Abstract
The mechanism of action of DNA methylation inhibitor 5-aza-2′-deoxycytidine (5-aza-CdR), a potential anticancer agent is believed to be activated by the demethylation of tumor suppressor genes. We tested here the hypothesis that demethylating agents also demethylate and activate genes involved in invasion and metastasis and therefore might increase the risk of developing tumor metastasis. The effect of 5-aza-CdR on noninvasive human breast cancer cells MCF-7 and ZR-75-1 was evaluated by cell proliferation, invasion, and migration assay. The ability of 5-aza-CdR to activate a panel of silenced prometastatic and tumor suppressor genes was evaluated using reverse transcription-polymerase chain reaction and bisulfite DNA sequence analysis in vitro and for change in tumor growth and gene expression in vivo. Treatment of MCF-7 and ZR-75-1 with 5-aza-CdR diminished cell proliferation, induced tumor suppressor RASSF1A, and altered cell cycle kinetics' G2/M-phase cell cycle arrest. While these effects of 5-aza-CdR slowed the growth of tumors in nude mice, it also induced a battery of prometastatic genes, namely, uPA, CXCR4, HEPARANASE, SYNUCLEIN γ, and transforming growth factor-beta (TGF-β), by demethylation of their promoters. These results draw attention to the critical role of demethylation as a potential mechanism that can promote the development and progression of tumor metastasis after demethylation therapy as an anticancer treatment.
Introduction
Hypermethylation of tumor suppressor genes is a common mechanism of gene silencing observed in cancer [1]. Knock down of DNA methyltransferase 1, the enzyme that is involved in the copying of DNA methylation during cell division, by either pharmacological [2,3] or genetic approaches [4], was shown to inhibit cancer in animal models. The DNA methyltransferase inhibitor, 5-aza-2′-deoxcytidine (5-aza-CdR), was shown to demethylate and induce the activity of methylated tumor suppressor genes [5]. 5-Aza-CdR is currently being tested in different clinical trials as an anticancer agent [6–8]. Breast cancer is the most common malignant disease affecting women of all age group globally; during this multistep process of tumor progression, the disease was initiated as a less-aggressive nonmetastatic hormone-responsive-type, which gradually develop as a highly invasive and metastatic hormone-insensitive malignancy. Most of the mortalities among these patients are due to delayed diagnosis, poor prognosis, and metastases to the distant organs especially lung and bones, which are more frequent sites of breast cancer metastases [9]. The early steps of the metastatic cascade involving the degradation of the extracellular matrix and subsequent invasion by cancer cells are accomplished by several proteinases, including among others, urokinase-type plasminogen activator (uPA), heparanase and matrix metalloproteinase (MMPs), and other proteins, by which different lines of data suggest to contribute to metastasis, such as transforming growth factor-beta (TGF-β), S100A4, SYNUCLEIN γ, and CXCR4 [10–20]. We have previously shown that uPA promoter region is hypomethylated, and its expression is upregulated in the invasive breast cancer cell line MDA-MB-231 (hormone insensitive), whereas the same promoter region is methylated in noninvasive breast cancer cells such as MCF-7 (hormone-responsive). Furthermore, 5-aza-CdR treatment induces the expression of uPA in noninvasive MCF-7 cells [21]. Similar effects were also observed in a hormone-responsive human prostate cancer cell line, LNCaP, after the treatment with 5-aza-CdR [22]. We have also shown that reverting the hypomethylated status of uPA promoter in MDA-MB-231 cells by either S-adenosyl-l-methionine treatment or knock down of methyl DNA binding domain protein 2 causes significant inhibition in uPA expression and invasion in vitro and tumor growth in vivo [23]. We previously hypothesized that reactivation of expression of prometastatic genes by DNA demethylation is a common contributing factor to promoting tumor metastasis [24]. Therefore, we suggested that although inhibition of DNA methylation results in decreasing tumor growth by the activation of tumor suppressor genes, it might, at the same time, result in the activation of prometastatic genes, suggesting divergence of growth control and invasion in breast cancer cells [24,25].
In the present study, we tested this hypothesis by examining the effects of 5-aza-CdR, a classic DNA methylation inhibitor, which is currently tested in clinical trials, on the expression and DNA methylation of the tumor suppressor gene, RASSF1A, and a set of genes involved in metastasis/angiogenesis of breast cancer, such as uPA, HEPARANASE, CXCR4 RECEPTOR, STROMAL CELL-DERIVED FACTOR 1α (CXCL12), BREAST CANCER-SPECIFIC GENE 1 or SYNUCLEIN γ (SNCG), VASCULAR ENDOTHELIAL GROWTH FACTOR (VEGF), and TGF-β as well as genes involved in cell cycle, such as cyclin-dependent kinase inhibitors (p21WAF1/Cip1, CYCLIN D1, and CYCLIN E1) and BAX, a proapoptotic gene of the Bcl-2 family [10,11,14,15,17,26–34]. We then tested whether 5-aza-CdR will convert nonmetastatic breast cancer cell lines MCF-7 and ZR- 75-1 to invasive and metastatic cells using in vitro and in vivo assays. Our data support a mechanism for the potential capacity of 5-aza-CdR to induce the expression of previously quiescent prometastatic genes and for the need to carefully evaluate and monitor patients receiving DNA methylation inhibitors.
Materials and Methods
Cell Culture and Treatment
Human breast cancer cell line, MCF-7 and ZR-75-1 cells were obtained from the American Type Culture Collection (Manassas, VA). MCF-7 cells were maintained in minimum Eagle's medium (Earle's salts; Invitrogen Life Technologies, Ontario, Canada) with 2 mM l-glutamine, 10% FBS, 10 µg/ml insulin, and 100 U/ml of penicillin-streptomycin sulfate. ZR-75-1 cells were grown in RPMI 1640 medium (Invitrogen Life Technologies) with 10% FBS, 2 mM l-glutamine adjusted to contain 1.5 g/l sodium bicarbonate, 4.5 g/l glucose, 10 mM HEPES, and 1.0 mM sodium pyruvate. Cells were incubated at 37°C in 5% CO2. In the 5-aza-CdR treatment group, MCF-7 and ZR-75-1 cells were treated for 7 days with 5-aza-CdR (Sigma Aldrich, Ontario, Canada) at final concentrations of 0.1, 1, and 5 µM in the regular medium. The culture medium was changed every second day in both control and treatment groups by maintaining the desired concentration of the drug.
Cell Proliferation Assay
MCF-7 and ZR-75-1 cells were plated in quadruplicate at a density of 50,000 cells/well in 2 ml of respective culture media in six-well plates. Cells were grown in the regular medium with 5% FBS in the control and treatment (5-aza-CdR) groups. Cell growth curve was analyzed at two different doses of 5-aza-CdR (1 and 5 µM). Cells were trypsinized, and the number of viable cells was determined by 0.4% trypan blue and counted at different time points starting from day 0 (without 5-aza-CdR) using a Coulter counter (Model ZF; Coulter Electronics, Hertfordshire, UK). Cell culture medium was replenished every second day along with the indicated dose of 5-aza-CdR in the treatment group.
Fluorescence-Activated Cell Sorting Analysis
The effect of 5-aza-CdR on cell cycle distribution was determined by flow cytometry after staining the cells with propidium iodide. Briefly, control and 5-aza-CdR-treated (5 µM for 7 days) MCF-7 and ZR-75-1 cells were trypsinized and washed with phosphate-buffered saline (PBS), and fixed with chilled 70% ethanol. The fixed cells were then centrifuged and washed with PBS. The cells were then treated with 1 U of DNase-free RNase to the cell suspension (106 cells in 1 ml), and incubated for 30 minutes at 37°C. Fifty microliters of 1 mg/ml propidium iodide was added directly to the cell suspension, and flow cytometry analysis was performed on a flow cytometer (FACS Calibur; BD Biosciences, Ontario, Canada). Results were analyzed further using the WinMDI software (version 2.8; Scripps Research Institute, La Jolla, CA).
Boyden Chamber Matrigel Invasion Assay
Cells' invasive capacity was determined using a two-compartment Boyden chamber Matrigel invasion assay (Transwell; Costar Corning Corporation, Burlington, MA) as described previously [21]. The 8-µm-pore polycarbonate filters were coated with basement membrane Matrigel (50 µg/filter). A total of 5 × 104 cells treated with or without 5-aza-CdR (1 and 5 µM) in 0.1 ml of medium were added to the upper chamber and placed on top of a lower chamber prefilled with 0.8 ml of serum-free medium supplemented with 25 µg/ml fibronectin (Sigma Aldrich) and then incubated at 37°C for 24 hours. After incubation, the medium was removed, and the polycarbonate filters with invaded cells were fixed in 2% paraformaldehyde, 0.5% glutaraldehyde (Sigma Aldrich) in 0.1 M phosphate buffer, pH 7.4, at room temperature for 30 minutes. They were then stained with 1.5% toluidine blue, washed, and mounted onto glass slides. The number of cells invaded was examined under a light microscope. Ten fields under a magnification of x400 were randomly selected, and the mean number of cells invading was calculated.
Cell Wounding Assay
Confluent MCF-7 monolayer was grown in six-well plates as described above with 2% fetal bovine serum. The cell monolayer was mechanically wounded in the center of each well with a sterile 200-µl pipette tip and maintained in the presence or absence of 5-aza-CdR (1 and 5 µM) for 48 hours where the culture medium was changed every 24 hours. Cells were then washed three times to get rid of the detached cells and debris. Wound healing was monitored at different time points with an inverted bright field microscopy under the 4x objective. Only cell-free area was selected, measured, and quantified using the Image Pro-Plus software (Media Cybernetics, Inc., Bethesda, MD) and calculated as percentage wound healing using the equation: %wound healing = [1 - (wound area at Tx hours/wound area at T0)], where the Tx is the respective time point and T0 is the time immediately after wounding.
Reverse Transcriptase-Polymerase Chain Reaction Analysis
Total cellular RNA from control and 5-aza-CdR-treated MCF-7 and ZR-75-1 cells were extracted using TRIzol (Invitrogen Life Technologies) according to the manufacturer's protocol. Two micrograms of total RNA was used for reverse transcription and amplification. The primers used for reverse transcription-polymerase chain reaction (RT-PCR) were designed so that there is an intron between the amplified regions to recognize any DNA contamination. Two sets of different RT-PCR primers were used in the present study; their sequence and annealing temperature are given in Table 1. The reactions were carried out using standard protocols. The DNA was amplified under the following conditions: 95°C for 5 minutes, 30 cycles at 95°C for 30 seconds, annealing temperature of the respective primer for 30 seconds as given in Table 1, and at 72°C for 30 seconds, and the final extension at 72°C for 5 minutes. Different numbers of cycles were used to ensure linearity of amplification. The gels were quantified by using the Scion Software (Scion Corporation, Frederick, MD) for the densitometry.
Table 1.
Different Sets of Primer Sequences and Annealing Temperatures Used for RT-PCR Analysis.
| Genes | Primer Sequence for RT-PCR | Tm Value (°C) |
| Tumor suppressor gene | ||
| RASSF1A | 5′-ACCTCTGTGGCGACTTCATCT-3′ | 52 |
| 5′-AGGTGAACTTGCAATGCGC-3′ | ||
| Involved in metastasis | ||
| uPA | 5′-ACATTCACTGGTGCAACTGC-3′ | 56 |
| 5′-CAAGCGTGTCAGCGCTGTAG-3′ | ||
| SNCG | 5′-CAAGAAGGGCTTCTCCATCGCCAAGG-3′ | 60 |
| 5′-CCTCTTTCTCTTTGGATGCCACACCC-3′ | ||
| HEPARANASE | 5′-TTCGATCCCAAGAAGGAATCAAC-3′ | 55 |
| 5′-GTAGTGATGCCATGTAACTGAATC-3′ | ||
| CXCR4 | 5′-GGAGGGGATCAGTATATACA-3′ | 55 |
| 5′-GAAGATGATGGAGTAGATGG-3′ | ||
| CXCL12 | 5′-AGAGCCAACGTCAAGCATCT-3′ | 55 |
| 5′-CGTCTTTGCCCTTTCATCTC-3′ | ||
| Involved in angiogenesis | ||
| VEGF | 5′-CCTGGTGGACATCTTCCAGGAGTA-3′ | 58 |
| 5′-CTCACCGCCTCGGCTTGTCACA-3′ | ||
| Involved in cell cycle | ||
| TGF-β | 5′-GTACCTGAACCCGTGTTGCT-3′ | 54 |
| 5′-TACAGCTGCCGCACGCAGCA-3′ | ||
| p21WAF1/Cip1 | 5′-CTCAGAGGAGGCGCCATG-3′ | 55 |
| 5′-GGGCGGATTAGGGCTTCC-3′ | ||
| CYCLIN D1 | 5′-TGGATGCTGGAGGTCTGCGAGGAA-3′ | 60 |
| 5′-GGCTTCGATCTGCTCCTGGCAGGC-3′ | ||
| CYCLIN E1 | 5′-AGTTCTCGGCTCGCTCCAGGAAGA-3 | 56 |
| 5′-TCTTGTGTCGCCATAATCCGGTCA-3′ | ||
| Involved in apoptosis | ||
| BAX | 5′-ATGGACGGCTCCGGGGAG-3′ | 51 |
| 5′-TGGAAGAAGATGGGCTGA-3′ | ||
| GAPDH | 5′-CCCTTCATTGACCTCAACTACATGGT-3′ | 56 |
| 3′-GAGGGGCCATCCACAGTCTTCTG-5′ |
Sodium Bisulfite Sequencing
Genomic DNA was extracted from control and 5-aza-CdR-treated (5 µM for 7 days) MCF-7 cells using DNazol (Invitrogen Life Technologies) following the manufacturer's instructions. Sodium bisulfite treatment of the genomic DNA was performed as described previously [21,22]. Two sets of primers were designed to amplify the modified DNA fragment within the uPA promoter region with outer primers (5′-GTAAGGGGGTTTGAGGTAGT-3′ and 5′-ATAACCAAACTCCCCAACTA-3′) and inner primers (5′-TTTAGGTAAGTTGGGGTTTAG-3′ and 5′-TCTCTCTCCTCTATAAACTC-3′) of the uPA promoter sequence. The amplification reaction was performed under the following conditions: 95°C for 5 minutes; 34 cycles at 95°C for 1 minute, at 51°C for 2.5 minutes, and at 72°C for 1 minute; final extension at 72°C for 5 minutes. For CXCR4, the bisulfite DNA was subjected to primary and nested PCR using two sets of primers from the promoter and exon 1 region; outer primers (5′-TTATTTATTTAGTAAGGATGGA-3′ and 5′-AATACCTCCAATATCCTAAC-3′) and inner primers (5′-AAATGTTTTTGGGAGGTTTTG-3′ and 5′-CACTAATCCCCTCCATAATA-3′) at 95°C for 5 minutes; 34 cycles at 95°C for 30 seconds, at 50°C for 30 seconds, and at 72°C for 30 seconds; final extension at 72°C for 5 minutes. For SNCG genomic bisulfite sequencing, first-step PCR was carried out using the outer set of primers (5′-GGTTGAGTTAGTAGGAGTTTA-3′ and 5′-CCTACCATACCCCACTTACCC-3′,) and nested PCR was carried out using the inner set of primers (5′-TATTTTGGAGGAAGGTGAGGTTG-3′ and 5′-CCACTTACCCACATACATAACC-3′) at 95°C for 5 minutes; 34 cycles at 95°C for 30 seconds, at 58°C for 30 seconds, and at 72°C for 30 seconds, and a final extension at 72°C for 5 minutes.
Finally, PCR products of the uPA, CXCR4, and SNCG were purified by using QIAquick PCR purification kit (Qiagen Inc., Ontario, Canada) and subcloned into pGEMT cloning vector (Promega, Madison, WI) according to the manufacturer's instruction, and plasmid DNA from different bacterial clones was prepared and analyzed for DNA sequences (Genome Centre, McGill University, Quebec, Canada). At least 10 clones for each gene were sequenced per treatment group.
In Vivo Tumor Growth in Breast Cancer Model
Six-week-old female Balb C nu/nu mice were obtained from Charles River, Inc. (St. Constant, Quebec, Canada). Before inoculation, MCF-7 cells were treated with 5 µM of 5-aza-CdR for 7 days in regular growth media. Growth medium and drug was changed every second day during treatment. On day 8, treated and untreated MCF-7 cells were trypsinized and washed with Hank's balanced buffer; 2.5 x 106 cells were resuspended in 100 µl of saline with 20% Matrigel (Becton Dickinson Labware, Ontario, Canada). An anaesthetic cocktail of ketamine (50 mg/kg), xylazine (5 mg/kg), and acepromazine (1 mg/kg) was injected intramuscularly, and 2.5 x 106 cells were inoculated using 30-gauge needles into the mammary fat pad of anaesthetized mice. All experimental animals (eight animals in each group) were implanted with 0.25 mg of estradiol pellet with 60-day release (Innovative Research of America, Sarasota, FL) through subcutaneous route using a trocar. All the experimental animals were taken care of in accord with the McGill University Animal Care Committee.
Both control and experimental animals were monitored at weekly intervals for the development of tumors for 6 weeks. Tumor volume was determined according to the formula: tumor volume = shorter diameter2 x longer diameter / 2. Results were presented as the mean of tumor volumes recorded from both groups. At the end of the study, control and experimental animals were sacrificed; primary tumors were removed and fixed in 10% buffered formalin and Bouin's solution, respectively. Before fixation, tumors were divided into two halves, one half was fixed in formalin and embedded in paraffin for histologic analyses by light microscopy. The other half was snap-frozen in liquid nitrogen to analyze the expression of various genes by RT-PCR.
Immunohistochemical Analyses
Bouin's solution-fixed tumor samples were processed for immunohistochemistry. The fixed tissues were embedded in paraffin. Paraffin-embedded tumor samples were cut into 5-µm-thick sections; for immunohistochemical analysis for uPA, CXCR4, and heparanase, a prometastatic marker was carried out as using the avidin-biotinperoxidase complex technique. Briefly, the sections were deparaffinized in xylene followed by rehydration through a series of ethanol to water gradients. The sections were treated with 1% normal goat serum (Vector Laboratories Inc., Burlingame, CA), to block the non-specific binding, for 30 minutes at room temperature before incubation with the primary antibodies: monoclonal antibody against uPA (American Diagnostics Inc., Stamford, CT) and monoclonal antibodies against CXCR4 and heparanase (Santa Cruz Biotechnologies, Santa Cruz, CA); primary anti-uPA, CXCR4, and heparanase antibodies were used at 1:50 dilution overnight at 4°C under the conditions described by manufacturer. Throughout the experiment, after each step, all sections were washed three times, 10 minutes each, with Tris buffer (pH 7.6). Biotinylated goat anti-mouse IgG (Vector Laboratories Inc., Burlingame, CA) and HRP donkey anti-goat or goat anti-mouse IgG (Bio-Rad Laboratories Ltd., Ontario, Canada) were used as the secondary antibodies at a dilution of 1:200 for 1 hour at room temperature. The sections for uPA staining were treated with vectastain ABC-AP kit (Vector Laboratories Inc., Burlingame, CA). The immunoreaction was visualized with Fast Red TR/Naphthol AS-MX phosphate (Sigma Aldrich) containing 1 mM levamisole (uPA) and DAB HRP substrate kit (Vector Laboratories Inc., Ontario, Canada) for CXCR4 and heparanase for 5 to 10 minutes. The sections were counterstained with hematoxylin (Fisher Scientific, Ltd., Ontario, Canada) and mounted with Kaiser's glycerol jelly or paramount. For all negative controls included in the study, the primary antibody was substituted with PBS. Ten random fields were selected from the previously coded slides and manually scored as arbitrary units for the expression level of these genes [35].
Statistical Analysis
Results were analyzed as the mean ± SEM, and comparisons of the experimental data were analyzed by a two-tailed independent-sample Student's t test at 0.05 level of significance.
Results
Effect of 5-Aza-CdR on Cell Proliferation, Cell Cycle, and Migration
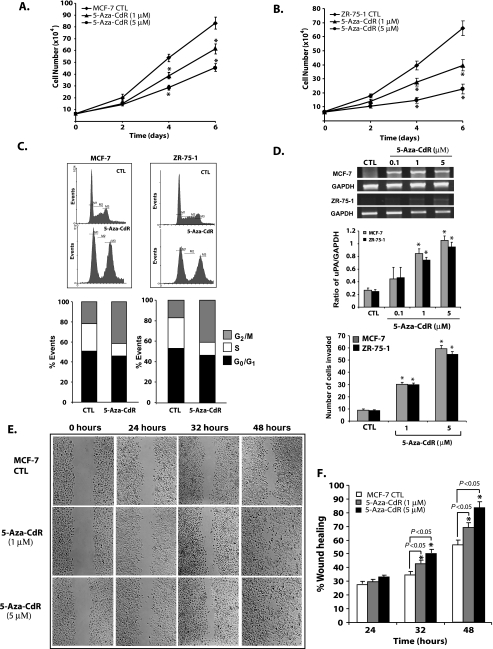
To verify that 5-aza-CdR exhibits the previously described effects on cell growth in vitro of the noninvasive breast cancer cell lines MCF-7 and ZR-75-1, we measured the effects of two doses of 5-aza-CdR on cell viability, cell doubling time, and cell cycle kinetics [36]. The results presented in Figure 1, A and B, show that 5-aza-CdR is increasing cell doubling time significantly at 1 and 5 µM doses. A FACS analysis of cell cycle distribution on control and 5-aza-CdR-treated cells (5 µM for 7 days) reveals a significant increase in the proportion of cells in the G2 phase, with a concomitant decline in S phase (Figure 1 C) in the treatment groups of both cell lines. Our results showed that 5-aza-CdR decreases cell proliferation under the conditions used in our study.
Figure 1.
Effect of 5-aza-CdR treatment on the proliferation, invasion, and migration of noninvasive cell lines. Noninvasive breast cancer cells, MCF-7 and ZR-75-1, were seeded at the same density and subjected to two different doses of 5-aza-CdR or with vehicle control alone (CTL) in a six-well plate, and growth curve analysis was carried out to determine the effect of 5-aza-CdR on the proliferation rate. At each time point, cells were trypsinized and counted as described in the Materials and Methods section (A and B). All data presented are expressed as mean ± SEM of quadruplicate wells for each time point. Similar results were obtained in two independent experiments. Cell cycle distribution analysis was carried out by FACS for control and 5-aza-CdR-treated MCF-7 and ZR-75-1 cells (5 µM for 7 days) after staining the cells with propidium iodide as described in the Materials and Methods section. Data were analyzed by WinMDI version 2.8 and plotted (C). Data are presented as mean of % events from three independent experiments by 100% stacked column graph for control and experimental groups. Expression of uPA, a protease involved in invasion, was analyzed by RT-PCR in MCF-7 and ZR-75-1 cells treated with different doses of 5-aza-CdR (D). Data were presented for control and experimental groups as mean ± SEM values from two independent experiments. The invasive capacity of MCF-7 and ZR-75-1 cells treated with different doses of 5-aza-CdR was determined using a Boyden chamber Matrigel invasion assay. The bar diagram represents the mean ± SEM as described in the Materials and Methods section. Wound healing migration assay was carried out on MCF-7 cells in the absence and presence of 5-aza-CdR (1 and 5µ M) in the culture medium with 2% fetal bovine serum as depicted in panels E and F. Cells were seeded at the same density and grown as monolayer and wounded as discussed in the Materials and Methods section. % Wound healing was recorded at different time points, and percentage of wound healing with respect to T0 was calculated using the described equation. Similar results were obtained from two different experiments. All data were presented as mean ± SEM of duplicate values from control and experimental groups from two independent experiments. Significant difference from the control is represented by an asterisk (*P < .05).
To determine whether 5-aza-CdR treatment affects the expression of uPA, a key protease involved in tumor cell invasion, MCF-7 and ZR-75-1 cells were treated with different doses of the 5-aza-CdR. Results from these studies showed a significant (P < .05) dose-dependent increase in the uPA expression at 1.0 and 5.0 µM concentrations of 5-aza-CdR (Figure 1 D). At these doses, more than 95% of tumor cells were viable. This change in the expression of uPA had a dosedependent increase in tumor cells' invasive capacity as determined by the Boyden chamber Matrigel invasion assay (Figure 1D). The effect of 5-aza-CdR on cell migration was analyzed using MCF-7 cells, rather than ZR-75-1 cells that grow in patches. A significant (P < .05) increase in wound healing (%) was observed in 5-aza-CdR-treated (5 µM) MCF-7 cells compared with control cells at 32 and 48 hours after wounding (Figure 1, E and F). Healing at the lower dose of 5-aza-CdR (1 µM) was also found to be significant (P < .05) compared with control MCF-7 cells. For both, control and 5-aza-CdR-treated groups, the rate of wound closure was highest at 24 and 32 hours after wounding. Migration assay on ZR-75-1 cells was not feasible, because these cells grow in patches and are unable to form a uniform monolayer.
Effect of 5-Aza-CdR on the Genes Involved in Invasion and Metastasis
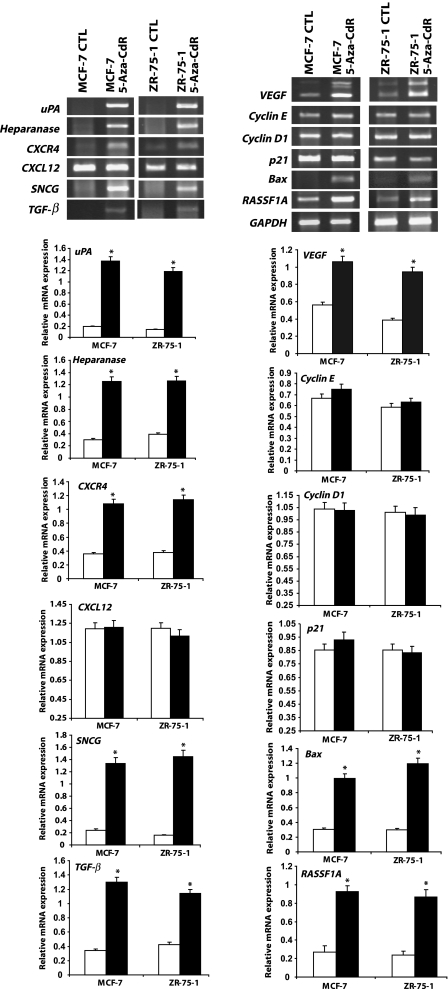
What is the mechanism by which 5-aza-CdR stimulates cell invasion and migration? We tested the hypothesis that 5-aza-CdR induces a battery of genes, which were previously reported to be involved in cell invasion and metastasis, including uPA, SNCG, HEPARANASE, CXCR4, and CXCL12. MCF-7 and ZR-75-1 cells were treated with 5 µM 5-aza-CdR for a duration of 7 days. At the end of the experiment, total cellular RNA was isolated and analyzed for the expression of mRNA levels of these genes as well as the tumor suppressor gene, RASSF1A, which is hypermethylated in 42% to 65% of primary breast tumors, including these cell lines [26]. We found that, as reported, 5-aza-CdR treatment upregulated the expression of the tumor suppressor gene, RASSF1A, in both cell lines. However, in addition to the tumor suppressor gene, a number of genes demonstrated to play a causal role in metastasis were induced as well, namely uPA, SNCG, HEPARANASE, the chemokine receptor CXCR4, the cytokine TGF-β, and the angiogenesis factor VEGF (Figure 2). There is no significant difference in CXCL12 or CYCLIN D1 and CYCLIN E1 mRNA expression in control and 5-aza-CdR-treated groups of either cell lines. Thus, our RT-PCR data confirm that 5-aza-CdR is responsible for the activation of quiescent genes or upregulation of a panel of prometastatic genes that are normally suppressed in these noninvasive cell lines concurrently with its activation of tumor suppressor genes.
Figure 2.
Treatment of MCF-7 and ZR-75-1 cells with 5-aza-CdR activates tumor suppressor and prometastatic genes. MCF-7 and ZR-75-1 cells were treated with 5µM 5-aza-CdR for 7 days, and at the end of the experiment, total cellular RNA was isolated with TRIzol. RNA from control and treatment groups were analyzed for the expression of genes involved in invasion, metastasis, cell cycle regulation, angiogenesis, and apoptosis by RT-PCR. The expression of tumor suppressor gene, RASSF1A, was also analyzed. Changes in the mRNA expression of the respective genes were determined by plotting the relative ratio against GAPDH of at least two different experiments, where white bars represent controls and solid black bars represent 5-aza-CdR-treated groups. Significant difference from the control is represented by an asterisk (*P < .05).
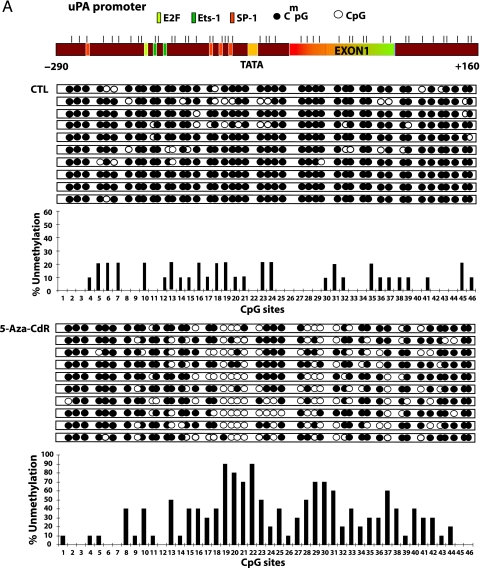
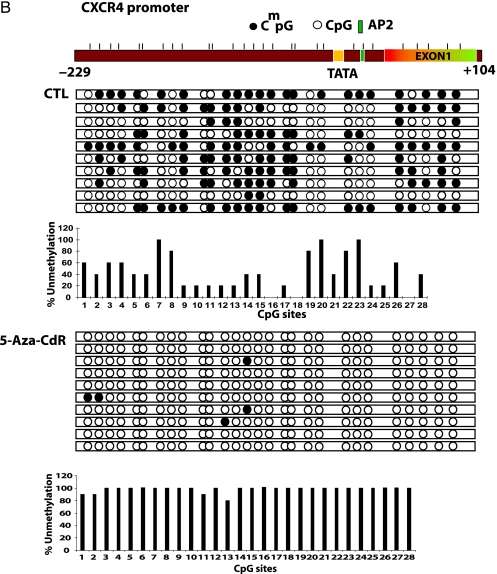
Effect of 5-Aza-CdR on the Methylation Status of the uPA, CXCR4, and SNCG Gene Promoters
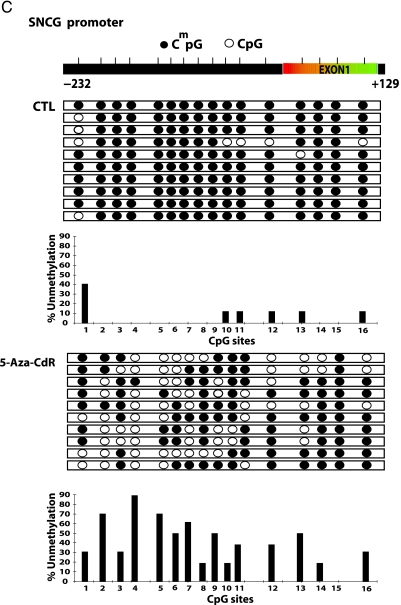
To test the hypothesis that the increase in mRNA expression of these genes is due to concurrent demethylation in the promoter and exon 1 region of these genes, we subjected genomic DNA from treated and untreated cells to sodium bisulfite mapping of the indicated regulatory regions of uPA, SNCG, and CXCR4 promoters (Figure 3 A). Demethylation of specific CpG sites after 5-aza-CdR treatment was noted for these three genes. The uPA promoter was highly methylated (81.8%) in the MCF-7 control; significant demethylation occurs at an SP1-binding site and four CpG sites located between TATA box and exon 1 as well as extensive demethylation of exon 1. CXCR4 regulatory region (-260 to +150) was almost totally demethylated by 5-aza-CdR (Figure 3 B). The CpG site numbers 2 to 14 and 16 on the SNCG promoter region are more prone to demethylation after 5-aza-CdR treatment, consistent with the possibility that demethylation of these sites may play an important role in the induction of this gene (Figure 3 C).
Figure 3.
Treatment with 5-aza-CdR causes demethylation of the promoter region of the uPA, CXCR4, and SNCG genes. MCF-7 cells were subjected to 5-aza-CdR treatment (5 µM) for 7 days, at which point genomic DNA was isolated with DNazol and subjected to bisulfite treatment as described in the Materials and Methods section. Genomic bisulfite-treated DNA was used for first PCR and nested PCR reactions to amplify uPA, CXCR4, and SNCG genes' 5′ regulatory regions and the first exon, followed by cloning in pGEMT vector. Ten clones for each gene were picked from control and treatment groups. A physical map of the uPA (A), CXCR4 (B), and SNCG (C) promoters is presented. Demethylated CpG site (empty circle) and methylated CpG site (filled circle). Beneath the clones is the bar diagram showing percent unmethylation for each CpG site present within the amplified region.
Effect of 5-Aza-CdR on In Vivo Tumor Growth
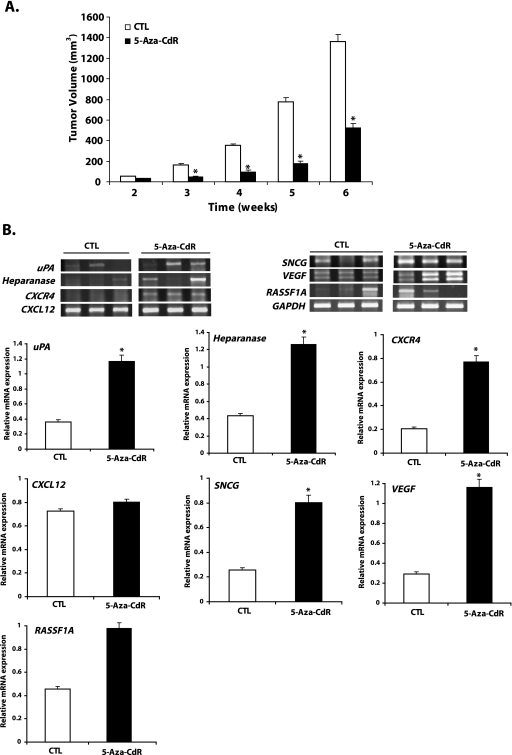
Following the in vitro data indicating that 5-aza-CdR induces a battery of genes involved in metastasis as well as tumor suppressor genes, we tested whether 5-aza-CdR-treated cells will exhibit this dual response; that is, reduced tumor growth and increased prometastasis gene expression in vivo. 5-Aza-CdR-treated (5 µM for 7 days in vitro) MCF-7 cells and control cells were inoculated in the mammary fat pad of female Balb C nu/nu mice in the presence of an estrogen pellet. As depicted in Figure 4 A, 5-aza-CdR-treated MCF-7 cells showed a reduced rate of increase in tumor volume compared with the control group of animals inoculated with MCF-7 cells treated with vehicle alone. Because 5-aza-CdR treatment was transient and the animals were not further treated with 5-aza-CdR after inoculation, we confirmed by RT-PCR that the genes (uPA, HEPARANASE, CXCR4, SNCG, and VEGF) induced by 5-aza-CdR in vitro were still induced in these tumors (Figure 4 B).
Figure 4.
5-Aza-CdR suppresses tumor growth and upregulates the expression of prometastatic, angiogenetic, and tumor suppressor genes in primary tumor. Female Balb C nu/nu mice were inoculated with either control MCF-7 cells or MCF-7 cells treated with 5 µM 5-aza-CdR for 7 days in the mammary fat pad region in the presence of 0.25 mg of estradiol pellet for a 60-day release. Tumors were measured weekly, and tumor volume was determined as described in the Materials and Methods section. Result represents the mean ± SEM of eight animals in each group. Significant difference from the control is represented by an asterisk (*P < .05) (A). At the termination of the in vivo experiment (week 6), primary tumors were excised and snap-frozen. Total cellular RNA was isolated from these tumors using TRIzol and was analyzed by RT-PCR for the expression of genes responsible for invasion and metastases (uPA, HEPARANASE, CXCR4, CXCL12, and SNCG) and angiogenesis (VEGF) and for the tumor suppressor gene (RASSF1A) as shown (representative three tumor samples from each group) in panel B. The bar diagram represents the mean ± SEM of tumors from three animals in each group (white bars represent mean ± SEM of controls and solid black bars represent mean ± SEM of the 5-aza-CdR-treated group). Significant difference from the control is represented by an asterisk (*P < .05).
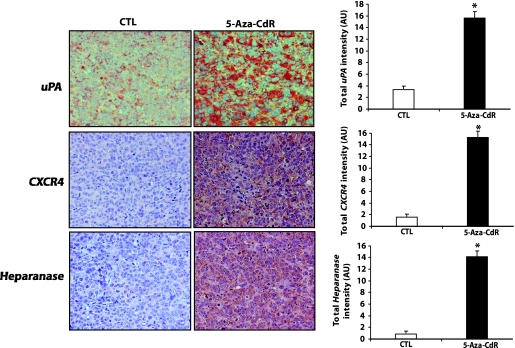
We then measured the levels of expression of uPA, CXCR4, and HEPARANASE, which are key players in tumor invasion and metastases, by immunohistochemistry in control tumors and tumors from animals inoculated with cells treated with 5-aza-CdR. We found that all tumors from the treatment group showed overexpression of uPA, CXCR4, and HEPARANASE both in terms of intensity and density compared with tumors from the control animals (Figure 5). Our data showed that genes induced by 5-aza-CdR in vitro are not changed after inoculating these cells in the in vivo model, both at the mRNA and protein levels.
Figure 5.
5-Aza-CdR alters the expression of invasive and prometastatic markers in the primary tumor tissues. At the termination of the in vivo experiment, primary tumors were excised and processed for immunohistochemical analyses as described in the Materials and Methods section to determine the levels of production of uPA, CXCR4, and HEPARANASE. A representative microphotograph for each group is shown; original magnification, x200. The intensity and density of positive staining was quantified as described in the Materials and Methods section. Ten random fields of observation were recorded in each group and were represented graphically as arbitrary units. Results represent the mean ± SEM of three animals from each group. Significant difference from the control is represented by an asterisk (*P < .05).
Discussion
Disruption in the DNA methylation machinery resulting in global hypomethylation [37] and regional hypermethylation [1] is well documented in most cancers. However, most of the focus of attention in the field has been on hypermethylation and silencing of tumor suppressor genes, leading to a number of clinical trials with the DNA methylation inhibitor 5-aza-CdR [7,38]. This approach is effective in inhibiting tumor growth, and it has shown promising results in the treatment of myelodysplastic syndromes [38,39]. However, this approach overlooks the other side of the methylation tale in cancer: global hypomethylation. If global hypomethylation plays a causal role in the cancer, then agents which globally inhibit DNA methylation, such as 5-aza-CdR, might have undesirable effects after their use, which can potentially affect cancer progression [24]. An important requirement in developing effective cancer therapeutics is to block not only primary tumor growth but also tumor metastases to distant organs, which is the major cause of cancer-associated morbidity and mortality [40]. We have previously shown that DNA methylation plays divergent roles in growth and metastasis in breast cancer cells [25]. Recent data implicating hypomethylation in the activation of certain prometastatic genes, such as uPA [22,23,25], in invasive breast cancer prompted us to test the hypothesis that inhibitors of DNA methylation would promote the transition of non-metastatic breast cancer cells into metastatic cancer by inducing a panel of genes required for metastasis.
Our study tested the hypothesis that hypomethylation coregulates a panel of genes involved in metastasis by a common activation mechanism rather than a specific gene. If this hypothesis is true, it has obvious important therapeutic implications. First, it draws attention to the potential danger in using demethylating agents in anticancer therapy because of their potential effects on activating a large number of quiescent genes including genes involved in promoting tumor metastases. Nevertheless, it puts forward the opportunity of an opposite approach, namely, inhibition of hypomethylation by hypermethylation therapy [23]. Such therapy might provide a unique opportunity for knocking out a whole panel of genes involved in metastasis, thus increasing the chances for successfully blocking metastasis in contrast to therapies, which target one protein at a time. Redundancy in activity of these proteins might require a therapeutic approach that coordinately targets a panel of genes involved in tumor progression.
Our data support this hypothesis. First, we show that 5-aza-CdR concurrently activates the tumor suppressor gene, RASSF1A, and a panel of prometastatic genes (Figure 2). We also show that the mechanism of action of 5-aza-CdR on three selected genes, namely, uPA, CXCR4, and SNCG, involves demethylation of their regulatory regions. These genes, uPA, CXCR4, and SNCG, were previously shown to be regulated by hypomethylation in other systems; however, here we show their simultaneous activation in the same noninvasive breast cancer cells with 5-aza-CdR [22,23,29,30,41,42].
Recently, a study has shown that overexpression of the metastatic gene, HEPARANASE, in MDA-MB-435 human breast carcinoma and rat C6 glioma cells resulted in a three- to six-fold increase in VEGF mRNA levels, which is mediated by the activation of Src family members [43]. In the present study, we also found the upregulation of HEPARANASE accompanied by the overexpression of VEGF mRNA level in 5-aza-CdR-treated MCF-7 cells. However, it is unclear whether this increase in VEGF expression is due to epigenetic regulation or is an outcome of HEPARANASE expression. It has been demonstrated that exogenous expression of SNCG in breast cancer cells (MDA-MB-435) led to a significant increase in cell motility and invasiveness in cell culture and to a profound augmentation of metastasis in nude mice [17]. The other genes of interest were CXCR4 and CXCL12; CXCR4 is a Gi protein-coupled receptor for the ligand CXCL12 and plays an important role in breast cancer metastasis. Chemokine receptors, such as CXCR4 and CXCR7, play an important role in cell migration and metastasis [42]. Recently, it has been shown that elevated secretion of CCL2 by bone marrow endothelial cells recruits prostate cancer epithelial cells to the bone microenvironment [44]. In parallel, Kollmer et al. [45] has also shown that the chemokine CXCL12 promotes solid metastatic tumor growth by pro-proliferative and antiapoptotic actions in an angiogenesis-dependent manner.
Another ubiquitous cytokine TGF-β that is found to be upregulated in our findings is involved in bone metastasis and inhibits growth of normal epithelia and early-stage tumors but stimulates invasion and metastasis of aggressive tumors [46]. Although in some cases, the loss of growth inhibition by TGF-β can arise because of mutational inactivation of the TGF-β receptors and SMAD signaling molecule [47] or might be due to hypermethylation. The effect of TGF-β on tumor growth is biphasic: carcinogenesis and early tumor growth are suppressed by TGF-β, whereas this growth factor apparently accelerates tumor progression in more advanced aggressive tumors. Thus, it presents another example of divergent effects on growth and metastasis in breast cancer progression. Most importantly, the high level of uPA in serum and extracts from the primary tumors are associated with poor prognosis and overall survival. In fact, uPA has been proven to be an important marker of the metastatic potential in human cancers [23,27,48,49]. uPA is required to generate the active zymogen plasmin necessary for the degradation of extracellular matrix and to promote metastasis, and there are reports where tumor metastasis was selectively reduced up to more than seven-fold in uPA-deficient mice [50].
The uPA promoter region we have evaluated has many transcription factor-binding sites, namely, ETS-1, AP-2, and SP-1, which may play important roles in the regulation of uPA expression. In previous studies, using mobility shift assay, we have shown that the transcription factor Ets-1 is involved in inducing uPA gene expression. This transcription factor-binding site coincides with methylation sites and is a candidate site that can be affected by DNA methylation [21]. In addition, uPA gene activation by 5-aza-CdR can also occur indirectly by additional transcription factors or by TGF-β which is known to regulate the production of uPA through the stabilization of uPA mRNA and through the SMAD-4 signaling pathway [51]. Furthermore, integrin-linked kinase, which is critical for TGF-β1-stimulated tumor cell-invasiveness, might also serve as a key signaling mediator of TGF-β regulation of the uPA/plasminogen activator inhibitor-1 system.
The main question with relevance to the clinical situation was whether 5-aza-CdR-treated breast cancer cells would metastasize in vivo. We opted to treat the cells in vitro and then inoculate them in vivo rather than treat systematically with 5-aza-CdR to measure the direct effect of the DNA-demethylating agent on the cancer cell itself without the confounding factors of the systemic effects of 5-aza-CdR and its potential effects on the stroma. In addition, this paradigm allows us to evaluate the long-term effects of transient 5-aza-CdR treatment. Because the action of 5-aza-CdR results in a stable change in the state of modification of the DNA molecule per se, we predicted that the early effects of 5-aza-CdR treatment would linger for the entire period of time of tumor growth in vivo well after the 5-aza-CdR was washed away. Our data show that while 5-aza-CdR decreases tumor growth in vivo, it also upregulates the expression of many prometastatic genes in the noninvasive breast cancer cells in vitro and in the primary tumors in vivo, which may result in metastases at the distant organ sites of these animals over time. The effects of 5-aza-CdR last long after the 5-aza-CdR is withdrawn, supporting the hypothesis that the effects of 5-aza-CdR are mediated through reprogramming of gene expression, which is consistent with a mechanism involving the change in DNA methylation rather than in an immediate nonspecific effect of the drug which is independent of DNA methylation. Because of the nonmetastatic behavior of MCF-7 cells, we were unable to conclusively demonstrate the ability of 5-aza-CdR to promote tumor metastases in this short-term in vivo model of breast cancer.
Altering the DNA methylation machinery is a potentially powerful approach to cancer therapy [52]. However, it is crucial to gain full understanding of the mechanisms involved in these two processes, namely, hypermethylation and demethylation, to properly use these drugs targeting the DNA methylation machinery in anticancer therapy. Because metastases is one of the most serious facets of cancer, and it plays a critical role in mortality from cancer, high vigilance is warranted when using demethylating agents in cancer therapy. Our challenge is to design agents that target the growth-promoting effect of DNA methyltransferases but are devoid of the hypomethylation action of critical prometastatic genes.
Footnotes
This work was supported by a grant from the Canadian Institutes for Health Research (MOP 12609). M.S. was supported by a grant from the National Cancer Institute of Canada.
References
- 1.Baylin SB, Esteller M, Rountree MR, Bachman KE, Schuebel K, Herman JG. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum Mol Genet. 2001;10:687–692. doi: 10.1093/hmg/10.7.687. [DOI] [PubMed] [Google Scholar]
- 2.MacLeod AR, Szyf M. Expression of antisense to DNA methyltransferase mRNA induces DNA demethylation and inhibits tumorigenesis. J Biol Chem. 1995;270:8037–8043. doi: 10.1074/jbc.270.14.8037. [DOI] [PubMed] [Google Scholar]
- 3.Ramchandani S, MacLeod AR, Pinard M, von Hofe E, Szyf M. Inhibition of tumorigenesis by a cytosine-DNA, methyltransferase, antisense oligodeoxynucleotide. Proc Natl Acad Sci USA. 1997;94:684–689. doi: 10.1073/pnas.94.2.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laird PW, Jackson-Grusby L, Fazeli A, Dickinson SL, Jung WE, Li E, Weinberg RA, Jaenisch R. Suppression of intestinal neoplasia by DNA hypomethylation. Cell. 1995;81:197–205. doi: 10.1016/0092-8674(95)90329-1. [DOI] [PubMed] [Google Scholar]
- 5.Merlo A, Herman JG, Mao L, Lee DJ, Gabrielson E, Burger PC, Baylin SB, Sidransky D. 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med. 1995;1:686–692. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- 6.Kuykendall JR. 5-Azacytidine and decitabine monotherapies of myelodysplastic disorders. Ann Pharmacother. 2005;39:1700–1709. doi: 10.1345/aph.1E612. [DOI] [PubMed] [Google Scholar]
- 7.Issa JP, Byrd JC. Decitabine in chronic leukemias. Semin Hematol. 2005;42:S43–S49. doi: 10.1053/j.seminhematol.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 8.Momparler RL. Epigenetic therapy of cancer with 5-aza-2′-deoxycytidine (decitabine) Semin Oncol. 2005;32:443–451. doi: 10.1053/j.seminoncol.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 9.Weigelt B, Peterse JL, van't Veer LJ. Breast cancer metastasis: markers and models. Nat Rev Cancer. 2005;5:591–602. doi: 10.1038/nrc1670. [DOI] [PubMed] [Google Scholar]
- 10.Andreasen PA, Kjoller L, Christensen L, Duffy MJ. The urokinasetype plasminogen activator system in cancer metastasis: a review. Int J Cancer. 1997;72:1–22. doi: 10.1002/(sici)1097-0215(19970703)72:1<1::aid-ijc1>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 11.Shteper PJ, Zcharia E, Ashhab Y, Peretz T, Vlodavsky I, Ben-Yehuda D. Role of promoter methylation in regulation of the mammalian heparanase gene. Oncogene. 2003;22:7737–7749. doi: 10.1038/sj.onc.1207056. [DOI] [PubMed] [Google Scholar]
- 12.Joyce JA, Freeman C, Meyer-Morse N, Parish CR, Hanahan D. A functional heparan sulfate mimetic implicates both heparanase and heparan sulfate in tumor angiogenesis and invasion in a mouse model of multistage cancer. Oncogene. 2005;24:4037–4051. doi: 10.1038/sj.onc.1208602. [DOI] [PubMed] [Google Scholar]
- 13.Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 14.Nawshad A, Lagamba D, Polad A, Hay ED. Transforming growth factor-beta signaling during epithelial-mesenchymal transformation: implications for embryogenesis and tumor metastasis. Cells Tissues Organs. 2005;179:11–23. doi: 10.1159/000084505. [DOI] [PubMed] [Google Scholar]
- 15.Elliott RL, Blobe GC. Role of transforming growth factor Beta in human cancer. J Clin Oncol. 2005;23:2078–2093. doi: 10.1200/JCO.2005.02.047. [DOI] [PubMed] [Google Scholar]
- 16.Jenkinson SR, Barraclough R, West CR, Rudland PS. S100A4 regulates cell motility and invasion in an in vitro model for breast cancer metastasis. Br J Cancer. 2004;90:253–262. doi: 10.1038/sj.bjc.6601483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jia T, Liu YE, Liu J, Shi YE. Stimulation of breast cancer invasion and metastasis by synuclein gamma. Cancer Res. 1999;59:742–747. [PubMed] [Google Scholar]
- 18.Burger JA, Kipps TJ. CXCR4: a key receptor in the crosstalk between tumor cells and their microenvironment. Blood. 2006;107:1761–1767. doi: 10.1182/blood-2005-08-3182. [DOI] [PubMed] [Google Scholar]
- 19.Kucia M, Jankowski K, Reca R, Wysoczynski M, Bandura L, Allendorf DJ, Zhang J, Ratajczak J, Ratajczak MZ. CXCR4-SDF-1 signalling, locomotion, chemotaxis and adhesion. J Mol Histol. 2004;35:233–245. doi: 10.1023/b:hijo.0000032355.66152.b8. [DOI] [PubMed] [Google Scholar]
- 20.Zlotnik A. Chemokines in neoplastic progression. Semin Cancer Biol. 2004;14:181–185. doi: 10.1016/j.semcancer.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 21.Guo Y, Pakneshan P, Gladu J, Slack A, Szyf M, Rabbani SA. Regulation of DNA methylation in human breast cancer. Effect on the urokinasetype plasminogen activator gene production and tumor invasion. J Biol Chem. 2002;277:41571–41579. doi: 10.1074/jbc.M201864200. [DOI] [PubMed] [Google Scholar]
- 22.Pakneshan P, Xing RH, Rabbani SA. Methylation status of uPA promoter as a molecular mechanism regulating prostate cancer invasion and growth in vitro and in vivo. FASEB J. 2003;17:1081–1088. doi: 10.1096/fj.02-0973com. [DOI] [PubMed] [Google Scholar]
- 23.Pakneshan P, Szyf M, Farias-Eisner R, Rabbani SA. Reversal of the hypomethylation status of urokinase (uPA) promoter blocks breast cancer growth and metastasis. J Biol Chem. 2004;279:31735–31744. doi: 10.1074/jbc.M401669200. [DOI] [PubMed] [Google Scholar]
- 24.Szyf M, Pakneshan P, Rabbani SA. DNA demethylation and cancer: therapeutic implications. Cancer Lett. 2004;211:133–143. doi: 10.1016/j.canlet.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 25.Pakneshan P, Szyf M, Rabbani SA. Methylation and inhibition expression of uPA by the RAS oncogene: divergence of growth control and invasion in breast cancer cells. Carcinogenesis. 2005;26:557–564. doi: 10.1093/carcin/bgi009. [DOI] [PubMed] [Google Scholar]
- 26.Agathanggelou A, Honorio S, Macartney DP, Martinez A, Dallol A, Rader J, Fullwood P, Chauhan A, Walker R, Shaw JA, et al. Methylation associated inactivation of RASSF1A from region 3p21.3 in lung, breast and ovarian tumours. Oncogene. 2001;20:1509–1518. doi: 10.1038/sj.onc.1204175. [DOI] [PubMed] [Google Scholar]
- 27.Duffy MJ. Urokinase-type plasminogen activator: a potent marker metastatic potential in human cancers. Biochem Soc Trans. 2002;30:207–210. doi: 10.1042/. [DOI] [PubMed] [Google Scholar]
- 28.Simizu S, Ishida K, Wierzba MK, Sato TA, Osada H. Expression heparanase in human tumor cell lines and human head and neck tumors. Cancer Lett. 2003;193:83–89. doi: 10.1016/s0304-3835(02)00719-x. [DOI] [PubMed] [Google Scholar]
- 29.Liu H, Liu W, Wu Y, Zhou Y, Xue R, Luo C, Wang L, Zhao W, Jiang JD, Liu J. Loss of epigenetic control of synuclein-gamma gene as a molecular indicator of metastasis in a wide range of human cancers. Cancer Res. 2005;65:7635–7643. doi: 10.1158/0008-5472.CAN-05-1089. [DOI] [PubMed] [Google Scholar]
- 30.Gupta A, Godwin AK, Vanderveer L, Lu A, Liu J. Hypomethylation of the synuclein gamma gene CpG island promotes its aberrant expression breast carcinoma and ovarian carcinoma. Cancer Res. 2003;63:664–673. [PubMed] [Google Scholar]
- 31.Tsutsui S, Inoue H, Yasuda K, Suzuki K, Takeuchi H, Nishizaki T, Higashi H, Era S, Mori M. Angiopoietin 2 expression in invasive ductal carcinoma of the breast: its relationship to the VEGF expression and microvessel density. Breast Cancer Res Treat. 2006;98:261–266. doi: 10.1007/s10549-005-9157-9. [DOI] [PubMed] [Google Scholar]
- 32.Cheng M, Olivier P, Diehl JA, Fero M, Roussel MF, Roberts JM, Sherr CJ. The p21(Cip1) and p27(Kip1) CDK “inhibitors” are essential activators of cyclin D-dependent kinases in murine fibroblasts. EMBO J. 1999;18:1571–1583. doi: 10.1093/emboj/18.6.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emi M, Kim R, Tanabe K, Uchida Y, Toge T. Targeted therapy against Bcl-2-related proteins in breast cancer cells. Breast Cancer Res. 2005;7:R940–R952. doi: 10.1186/bcr1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vos MD, Dallol A, Eckfeld K, Allen NP, Donninger H, Hesson LB, Calvisi D, Latif F, Clark GJ. The RASSF1A tumor suppressor activates Bax via MOAP-1. J Biol Chem. 2006;281:4557–4563. doi: 10.1074/jbc.M512128200. [DOI] [PubMed] [Google Scholar]
- 35.Inoue S, Hartman A, Branch CD, Bucana CD, Bekele BN, Stephens LC, Chada S, Ramesh R. mda-7 In combination with bevacizumab treatment produces a synergistic and complete inhibitory effect on lung tumor xenograft. Mol Ther. 2007;15:287–294. doi: 10.1038/sj.mt.6300035. [DOI] [PubMed] [Google Scholar]
- 36.Schmelz K, Wagner M, Dorken B, Tamm I. 5-Aza-2′-deoxycytidine induces p21WAF expression by demethylation of p73 leading to p53-independent apoptosis in myeloid leukemia. Int J Cancer. 2005;114:683–695. doi: 10.1002/ijc.20797. [DOI] [PubMed] [Google Scholar]
- 37.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 38.Fenaux P. Inhibitors of DNA methylation: beyond myelodysplastic syndromes. Nat Clin Pract Oncol. 2005;2(Suppl 1):S36–S44. doi: 10.1038/ncponc0351. [DOI] [PubMed] [Google Scholar]
- 39.Rosenfeld CS. Clinical development of decitabine as a prototype for an epigenetic drug program. Semin Oncol. 2005;32:465–472. doi: 10.1053/j.seminoncol.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 40.van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 41.Zhao W, Liu H, Liu W, Wu Y, Chen W, Jiang B, Zhou Y, Xue R, Luo C, Wang L, et al. Abnormal activation of the synuclein-gamma gene in hepatocellular carcinomas by epigenetic alteration. Int J Oncol. 2006;28:1081–1088. [PubMed] [Google Scholar]
- 42.Mori T, Kim J, Yamano T, Takeuchi H, Huang S, Umetani N, Koyanagi K, Hoon DS. Epigenetic up-regulation of C-C chemokine receptor 7 and C-X-C chemokine receptor 4 expression in melanoma cells. Cancer Res. 2005;65:1800–1807. doi: 10.1158/0008-5472.CAN-04-3531. [DOI] [PubMed] [Google Scholar]
- 43.Zetser A, Bashenko Y, Edovitsky E, Levy-Adam F, Vlodavsky I, Ilan N. Heparanase induces vascular endothelial growth factor expression: correlation with p38 phosphorylation levels and Src activation. Cancer Res. 2006;66:1455–1463. doi: 10.1158/0008-5472.CAN-05-1811. [DOI] [PubMed] [Google Scholar]
- 44.Loberg RD, Day LL, Harwood J, Ying C, St John LN, Giles R, Neeley CK, Pienta KJ. CCL2 is a potent regulator of prostate cancer cell migration and proliferation. Neoplasia. 2006;8:578–586. doi: 10.1593/neo.06280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kollmar O, Rupertus K, Scheuer C, Junker B, Tilton B, Schilling MK, Menger MD. Stromal Cell-Derived Factor-1 promotes cell migration and tumor growth of colorectal metastasis. Neoplasia. 2007;9:862–870. doi: 10.1593/neo.07559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yin JJ, Selander K, Chirgwin JM, Dallas M, Grubbs BG, Wieser R, Massague J, Mundy GR, Guise TA. TGF-beta signaling blockade inhibits PTHrP secretion by breast cancer cells and bone metastases development. J Clin Invest. 1999;103:197–206. doi: 10.1172/JCI3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kretzschmar M. Transforming growth factor-beta and breast cancer: transforming growth factor-beta/SMAD signaling defects and cancer. Breast Cancer Res. 2000;2:107–115. doi: 10.1186/bcr42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duffy MJ, Maguire TM, McDermott EW, O'Higgins N. Urokinase plasminogen activator: a prognostic marker in multiple types of cancer. J Surg Oncol. 1999;71:130–135. doi: 10.1002/(sici)1096-9098(199906)71:2<130::aid-jso14>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 49.Pakneshan P, Tetu B, Rabbani SA. Demethylation of urokinase promoter as a prognostic marker in patients with breast carcinoma. Clin Cancer Res. 2004;10:3035–3041. doi: 10.1158/1078-0432.ccr-03-0545. [DOI] [PubMed] [Google Scholar]
- 50.Almholt K, Lund LR, Rygaard J, Nielsen BS, Dano K, Romer J, Johnsen M. Reduced metastasis of transgenic mammary cancer in urokinase-deficient mice. Int J Cancer. 2005;113:525–532. doi: 10.1002/ijc.20631. [DOI] [PubMed] [Google Scholar]
- 51.Shiou SR, Datta PK, Dhawan P, Law BK, Yingling JM, Dixon DA, Beauchamp RD. Smad4-dependent regulation of urokinase plasminogen activator secretion and RNA stability associated with invasiveness by autocrine and paracrine transforming growth factor-beta. J Biol Chem. 2006;281:33971–33981. doi: 10.1074/jbc.M607010200. [DOI] [PubMed] [Google Scholar]
- 52.Szyf M. Towards a pharmacology of DNA methylation. Trends Pharmacol Sci. 2001;22:350–354. doi: 10.1016/s0165-6147(00)01713-2. [DOI] [PubMed] [Google Scholar]