Abstract
The transactivation activity of the p53 tumor suppressor protein is critical for regulating cell growth and apoptosis. We describe the identification of a transcription factor that is functionally similar to p53 and contains the same DNA binding and transcription activities specific for the p53 responsive DNA element (p53RE). This protein was highly purified through chromatography from HeLa cell extracts. The purified protein was able to bind specifically to the p53RE derived from a p21waf1 promoter and to stimulate p53RE-dependent transcription but not basal transcription in vitro. Its DNA-binding activity was inhibited by the wild type but not mutant p53RE-containing DNA oligomers. Also, this p53RE-binding activity was found in human p53 null Saos-2 osteosarcoma and H1299 small cell lung carcinoma cells. Interestingly, this activity exhibited a p53RE sequence preference that was distinct from the p53 protein. The activity is neither p53 nor p73, because anti-p53 or anti-73 antibodies were unable to detect this purified protein nor were the antibodies able to alter the p53-like activity, the p53RE-protein complex. These results demonstrate that, besides p73, an additional p53-like protein exists in cells, which is named NBP for non-p53, p53RE binding protein.
Regulation of cell growth and apoptosis by the p53 tumor suppressor protein is primarily executed through its transactivation capability (1–3). Numerous studies have demonstrated that the wild-type p53 nuclear phosphoprotein acts as a transcriptional activator with a sequence-specific DNA-binding activity (1–3). This protein contains three major functional domains: (i) the N-terminal transactivation domain (amino acids 1–45) (4, 5), (ii) the central sequence-specific DNA binding domain (aa 113–290) (6–9), and (iii) the C-terminal regulatory region (1, 10). The N terminus contacts several transcription regulators including the components of RNA polymerase II transcription machinery, such as TAFII31 and TAFII70 (11, 12), and the coactivator proteins p300 and CBP (13–15). These interactions are crucial for the transcription function of p53, although the detailed mechanisms remain elusive. The C-terminus also plays a role in regulating the p53-dependent transcription (1–3 and references therein). Posttranslational modification such as phosphorylation of this region and the N terminus has been postulated to activate p53’s DNA binding (16) and transcription activities in response to DNA damage (17–19).
The importance of the p53 transcription activity is also evident in the fact that p53 can transactivate the expression of many target genes. The p53 central domain specifically binds to a p53 responsive DNA element (p53RE) with two copies of the 10-mer (5′RRRCA/TT/AGYYY3′) (20, 21). The p53RE with some variations among the p53 target genes (for example see Fig. 4A) is usually localized at 300–2,000 bases upstream from the transcriptional initiation site. Many putative p53 responsive elements have been identified (22) and some of them well characterized as downstream genes in the p53 pathway (1–3). For example, the p21waf1 gene (23, 24) is transcriptionally stimulated by p53 and mediates in part a G1 arrest, by binding to and inhibiting CDK family kinases (25). Also, the expression of the cellular p53 negative regulator, MDM2 (26, 27), is elevated by p53, forming a negative regulatory feedback loop (28, 29). Moreover, p53 stimulates the expression of several apoptosis regulatory genes (30) including Bax1 (31), which may serve as downstream mediators of p53-induced apoptosis (30, 31). Thus, the transcription activity of p53 is important for its function to protect cells from undergoing tumorigenesis. The central DNA-binding domain is evolutionarily conserved from Xenopus to human (32) and is the region that sustains the mutations during carcinogenesis (33).
Figure 4.
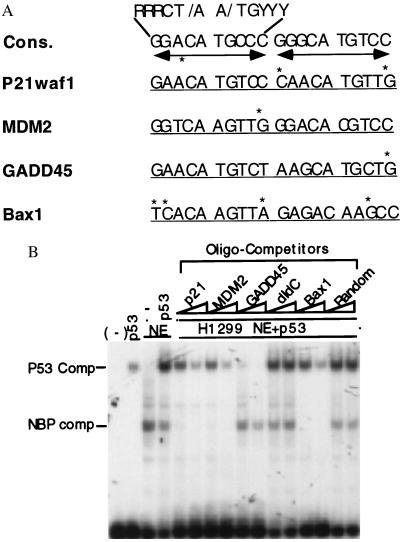
NBP and p53 display differential p53RE-binding affinities. (A) Alignments of the p53RE motif 20 mers derived from different p53 responsive promoters (20, 21, 23, 24, 28, 31, 48). Cons., The consensus p53 binding sequence. Asterisks are placed above nucleotides to indicate deviations from the consensus sequence. The dual arrow bars denote the palindrome sequences. (B) NBP exhibits a differential sequence preference distinct from p53. The EMSA was performed as described in Fig. 1B. p53 (40 ng) was used in lanes 2, 4–16. One microliter (≈4 μg of protein) of the H1299 cell nuclear extracts was used in lanes 3–16 as indicated on top. All the nonlabeled oligomer competitors (10 and 30 ng) were used in the lanes as indicated on top.
Given the central role that p53 mutations play in the origins of cancer, it was perhaps surprising that related transcriptional activities have not been reported until recently. A p53-related gene product, p73, containing ≈60% identity of amino acid sequences in the DNA binding domain has been described (34). P73 can transcriptionally activate p53 target genes such as p21 or bax-1, and induce growth suppression or apoptosis (34, 35). These studies suggested the possibility that there may be other p53 homologs in cells.
As a first step in investigating these possibilities, a search was initiated for p53-independent transcription and DNA-binding activities that were specific to the p53RE derived from the p21waf1 promoter (23, 24). Using a conventional fractionation procedure with HeLa cell nuclear extracts, a p53-like transcriptional factor was identified. This factor has been highly purified. It exhibits a p53RE-specific DNA binding and transactivation activity in vitro. This protein was neither p53 nor p73, as anti-p53 and anti-p73 antibodies failed to alter the DNA-protein complexes nor detected any polypeptide in the purified protein sample. This activity was also found in p53-deficient Saos-2 and H1299 cell lines. Interestingly, this protein displayed a p53RE sequence preference distinct from p53 itself. These results demonstrate the existence of another p53 functional homolog in cells. Because of its functional similarity to p53, this activity has been named non-p53 p53RE binding protein (NBP).
MATERIALS AND METHODS
Cell Culture.
Human cervical carcinoma HeLa, human osteosarcoma Saos-2, and small cell lung carcinoma H1299 cells were cultured as described (11, 36, 37).
Preparation of Nuclear Extracts.
Nuclear extracts were prepared from Soas-2, H1299 and HeLa cells by using a method described previously (38).
Purification of the p53-Like Transcriptional Factor NBP.
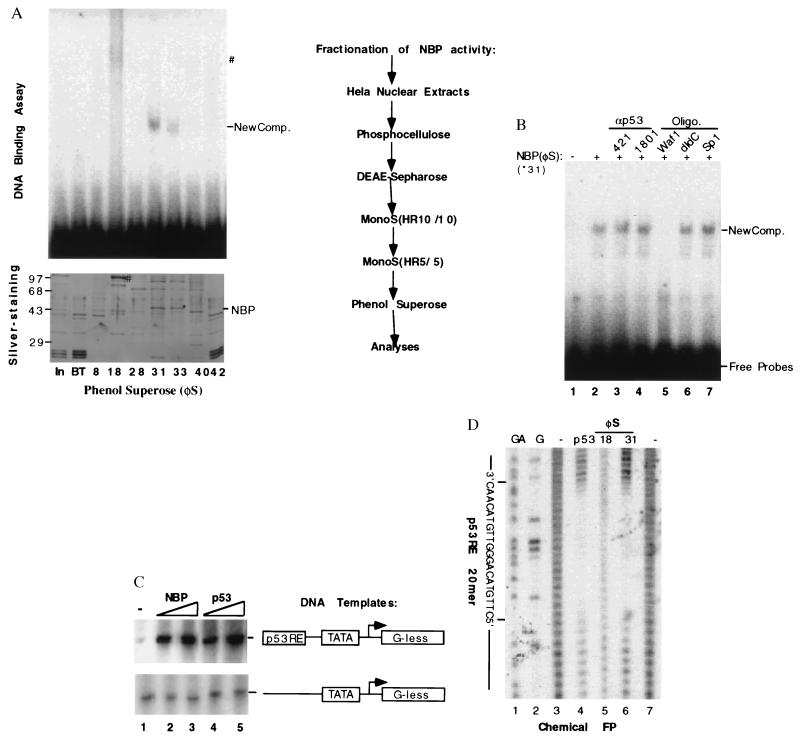
Nuclear extracts [0.6 g (in 60 ml)] from ≈1010 HeLa cells was used as starting material and was fractionated through phosphocellulose (P11) and DEAE Sepharose columns as described (39). The p53RE-binding and transcriptional activities of the fractions were assayed by using electrophoretic mobility-shift assay (EMSA) and in vitro transcription reaction as described before (11, 40). P53RE DNA binding and transcription activities were found in the 0.5 M wash of the first column and the flow through fraction from the second column (Fig. 2A). This active flow through material (6 mg) was further fractionated on a MonoS (HR10/10, Pharmacia) column that was equilibrated with buffer C 100 (BC100) containing 20 mM Tris⋅HCl (pH 7.9), 0.1 mM EDTA, 10% glycerol, 100 mM KCl, 4 mM MgCl2, 0.2 mM phenylmethylsulfonyl fluoride, 1 mM DTT, and 0.25 μg/ml of pepstatin A. Proteins were eluted with a linear gradient of KCl from 0.1–0.8 M. The pool (1.2 mg) of fractions containing the p53RE-binding activity was dialyzed against BC100 and further purified on a smaller MonoS (HR5/5, Pharmacia) column with a different linear gradient range of KCl from 0.1 to 0.5 M. The pool (0.7 mg) of the p53RE-binding activity containing fractions was dialyzed against a buffer identical to BC100 except 1.3 M ammonium sulfate used and loaded onto a micro phenol Superose column (Pharmacia) for further purification. The active protein fractions were dialyzed against BC50 (50 mM KCl) and stored at −80°C for further analyses.
Figure 2.
Purification and functional characterization of NBP. (A) Purification steps of the NBP protein from HeLa cell nuclear extracts and analyses of the fractions from the last phenol Superose (φS) by EMSA and silver-staining SDS-PAGE. The columns used for purification of the p53RE-mediated transcription activity were listed on right. On left are shown the results from EMSA (Upper) and silver-stained SDS/10% polyacrylamide gel analyses of the peak fractions from the last column. For EMSA, 2 μl of each fraction was used & for silver-staining gel, 10 μl of each fraction was directly loaded. NBP and ∗ indicate the polypeptide band on the denatured gel, which is corresponding to the p53RE binding complexes on the EMSA gel. #, The polypeptide (Lower) that may be responsible for the smeared DNA-protein complex on the native gel (Upper). (B) The effect of anti-p53 antibodies and DNA oligomers on the p53RE-binding activity of the purified NBP. Fraction 31 (60 ng) was used in this EMSA. 0.5 μg of 421 and 1801, 0.1 μg of each oligomer as indicated on top were incubated with the purified protein 15 min before addition of the 32P-labeled p53RE DNA probe mixture into the mixture. Similar assays were conducted with anti-p73 antibodies and the mutant p53RE DNA oligomers derived from the p21 promoter. Anti-p73 antibodies and the mutant oligomers did not affect the NBP-p53RE complex (data not shown). New Comp., The NBP-p53RE complex. (C) The transcription activity of NBP in vitro. The in vitro transcription assay was conducted by using the same conditions as for Fig. 1A. Fraction 31 (60 and 120 ng) of the phenol Superose (φS) column and purified p53 (40 and 80 ng) were used in the reaction, respectively. (D) The chemical footprinting analysis of the purified NBP protein. For each lane, the DNA probes were pooled from quadruple DNA-binding reactions.
EMSA.
This assay was performed based on a published method (20, 40). The protein components, as indicated in figure legends, were incubated with 3′end-labeled DNA fragments harboring two copies of the p53RE sequence (5,000 cpm, 1.0 ng DNA per assay) for 30 min at room temperature. The reaction mixture (20 μl) contained 10 mM Hepes buffer, pH 7.5, 4 mM MgCl2, 60 mM NaCl, 0.1 μg poly(dIdC), 0.1% Nonidet P-40, and 0.1 mM EDTA. The complexes formed were separated by electrophoresis on a 4% native gel.
In Vitro Transcriptional Assay.
Mixtures (40 μl) for the specific transcription reaction as described (11, 41) were incubated at 30°C for 60 min. and contained 20 mM Hepes buffer (pH 7.9), 8 mM MgCl2, 60 mM KCl, 10 mM Ammonium sulfate, 12% glycerol, 10 mM 2-mercaptoethanol, 2% (wt/vol) PEG 8,000, 20 units RNase T1, 0.6 μM ATP and CTP, 15 mM [α-32P]UTP (10,000 cpm/pmol), and 0.5 μg of p53RE-adenovirus major late promoter (AdMLP) or AdMLP plasmid DNAs (11). The transcription factors purified by published methods (41 and references therein) were: TFIIA (DEAE-5PW fraction, 250 ng), rTFIIB (P11 fraction, 30 ng), purified eTFIID (12CA5 immunoaffinity column, 60 ng), rTFIIE (Sephacryl 200, 30 ng), rTFIIF (40 ng), TFIIH (μMono Q, 40 ng), and RNA polymerase II (DEAE-5PW, 50 ng). RNA transcripts of the reactions were separated by a urea-PAGE and visualized by autoradiography.
Chemical Footprinting Analysis.
A chemical footprinting assay was performed as described (42). DNA-protein complexes were formed as described above. Before electrophoresis, the p53RE-containing DNA templates were digested with a chemical mixture of methidiumpropyl-EDTA-(Fe)II and Fe(NH4)2(SO4)2. The reactions were halted by addition of 1 μl of 0.5 M EDTA and the complexes were separated by a native PAGE. The DNA fragments in the complexes were electroeluted from the gel and subjected to electrophoresis on an 8% polyacrylamide sequencing gel. The G and G + A DNA ladders were prepared by the Maxam–Gilbert method (43).
Western Blot Analysis.
The protein fractions from different columns and nuclear extracts were subject to SDS/PAGE. Western blot analysis was carried out as described (11) using anti-p73 (anti-p73-α and p73-β mAbs provided by William Kaelin) and anti-p53 antibodies and proteins were detected with enhanced chemiluminescence reagents (Amersham).
RESULTS AND DISCUSSION
Identification of a p53RE-Dependent DNA-Binding and Transcription Activity from HeLa Cells.
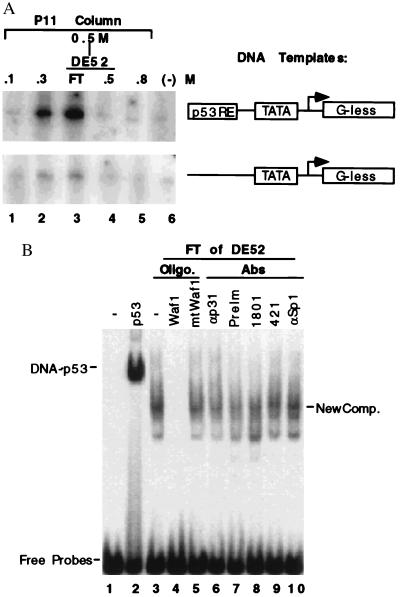
During the purification of general transcription factors specific for RNA polymerase II from HeLa cell nuclear extracts (39–41), the fractions from the first two columns (see the purification chart in Fig. 2A) were examined to determine whether a transcription activity driven by the p53RE-containing promoter was present by using an in vitro specific transcription run-off assay as described in Materials and Methods (11, 41). Two DNA templates (Fig. 1A) used in this reaction were the AdMLP, which is linked to a G-less cassette of 400 bases located 3′ to the transcriptional initiation site (44), with or without two copies of a synthetic p53RE sequence derived from either the p21waf1 (20, 21) or the MDM2 (26, 27) promoters as described (11). The proteins (39–41) used in the reaction included RNA polymerase II, TFIIA, eTFIID, and TFIIH purified from HeLa nuclear extracts, recombinant (r) TFIIB, rTFIIE, and rTFIIF purified from Escherichia coli. A p53RE-dependent transcription activity was detected in two fractions, the 0.3 M salt wash of the P11 column and the flowthrough (FT) of DE-52 column, which derived from the 0.5 M wash of P11. This transcription activity was specific to the p53 binding site (Upper), but rarely detectable when a p53RE-less template was used (Lower). The 0.3 M wash was later found to contain anti-p73 antibody-reacting materials determined by Western blot analysis (Fig. 3) and thus that transcription activity may be mediated by the p73 protein. The FT of DE-52 was chosen for further purification because this fraction appeared to possess a higher activity than the 0.3 M fraction of P11 (compare lane 3 with lane 2). This result indicates that there is a p53RE-dependent transcription activity in HeLa cells, which may not be due to p53. p53 is hardly detectable in the HeLa cells due its proteolytic degradation enhanced by the papillomavirus E6 oncoprotein (45).
Figure 1.
Identification of a p53RE-dependent DNA binding and transcription activity. (A) A p53RE-dependent transcription activity in HeLa nuclear extracts. The fractions (2 μl) from P11 column and DE-52 column were assayed for the transcriptional activation by using AdMLP promoter-derived templates with or without the p53RE as indicated on right, as described in Materials and Methods. The fractions tested are indicated on top. (B) The effect of anti-p53 antibodies and DNA oligomers on the p53RE-binding activity in the flow through (FT) of DE-52 column. 1 μl of the FT sample was used in the assay containing 5,000 cpm of the p53RE probes (1 ng) digested from the p53RE-AdMLP DNA template used in the above transcription reaction. 0.5 μg of poly(dIdC) as nonspecific competitors in all the reactions and additionally, 0.3 μg of specific DNA competitors as indicated on top were used in lanes 4 and 5. 0.5 μg of antibodies as indicated on top was used. On lane 2, 100 ng of p53 purified from baculovirus-infected insect SF9 cells was used.
Figure 3.
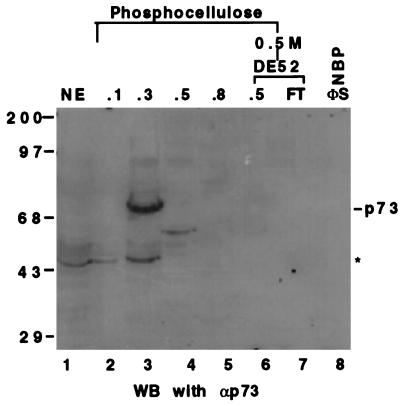
The Western blot analysis of the fractions from P11 and DE-52 columns using anti-p73 mAbs. Twenty-five microliters of each fraction or HeLa cell nuclear extracts as indicated (Top) was directly loaded onto a SDS/10% polyacrylamide gel. The antibodies recognize both the α and β forms of the p73 protein (35). The asterisk denotes that those signals picked up by the antibodies could be due to nonspecific cross reaction or to the proteolytically degraded products of p73. The band on lane 4 (fraction 0.5 M of P11) may be p73β. The numbers on top indicate the salt concentration that was used to wash the proteins from the columns.
To test whether the transcriptionally active FT fractions possess a p53RE binding activity, EMSA was conducted with the p53RE-containing DNA fragment derived from the p21 promoter (20, 21). The DE-52 FT exhibited a DNA-protein complex (lane 3, Fig. 1B) distinct from the p53-DNA complex (lane 2). This complex was competed specifically by wild-type (lane 4) but not mutant (lane 5) p53RE-containing DNA oligomers, indicating that this is a p53RE specific DNA complex. The two anti-p53 mAbs Pab1801 and Pab421 (lanes 8 and 9) did not affect (i.e., gel shift) the new DNA-protein complexes. The p53 protein was not detected either in the fractions by immunoprecipitation with another anti-p53 mAb Pab 1621 (its epitope locates at aa 88–103) followed by Western blot analysis with 1801 recognizing the aa 32–79 or 421 interacting with the aa 370–378 (data not shown). As controls, preimmune serum, anti-TAFII31 and anti-Sp1 antibodies did not affect these complexes. Thus, these results suggest that a human transcription factor that binds to the p53RE, but is not itself p53, might be detected in these fractions.
Purification of the p53RE-Binding Activity.
To learn the nature of the activity and to further characterize it, we decided to purify this new factor by monitoring the p53RE-binding activity using EMSA through conventional chromatography. This p53RE binding activity has been extensively purified through five columns, P11, DE-52, MonoS (large), MonoS (small), and phenol Superose (Fig. 2A). Starting with 0.6 g of HeLa nuclear extracts, we ended with 5 μg of proteins that contain the p53RE-binding activity. A representative result in Fig. 2A displays the silver-staining and EMSA of some protein peak fractions from the last column. As it reveals, the fractions 31 and 33 of the phenol Superose column exhibited sharp p53RE-protein complexes (Fig. 2A Upper). Noticeably, the fraction #18 also showed a smeared, slowly migrating DNA-protein complex that copurified with a predominant polypeptide of ≈90 Kd. This protein was excluded as a candidate because we determined that it is the poly(ADP ribose) polymerase by micropeptide sequencing. Poly(ADP ribose) polymerase modifies nuclear proteins by poly(ADP ribosyl) action (46) and binds to DNA oligomers nonspecifically as confirmed by a chemical footprinting analysis (Fig. 2D, lane 5). Hence, the active fractions 31–33 were further analyzed as described below.
NBP Is Neither p53 Nor p73.
As shown in Fig. 2A, several polypeptides appeared to cofractionate with this DNA binding activity as analyzed by silver-staining after SDS/PAGE (Fig. 2A Lower). The polypeptide migrating at the ≈44 Kd position appears to be a good candidate for the following reasons: (i) The protein density of this band correlates with the strength of the p53RE-binding activity (compare fraction 31 with fraction 33); (ii) The DNA-protein complex formed with this protein migrates faster than the p53-DNA complex (Fig. 2A), consistent with the size of this polypeptide (≈44 Kd). Hence, we believe that the 44 Kd polypeptide may represent NBP.
P53 was not present in the active fractions as none of the anti-p53 antibodies either reacted with proteins in the fractions by immunoprecipitation-Western blot analysis analyses (data not shown) or altered the DNA-protein complex formed with the fractions (Fig. 2B). Also, a band that comigrates with p53 was not visualized on the silver-stained SDS-gel (Fig. 2A Lower). The 44 Kd protein is not the proteolytic product of p53 for two reasons: (i) Neither 1801 nor 421 antibodies detected a protein from the fractions 31 and 33, indicating that none of the p53 fragments containing the N or C terminus exists in the fractions; and (ii) The fraction 31 was active in the in vitro p53RE-dependent transcription assay (Fig. 2C). P53 with a deleted amino terminus is not functional in a transcription assay (4–8). Thus, these results suggest that this transcription activity is not p53.
P73 is not likely NBP either (Fig. 2A) as evident from these observations: (i) The new DNA-protein complexes migrated faster than the p53- or p73-DNA complex (Fig. 2A and data not shown), suggesting that a polypeptide smaller than p53 or p73 may be the candidate; (ii) p73 was hardly detectable in the active fractions using Western blot analysis with anti-p73 mAbs (Fig. 3, lanes 7 and 8). Instead, a significant amount of the p73 protein was detected in the 0.3 M fraction of the P11 column (Fig. 3, lane 3), which contains a p53-like transcription activity (Fig. 1A). Although the real native molecular mass of NBP remains to be determined, its DNA complex would not be as small as was detected (Figs. 1B and 2B) if it was composed of a polypeptide >70 Kd. It remains formally possible that NBP could be an as yet undetected splice variant of p73 that fails to react with the available antibodies.
The Partially Purified NBP Is Active in p53RE-Binding and Transcription Reactions in Vitro.
To verify whether NBP truly binds to the p53 responsive DNA site, oligomer competition EMSA and footprinting analyses were utilized. The p53RE-NBP complex was reduced (Fig. 2B, lane 5) in the presence of the p53RE-containing oligomer from the p21waf1 promoter but not of nonspecific DNA oligomers (lanes 6 and 7). A chemical methidiumpropyl-EDTA/(Fe)II method was employed for the footprinting analysis, as described (42). The chemical footprinting analysis showed that NBP from the fraction 31 of phenol Superose, like p53, protected the exact p53RE 20 mer from digestion by the methidiumpropyl-EDTA reagent (Fig. 2D, compare lanes 4 with 6). In contrast, the proteins in the fraction 18 was unable to protect this sequence specifically (lane 5). This clearly proves that NBP possesses a bona fide p53RE-binding activity. Accordingly, the purified NBP protein, like p53 (Fig. 2C Upper, lanes 4 and 5), also stimulated the p53RE-driven transcription in vitro (Fig. 2C Upper, lanes 2 and 3) in a dose-dependent fashion. By contrast, in the absence of the p53RE motif, NBP was without any effect on the basal transcription (bottom, Fig. 2C). Thus, the purified NBP is active in both p53RE-dependent DNA binding and transcription reactions in vitro.
The Differential Binding Affinities of NBP and p53 to Different p53RE Motifs.
Consistent with the above results, an NBP-like p53RE binding activity was also identified in two human p53 null cell lines: Saos-2 and H1299 cells (data not shown), confirming the existence of the NBP activity in cells. It was observed that the NBPp53RE complexes from these cell extracts were readily abolished by the p53RE-containing oligomers derived from the p21 promoter but not from the GADD45 promoter (data not shown). This suggests that the activity may have a different affinity to the different p53RE motifs. Supporting this idea is the fact that the p53RE sequence from one p53 target promoter always differs from that derived from other p53 target genes. For instance, the Bax1-derived p53RE motif has four mismatches, whereas the GADD45-derived p53RE only possesses a single mismatch, compared with the p53RE consensus sequence (Fig. 4A). It is also possible that NBP and p53 may have different affinities for the same p53RE motif. To test these ideas, the following oligomer competition experiment was conducted (Fig. 4B). P53 was preincubated together with H1299 nuclear extracts in the presence of increasing amounts of the nonlabeled oligomer competitors at room temperature for 10 minutes before addition of the 32P-labeled p53RE oligomer probes derived from the p21 promoter. Interestingly, 10 ng (10-fold more in molar ratio than the probe) of the p21-, MDM2-, and Bax1-derived p53RE motifs completely abolished the NBP-DNA complex but not the p53-DNA complex (compare lane 4 with lanes 5, 7, and 13). In contrast, the same amount of the GADD45-derived p53RE sequence conversely competed out the p53-DNA complex but not the complexes formed with NBP (compare lane 4 with lane 9). These differential effects of the distinct p53RE motifs on the two types of p53RE-protein complexes were dose-dependent. The more oligomer competitors were added, the fewer DNA-protein complexes were formed (lanes 6, 8, 10, and 14). These complexes are specific to the p53 binding DNA motif, as neither random sequences nor dIdC oligomers at the same concentration blocked any of the DNA-protein complexes (compare lane 4 with lanes 11, 12, 15, and 16). Also, p53 appeared to compete with the NBP activity for the p53RE probes, as lower levels of the NBP-DNA complex were observed when the p53 protein was mixed with the H1299 nuclear extracts (comparing lane 4 with lanes 5 and 6 Left). Notably, the H1299 nuclear extracts may possess a factor that activates the DNA-binding activity of p53 as the p53-DNA complex increased when p53 was blended with the nuclear extracts (compare lanes 2 with 4). Taken together, these results, which were reproducible, suggest that p53 has a higher affinity for the p53RE motifs with fewer mismatched nucleotides, such as GADD45, while the NBP-like protein preferentially binds to the p53RE motifs that harbor more mismatched nucleotides, such as p21waf1 and Bax1.
In summary, these results demonstrate that there is an NBP-like activity in cells and also suggest that p53 and NBP may differentially target a subset of the p53 responsive genes under selected physiological circumstances. The physiological level of or possible interaction between the two proteins may play a role in determining which is primarily responsible for activation of certain p53 target genes in response to a variety of intracellular or extracellular signals or at certain stages of cell cycle (1–3). This remains to be addressed when the NBP cDNA becomes available. It would also be interesting and important to learn how differently p73 binds to the p53RE motifs by comparing it with NBP and p53.
Most recently, a murine cellular p53RE-binding activity called p53 competing protein was identified by searching for a regulatory activity specific for the putative p53 binding sequence derived from the TIMP-3 promoter (47). It is unclear at present whether p53 competing protein and NBP are related proteins from the mouse and human respectively. Both p53 competing protein and NBP support the idea that additional p53 homologs exist in cells. Elucidation of each member of the p53 family (34, 35, 47) should be important for a better understanding of the networking among these components in regulating cell growth and controlling tumorigenicity.
Acknowledgments
We thanks Ms. Ashleigh Miller for preparing some of the reagents for this study, William G. Kaelin for providing the p73 mAb, Ms. Crissa Elliot for her secretary expertise, and Drs. Richard Goodman and Richard Maurer for their valuable discussion. H.L. was supported by a junior faculty start fund granted by the Department of Biochemistry and Molecular Biology, Oregon Health Science University. A.J.L. was supported by a National Institutes of Health grant.
ABBREVIATIONS
- AdMLP
adenovirus major late promoter
- EMSA
electrophoretic mobility-shift assay
- FT
flow through
- NBP
non-p53 p53RE binding protein
- p53RE
p53 responsive DNA element
- P11
phosphocellulose
References
- 1.Ko J L, Prives C. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 2.Levine A J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 3.Gottlieb T M, Oren M. Biochim Biophys Acta. 1996;1287:77–102. doi: 10.1016/0304-419x(95)00019-c. [DOI] [PubMed] [Google Scholar]
- 4.Raycroft L, Wu H, Lozano G. Science. 1990;249:1049–1051. doi: 10.1126/science.2144364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fields S, Jang S K. Science. 1990;249:1046–1049. doi: 10.1126/science.2144363. [DOI] [PubMed] [Google Scholar]
- 6.Pavletich N P, Chambers K A, Pabo C O. Genes Dev. 1993;7:2556–2564. doi: 10.1101/gad.7.12b.2556. [DOI] [PubMed] [Google Scholar]
- 7.Bargonetti J, Manfredi J J, Chen X, Marshak D R, Prives C. Genes Dev. 1994;7:2565–2574. doi: 10.1101/gad.7.12b.2565. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y, Reed M, Wang P, Stenger J E, Mayr G, Anderson M E, Schwedes J F, Tegtmeyer P. Genes Dev. 1994;7:2575–2586. doi: 10.1101/gad.7.12b.2575. [DOI] [PubMed] [Google Scholar]
- 9.Cho Y, Gorina S, Jeffrey P D, Pavletich N P. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 10.Balkalkin G, Yakovleva T, Selivanova G, Magnusson K P, Szekely L, Kisaleva E, Klein G, Terenius L, Wiman K G. Proc Natl Acad Sci USA. 1994;91:413–417. doi: 10.1073/pnas.91.1.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu H, Levine A J. Proc Natl Acad Sci USA. 1995;92:5154–5158. doi: 10.1073/pnas.92.11.5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thut C J, Chen J L, Klemin R, Tjian R. Science. 1995;267:100–104. doi: 10.1126/science.7809597. [DOI] [PubMed] [Google Scholar]
- 13.Avantaggiati M L, Ogryzko V, Gardner K, Giordano A, Levine A S, Kelly K. Cell. 1997;89:1175–1164. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- 14.Gu W, She X, Roeder R G. Nature (London) 1997;387:819–823. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- 15.Lill N L, Grossman S R, Ginsberg D, DeCaprio J, Livingston D M. Nature (London) 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 16.Hupp T R, Lane D P. Curr Biol. 1994;4:865–875. doi: 10.1016/s0960-9822(00)00195-0. [DOI] [PubMed] [Google Scholar]
- 17.Shieh S, Ikeda M, Taya Y, Prives C. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 18.Siliciano J D, Canman C E, Taya Y, Sakaguchi K, Appella E, Kastan M B. Genes Dev. 1998;11:3471–3481. doi: 10.1101/gad.11.24.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu, H., Taya, Y., Ikeda, M. & Levine, A. J. (1998) Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 20.Kern S E, Kinzler K W, Bruskin A, Jarosz D, Friedman P, Prives C, Vogelstein B. Science. 1991;252:1708–1711. doi: 10.1126/science.2047879. [DOI] [PubMed] [Google Scholar]
- 21.Bargonetti J, Friedman P N, Kern S E, Vogelstein B, Prives C. Cell. 1991;65:1083–1091. doi: 10.1016/0092-8674(91)90560-l. [DOI] [PubMed] [Google Scholar]
- 22.Tokino T, Thiagalingam S, El-Deiry W S, Waldman T, Kinzler K W, Vogelstein B. Hum Mol Genet. 1994;3:1537–1542. doi: 10.1093/hmg/3.9.1537. [DOI] [PubMed] [Google Scholar]
- 23.El-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 24.Dulic V, Kaufmann W K, Lees S J, Tisty T D, Lees E, Harper J W, Elledge S J, Reed S I. Cell. 1994;76:1013–1023. doi: 10.1016/0092-8674(94)90379-4. [DOI] [PubMed] [Google Scholar]
- 25.Xiong Y, Hannon G J, Zhang H, Casso D, Kobayashi R, Beach D. Nature (London) 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 26.Momand J, Zambetti G P, Olson D C, George D, Levine A J. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 27.Chen J, Marechal V, Levine A J. Mol Cell Biol. 1993;13:4107–4114. doi: 10.1128/mcb.13.7.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu X, Bayle J H, Olson D, Levine A J. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 29.Perry M E, Piette J, Zawadzki J, Harvey D, Levine A J. Proc Natl Acad Sci USA. 1993;90:11623–11627. doi: 10.1073/pnas.90.24.11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Polyak K, Xia Y, Zweier J L, Kinsler K W, Vogelstein B. Nature (London) 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 31.Miyashita T, Reed J C. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 32.Hollstein M, Sidransky D, Vogelstein B, Harris C C. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 33.Levine A J, Momand J, Finlay C A. Nature (London) 1991;351:453–455. doi: 10.1038/351453a0. [DOI] [PubMed] [Google Scholar]
- 34.Kaghad M, Bonnet H, Yang A, Creancier L, Biscan J, Valent A, Minty A, Chalon P, Lelias J, Dumont X, et al. Cell. 1997;90:809–819. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- 35.Christine A J, Maria C M, Kaelin W G., Jr Nature (London) 1997;389:191–194. doi: 10.1038/38298. [DOI] [PubMed] [Google Scholar]
- 36.Chen J, Wu X, Lin J, Levine A J. Mol Cell Biol. 1996;16:2445–2452. doi: 10.1128/mcb.16.5.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chiba I, Takahashi T, Nau M M, D’Amico D, Curiel D T, Mitsudomi T, Buchhagen D L, Carbone D, Piantadosi S, Koga H. Oncogene. 1990;10:1603–1610. [PubMed] [Google Scholar]
- 38.Dignam J D, Lebovitz R M, Roeder R G. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flores O, Lu H, Reinberg D. J Biol Chem. 1992;267:2786–2793. [PubMed] [Google Scholar]
- 40.Lu H, Fisher R, Bailey P, Levine A J. Mol Cell Biol. 1997;17:5923–5934. doi: 10.1128/mcb.17.10.5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu H, Zawel L, Fisher L, Egly J M, Reinberg D. Nature (London) 1992;358:641–645. doi: 10.1038/358641a0. [DOI] [PubMed] [Google Scholar]
- 42.Van Dyke M W, Dervan P B. Nucleic Acids Res. 1983;11:5555–5567. doi: 10.1093/nar/11.16.5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. p. 13. : 11. [Google Scholar]
- 44.Sawadogo M, Roeder R G. Proc Natl Acad Sci USA. 1985;82:4394–4398. doi: 10.1073/pnas.82.13.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 46.Gradwohl G, Menissier de Murcia J M, Molinete M, Simonin F, Koken M, Hoeijmakers J H, de Murcia G. Proc Natl Acad Sci USA. 1990;87:2990–2994. doi: 10.1073/pnas.87.8.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bian J, Sun Y. Proc Natl Acad Sci USA. 1997;94:14753–14758. doi: 10.1073/pnas.94.26.14753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kastan M B, Zhan Q, El-Deiry W S, Carrier F, Jacks T, Walsh W V, Plunkett B S, Vogelstein B, Fornace A J., Jr Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]