL, Ulrich CM, Potter JD. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11:1513–1530. [PubMed] [Google Scholar]
- 8.Lunn RM, Helzlsouer KJ, Parshad R, Umbach DM, Harris EL, Sanford KK, Bell DA. XPD polymorphisms: effects on DNA repair proficiency. Carcinogenesis. 2000;1:551–555. doi: 10.1093/carcin/21.4.551. [DOI] [PubMed] [Google Scholar]
- 9.Spitz MR, Wu X, Wang Y, Wang LE, Shete S, Amos CI, Guo Z, Lei L, Mohrenweiser H, Wei Q. Modulation of nucleotide excision repair capacity by XPD polymorphisms in lung cancer patients. Cancer Res. 2001;61:1354–1357. [PubMed] [Google Scholar]
- 10.Cornetta T, Festa F, Testa A, Cozzi R. DNA damage repair and genetic polymorphisms: assessment of individual sensitivity and repair capacity. Int J Radiat Oncol Biol Phys. 2006;66:537–545. doi: 10.1016/j.ijrobp.2006.06.037. [DOI] [PubMed] [Google Scholar]
- 11.Naccarati A, Soucek P, Stetina R, Haufroid V, Kumar R, Vodickova L, Trtkova K, Dusinska M, Hemminki K, Vodicka P. Genetic polymorphisms and possible gene-gene interactions in metabolic and DNA repair genes: effects on DNA damage. Mutat Res. 2006;593:22–31. doi: 10.1016/j.mrfmmm.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 12.Vodicka P, Kumar R, Stetina R, Sanyal S, Soucek P, Haufroid V, Dusinska M, Kuricova M, Zamecnikova M, Musak L, et al. Genetic polymorphisms in DNA repair genes and possible links with DNA repair rates, chromosomal aberrations and single-strand breaks in DNA. Carcinogenesis. 2004;25:757–763. doi: 10.1093/carcin/bgh064. [DOI] [PubMed] [Google Scholar]
- 13.Matullo G, Palli D, Peluso M, Guarrera S, Carturan S, Celentano E, Krogh V, Munnia A, Tumino R, Polidoro S, et al. XRCC1, XRCC3, XPD gene polymorphisms, smoking and (32)P-DNA adducts in a sample of healthy subjects. Carcinogenesis. 2001;22:1437–1445. doi: 10.1093/carcin/22.9.1437. [DOI] [PubMed] [Google Scholar]
- 14.Matullo G, Guarrera S, Sacerdote C, Polidoro S, Davico L, Gamberini S, Karagas M, Casetta G, Rolle L, Piazza A. Polymorphisms/haplotypes in DNA repair genes and smoking: a bladder cancer case-control study. Cancer Epidemiol Biomarkers Prev. 2005;14:569–2578. doi: 10.1158/1055-9965.EPI-05-0189. [DOI] [PubMed] [Google Scholar]
- 15.Wu X, Gu J, Grossman HB, Amos CI, Etzel C, Huang M, Zhang Q, Millikan RE, Lerner S, Dinney CP, et al. Bladder cancer predisposition: a multigenic approach to DNA-repair and cell-cycle-control genes. Am J Hum Genet. 2006;78:464–479. doi: 10.1086/500848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanyal S, Festa F, Sakano S, Zhang Z, Steineck G, Norming U, Wijkstrom H, Larsson P, Kumar R, Hemminki K. Polymorphisms in DNA repair and metabolic genes in bladder cancer. Carcinogenesis. 2004;25:729–734. doi: 10.1093/carcin/bgh058. [DOI] [PubMed] [Google Scholar]
- 17.Sakano S, Kumar R, Larsson P, Onelov E, Adolfsson J, Steineck G, Hemminki K. A single-nucleotide polymorphism in the XPG gene, and tumour stage, grade, and clinical course in patients with nonmuscle-invasive neoplasms of the urinary bladder. BJU Int. 2006;97:847–851. doi: 10.1111/j.1464-410X.2005.05994.x. [DOI] [PubMed] [Google Scholar]
- 18.Sakano S, Wada T, Matsumoto H, Sugiyama S, Inoue R, Eguchi S, Ito H, Ohmi C, Matsuyama H, Naito K. Single nucleotide polymorphisms in DNA repair genes might be prognostic factors in muscle-invasive bladder cancer patients treated with chemoradiotherapy. Br J Cancer. 2006;95:561–570. doi: 10.1038/sj.bjc.6603290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu J, Zhao H, Dinney CP, Zhu Y, Leibovici D, Bermejo CE, Grossman HB, Wu X. Nucleotide excision repair gene polymorphisms and recurrence after treatment for superficial bladder cancer. Clin Cancer Res. 2005;11:1408–1415. doi: 10.1158/1078-0432.CCR-04-1101. [DOI] [PubMed] [Google Scholar]
- 20.Sanyal S, De Verdier PJ, Steineck G, Larsson P, Onelov E, Hemminki K, Kumar R. Polymorphisms in XPD, XPC and the risk of death in patients with urinary bladder neoplasms. Acta Oncol. 2007;46:31–41. doi: 10.1080/02841860600812693. [DOI] [PubMed] [Google Scholar]
- 21.Jensen OM, Knudsen JB, McLaughlin JK, Sorensen BL. The Copenhagen case-control study of renal pelvis and ureter cancer: role of smoking and occupational exposures. Int J Cancer. 1988;41:557–561. doi: 10.1002/ijc.2910410414. [DOI] [PubMed] [Google Scholar]
- 22.Kirkali Z, Tuzel E. Transitional cell carcinoma of the ureter and renal pelvis. Crit Rev Oncol Hematol. 2003;47:155–169. doi: 10.1016/s1040-8428(03)00079-9. [DOI] [PubMed] [Google Scholar]
- 23.Rigola MA, Fuster C, Casadevall C, Bernues M, Caballin MR, Gelabert A, Egozcue J, Miro R. Comparative genomic hybridization analysis of transitional cell carcinomas of the renal pelvis. Cancer Genet Cytogenet. 2001;127:59–63. doi: 10.1016/s0165-4608(00)00426-x. [DOI] [PubMed] [Google Scholar]
- 24.Yao R, Yi Y, Grubbs CJ, Lubet RA, You M. Gene expression profiling of chemically induced rat bladder tumors. Neoplasia. 2007;9:207–221. doi: 10.1593/neo.06814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakano S, Matsumoto H, Yamamoto Y, Kawai Y, Eguchi S, Ohmi C, Matsuyama H, Naito K. Association between DNA repair gene polymorphisms and p53 alterations in Japanese patients with muscle-invasive bladder cancer. Pathobiology. 2006;73:295–303. doi: 10.1159/000099124. [DOI] [PubMed] [Google Scholar]
- 26.de las Penas R, Sanchez-Ronco M, Alberola V, Taron M, Camps C, Garcia-Carbonero R, Massuti B, Queralt C, Botia M, Garcia-Gomez R, et al. Polymorphisms in DNA repair genes modulate survival in cisplatin/gemcitabine-treated non-small-cell lung cancer patients. Ann Oncol. 2006;17:668–675. doi: 10.1093/annonc/mdj135. [DOI] [PubMed] [Google Scholar]
- 27.Sakano S, Hinoda Y, Okayama N, Kawai Y, Korenaga Y, Eguchi S, Nagao K, Ohmi C, Naito K. The association of DNA repair gene polymorphisms with the development and progression of renal cell carcinoma. Ann Oncol. 2007;18:1817–1827. doi: 10.1093/annonc/mdm337. [DOI] [PubMed] [Google Scholar]
- 28.Greene FL, Page DL, Fleming ID, Fritz A, Balch CM, Haller DG, Morrow M. American Joint Committee on Cancer Staging Manual. 6th ed. Philadelphia: PA Springer; 2002. [Google Scholar]
- 29.Seaman SR, Muller-Myhsok B. Rapid simulation of P values for product methods and multiple-testing adjustment in association studies. Am J Hum Genet. 2005;76:399–408. doi: 10.1086/428140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inoue K, Kamada M, Slaton JW, Fukata S, Yoshikawa C, Tamboli P, Dinney CP, Shuin T. The prognostic value of angiogenesis and metastasis-related genes for progression of transitional cell carcinoma of the renal pelvis and ureter. Clin Cancer Res. 2002;8:1863–1870. [PubMed] [Google Scholar]
- 31.Ohtsuka Y, Kawakami S, Fujii Y, Koga F, Saito K, Ando N, Takizawa T, Kageyama Y, Kihara K. Loss of uroplakin III expression is associated with a poor prognosis in patients with urothelial carcinoma of the upper urinary tract. BJU Int. 2006;97:1322–1326. doi: 10.1111/j.1464-410X.2006.06158.x. [DOI] [PubMed] [Google Scholar]
- 32.Fromont G, Roupret M, Amira N, Sibony M, Vallancien G, Validire P, Cussenot O. Tissue microarray analysis of the prognostic value of E-cadherin, Ki67, p53, p27, survivin and MSH2 expression in upper urinary tract transitional cell carcinoma. Eur Urol. 2005;48:764–770. doi: 10.1016/j.eururo.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 33.Gurubhagavatula S, Liu G, Park S, Zhou W, Su L, Wain JC, Lynch TJ, Neuberg DS, Christiani DC. XPD and XRCC1 genetic polymorphisms are prognostic factors in advanced non-small-cell lung cancer patients treated with platinum chemotherapy. J Clin Oncol. 2004;22:2594–2601. doi: 10.1200/JCO.2004.08.067. [DOI] [PubMed] [Google Scholar]
- 34.Brown GA, Busby JE, Wood CG, Pisters LL, Dinney CP, Swanson DA, Grossman HB, Pettaway CA, Munsell MF, Kamat AM, et al. Nephroureterectomy for treating upper urinary tract transitional cell carcinoma: time to change the treatment paradigm? BJU Int. 2006;98:1176–1180. doi: 10.1111/j.1464-410X.2006.06524.x. [DOI] [PubMed] [Google Scholar]
- 35.Shen J, Desai M, Agrawal M, Kennedy DO, Senie RT, Santella RM, Terry MB. Polymorphisms in nucleotide excision repair genes and DNA repair capacity phenotype in sisters discordant for breast cancer. Cancer Epidemiol Biomarkers Prev. 2006;15:1614–1619. doi: 10.1158/1055-9965.EPI-06-0218. [DOI] [PubMed] [Google Scholar]
- 36.Loktionov A. Common gene polymorphisms, cancer progression and prognosis. Cancer Lett. 2004;208:1–33. doi: 10.1016/j.canlet.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 37.Hunter K. Host genetics influence tumour metastasis. Nat Rev Cancer. 2006;6:141–146. doi: 10.1038/nrc1803. [DOI] [PubMed] [Google Scholar]
- 38.Stern MC, Umbach DM, Lunn RM, Taylor JA. DNA repair gene XRCC3 codon 241 polymorphism, its interaction with smoking and XRCC1 polymorphisms, and bladder cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11:939–943. [PubMed] [Google Scholar]
- 39.Neumann AS, Sturgis EM, Wei Q. Nucleotide excision repair as a marker for susceptibility to tobacco-related cancers: a review of molecular epidemiological studies. Mol Carcinog. 2005;42:65–92. doi: 10.1002/mc.20069. [DOI] [PubMed] [Google Scholar]
- 40.McCredie M, Stewart JH, Day NE. Different roles for phenacetin and paracetamol in cancer of the kidney and renal pelvis. Int J Cancer. 1993;53:245–249. doi: 10.1002/ijc.2910530212. [DOI] [PubMed] [Google Scholar]
- 41.Radovanovic Z, Jankovic S, Jevremovic I. Incidence of tumors of urinary organs in a focus of Balkan endemic nephropathy. Kidney Int Suppl. 1991;34:S75–S76. [PubMed] [Google Scholar]
- 42.Watson P, Lynch HT. The tumor spectrum in HNPCC. Anticancer Res. 1994;14:1635–1639. [PubMed] [Google Scholar]
- 43.Amira N, Rivet J, Soliman H, Cancel-Tassin G, Le Duc A, Janin A, Cussenot O. Microsatellite instability in urothelial carcinoma of the upper urinary tract. J Urol. 2003;170:1151–1154. doi: 10.1097/01.ju.0000086551.22844.cd. [DOI] [PubMed] [Google Scholar]
- 44.Aaltonen LA, Peltomaki P, Leach FS, Sistonen P, Pylkkanen L, Mecklin JP, Jarvinen H, Powell SM, Jen J, Hamilton SR, et al. Clues to the pathogenesis of familial colorectal cancer. Science. 1993;260:812–816. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- 45.Gonzalez-Zulueta M, Ruppert JM, Tokino K, Tsai YC, Spruck CH, Miyao N, Nichols PW, Hermann GG, Horn T, Steven K, et al. Microsatellite instability in bladder cancer. Cancer Res. 1993;53:5620–5623. [PubMed] [Google Scholar]
- 46.Bonnal C, Ravery V, Toublanc M, Bertrand G, Boccon-Gibod L, Henin D, Grandchamp B. Absence of microsatellite instability in transitional cell carcinoma of the bladder. Urology. 2000;55:287–291. doi: 10.1016/s0090-4295(99)00399-4. [DOI] [PubMed] [Google Scholar]