Abstract
The purpose of this study was to investigate whether detectable protein biomarker overexpression is a prerequisite for the presence of increased gene copy number or activating mutations and responsiveness to the epidermal growth factor receptor (EGFR) inhibitors gefitinib and erlotinib in patients with lung adenocarcinomas. EGFR status was prospectively analyzed in tumor biopsy samples by three methods: protein expression (n = 117) by standardized immunohistochemistry (IHC), gene copy number (n = 97) by fluorescent in situ hybridization (FISH), and mutation analysis by sequencing (n = 126). Fifty-nine percent of the samples were positive by IHC, 40% were positive by FISH, and 13.5% contained activating kinase domain mutations. Thirty-four percent of the FISH-positive and 27% of the mutant samples were also IHC-negative. All EGFR mutant patients had major clinical responses (five complete response and five partial response) to gefitinib or erlotinib treatment, although three of these tumors were IHC-negative and four were FISH-negative. In a retrospective analysis of samples from nine patients with excellent therapeutic responses (three complete response, five partial response, one stable disease) to erlotinib or gefitinib, mutations were identified in eight cases, but IHC was negative in four of these tumors. These results indicate that molecular diagnostic methods appear to be most important for the identification of lung adenocarcinoma patients who may benefit from EGFR inhibitor treatments.
The detectable expression of the target protein of novel molecular targeted drugs is often assumed to both sufficient and necessary for predicting the efficacy of these novel drugs. The clinical experience with anti-epidermal growth factor receptor (EGFR) antibody therapy cetuximab in the treatment of colon carcinoma patients revealed that not only not all EGFR-expressing tumors responded, but more surprisingly, tumors without detectable EGFR expression by standard immunohistochemistry (IHC) had similar clinical response to anti-EGFR therapy.1 The oncogenic dependence on a signal transduction molecule may be more dependent on the genetic changes in the target gene itself or downstream signal proteins like RAS, in the case of anti-EGFR therapies.2
Gefitinib and erlotinib are small-molecule inhibitors of the tyrosine kinase domain (TKI) of the EGFR. These EGFR TKIs have an objective response rate of 9 to 19%, mild side effects, and in some patients there was rapid and dramatic tumor shrinkage.3,4,5,6,7,8,9 Biomarkers and clinical characteristics with reliable predictive value remain the focus of several investigations. Adenocarcinoma histology, nonsmoking history, Asian race, and female gender were the characteristics that were associated with increased response to both EGFR TKIs.3,4,5,6,8 Mutations in the tyrosine kinase domain of EGFR were reported in the majority of tumors with dramatic responses to gefitinib and erlotinib,10,11,12 and in some series, the presence of mutations was associated with improved survival.13,14,15,16,17,18 EGFR mutations were more common in patients with the same clinical characteristics as those associated with better treatment response. The latest advances in research of biological and clinical relevance of activating mutations have been reviewed recently.3 The frequencies of mutations in lung adenocarcinomas were 22 to 67% in Asia, 3 to 25% in North America, and 10 to 24% in South Europe.11,13,14,15,17,18,19,20,21 The prevalence of EGFR gene mutations and copy number alterations in Eastern and Central Europe has not been published.
EGFR gene copy number, detected by fluorescent in situ hybridization (FISH), is also associated with response to gefitinib. Gefitinib-treated patients carrying EGFR gene amplification or high polysomy (FISH+) had a statistically significant improvement in response, time to progression, and survival compared with patients with no or low genomic gain for EGFR.22 The efficiency of this molecular predictive marker was confirmed on a subgroup of samples of phase II study of gefitinib (S0126) and on subgroup of specimens of phase III study of erlotinib (BR.21).23,24
Although initial retrospective studies had suggested that the protein expression is not associated with gefitinib response,25,26,27 two subsequent studies reported longer survival among TKI-treated patients with protein overexpression detected by IHC.23,24 In the BR.21 study, survival among patients with protein overexpression (50–55%) was longer in the erlotinib group than in the placebo group, but there was no survival advantage among patients with EGFR IHC-negative tumors, although the P value for interaction was 0.25, which indicates very low level of statistical significance.24 In addition, recent preclinical studies in cell lines did not find correlation between EGFR sensitivity and EGFR protein expression.28 In 2004 our group identified the simultaneous presence of amplification and mutation of the EGFR gene and overexpression of the EGFR protein in primary non-small cell lung cancer (NSCLC) with complete regression of brain and lung metastases in response to gefitinib.7
Due to the lack of consensus on the significance of predictive diagnostic tests, in particular the mutation tests, clinical oncologists both in the United States and the European Union most often rely on the IHC of EGFR for patient selection. This decision is based on the assumption that detection of the molecular target protein is the most reliable way to use a molecular targeted therapeutic drug. In addition, it is also assumed that the IHC-positive population includes the smaller patient populations of FISH-positive and tumors with activating EGFR mutations.
In this study we used the standardized PharmDx (Dako) IHC kit to analyze EGFR expression by IHC in large set of NSCLC samples. This is the most commonly used IHC test for EGFR, and this was the clinical trial assay in the BR21 study that led to the market authorization of erlotinib. We also analyzed gene copy number by FISH using the most standard probes (Vysis) and the presence of activating mutations by the gold standard method of bidirectional sequencing. The clinical significance of EGFR mutations in response to gefitinib has been far more studied (over 20 publications) than in response to erlotinib (three publications).3 The BR21 clinical trial included only eight erlotinib-treated patients with classic EGFR mutations. In this study we provide clinical data of 14 erlotinib-treated patients with EGFR mutations both IHC-positive and -negative.
Our study revealed that the positive patient populations of these common EGFR biomarkers do not overlap. Almost half of the FISH-positive and mutant samples were IHC-negative. We also found examples of NSCLC patients with IHC-negative but EGFR mutant NSCLC tumors who had complete responses to erlotinib treatment.
Materials and Methods
Tumor Specimens
One hundred twenty-seven primary NSCLC tissues prospectively before and a further nine samples retrospectively after EGFR TKI therapy were examined in the same laboratory throughout a 1-year period. Each specimen was reviewed by a pathologist (P.J.), and only those with ≥30% tumor component were used for DNA mutation analysis in the prospective examination. In retrospective analysis two specimens with <30% tumor component were reanalyzed by mutant-enriched polymerase chain reaction (PCR) and sequencing. For treatment, 250 mg of gefitinib or 150 mg of erlotinib was administered daily. Tumor response was evaluated in accordance with Response Evaluation Criteria in Solid Tumors (RECIST).
PCR and Sequencing
After DNA extraction from the paraffin-embedded specimens and biopsy smears, exons 18, 19, and 21 of the EGFR gene were amplified using nested PCR with “touch-down” protocol. Primers are shown in Table 1. The universal amplification protocol was as follows: 95°C for 2 minutes, 10 cycles of 94°C for 30 seconds, 62°C decreased with 1°C in each cycle for 30 seconds and 72°C for 45 seconds, 30 cycles of 94°C for 30 seconds, 52°C for 30 seconds and 72°C for 45 seconds, closing cycle of 72°C for 10 minutes. After purification and bidirectional sequencing reactions using the second step primers, sequencing fragments were detected with ABI Prism 310 Genetic Analyzer (Applied Biosystems, Foster City, CA). High-quality sequence variations were confirmed from both directions in two independent PCR reactions of the original DNA samples.
Table 1.
Primers Used for PCR Amplification of Exons 18, 19, and 21 of the EGFR Gene
| Name | Sequence |
|---|---|
| 18externalF* | 5′-CAAGTGCCGTGTCCTGGCACCCAAGC-3′ |
| 18externalR* | 5′-CCAAACACTCAGTGAAACAAAGAG-3′ |
| 18nestedF | 5′-GCCATGTCTGGCACTGCTTT-3′ |
| 18nestedR | 5′-AGTAGATGATGGAAATATACAGCTTGC-3′ |
| 19externalF | 5′-CTGGTAACATCCACCCAGATCACTG-3′ |
| 19externalR | 5′-GAGATGAGCAGGGTCTAGAGCAGAG-3′ |
| 19nestedF | 5′-CAGATCACTGGGCAGCATGTGG-3′ |
| 19nestedR | 5′-CTAGAGCAGAGCAGCTGCCAGACAT-3′ |
| 21externalF | 5′-CTGAATTCGGATGCAGAGCTTCTT-3′ |
| 21externalR | 5′-CATCCTCCCCTGCATGTGTTAAA-3′ |
| 21nestedF | 5′-GATGCAGAGCTTCTTCCCATGAT-3′ |
| 21nestedR | 5′-GCATGTGTTAAACAATACAGCTAGTGG-3′ |
F, forward; R, reverse.
See Lynch et al.10
Mutant-Enriched PCR and Sequencing
Mutant-enriched PCR is a multistep PCR with intermittent restriction digestion to eliminate wild-type genes selectively, thus enriching the genes with exon 19 deletion or L858R exon 21 point mutation. We used a modified protocol described by Asano et al.29
Fluorescent in Situ Hybridization
Gene copy number per cell was investigated by FISH using the Vysis EGFR probe (Abbott Laboratories, Des Plaines, IL) and semiautomated or manual procedure. In the former procedure the tissue was pretreated by a Discovery Automatic Hybridizator (Ventana, Tucson, AZ). The classification was done according to the six FISH categories defined by Cappuzzo et al22 and was also used in the BR.21 study.24 Samples with a high EGFR gene copy number (high polysomy or amplification) were considered to be FISH-positive.
Immunohistochemistry
The expression of EGFR protein was determined by IHC using Dako EGFR PharmDx kits (DakoCytomation), and the slides were counterstained with hematoxylin. For evaluation the same categorization was used as in the BR.21 study; samples with more than 10% tumor cells showing membranous (partial or complete) staining of any intensity were stated as positive for EGFR.21 For semiquantitation we also used the scoring system defined by Capuzzo et al22 with the modification that we evaluated only membranous staining and determined four levels of intensity (0, 1+, 2+ = control slide of the kit, 3+) according to the vendor (Dako). The IHC score was calculated by multiplying the staining intensity and the fraction of the positive cells (0–100%).
Statistical Methods
Relationships between EGFR status and clinical characteristics and between mutation type and complete response/partial response and stable disease rate were analyzed by χ2 or Fisher's exact test. Age differences of various subpopulations were compared with the use of t-test for independent samples. Correlation between IHC score and other EGFR status was analyzed using the Mann-Whitney U-test. All reported P values are two-sided.
Results
The EGFR status in paraffin-embedded lung adenocarcinoma samples from 127 Hungarian patients was evaluated. The results of the prospective EGFR status analysis are shown in Table 2.
Table 2.
Prospective Analysis of the Association between EGFR Mutation, FISH, and IHC Status and Characteristics of Lung Adenocarcinoma Patients
| Mutation+ | FISH+ | Amplified* | IHC+ | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Parameter Y | All | % | (Nmut+/N Y− examined) | P | % | (N FISH+/N Y− examined) | P | % | (Namp+/N Y− examined) | P | % | (N IHC+/N Y− examined) | P |
| Total | 127 | 13.5 | (17/126) | 40 | (39/97) | 6 | (6/97) | 59 | (68/116) | ||||
| Sex | |||||||||||||
| Female | 74 | 16 | (12/74) | 0.286 | 42 | (24/57) | 0.649 | 7 | (4/57) | 1.000 | 58 | (39/67) | 0.916 |
| Male | 53 | 10 | (5/52) | 38 | (15/40) | 5 | (2/40) | 59 | (29/49) | ||||
| Smoking status | |||||||||||||
| Current/former | 52 | 4 | (2/51) | <0.0001 | 42 | (16/38) | 0.354 | 5 | (2/38) | 0.366 | 64 | (32/50) | 0.388 |
| Never | 31 | 42 | (13/31) | 54 | (13/24) | 13 | (3/24) | 73 | (22/30) | ||||
| Female | 25 | 40 | (10/25) | 0.676 | 47 | (9/19) | 0.327 | 11 | (2/19) | 0.521 | 71 | (17/24) | 1.000 |
| Male | 6 | 50 | (3/6) | 80 | (4/5) | 20 | (1/5) | 83 | (5/6) | ||||
| Unknown | 44 | 5 | (2/44) | 71 | (25/35) | 3 | (1/35) | 39 | (14/36) | ||||
| Age (year) +/− | 54.9 | 58.3/54.0 | 0.086 | 54.1/56.7 | 0.163 | 50.3/56.0 | 0.135 | 55.0/54.6 | 0.795 | ||||
| Mutant | 17 | 63 | (10/16) | 0.051 | 25 | (4/16) | 0.007 | 73 | (11/15) | 0.204 | |||
| Wild type | 109 | 36 | (29/80) | 3 | (2/80) | 56 | (56/100) | ||||||
| FISH+ | 39 | 26 | (10/39) | 0.051 | 66 | (25/38) | 0.377 | ||||||
| Amplified* | 6 | 67 | (4/6) | 0.007 | 100 | (6/6) | 0.078 | ||||||
| FISH− | 58 | 10.5 | (6/57) | 57 | (30/53) | ||||||||
| IHC+ | 68 | 16 | (11/67) | 0.204 | 45 | (25/55) | 0.377 | 11 | (6/55) | 0.078 | |||
| IHC− | 48 | 8 | (4/48) | 36 | (13/36) | 0 | (0/36) | ||||||
| IHC score | |||||||||||||
| 0–99 | 74 | 8.1 | (6/74) | 0.025 | 31 | (17/55) | 0.077 | 0 | (0/55) | <0.0001 | 35 | (26/74) | <0.0001 |
| 100–199 | 25 | 16 | (4/25) | 57 | (12/21) | 5 | (1/21) | 100 | (25/25) | ||||
| 200–300 | 17 | 31 | (5/16) | 60 | (9/15) | 33 | (5/15) | 100 | (17/17) | ||||
Specimens of 90 primary tumors and 37 metastases were obtained from 21 pathology departments. There were seven bronchoscopic biopsies and two small brain biopsies. The others were samples of surgical resections of the standard size. amp, gene amplification; mut, mutation.
Subgroup of the FISH+ group.
EGFR Mutation Analysis
The evaluation was successful in almost all (99%) cases (126). EGFR kinase domain mutation was identified in 17 cases (13.5%). Characteristics of the patients with mutations are shown in Table 3. Nine mutations were 19 exon deletions and seven were exon 21 point mutations (6x T2573G>L858R, 1x T2582A>L861Q) (Figure 1, A and B). Besides these mutations that have been reported to be associated with responsiveness to EGFR TKIs, we also identified a new point mutation of exon 19 (G2227A>A743T).
Table 3.
Characteristics of Patients with EGFR TK Mutations Designated by Prospective Analysis and Patients Who Responded to EGFR TKI Treatment
| EGFR status |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Analysis | Case | Histology | Age | Sex | Smoking status | Phenotype | FISH | IHC (score) | TKI | Response |
| Prospective | Pr 1 | ADC | 61 | F | Never | del747-750insP | + | + (150) | Gefitinib | CR |
| Pr 2 | ADC | 45 | F | Never | del746-752insV | + (ampl) | + (240) | Gefitinib | CR | |
| Pr 3 | ADC | 50 | M | Former | del747-753insS | − | + (60) | Erlotinib | CR | |
| Pr 4 | ADC | 55 | F | Never | del746-750 | + | − (0) | Erlotinib | CR | |
| Pr 5 | ADC | 59 | F | Never | L858R | ND | + (120) | Gefitinib | CR | |
| Pr 6 | ADC | 67 | F | Never | del746-750 | − | + (15) | Erlotinib | PR | |
| Pr 7 | ADC | 53 | F | Never | L858R | − | ND | Erlotinib | PR | |
| Pr 8 | ADC | 67 | M | Never | L858R | + | − (0) | Erlotinib | PR | |
| Pr 9 | ADC | 64 | F | Never | L858R | − | − (0) | Erlotinib | PR | |
| Pr 10 | ADC | 55 | M | Never | L858R | + (ampl) | + (270) | Erlotinib | Primar: PR, meta: SD | |
| Pr 11 | ADC | 62 | M | Never | L858R | + (150) | + (270) | Erlotinib, ceased* | ||
| Pr 12 | ADC | 57 | F | Never | del746-750 | + (ampl) | + (180) | – | – | |
| Pr 13 | ADC | 54 | F | ? | del747-751 | + (ampl) | + (270) | – | – | |
| Pr 14 | ADC | 43 | F | Never | del746-750 | + | − (5) | – | – | |
| Pr 15 | ADC | 56 | F | Smoker | del747-753insS | − | + (180) | – | – | |
| Pr 16 | ADC | 62 | F | Never | L861Q | + | + (240) | – | – | |
| Pr 17 | ADC | 81 | M | ? | A743T | − | ND | – | – | |
| Retrospective | Rtr 1 | ADC | 58 | F | Never | del747-753insS | + | + (150) | Gefitinib | CR |
| Rtr 2 | ADC | 68 | F | Never | del746-750 | ND (smear) | − (0) | Erlotinib | CR | |
| Rtr 3 | adsq. c. | 62 | F | Former | L858R | + (ampl) | + (160) | Erlotinib | CR | |
| Rtr 4 | ADC | 66 | F | Never | del746-750 | ND (smear) | − (5) | Erlotinib | PR | |
| Rtr 5 | ADC | 50 | M | Never | del746-751insA | + (ampl) | + (120) | Erlotinib | PR | |
| Rtr 6 | ADC | 65 | F | Never | L858R | + | + (70) | Erlotinib | PR | |
| Rtr 7 | ADC | 72 | F | Never | del746-750 | ND | − (5) | Erlotinib | PR | |
| Rtr 8 | anap. c. | 54 | M | Former | del746-750† | + | − (0) | Erlotinib | SD | |
| Rtr 9 | anap. c. | 60 | F | ? | wild type‡ | + | + (200) | Erlotinib | PR | |
Each mutant sample contained only one confirmed mutation. All samples were taken before EGFR TKI therapy. ADC, adenocarcinoma; adsq. c., adenosquamous carcinoma; anap. c., anaplastic carcinoma; F, female; M, male; meta, metastasis; ?, not known; ND, not determined; ampl, gene amplification; CR, complete response; PR, partial response; Retrosp, retrospective; SD, stable disease.
Cessation due to skin rash.
Bronchoscopic biopsy specimen with 25% tumor cell proportion. Mutant-enriched PCR assay was applied.
Bronchoscopic biopsy specimen with <10% tumor cell proportion. Mutant-enriched PCR assay was applied.
Figure 1.
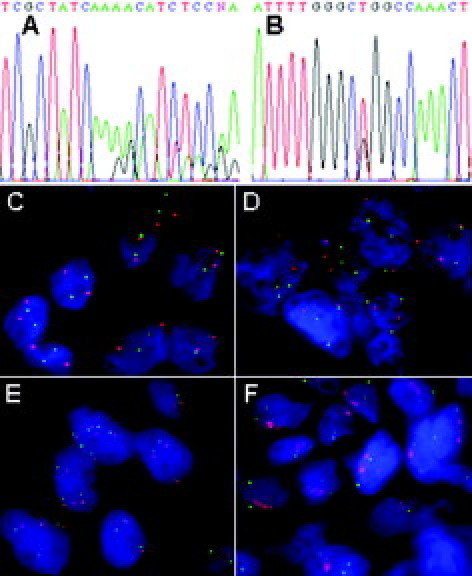
Chromatogram of the del2235-2249>del746-750 exon 19 deletion (A) and T2573G>L858R exon 21 point mutation (B). FISH analysis of disomy (C), high trisomy (D), high polysomy (E), and gene amplification (F). The EGFR gene signal is red; the chromosome 7 signal is green.
There was a conspicuously higher mutation rate in those who had never smoked than in current or former smokers (42% versus 4%, P < 0.001) and in female patients (16% versus 10%, not significant). The frequency of mutations was higher, but not significantly, in FISH-positive cases (26% mutation rate in FISH+ versus 10.5% in FISH−, P = 0.051; 67% in amplified versus 13.3% in nonamplified).
Using the 10% as a cut-off criteria for IHC positivity, the frequency of mutations was higher (16%) in IHC+ than in IHC− tumors (8%), but this difference was not significant (P = 0.204). The frequency of mutations was 8.1% in the tumors with low IHC score (0–99), 16% in the middle range (100–199), and 31% in tumors with the highest IHC score (200–299) by semiquantitative score analysis. The correlation between the IHC score and the frequency of mutations is significant (P = 0.025).
An additional 36, most not published, sequence variations were found in 28 (22%) samples. However, these sequence alterations could not be confirmed from the repeated second and third independent PCR reactions, and therefore these alterations were not considered valid mutations.
FISH Analysis
EGFR gene copy number was assessed by FISH analysis in 118 patients and was successful in 97 cases (82%). It was increased in 39 samples (40% FISH+). In six (6%) of them the EGFR gene was amplified. One sample showed both EGFR amplification and polysomy (≥4 chromosome 7 centromeres). The other 33 (34%) FISH+ samples had high polysomy without amplification. Disomy for the EGFR gene was present in 42 (43%), low trisomy in one, high trisomy in 14, and low polysomy in one specimen. These populations were categorized as FISH− (60%). The four major FISH patterns are illustrated in Figure 1, C–F.
There were no significant differences in the rate of FISH positivity and gene amplification between groups with different clinical characteristics (age, gender, smoking status). A significant association was observed between gene amplification and mutation (25% EGFR gene amplification in patients with mutants versus 2.5% in patients with wild type). Although the prevalence of amplification and FISH+ were higher in case of IHC positivity (11% amplification/45% FISH+) than in the case of IHC negativity (0% amplification/36% FISH+), these associations did not reach significance.
IHC Analysis
EGFR protein expression was successfully evaluated in 116 patients (success rate, 99%). Overexpression was found in 68 cases (59%). There were no significant differences in the rate of IHC positivity in patients of different age, gender, and smoking status. Only the semiquantitatively estimated protein expression using the IHC score showed significant association with the frequency of mutation. Although all gene amplification caused a very strong immunohistochemical membrane staining, statistically the protein expression was unrelated to gene copy number (Figure 2).
Figure 2.
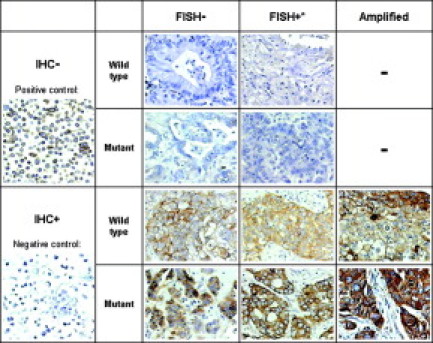
EGFR protein expression of the samples with different genotypes. The expression of EGFR protein was determined by immunohistochemistry using Dako EGFR PharmDx kits (DakoCytomation), and the slides were counterstained with hematoxylin. *High polysomy, not amplified.
Samples Analyzed by All Three Methods
In case of 90 patients' samples all three analyses were successfully performed, so this population can be divided into eight subgroups based on the EGFR status (Figure 3). The independence of the three methods in the analysis of the EGFR status manifests again. None of the subgroups of the mutant, FISH+, and IHC+ patients fully overlapped the other, so neither method can serve as a substitution or even preselection of the other. Only 24% of the lung adenocarcinomas had a totally negative EGFR status by all three methods.
Figure 3.
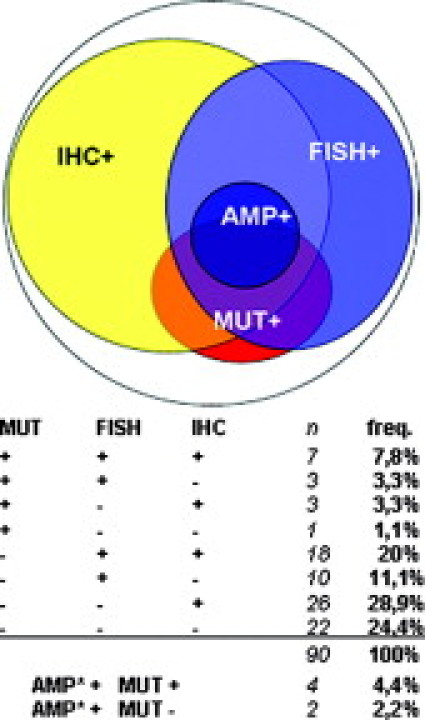
Subgroups of lung adenocarcinomas designated by the three EGFR status analyses. Since the population of this three-sided analysis has been only a subgroup of the population of the two-sided analyses, the analogous frequencies are slightly different (mutation+, 15.6%; FISH+, 42.2%; amplification+, 6.7%; IHC+, 60%). AMP, amplification; freq: frequency; MUT, mutation. *Subgroup of the FISH+ group.
Clinical Responses of EGFR Mutant Tumors to Gefitinib or Erlotinib
Ten patients were treated with either gefitinib or erlotinib after detection of the EGFR TK domain mutation (Table 3). All (100%) permanently treated patients with EGFR mutant responded to therapy. Five patients showed complete response, and five had partial response. Patients with exon 19 deletion were more sensitive to TKI treatment than L858R mutants. While four of the five patients with exon 19 mutations showed complete response, only one of the five L858R mutants showed complete response and the other four had partial response.
Five EGFR mutant responders were FISH-positive (two had EGFR amplification) and four were FISH-negative. In one case the FISH analysis was not successful. The EGFR protein was overexpressed in six cases. In three cases, however, mutant responders were negative by IHC. One sample from a partial responder patient was not determined by IHC due to technical failure. This tumor had also a negative FISH status. Another patient who had a partial response as a result of erlotinib treatment had normal EGFR gene copy number (disomy, FISH−) and protein expression (IHC−) but showed T2573G>L858R mutation as the only EGFR abnormality.
Retrospective Biomarker Analysis of Tumor Samples from NSCLC Patients with Good Clinical Response to EGFR TKI Treatment
To further evaluate which biomarker analysis is associated most with the responsiveness to EGFR TKIs, we analyzed the EGFR status in tumors of nine Hungarian lung cancer patients who had responded on erlotinib or gefitinib therapy (Table 3). Eight patients were treated with erlotinib and one patient was a gefitinib responder. Three patients showed complete response, five had partial response, and one patient had stable disease.
Seven samples were formalin-fixed and paraffin-embedded, but only smears were available from two patients. After IHC the smears were scraped for mutation analysis, and therefore FISH could not be performed in these cases. The mutation analysis was successful in all cases. Eight (89%) patients had mutant and only one had wild-type tyrosine kinase domain. Six mutations were exon 19 deletions and two were point mutations of exon 21. In bronchoscopic biopsy specimens (two cases) the proportion of tumor cells was low. We could not identify mutations in these samples by the standard PCR and sequencing method. However after mutant-enriched PCR of exons 19 and 21, an exon 19 deletion was found in the sample, which contained about 25% tumor cells. In the specimen where tumor cell content was less than 10% we were not able to identify any mutation.
Gene copy number was successfully assessed in six specimens. All of them were FISH-positive. Two had EGFR amplification, and four had polysomy. The bronchoscopic sample from a partial responder showed no mutation, but polysomy. Five responders had protein overexpression detected by IHC and four (44%) responders were IHC-negative.
Discussion
Prospective, randomized, placebo-controlled clinical trials are currently underway to determine the exact role of different predictive biomarkers in selection of non-small-cell lung cancer patients for EGFR TKI treatment, in particular, erlotinib therapy. It will take probably years to complete these trials. At the present, oncologists can only rely on the existing reports of clinical benefit in patient populations positive for different biomarkers related to the increased activity of EGFR, protein overexpression, increased gene copy number, and presence of activating mutations.
This study provides evidence that patient populations positive by these different EGFR diagnostics only partly overlap. Most importantly, patients who do not overexpress EGFR protein, therefore presenting negative EGFR immunohistochemical reaction, can carry activating mutations and have excellent response to EGFR TKI therapy, including erlotinib.
Although no conclusion can be reached about the negative predictive value of the absence of activating mutations due to lack of clinical information of all patients, the 100% (14/14) response rate to erlotinib in patients carrying activating mutations is remarkably higher than reported in the study published by Tsao et al. Tsao et al found a higher, but only two of eight (25%), response rate in patients with EGFR (classical mutations) mutants, in comparison to the 9% response rate in the whole population.24
In the BR21 trial, only EGFR IHC-positive patients gained a significant survival benefit from erlotinib treatment, although the P value for interaction was not significant (0.25), indicating that the diminished therapeutic effect of erlotinib in EGFR IHC-negative patients could be attributed to the smaller population size. The gene copy number analysis by FISH proved to be a better marker to exclude patients who do not benefit from erlotinib but not significantly (P = 0.1). Based on this study, erlotinib was registered by the Food and Drug Administration for the second- and third-line treatment of NSCLC independently from any biomarker analysis. However, the registration by the European Medicines Agency in the European Union included an important remark: “The survival benefit of erlotinib is not proven in EGFR negative patients.”
The advantages of IHC analysis are that 1) it is the most cost-efficient and easiest method; 2) most in vitro diagnostic units can perform this test; and 3) this test is the most frequently positive (in our study 59%), therefore excluding the fewest patients from therapy. The disadvantage of IHC lies in the difficulties of standardization. In our laboratory, we use the same standardized IHC protocol used by Tsao et al and the same evaluation criteria of 10% positivity as the threshold for positive EGFR IHC status.24 The sensitivity of the IHC reaction can be increased, but the altering the standardized protocol result abolishes the quantitative information of expression levels and interlaboratory comparability. These technical problems are discussed for HER2 testing.30,31 However there is no similar consensus for the guidelines of EGFR IHC and FISH testing in lung cancer.
The FISH analysis is a more expensive and difficult method. In 61 samples the FISH signals were already valuable after the first FISH procedure. However, in case of 36 samples we had to repeat the reaction with a different extent of digestion. In the case of 21 specimens we could not gain sufficient signal intensity for the adequate diagnosis, even after multiple attempts with modified FISH protocol. After extensive optimization of the protocol we reached an 82% success rate, but it is still less then the 99% success of the IHC analysis. However, FISH analysis is a more reliable method than IHC, since the presence of the normal signals of the normal karyotype (disomy) exclude the possibility of false negativity. The quantitative evaluation of FISH is also more objective. These technological differences may have contributed to the better predictive value of FISH in comparison to IHC in the retrospective analysis of BR2124 similarly to results found previously with gefitinib.23
In our study FISH was less frequently positive (40%) than IHC (59%). Since the survival benefit was significant in the larger population of IHC-positive patients of the BR21 trial, it is not warranted to exclude FISH-negative but IHC-positive patients from erlotinib treatment. All of the six patients with intrachromosomal gene amplifications were also 3+ IHC-positive, similar to HER-2 in breast cancer, but 41% of the FISH defined by polysomy were IHC-negative. The FISH-positive patients significantly benefited from erlotinib regardless of the IHC status in BR21. Therefore, IHC-negative but FISH-positive patients cannot be regarded as EGFR-negative.
The sequence analysis of archived tissue samples by automatic sequencing is considered the gold standard for mutation detection but is also a very labor intensive and difficult assay. However, in our laboratory EGFR DNA sequence could be determined in 99% of surgical biopsies.
Activating mutations of the EGFR may increase the receptor activity even in the absence of protein overexpression that can lead to oncogene dependence. To test this hypothesis and to explore the potential clinical significance of these mutations in the absence of IHC-positive protein, we obtained clinical information of 10 patients with mutations who received either gefitinib or erlotinib treatment following our diagnosis. Remarkably, there were five complete responses and five partial responses in this group of patients regardless of IHC positivity. In addition, we retrospectively analyzed the samples of nine patients reportedly having an exceptionally good response (complete or partial) to EGFR TKI treatment. We found activating mutations in all except one of these tumors. We have no clinical data for the patients without mutations, but these results can be evaluated in consideration of the low frequency (13.5%) of mutant tumors in our set of 127 tumor samples and in comparison to the 9% response rate in the unselected population of the BR21 trial.6
Most importantly, seven of the patients with mutations who responded to EGFR TKI treatment were EGFR IHC-negative. Five of these samples were completely IHC-negative (0%) and two did not meet the 10% threshold criteria. Alterations from the standard IHC protocols can increase the ratio of IHC-positive samples but it is already 59%, and further increase in sensitivity would diminish the quantitative information on EGFR expression. Previous studies have reported lack of correlation between EGFR mutation and IHC positivity22 and EGFR mutant tumors with absent IHC positivity32 in gefitinib-treated NSCLC patients, but this is the first study that focuses on this phenomenon in lung adenocarcinomas treated with both erlotinib and gefitinib.
The evaluation of the clinical significance of EGFR mutations always suffers from the low number of patients that underpower the statistical analysis. The BR21 trial that led to the market authorization of erlotinib included only eight erlotinib-treated patients with classic activating EGFR mutations. There were only one complete response and one partial response (25%) in those patients, far below the 65 to 100% response others reported with EGFR TKI-treated patients with EGFR mutations.10,11,12,13,14,15,16,17,18,19 In this study we provide clinical data of 18 (14 erlotinib-treated, four gefitinib-treated) patients with EGFR mutations. We found a 100% partial response or complete response rate in these patients. The reason for this difference may lay in the different laboratory practices in different laboratories.
The BR21 study also concluded that patients carrying EGFR mutations have better prognosis independently from EGFR TKI treatments.24 The median survival of patients with EGFR mutations in the control arm was 9.1 months (n = 20), and the median survival in control arm of the unselected population was 4.7 months.24 In contrast, several others have reported the average survival of patients with EGFR mutations receiving EGFR-TKI treatment to be over 30 months.13,15,16 These patient populations cannot be directly compared and new randomized prospective studies will be necessary to draw a final conclusion. Our observation is that patients with EGFR mutations in this study received the EGFR TKI treatment at a very late stage of their disease, and therefore the major reduction, often complete elimination of the tumor burden, most evidently prolongs their survival, although further follow-up (probably years) is required to determine median survival time. The first patient with EGFR mutation in this study started the gefitinib treatment 2 years ago against multiple brain metastases in a moribund clinical stage, and she is still tumor-free and enjoys a good quality of life.
In our view, although further studies will be required to decide which biomarker analysis is the most suitable for individualized EGFR TKI treatment, at the present, this important clinical decision cannot solely be based on a single method, immunohistochemistry, but molecular diagnostic methods, particularly DNA sequence analysis, should be part of the biomarker analysis of EGFR status of NSCLC patients. This report also includes three patients with EGFR mutations who formerly smoked who had excellent therapeutic response. Most importantly, this study does not suggest that only patients with EGFR mutations should be exclusively selected for EGFR TKI therapy, but provides strong evidence that all patients with EGFR mutations with lung adenocarcinoma should be treated regardless of other biomarkers or smoking status.
Acknowledgements
We are grateful for the valuable comments of Professor Gabriel N. Hortobagyi, University of Texas M. D. Anderson Cancer Center, Houston, TX, in the preparation of this manuscript.
Footnotes
I.P. was supported by Terry Fox Run for Cancer Research Foundation (Hungary), grants ETT 574/2006, OTKA-T046665, OTKA TS 049887, NFKP-07-A2-NANODRUG, and OMFB-00287/KKKII-05/2005, and by the National Office for Research and Technology (NKTH), Hungary.
References
- 1.Chung KY, Shia J, Kemeny NE, Shah M, Schwartz GK, Tse A, Hamilton A, Pan D, Schrag D, Schwartz L, Klimstra DS, Fridman D, Kelsen DP, Saltz LB. Cetuximab shows activity in colorectal cancer patients with tumors that do not express the epidermal growth factor receptor by immunohistochemistry. J Clin Oncol. 2005;23:1803–1810. doi: 10.1200/JCO.2005.08.037. [DOI] [PubMed] [Google Scholar]
- 2.Lièvre A, Bachet JB, Le Corre D, Boige V, Landi B, Emile JF, Côté JF, Tomasic G, Penna C, Ducreux M, Rougier P, Penault-Llorca F, Laurent-Puig P. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992–3995. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 3.Uramoto H, Mitsudomi T. Which biomarker predicts benefit from EGFR-TKI treatment for patients with lung cancer. Br J Cancer. 2007;96:857–863. doi: 10.1038/sj.bjc.6603665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kris MG, Natale RB, Herbst RS, Lynch TJ, Jr, Prager D, Belani CP, Schiller JH, Kelly K, Spiridonidis H, Sandler A, Albain KS, Cella D, Wolf MK, Averbuch SD, Ochs JJ, Kay AC. Efficacy of gefitinib, an inhibitor of the epidermal growth factor receptor tyrosine kinase, in symptomatic patients with non-small cell lung cancer: a randomized trial. JAMA. 2003;290:2149–2158. doi: 10.1001/jama.290.16.2149. [DOI] [PubMed] [Google Scholar]
- 5.Fukuoka M, Yano S, Giaccone G, Tamura T, Nakagawa K, Douillard JY, Nishiwaki Y, Vansteenkiste J, Kudoh S, Rischin D, Eek R, Horai T, Noda K, Takata I, Smit E, Averbuch S, Macleod A, Feyereislova A, Dong RP, Baselga J. Multi-institutional randomized phase II trial of gefitinib for previously treated patients with advanced non-small-cell lung cancer (The IDEAL 1 Trial) J Clin Oncol. 2003;21:2237–2246. doi: 10.1200/JCO.2003.10.038. [DOI] [PubMed] [Google Scholar]
- 6.Shepherd FA, Rodrigues Pereira J, Ciuleanu T, Tan EH, Hirsh V, Thongprasert S, Campos D, Maoleekoonpiroj S, Smylie M, Martins R, van Kooten M, Dediu M, Findlay B, Tu D, Johnston D, Bezjak A, Clark G, Santabarbara P, Seymour L, National Cancer Institute of Canada Clinical Trials Group Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med. 2005;353:123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 7.Schwab R, Pinter F, Moldavy J, Papay J, Strausz J, Kopper L, Keri G, Pap A, Petak I, Oreskovich K, Mangel L. Amplification and mutation of the epidermal growth factor receptor in metastatic lung cancer with remission from gefitinib. J Clin Oncol. 2005;23:7736–7738. doi: 10.1200/JCO.2005.02.4760. [DOI] [PubMed] [Google Scholar]
- 8.Thatcher N, Chang A, Parikh P, Rodrigues Pereira J, Ciuleanu T, von Pawel J, Thongprasert S, Tan EH, Pemberton K, Archer V, Carroll K. Gefitinib plus best supportive care in previously treated patients with refractory advanced non-small-cell lung cancer: results from a randomised, placebo-controlled, multicentre study (Iressa Survival Evaluation in Lung Cancer) Lancet. 2005;366:1527–1537. doi: 10.1016/S0140-6736(05)67625-8. [DOI] [PubMed] [Google Scholar]
- 9.US Food and Drug Administration . FDA public health advisory: new labeling and distribution program for gefitinib (Iressa) US Food and Drug Administration; Washington, DC: 2005 June 17. [Google Scholar]
- 10.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 11.Paez JG, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE, Meyerson M. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 12.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, Mardis E, Kupfer D, Wilson R, Kris M, Varmus H. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han SW, Kim TY, Hwang PG, Jeong S, Kim J, Choi IS, Oh DY, Kim JH, Kim DW, Chung DH, Im SA, Kim YT, Lee JS, Heo DS, Bang YJ, Kim NK. Predictive and prognostic impact of epidermal growth factor receptor mutation in non-small-cell lung cancer patients treated with gefitinib. J Clin Oncol. 2005;23:2493–2501. doi: 10.1200/JCO.2005.01.388. [DOI] [PubMed] [Google Scholar]
- 14.Cortes-Funes H, Gomez C, Rosell R, Valero P, Garcia-Giron C, Velasco A, Izquierdo A, Diz P, Camps C, Castellanos D, Alberola V, Cardenal F, Gonzalez-Larriba JL, Vieitez JM, Maeztu I, Sanchez JJ, Queralt C, Mayo C, Mendez P, Moran T, Taron M. Epidermal growth factor receptor activating mutations in Spanish gefitinib-treated non-small-cell lung cancer patients. Ann Oncol. 2005;16:1081–1086. doi: 10.1093/annonc/mdi221. [DOI] [PubMed] [Google Scholar]
- 15.Mitsudomi T, Kosaka T, Endoh H, Horio Y, Hida T, Mori S, Hatooka S, Shinoda M, Takahashi T, Yatabe Y. Mutations of the epidermal growth factor receptor gene predict prolonged survival after gefitinib treatment in patients with non-small-cell lung cancer with postoperative recurrence. J Clin Oncol. 2005;23:2513–2520. doi: 10.1200/JCO.2005.00.992. [DOI] [PubMed] [Google Scholar]
- 16.Taron M, Ichinose Y, Rosell R, Mok T, Massuti B, Zamora L, Mate JL, Manegold C, Ono M, Queralt C, Jahan T, Sanchez JJ, Sanchez-Ronco M, Hsue V, Jablons D, Sanchez JM, Moran T. Activating mutations in the tyrosine kinase domain of the epidermal growth factor receptor are associated with improved survival in gefitinib-treated chemorefractory lung adenocarcinomas. Clin Cancer Res. 2005;11:5785–5878. doi: 10.1158/1078-0432.CCR-04-2618. [DOI] [PubMed] [Google Scholar]
- 17.Chou TY, Chiu CH, Li LH, Hsiao CY, Tzen CY, Chang KT, Chen YM, Perng RP, Tsai SF, Tsai CM. Mutation in the tyrosine kinase domain of epidermal growth factor receptor is a predictive and prognostic factor for gefitinib treatment in patients with non-small cell lung cancer. Clin Cancer Res. 2005;11:3750–3757. doi: 10.1158/1078-0432.CCR-04-1981. [DOI] [PubMed] [Google Scholar]
- 18.Takano T, Ohe Y, Tsuta K, Fukui T, Sakamoto H, Yoshida T, Tateishi U, Nokihara H, Yamamoto N, Sekine I, Kunitoh H, Matsuno Y, Furuta K, Tamura T. Epidermal growth factor receptor gene mutations and increased copy numbers predict gefitinib sensitivity in patients with recurrent non-small-cell lung cancer. J Clin Oncol. 2005;28:6829–6837. doi: 10.1200/JCO.2005.01.0793. [DOI] [PubMed] [Google Scholar]
- 19.Tokumo M, Toyooka S, Kiura K, Shigematsu H, Tomii K, Aoe M, Ichimura K, Tsuda T, Yano M, Tsukuda K, Tabata M, Ueoka H, Tanimoto M, Date H, Gazdar AF, Shimizu N. The relationship between epidermal growth factor receptor mutations and clinicopathologic features in non-small cell lung cancers. Clin Cancer Res. 2005;11:1167–1173. [PubMed] [Google Scholar]
- 20.Shigematsu H, Lin L, Takahashi T, Nomura M, Suzuki M, Wistuba II, Fong KM, Lee H, Toyooka S, Shimizu N, Fujisawa T, Feng Z, Roth JA, Herz J, Minna JD, Gazdar AF. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97:339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 21.Marchetti A, Martella C, Felicioni L, Barassi F, Salvatore S, Chella A, Camplese PP, Iarussi T, Mucilli F, Mezzetti A, Cuccurullo F, Sacco R, Buttitta F. EGFR mutations in non-small-cell lung cancer: analysis of a large series of cases and development of a rapid and sensitive method for diagnostic screening with potential implications on pharmacologic treatment. J Clin Oncol. 2005;23:857–865. doi: 10.1200/JCO.2005.08.043. [DOI] [PubMed] [Google Scholar]
- 22.Cappuzzo F, Hirsch FR, Rossi E, Bartolini S, Ceresoli GL, Bemis L, Haney J, Witta S, Danenberg K, Domenichini I, Ludovini V, Magrini E, Gregorc V, Doglioni C, Sidoni A, Tonato M, Franklin WA, Crino L, Bunn PA, Jr, Varella-Garcia M. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst. 2005;97:643–655. doi: 10.1093/jnci/dji112. [DOI] [PubMed] [Google Scholar]
- 23.Hirsch FR, Varella-Garcia M, McCoy J, West H, Xavier AC, Gumerlock P, Bunn PA, Jr, Franklin WA, Crowley J, Gandara DR, Southwest Oncology Group Increased epidermal growth factor receptor gene copy number detected by fluorescence in situ hybridization associates with increased sensitivity to gefitinib in patients with bronchoalveolar carcinoma subtypes: a Southwest Oncology Group Study. J Clin Oncol. 2005;23:6838–6845. doi: 10.1200/JCO.2005.01.2823. [DOI] [PubMed] [Google Scholar]
- 24.Tsao MS, Sakurada A, Cutz JC, Zhu CQ, Kamel-Reid S, Squire J, Lorimer I, Zhang T, Liu N, Daneshmand M, Marrano P, da Cunha Santos G, Lagarde A, Richardson F, Seymour L, Whitehead M, Ding K, Pater J, Shepherd FA. Erlotinib in lung cancer: molecular and clinical predictors of outcome. N Engl J Med. 2005;353:133–144. doi: 10.1056/NEJMoa050736. [DOI] [PubMed] [Google Scholar]
- 25.Bailey LR, Kris M, Wolf M, Kay A, Averbuch S, Askaa J, Jans M, Schmidt K, Fukuoka M. Tumor EGFR membrane staining is not clinically relevant for predicting response in patients receiving gefitinib (“Iressa” ZD1839) monotherapy for pretreated advanced non-small-cell lung cancer: iDEAL 1 and 2 (abstr LB-170) Proc Am Assoc Cancer Res. 2003;44:1362. [Google Scholar]
- 26.Cappuzzo F, Gregorc V, Rossi E, Cancellieri A, Magrini E, Paties CT, Ceresoli G, Lombardo L, Bartolini S, Calandri C, de Rosa M, Villa E, Crino L. Gefitinib in pretreated non-small-cell lung cancer (NSCLC): analysis of efficacy and correlation with HER2 and epidermal growth factor receptor expression in locally advanced or metastatic NSCLC. J Clin Oncol. 2003;21:2658–2663. doi: 10.1200/JCO.2003.01.039. [DOI] [PubMed] [Google Scholar]
- 27.Parra HS, Cavina R, Latteri F, Campagnoli E, Morenghi E, Torri W, Brambilla G, Alloisio M, Santoro A. Analysis of epidermal growth factor receptor expression as a predictive factor for response to gefitinib (“Iressa” ZD1839) in non-small-cell lung cancer. Br J Cancer. 2004;91:208–212. doi: 10.1038/sj.bjc.6601923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Helfrich BA, Raben D, Varella-Garcia M, Gustafson D, Chan DC, Bemis L, Coldren C, Barón A, Zeng C, Franklin WA, Hirsch FR, Gazdar A, Minna J, Bunn PA., Jr Antitumor activity of the epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor gefitinib (ZD1839, Iressa) in non-small cell lung cancer cell lines correlates with gene copy number and EGFR mutations but not EGFR protein levels. Clin Cancer Res. 2006;12:7117–7125. doi: 10.1158/1078-0432.CCR-06-0760. [DOI] [PubMed] [Google Scholar]
- 29.Asano H, Toyooka S, Tokumo M, Ichimura K, Aoe K, Ito S, Tsukuda K, Ouchida M, Aoe M, Katayama H, Hiraki A, Sugi K, Kiura K, Date H, Shimizu N. Detection of EGFR gene mutation in lung cancer by mutant-enriched polymerase chain reaction assay. Clin Cancer Res. 2006;12:43–48. doi: 10.1158/1078-0432.CCR-05-0934. [DOI] [PubMed] [Google Scholar]
- 30.Ross JS, Fletcher JA, Bloom KJ, Linette GP, Stec J, Symmans WF, Pusztai L, Hortobagyi GN. Targeted therapy in breast cancer: the HER-2/neu gene and protein. Mol Cell Proteomics. 2004;4:379–398. doi: 10.1074/mcp.R400001-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, Dowsett M, Fitzgibbons PL, Hanna WM, Langer A, McShane LM, Paik S, Pegram MD, Perez EA, Press MF, Rhodes A, Sturgeon C, Taube SE, Tubbs R, Vance GH, van de Vijver M, Wheeler TM, Hayes DF, American Society of Clinical Oncology, College of American Pathologists American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007;25:118–145. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 32.Hirsch FR, Varella-Garcia M, Bunn PA, Jr, Franklin WA, Dziadziuszko R, Thatcher N, Chang A, Parikh P, Pereira JR, Ciuleanu T, von Pawel J, Watkins C, Flannery A, Ellison G, Donald E, Knight L, Parums D, Botwood N, Holloway B. Molecular predictors of outcome with gefitinib in a phase III placebo-controlled study in advanced non-small-cell lung cancer. J Clin Oncol. 2006;24:5034–5042. doi: 10.1200/JCO.2006.06.3958. [DOI] [PubMed] [Google Scholar]


