Abstract
We have used electron cryo-microscopy and image analysis to examine the native structure of immature, protease-deficient (PR−) and mature, wild-type (WT) Moloney murine leukemia virus (MuLV). Maturational cleavage of the Gag polyprotein by the viral protease is associated with striking morphological changes. The PR− MuLV particles exhibit a rounded central core, which has a characteristic track-like shell on its surface, whereas the WT MuLV cores display a polygonal surface with loss of the track-like feature. The pleomorphic shape and inability to refine unique orientation angles suggest that neither the PR− nor the WT MuLV adheres to strict icosahedral symmetry. Nevertheless, the PR− MuLV particles do exhibit paracrystalline order with a spacing between Gag molecules of ≈45 Å and a length of ≈200 Å. Because of the pleomorphic shape and paracrystalline packing of the Gag–RNA complexes, we raise the possibility that assembly of MuLV is driven by protein–RNA, as well as protein–protein, interactions. The maturation process involves a dramatic reorganization of the packing arrangements within the ribonucleoprotein core with disordering and loosening of the individual protein components.
Keywords: image processing, retroviruses, virus assembly
There is a wealth of information about the biochemistry and genetics of retroviruses (reviewed in refs. 1 and 2). In addition, x-ray crystallography and NMR spectroscopy have provided high resolution structures for key virion proteins: HIV protease (3), HIV reverse transcriptase (4, 5), the HIV matrix protein (MA) (6), the HIV capsid protein (CA) (7–10), the catalytic and terminal domains of the HIV integrase (11), the HIV nucleocapsid protein (12–14), and fragments of the HIV gp41 envelope protein (15–17) and the MuLV envelope (Env) protein (18, 19). However, comparatively little is known about the supramolecular organization of retroviruses.
Moloney murine leukemia virus (MuLV) is a member of the mammalian oncoviridae and has long been an important model system for the analysis of retrovirus biology. Like other type C retroviruses, it seems to assemble only at the plasma membrane. Expression of the Gag polyprotein in a eukaryotic cell is sufficient for MuLV assembly (20, 21); therefore, this molecular species is capable of self-assembly into a budding virus particle and of incorporating into the particle the other normal constituents, including the Gag–Pol polyprotein, the Env protein complex, and the genomic RNA. Nevertheless, very little is known regarding the mechanisms of retrovirus assembly.
After a retrovirus particle has budded from the cell, the Gag and Gag–Pol polyproteins are cleaved by the viral protease (PR) into a series of smaller proteins. This process, which has been called virus maturation, is a prerequisite for infectivity. It is accompanied by a profound change in the morphology of the particle, as viewed by conventional electron microscopy (22–24). Thus, the immature particles have a dense ring of stain with a hollow or doughnut-shaped interior, thought to contain the viral ribonucleoprotein. In contrast, the mature form displays a densely stained, condensed ribonucleoprotein core at the center of the particle. Some investigators have proposed that the cores have icosahedral symmetry (22, 25–28). However, conventional thin-section and negative stain electron microscopy have revealed that retroviridae are pleomorphic, an observation confirmed recently by electron cryo-microscopy (26).
Here, we report an analysis of the supramolecular design of native wild-type (WT) MuLV and protease-defective (PR−), immature MuLV particles by electron cryo-microscopy. This technique avoids many artifacts that can appear in other electron-microscopic methods, as it does not require fixation, dehydration, embedding, shadowing, or staining. We found that both the WT and PR− MuLV particles are pleomorphic, but they can be readily distinguished by electron cryo-microscopy. Although the core of the immature particles manifests paracrystalline packing, we found no evidence that the subunits in either the immature or mature particles adhere to strict icosahedral symmetry. The implications of these findings for mechanisms of MuLV assembly and maturation are discussed.
MATERIALS AND METHODS
Virus Preparation.
The parental clone of WT MuLV is a plasmid designated pRR88, which contains an infectious Moloney MuLV genome in a plasmid vector derived from pSV2neo (29). The PR− MuLV, designated, D32L, was identical to pRR88 except that the aspartate codon at the active site of the viral protease had been changed to leucine, thereby abolishing protease activity (30). WT MuLV was grown in NIHT3 cells or was produced by transient transfection of 293 cells expressing SV40 T antigen (a gift of Nancy Rice, National Cancer Institute—Frederick Cancer Research Facility). PR− MuLV was produced by transient transfection of 293 cells containing T antigen or by stable transfection of Chinese hamster ovary cells (30). All transfections were performed using calcium phosphate (31). When transient transfection was used, 1-day harvests of culture fluids were collected 2, 3, and 4 days after the addition of DNA. All supernatants were passed through a 0.45-μm filter before further manipulations. Virus particles were purified before cryo-microscopy in either of two ways. In some experiments, 30 ml of culture fluid was layered on a 5-ml sucrose cushion (20% wt/wt in 0.05 M NaCl/0.01 M Tris, pH 7.4/0.001 M EDTA) and centrifuged for 35 min at 25,000 rpm in a Beckman SW27 rotor at 4°C. The particles were then collected by resuspending the pellet in 0.1 M NaCl, 0.01 M Tris, pH 7.4, 0.001 M EDTA. In other experiments, virus particles were purified by sucrose gradient centrifugation in the same buffer and collecting the virus fraction at 35–40% sucrose (wt/wt). Some MuLV samples were mixed with purified rhesus rotavirus (kindly provided by Harry Greenberg, Stanford University Medical Center) and centrifuged as described above. We found that appearance of the particles was independent of the cell line used to produce the virus or the procedure used to purify it before examination.
Electron Cryo-Microscopy and Image Analysis.
Frozen-hydrated specimens and electron cryo-microscopy were prepared as described (32, 33). Micrographs were digitized at 25-μm steps, corresponding to a ≈7.14-Å sampling interval at the specimen. Images of 67 mutant and 32 wild-type particles that did not appear to be distorted, disrupted, or overlapped by neighboring particles were extracted from the digitized area using a circular mask. Density attributable to neighboring particles not excluded by the mask was manually adjusted to the average density of the surrounding vitreous water, and the background was subtracted as described (32). Fourier transforms and radial density plots were computed as described (32).
Diameter Measurements of WT and PR− MuLV Particles.
The outer diameter (Do) of 67 mutant and 32 wild-type particles was measured using a circular mask. The core of the mutant particles was also measured, as it displayed a well defined inner boundary (Di). The irregular perimeter of the core boundary in the wild-type particles precluded accurate measurement of a mean inner diameter.
Icosahedral Analysis.
The translation (center or origin point) and view orientation (θ, φ, ω) (34) parameters for each virion image were determined as previously described (32, 33) using common lines procedures (35–37). The Fourier transform was truncated at low resolution (1/280 Å−1) to exclude spherically symmetric information (32). The maximum resolution for refinement of the orientation parameters ranged from 1/250 to 1/45 Å−1, and the mean weighted phase error was computed at each resolution.
Quantitation of Particle Features.
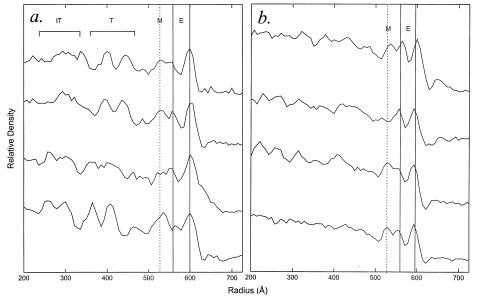
The particles in Fig. 3 were circularly averaged to determine the mean radial location and thickness of the membrane envelope (M) and the track-like density (T). Particles were centered using cross-correlation techniques (32), and the radial density was circularly averaged in annuli with a width of 3 pixels (21.4 Å).
Figure 3.
Montage of frozen-hydrated images of immature PR− MuLV (a) and mature WT MuLV (b) that have been masked from the original negative, centered, and background subtracted. In rare instances, PR− MuLV particles were identified that displayed a core layer manifesting discrete diffraction spots (Inset in a).
For several PR− MuLV particles, the core was surrounded by a well defined fringe with track-like densities (T) (Fig. 3a). For six particles, 90° sectors were extracted in which the fringe was circular (Fig. 5 Left), and rotational correlation analysis (38) revealed 56- to 64-fold symmetry (Fig. 5 Right), corresponding to the repeat pattern of the striations in the track pattern.
Figure 5.
Rotational correlation analysis of PR− MuLV images. Sectors were extracted from PR− MuLV that displayed a well defined track pattern. Image analysis revealed 56- to 64-fold rotational symmetry, corresponding to the repeat pattern for the striations in the track pattern (Right). This rotational symmetry gives a spacing between the ties on the tracks of 44 ± 4 Å, and the peak-to-peak separation of the tracks was also ≈44 Å. Note the variation in the radial thickness of the region between the tracks and the lipid envelope.
RESULTS
Morphological Features of Immature (PR−) and Mature (WT) MuLV.
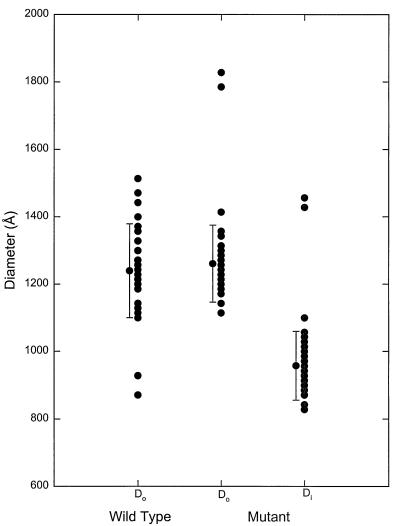
PR− and WT MuLV particles were prepared as described; images of these frozen-hydrated virions are shown in Fig. 1. Several common features are evident, including a well defined lipid bilayer envelope, a pleomorphic shape, and a central core with a higher density than the region between it and the membrane. The diameters of the WT and PR− particles are both ≈1,200 Å but show a significant (≈10%) variation around this average value (Fig. 2). The extremely large diameters represent particles with an oblong shape or with “blebs” on their surfaces.
Figure 1.
Frozen-hydrated images of immature, protease-deficient (PR−) (a, b) and mature, wild-type (WT) (c) MuLV particles. Both particle types are bounded by a lipid bilayer envelope (E) and display a variation in diameter. The inner membrane leaflet was thicker than the outer leaflet, which we attribute at least in part to the matrix protein (M). Both particles displayed a core region (C), which was rounded in the PR− MuLV and polygonal in the WT MuLV. The surface of the core in the PR− MuLV displayed a characteristic track-like structure (T). Occasionally, a second inner track (IT) could be identified that was concentric or continuous with the outer fringe. Rotavirus particles (labeled RV) (≈1,000 Å diameter) retained their constant diameter and spherical shape during ultracentrifugation with the PR− MuLV (d) and WT MuLV (e).
Figure 2.
Size distribution for 67 PR− MuLV and 32 WT-MuLV particles. PR− MuLV and WT MuLV have comparable diameters (Do) (PR− MuLV 1,260 ± 114 Å, WT MuLV 1,239 ± 139 Å). The irregular polygonal shape of the core in the WT MuLV precluded measurement of the diameter. The more rounded core in the PR− MuLV was readily measured and yielded a core diameter (Di) of 958 ± 102 Å.
It seemed possible that the pleomorphism may have resulted from particle disruption or deformation during isolation. As one approach to testing this possibility, we mixed rotavirus particles with the MuLV samples and subjected the mixture to ultracentrifugation through 20% sucrose (the final step in MuLV preparation). Rotavirus is susceptible to degradation during isolation (32, 33), and the lack of deformation and breakage in the rotavirus particles in Fig. 1, d and e, suggests that the pleomorphism of MuLV is an intrinsic property rather than an artifact of specimen preparation.
Although the PR− and WT MuLV particles share a pleomorphic rounded shape, they also exhibit dramatic differences. The WT MuLV virions have a polygonal, dense core with an irregular boundary. In contrast, the PR− MuLV particles have a rounded core, with a diameter of 958 ± 102 Å (Fig. 2). A striking feature of the PR− particles is a track-like striation or “railroad-track” pattern on the surface of the core. Occasionally, a second, inner track, possibly continuous with the outer track, could be seen.
Fine Structure of PR− and WT MuLV.
The images of PR− and WT MuLV were first analyzed by calculating circularly averaged radial density profiles from the most rounded particles (Figs. 3 and 4). These profiles delineated the high density phosphate headgroup region of the membrane envelope (E) and a dense layer on the inner leaflet of the lipid bilayer, which we designated “M.” In addition, the PR− profiles exhibited the track region (T) at the outer surface of the virion core. The PR− images displayed in Fig. 3a were then subjected to rotational correlation analysis. Depending on the diameter of the particles, the track-like feature was reinforced when 56- to 64-fold rotational symmetry was applied (Fig. 5), yielding a spacing between the ties in the track pattern of 44 ± 4 Å. The spacing between the two peaks of density in the tracks was also similar to this value (Fig. 4a). Additional density could also be detected by this analysis at a lower radius within the viral core (termed IT for inner track) (Fig. 4a).
Figure 4.
Circularly averaged radial density profiles for the most rounded PR− MuLV (a) and WT MuLV (b) displayed in Fig. 3. The locations of the membrane envelope (E), the track pattern (T), and the inner track pattern (IT) are indicated. The lipid bilayer envelope (E) has a constant distance between the phosphate head groups (peak-to-peak distance 37.5 ± 3.3 Å; n = 8) for both particles, but the inner leaflet seems to have a lower density in the PR− MuLV compared with the WT MuLV. We attribute the shoulder of density on the inner leaflet of the bilayer to the region occupied by the matrix protein (M), which has a thickness of 40 ± 9 Å. The track region (T) corresponds to the outer surface of the core and is not well defined in the WT MuLV particles. For the PR− MuLV, there was considerable variation in the lucent region between the track and matrix regions (43 ± 16 Å, n = 4). Consequently, the radial location of the track pattern varies from particle to particle.
Packing Within the MuLV Core.
The core region of some PR− MuLV particles displayed hexagonal diffraction (see Fig. 3b Insets). Presumably, the orientation of these particles afforded an end-on view of the core subunits in a sufficiently large planar sheet so that the Fourier transform displayed the packing of the subunits. We reasoned that most particles would have randomly oriented paracrystalline sheets within the core so that the diffraction pattern would be spherically averaged. Hence, we analyzed the core regions of the particles in Fig. 3 by computing circularly averaged Fourier transforms (FFT) (Fig. 6). In the PR− MuLV particles, the FFT displayed peaks centered at 0.015 ± 0.003 and 0.022 ± 0.006 Å−1, corresponding to spacings of 67 ± 11 and 45 ± 10 Å in real space. For a hexagonal paracrystal with a unit cell spacing of ≈80 Å, these peaks could be indexed as the (1,0) and (1,1) reflections, respectively. The lattice could also be orthorhombic (a, ≈80 Å; b, ≈140 Å), in which case these peaks would correspond to the (1,1) and (0,2) reflections. These predicted unit cell parameters are in very good agreement for the two-dimensional crystalline packing of expressed, N-terminal, histidine-tagged capsid molecules of MuLV on lipid monolayers (39). The FFT for the WT particles also showed two peaks, but these were far broader and shallower than those in the PR− patterns. These broad peaks were roughly centered at 0.010 and 0.018 Å−1, corresponding to spacings of ≈100 and ≈55 Å, respectively, in real space. If these two broad peaks similarly arise from the (1,0) and (1,1) reflections of a hexagonal lattice, then the unit cell spacing is ≈115 Å. The broadness and flatness of the peaks suggest that the lattice must be quite loosely packed.
Figure 6.
Circularly averaged Fourier transforms (FFTs) for the core regions of PR− MuLV (solid line) and WT MuLV (dotted line). The FFT for the PR− MuLV particles displays well defined peaks centered at 0.022 and 0.032 Å−1, corresponding to spacings of 45.4, and 31.8 Å. In contrast, the FFT for the WT MuLV displays two broad peaks centered at 0.015 Å−1 and 0.025 Å, corresponding to spacings of 67 and 40 Å, respectively, in real space.
Assessment of Icosahedral Symmetry.
To quantitatively evaluate whether MuLV particles manifest icosahedral symmetry, we computed the mean weighted phase error for selected circular particles that would be more likely to manifest icosahedral symmetry. For icosahedral particles, one would expect that the orientation angles could be refined to a single set of values with phase errors < about 50° (32). In fact, the orientation angles varied dramatically, and the 10 best sets of solutions gave phase residuals >50° (data not shown). These phase residuals were comparable with those obtained for refinement of random noise in the vitrified ice background. Hence, even the most spherical PR− MuLV did not exhibit the phase relationships expected for icosahedral particles.
DISCUSSION
In this study, we have recorded images of frozen-hydrated immature (PR−) and mature (WT) MuLV particles using electron cryo-microscopy. This technique avoids many of the potential artifacts associated with other ultrastructural techniques because it does not involve chemical fixation, dehydration, embedding, shadowing, or staining. We have also subjected the images to detailed image analysis in the hope of gaining new insights into the underlying structure and mechanisms of assembly of these particles. The two important results to emerge from this analysis are that 1) there are striking morphological differences between the immature and the mature, infectious particles and 2) neither the immature nor the mature particles exhibit icosahedral symmetry. Instead, the immature particles display paracrystalline packing of subunits that becomes much looser in the mature particle form.
Lack of Icosahedral Symmetry in MuLV.
The PR− and WT MuLV images failed several tests for strict icosahedral symmetry. The pleomorphism in size and shape of the MuLV particle images contrasts with the circular shape and constant diameter for images of icosahedral particles, as is evident in mixtures of MuLV and rotavirus particles (Fig. 1). Furthermore, rotational correlation analysis (Fig. 5) revealed a variable symmetry in the PR− particles (56- to 64-fold), corresponding to the spacing between the tracks in the core surface; this variation in the rotational symmetry was simply due to variation in the particle diameters. Finally, an attempt to refine the θ and φ orientation angles for an expected icosahedral lattice in the most circular particles gave rise to phase errors that were comparable with the errors in refining background noise to an icosahedral lattice. A lack of icosahedral symmetry has also been observed for virus-like particles formed by HIV-1 Gag expressed in insect cells (40).
Previous image analysis of retrovirus particles examined by conventional electron microscopic techniques have suggested that retroviral cores exhibit icosahedral symmetry (22, 26, 27, 28, 41). Although we cannot definitively exclude the existence of such symmetry, our analysis of frozen-hydrated MuLV particles that are presumably in a native state argues strongly against it.
A Model for the Packing of Gag Molecules in PR− MuLV Particles.
Although no icosahedral symmetry could be found in the particles, several lines of evidence are all consistent with the hypothesis that a PR− MuLV particle is composed of subunits arranged side-by-side in a paracrystalline lattice with an average center-to-center distance between these subunits of ≈45 Å. The evidence includes the diffraction pattern of individual particles (Fig. 3a Insets); the circularly averaged FFTs of multiple particles, showing diffraction fringes consistent with a hexagonal paracrystal with a unit cell spacing of ≈80 Å for the PR− MuLV and ≈115 Å for the WT MuLV (Fig. 6); and the results of rotational correlation analysis (Fig. 5). We suggest that the subunits observed in our analysis are the individual Gag polyprotein monomers. Furthermore, it seems likely that they are arranged radially with their N termini embedded in the viral membrane and their C termini in the interior of the particle, as originally suggested by Bolognesi et al. (42).
The track-like striation is the dominant feature in the PR− MuLV particles and can be used to estimate the stoichiometry of Gag molecules per particle. The PR− MuLV particles have a core diameter that ranges from 850 to 1,060 Å (Fig. 2). Using a value of 44 Å as the separation between the striations in Fig. 5 (and hence Gag monomers), the predicted number of Gag molecules per particle is 1,100–1,800, in reasonable agreement with other estimates (43). It seems likely that the individual domains of the Gag polyprotein fold independently, as (at least in HIV) each of them has a well defined tertiary structure when they are expressed as separate proteins (6–10, 44). Therefore, assuming that the Gag monomers are arranged as suggested above, we can tentatively assign the densities in the circularly averaged radial density profiles and the rotationally filtered images (Figs. 4 and 5) to individual domains. These assignments are shown in Fig. 7.
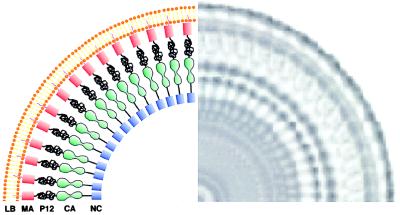
Figure 7.
Schematic model for the packing of the Gag polyprotein in immature murine leukemia virus. The rotationally averaged image of a single particle is shown to the right. The thicker inner leaflet of the lipid bilayer envelope is attributed to the matrix protein (MA), which is known to be anchored to the membrane via a myristoyl group. The low density zone between the MA region and the tracks is assigned to the location of the pp12 protein, which is known to be rich in proline and is likely to be disordered. The ordered track-like structure is assigned to the location of the capsid protein but could also include contributions from the nucleocapsid protein together with bound RNA.
The outer peaks in the radial density profiles (Fig. 4 a and b) are interpreted as the electron dense phosphate headgroups of the lipid bilayer (center-to-center distance 37.5 ± 3.3 Å). The inner leaflet of the bilayer displays a shoulder of density (thickness 40 ± 9 Å) that we attribute to the matrix (MA) protein, which is known to be anchored in the bilayer, in part by its N-terminal myristoyl group (45, 46) that intercalates into the bilayer as well as electrostatic interactions involving positively charged residues at the N terminus of MA and the phospholipid headgroups (47). The “railroad track” probably corresponds to the CA domain. HIV-1 CA is known to consist of an amino-terminal cyclophilin A binding domain (residues 1 to ≈151) and a carboxyl-terminal dimerization domain (≈151 to 231) (7–10, 44). Therefore, it seems reasonable that MuLV CA also consists of two domains. If the long axis of the CA molecules is radially aligned, then the ties of the tracks would correspond to the two domains within each monomer, and the tracks are a manifestation of the CA–CA interactions between adjacent Gag molecules. The lucent region between MA and CA, which is the most variable in length in our images (43 ± 16 Å, measured from the inner minimum of the bilayer density to the outer minimum of the track density), presumably corresponds to the pp12 protein, which is proline-rich (48) and probably quite flexible. This predicted flexibility is consistent with the striking variation in length of this domain. Finally, the inner track-like feature, within the interior of the particle, presumably contains the NC domain in association with the viral RNA.
A recent paper described a similar analysis of “immature” virus-like particles (VLPs) formed by expression of the HIV-1 Gag polyprotein in insect cells (40). In general, the results and conclusions are completely consistent with our results. However, one striking difference between HIV-1 VLPs and immature MuLV particles is in the radial distance between the outer border of the “railroad track” and the inner border of the matrix region. In the HIV-1 VLPs, this low density zone was ≈20 Å, whereas in the immature MuLV, it was quite variable (see Fig. 4a), ranging between 25 and 60 Å. As noted, we propose that this zone in MuLV includes the pp12 domain of Gag. This difference in morphology between immature MuLV particles and HIV-1 VLPs is consistent with the fact that HIV-1 Gag does not have a domain corresponding to pp12; taken together, these data lend support to the assignment of morphological features to the domains of Gag given in Fig. 7.
Yet another way to assess the plausibility of this model for the PR− MuLV particles is to calculate the predicted diameters of each of these domains. For instance, assuming a protein partial specific volume of 0.77 cm3/g, then the MA domain (15 kDa) should occupy 19,186 Å3. If this were a cylinder of length 40 Å, then the diameter would be 25 Å. In a similar fashion, we can use the lengths assigned to each of the Gag domains based on the radial density plots in Fig. 4 and the respective protein molecular weights to calculate that these other domains will also have diameters of ≈20–25 Å (Table 1). This diameter estimate is in reasonable agreement with the sizes observed for the MA (6) and CA fragments (8–10) of Gag solved by x-ray crystallography. This conclusion is also consistent with the hypothesis that the immature MuLV particle is composed of Gag monomers of roughly cylindrical shape and a mean diameter of ≈25 Å that are packed side-by-side with a center-to-center spacing of ≈45 Å, comparable with that observed for HIV-1 Gag VLPs (40). Notably, the published crystal structures of HIV-1 MA and CA are, respectively, trimeric and dimeric (6–10). Our data do not address whether the Gag proteins are oligomeric in either the immature or mature particles.
Table 1.
PR− MuLV composition and estimated protein dimensions
| Protein | Size | Volume, Å3 | Length, Å | Diameter, Å |
|---|---|---|---|---|
| Matrix (MA) | p15 | 19,186 | ≈40 | ≈25 |
| Connector (pp12) | p12 | 15,349 | ≈25–60 | ≈18–28 |
| Capsid (CA) | p30 | 38,372 | ≈100 | ≈20 |
| Nucleocapsid (NC) | p10 | 12,791 | ≈25 | ≈25 |
| Total length | ≈200 |
Implications for Viral Assembly.
A wide variety of plant and animal viruses have been shown to possess icosahedral symmetry. The hallmark of these symmetrical particles is an extremely regular arrangement of protein subunits, in which the only points of deviation from 6-fold symmetry are 12 axes of 5-fold symmetry at the vertices of the icosahedron (49, 50).
We now report that immature retrovirus particles apparently lack icosahedral symmetry. Thus, the geometric arrangement of the protein subunits in these particles is less regular than that in symmetrical particles. One explanation for the lack of strict regularity in the arrangement of the Gag protein subunits is that other interactions, in addition to Gag–Gag interactions, contribute to the assembly process. Intermolecular interactions that may play a role in assembly could, in principle, occur between Gag monomers and any of the other viral constituents. These constituents include a lipid bilayer derived from the plasma membrane of the host cell, the Env glycoprotein complex, the products of the Pol gene, the viral genomic RNA, and a significant number of low molecular weight RNA molecules. Which of these constituents might be significant in facilitating the self-assembly of Gag polyproteins?
In the type-C retroviruses, assembly is apparently initiated at the plasma membrane, and a strong plasma membrane-targeting signal is present at the N terminus of the Gag polyprotein. Thus, it is likely that membrane association facilitates Gag–Gag interactions by concentrating and aligning the Gag monomers at the membrane. There is strong support for this hypothesis in the case of MuLV, as mutant Gag molecules lacking the myristate portion of the membrane-targeting signal fail to be incorporated into virus particles, even in the presence of wild-type Gag (51). It should be noted, however, that MuLV differs from HIV-1 (52) and Rous sarcoma virus (53) in this regard.
In the type-B and -D retroviruses, core particles assemble in the cytoplasm without any prior membrane association of the Gag monomers (2). It has also been demonstrated that unmyristoylated HIV Gag, lacking a portion of the normal membrane-targeting signal, can assemble when expressed at high levels (54, 55). This suggests that these viruses follow a different assembly pathway from other retroviruses. However, it seems likely that the pathways are not qualitatively or fundamentally different because a single amino acid change in a type-D Gag polyprotein converts its morphogenesis pattern into that of a type-C virus (56). If this were correct, then interactions with the membrane would not play a fundamental role in type-C virus assembly; on the other hand, it seems that the basic distinction between type-C and type-D viruses is precisely the degree of dependence of assembly on membrane association.
Interactions with the Pol and Env gene products can both be excluded as essential for retroviral particle assembly because, as noted above, assembly occurs in the absence of these constituents (20, 21, 57). Particle assembly is also not dependent on genomic RNA, as was first demonstrated by treatment of cells producing wild-type MuLV with actinomycin D (58) and is also evident in packaging cells, in which wild-type viral proteins are encoded by mRNAs lacking packaging signals (ψ -RNA) (59) and in cells in which the zinc fingers of the NC domain have been eliminated by mutation (60–63).
As noted previously, retrovirus particles contain a substantial number of small RNA molecules in addition to the genomic RNA. These additional RNAs are presumably present in the particles lacking genomic RNA mentioned above (62, 64). The significance of these additional RNAs is completely unknown, except that if the Pol gene product is present, they are enriched for the tRNA species, which acts as the primer for reverse transcription. The mass of these RNAs in the particle is only a few times lower than that of the dimeric genomic RNA; thus, they are present at a ratio of several nucleotides per Gag monomer. This approaches the ratio at which free NC is saturated with nucleic acid (65). Surprisingly, whereas Rous sarcoma virus and HIV Gag proteins (or truncated Gag proteins) can assemble in vitro into virus-like particles without membranes or other proteins, this assembly, at least under certain conditions, is dependent on the presence of nucleic acid (66–68). This observation lends significant support for the possibility that the nucleic acid acts as a scaffold upon which Gag monomers assemble and that Gag–RNA interactions are critical for retrovirus assembly. Genomic RNA is not necessary for assembly, so it seems possible that the small RNAs can fulfill this role in the particle. Assembly of MuLV is presumably driven by a balance of protein–protein and protein–RNA interactions in which the protein contacts are not as rigidly fixed as in strictly icosahedral particles.
Implications for Budding.
The morphological signature of budding is the formation of a convex curvature in the plasma membrane. However, because type-B and -D retroviruses form roughly spherical cores before they reach the membrane, the lipid bilayer evidently plays no role in the formation of these curved structures during budding. Because, as noted above, type-C assembly pathways are probably not fundamentally different from those of type-B and -D viruses, the same is very likely true of Moloney MuLV.
To form a curved structure, the paracrystalline sheets of Gag monomers must have some curvature. As noted by Nermut and Hockley (69), if a Gag molecule deviated from a purely cylindrical shape by having a slight taper, then packing them together could give rise to spherical particles with diameters of ≈1,000 Å. It is also conceivable that curvature arises because the C termini of the monomers are held together more tightly by intermolecular interactions than the N termini.
Implications for Maturation.
Cleavage of the Gag polyprotein is associated with striking structural rearrangements of MuLV. Thus, as noted above, the paracrystalline packing in the PR− MuLV particles, manifested as first-order reflections in diffraction patterns (Fig. 3a Insets) and sharp peaks in rotationally averaged Fourier transforms (Fig. 6), is almost entirely absent in the WT MuLV particles. These changes suggest a loss of paracrystalline order upon polyprotein cleavage and a loosening of the packing of the individual proteins that were covalently attached as domains in the Gag polyprotein.
It seems reasonable that cleavage of the Gag precursor would lead to a substantial loss of paracrystalline order. For example, the MA molecules would no longer be covalently connected to the pp12 molecules, allowing rotational slippage between the MA and pp12 layers. The same would hold true for the other cleavage products. The loss of radial register would lead to rotational and radial disordering, as suggested by the Fourier analysis in Fig. 6. The angular appearance of the WT MuLV cores (Figs. 1 and 3b) may be a remnant of the paracrystalline packing of the CA domains in the immature particles.
Further clarification of these possibilities may require a combination of site-specific mutagenesis, electron cryo-microscopy, and image processing. Although our results only provide a low-resolution snapshot of two dynamic states of MuLV, the ultimate goal of our studies is to provide particle structures with sufficient detail to allow the docking of atomic-resolution structures of the component proteins into a model of the entire particle. Given the pleomorphic nature of retroviruses, we anticipate that this approach will provide constraints for a family of structures that are in agreement with the data.
Acknowledgments
We thank Steven Fuller and Volker Vogt and their colleagues for sharing their results with us prior to publication, John E. Johnson for helpful discussions, Stephen Campbell for a thoughtful reading of the manuscript, and Ron Milligan for assistance with preliminary experiments. We also acknowledge the technical assistance of Jane Mirro in the preparation of the MuLV and Brian Sheehan for preparing Fig. 7. A.R. has been supported in part by the National Cancer Institute, Department of Health and Human Services, under contract to Advanced BioScience Laboratories. P.O.B. is an Associate Investigator of the Howard Hughes Medical Institute. M.Y. is an Established Investigator of the American Heart Association and Bristol-Myers Squibb and has been supported by the National Institutes of Health (RO1AI31535, RO1HL48908), the Gustavus and Louise Pfeiffer Research Foundation, and the Donald E. and Delia B. Baxter Research Foundation.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: PR−, protease-deficient; WT, wild type; MuLV, murine leukemia virus; VLPs, virus-like particles; FFT, Fourier transforms.
References
- 1.Varmus H, Brown P. In: Mobile DNA. Howe M M, Berg D E, editors. Washington, DC: Am. Soc. Microbiol.; 1989. pp. 53–108. [Google Scholar]
- 2.Coffin J M. In: Fields Virology. Fields B N, Knipe D M, Howley P M, et al., editors. Philadelphia: Lippincott; 1996. pp. 1767–1847. [Google Scholar]
- 3.Ringe D. Methods Enzymol. 1994;241:157–177. doi: 10.1016/0076-6879(94)41064-x. [DOI] [PubMed] [Google Scholar]
- 4.Kohlstaedt L A, Wang J, Friedman J M, Rice P A, Steitz T A. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 5.Jacobo-Molina A, Ding J, Nanni R G, Clark A D, Jr, Lu X, Tantillo C, Williams R L, Kamer G, Ferris A L, Clark P, Hizi A, Hughes S H, Arnold E. Proc Natl Acad Sci USA. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hill C P, Worthylake D, Bancroft D P, Christensen A M, Sundquist W I. Proc Natl Acad Sci USA. 1996;93:3099–3104. doi: 10.1073/pnas.93.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Momany C, Kovari L C, Prongay A J, Keller W, Gitti R K, Lee B M, Gorbalenya A E, Tong L, McClure J, Ehrlich L S, Summers M F, Carter C, Rossmann M G. Nat Struct Biol. 1996;3:763–770. doi: 10.1038/nsb0996-763. [DOI] [PubMed] [Google Scholar]
- 8.Gitti R K, Lee B M, Walker J, Summers M F, Yoo S, Sundquist W I. Science. 1996;273:231–235. doi: 10.1126/science.273.5272.231. [DOI] [PubMed] [Google Scholar]
- 9.Gamble T R, Vajdos F F, Yoo S, Worthylake D K, Houseweart M, Sundquist W I, Hill C P. Cell. 1996;87:1285–1294. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- 10.Gamble T R, Yoo S, Vajdos F F, von Schwedler U K, Worthylake D K, Wang H, McCutcheon J P, Sundquist W I, Hill C P. Science. 1997;278:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 11.Dyda F, Hickman A B, Jenkins T M, Engelman A, Craigie R, Davies D R. Science. 1994;266:1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- 12.Summers M F, Henderson L E, Chance M R, Bess J W, Jr, South T L, Blake P R, Sagi I, Perez-Alvarado G, Sowder R C, III, Hare D R, et al. Protein Sci. 1992;1:563–574. doi: 10.1002/pro.5560010502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morellet N, Jullian N, de Rocquigny H, Maigret B, Darlix J-L, Roques B P. EMBO J. 1992;11:3059–3065. doi: 10.1002/j.1460-2075.1992.tb05377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morellet N, de Rocquigny H, Mély Y, Jullian N, Déméné H, Ottmann M, Gérard D, Darlix J L, Fournie-Zaluski M C, Roques B P. J Mol Biol. 1994;235:287–301. doi: 10.1016/s0022-2836(05)80033-6. [DOI] [PubMed] [Google Scholar]
- 15.Lu M, Blacklow S C, Kim P S. Nat Struct Biol. 1995;2:1075–1082. doi: 10.1038/nsb1295-1075. [DOI] [PubMed] [Google Scholar]
- 16.Chan D C, Fass D, Berger J M, Kim P S. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 17.Weissenhorn W, Dessen A, Harrison S C, Skehel J J, Wiley D C. Nature (London) 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 18.Fass D, Harrison S C, Kim P S. Nat Struct Biol. 1996;3:465–469. doi: 10.1038/nsb0596-465. [DOI] [PubMed] [Google Scholar]
- 19.Fass D, Davey R A, Hamson C A, Kim P S, Cunningham J M, Berger J M. Science. 1997;277:1662–1666. doi: 10.1126/science.277.5332.1662. [DOI] [PubMed] [Google Scholar]
- 20.Shields A, Witte O N, Rothenberg E, Baltimore D. Cell. 1978;14:601–609. doi: 10.1016/0092-8674(78)90245-3. [DOI] [PubMed] [Google Scholar]
- 21.Wills J W, Craven R C. AIDS. 1991;5:639–654. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]
- 22.Nermut M V, Frank H, Schäfer W. Virology. 1972;49:345–358. doi: 10.1016/0042-6822(72)90487-4. [DOI] [PubMed] [Google Scholar]
- 23.Yoshinaka Y, Luftig R B. Proc Natl Acad Sci USA. 1977;74:3446–3450. doi: 10.1073/pnas.74.8.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schätzl H, Gelderblom H R, Nitschko H, von der Helm K. Arch Virol. 1991;120:71–81. doi: 10.1007/BF01310950. [DOI] [PubMed] [Google Scholar]
- 25.Gelderblom H R, Hausmann E H S, Özel M, Pauli G, Koch M A. Virology. 1987;156:171–176. doi: 10.1016/0042-6822(87)90449-1. [DOI] [PubMed] [Google Scholar]
- 26.Nermut M V, Grief C, Hashmi S, Hockley D J. AIDS Res Hum Retroviruses. 1993;9:929–938. doi: 10.1089/aid.1993.9.929. [DOI] [PubMed] [Google Scholar]
- 27.Hockley D J, Nermut M V, Grief C, Jowett J B M, Jones I M. J Gen Virol. 1994;75:2985–2997. doi: 10.1099/0022-1317-75-11-2985. [DOI] [PubMed] [Google Scholar]
- 28.Nermut M V, Hockley D J, Jowett J B M, Jones I M, Garreau M, Thomas D. Virology. 1994;198:288–296. doi: 10.1006/viro.1994.1032. [DOI] [PubMed] [Google Scholar]
- 29.Southern P J, Berg P. J Mol Appl Genet. 1982;1:327–341. [PubMed] [Google Scholar]
- 30.Fu W, Rein A. J Virol. 1993;67:5443–5449. doi: 10.1128/jvi.67.9.5443-5449.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graham F L, van der Eb A J. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 32.Yeager M, Dryden K A, Olson N H, Greenberg H B, Baker T S. J Cell Biol. 1990;110:2133–2144. doi: 10.1083/jcb.110.6.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yeager M, Berriman J A, Baker T S, Bellamy A R. EMBO J. 1994;13:1011–1018. doi: 10.1002/j.1460-2075.1994.tb06349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klug A, Finch J T. J Mol Biol. 1968;31:1–12. doi: 10.1016/0022-2836(68)90050-8. [DOI] [PubMed] [Google Scholar]
- 35.Crowther R A. Phil Trans R Soc Lond B. 1971;261:221–230. doi: 10.1098/rstb.1971.0054. [DOI] [PubMed] [Google Scholar]
- 36.Fuller S D. Cell. 1987;48:923–934. doi: 10.1016/0092-8674(87)90701-x. [DOI] [PubMed] [Google Scholar]
- 37.Baker T S, Drak J, Bina M. Proc Natl Acad Sci USA. 1988;85:422–426. doi: 10.1073/pnas.85.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crowther R A, Amos L A. J Mol Biol. 1971;60:123–130. doi: 10.1016/0022-2836(71)90452-9. [DOI] [PubMed] [Google Scholar]
- 39.Barklis E, McDermott J, Wilkens S, Schabtach E, Schmid M F, Fuller S, Karanjia S, Love Z, Jones R, Rui Y, Zhao X, Thompson D. EMBO J. 1997;16:1199–1213. doi: 10.1093/emboj/16.6.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fuller S D, Wilk T, Gowen B E, Kräusslich H-G, Vogt V M. Curr Biol. 1997;7:729–738. doi: 10.1016/s0960-9822(06)00331-9. [DOI] [PubMed] [Google Scholar]
- 41.Gelderblom H R. AIDS. 1991;5:617–638. [PubMed] [Google Scholar]
- 42.Bolognesi D P, Montelaro R C, Frank H, Schäfer W. Science. 1978;199:183–186. doi: 10.1126/science.202022. [DOI] [PubMed] [Google Scholar]
- 43.Davis N L, Rueckert R R. J Virol. 1972;10:1010–1020. doi: 10.1128/jvi.10.5.1010-1020.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoo S, Myszka D G, Yeh C-y, McMurray M, Hill C P, Sundquist W I. J Mol Biol. 1997;269:780–795. doi: 10.1006/jmbi.1997.1051. [DOI] [PubMed] [Google Scholar]
- 45.Henderson L E, Krutzsch H C, Oroszlan S. Proc Natl Acad, Sci USA. 1983;80:339–343. doi: 10.1073/pnas.80.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rein A, McClure M R, Rice N R, Luftig R B, Schultz A M. Proc Natl Acad Sci USA. 1986;83:7246–7250. doi: 10.1073/pnas.83.19.7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Craven R C, Parent L J. Curr Top Microbiol Immunol. 1996;214:65–94. doi: 10.1007/978-3-642-80145-7_3. [DOI] [PubMed] [Google Scholar]
- 48.Shinnick T M, Lerner R A, Sutcliffe J G. Nature (London) 1981;293:543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- 49.Caspar D L D, Klug A. Cold Spring Harbor Symp Quant Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 50.Johnson J E, Speir J A. J Mol Biol. 1997;269:665–675. doi: 10.1006/jmbi.1997.1068. [DOI] [PubMed] [Google Scholar]
- 51.Schultz A M, Rein A. J Virol. 1989;63:2370–2373. doi: 10.1128/jvi.63.5.2370-2373.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morikawa Y, Hinata S, Tomoda H, Goto T, Nakai M, Aizawa C, Tanaka H, Omura S. J Biol Chem. 1996;271:2868–2873. doi: 10.1074/jbc.271.5.2868. [DOI] [PubMed] [Google Scholar]
- 53.Wills J W, Craven R C, Weldon R A, Jr, Nelle T D, Erdie C R. J Virol. 1991;65:3804–3812. doi: 10.1128/jvi.65.7.3804-3812.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee P P, Linial M L. J Virol. 1994;68:6644–6654. doi: 10.1128/jvi.68.10.6644-6654.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spearman P, Wang J-J, Vander Heyden N, Ratner L. J Virol. 1994;68:3232–3242. doi: 10.1128/jvi.68.5.3232-3242.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rhee S S, Hunter E. Cell. 1990;63:77–86. doi: 10.1016/0092-8674(90)90289-q. [DOI] [PubMed] [Google Scholar]
- 57.Scheele C M, Hanafusa H. Virology. 1971;45:401–410. doi: 10.1016/0042-6822(71)90341-2. [DOI] [PubMed] [Google Scholar]
- 58.Levin J G, Grimley P M, Ramseur J M, Berezesky I K. J Virol. 1974;14:152–161. doi: 10.1128/jvi.14.1.152-161.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mann R, Mulligan R C, Baltimore D. Cell. 1983;33:153–159. doi: 10.1016/0092-8674(83)90344-6. [DOI] [PubMed] [Google Scholar]
- 60.Méric C, Spahr P-F. J Virol. 1986;60:450–459. doi: 10.1128/jvi.60.2.450-459.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gorelick R J, Henderson L E, Hanser J P, Rein A. Proc Natl Acad Sci USA. 1988;85:8420–8424. doi: 10.1073/pnas.85.22.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Méric C, Goff S P. J Virol. 1989;63:1558–1568. doi: 10.1128/jvi.63.4.1558-1568.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Berkowitz R, Fisher J, Goff S P. Curr Top Microbiol Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 64.Levin T G, Seidman J G. J Virol. 1979;29:328–335. doi: 10.1128/jvi.29.1.328-335.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Karpel R L, Henderson L E, Oroszlan S. J Biol Chem. 1987;262:4961–4967. [PubMed] [Google Scholar]
- 66.Campbell S, Vogt V M. J Virol. 1995;69:6487–6497. doi: 10.1128/jvi.69.10.6487-6497.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Campbell S, Vogt V M. J Virol. 1997;71:4425–4435. doi: 10.1128/jvi.71.6.4425-4435.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gross I, Hohenberg H, Kräusslich H-G. Eur J Biochem. 1997;249:592–600. doi: 10.1111/j.1432-1033.1997.t01-1-00592.x. [DOI] [PubMed] [Google Scholar]
- 69.Nermut M V, Hockley D J. Curr Top Microbiol Immunol. 1996;214:1–24. doi: 10.1007/978-3-642-80145-7_1. [DOI] [PubMed] [Google Scholar]