Abstract
The Drosophila HMG1-like protein DSP1 was identified by its ability to inhibit the transcriptional activating function of Dorsal in a promoter-specific fashion in yeast. We show here that DSP1 as well as its mammalian homolog hHMG2 bind to the mammalian protein SP100B and that SP100B in turn binds to human homologs of HP1. The latter is a Drosophila protein that is involved in transcriptional silencing. Each of these proteins represses transcription when tethered to DNA in mammalian cells. These results suggest how heterochromatin proteins might be recruited to specific sites on DNA with resultant specific effects on gene expression.
The Drosophila protein Dorsal can act as a transcriptional activator or repressor depending on the promoter context. For example, in Drosophila, Dorsal activates the twist promoter but represses the zen promoter. A Dorsal-binding element taken from the zen promoter, called the ventral repression element, and placed upstream of an activated gene in a Drosophila embryo, mediates Dorsal-dependent repression of that gene (1). In Saccharomyces cerevisiae, however, Dorsal activates transcription from both the twist and zen promoters. DSP1, a member of the high mobility group1/2 (HMG) family of non-histone chromosomal DNA-binding proteins, was isolated as a putative corepressor that inhibits Dorsal from activating the zen promoter but has no effect on Dorsal activation of a reporter bearing certain isolated Dorsal-binding sites (2). DSP1 interacts with Dorsal and with p50/p65 heterodimer NF-κB and binds cooperatively with these proteins to DNA (ref. 2 and J. Brickman and M.P., unpublished data).
In Drosophila, various proteins bearing “chromo domains” (CDs) or closely related sequences have been implicated in gene repression (3). For example, Polycomb, which contains one such domain, is required to maintain repression of genes of the antennapedia and bithorax complexes in certain tissues (4, 5). A variety of experiments suggest that Polycomb, which evidently does not bind DNA directly (6), is found in complexes at specific sites associated with repressed genes (7, 8). The CD of Polycomb, a sequence of some 53 amino acids, is required for proper localization of the protein (9). Another example of a repressing non-histone chromosomal protein that bears a CD is HP1, also known as Su(var)205 (10). This protein, which bears in addition to a CD a related sequence called a chromo shadow domain (CSD) (see Discussion), is required for the form of gene repression called position effect variegation. Position effect variegation is the partially repressed expression of a gene that, by chromosomal rearrangement, has been placed into or near heterochromatic regions (11). Mutation of HP1 restores expression of such a gene (10). HP1 is found predominantly but not exclusively in heterochromatin as visualized by in situ staining of polytene chromosomes (12). Either the CD or the CSD suffices to direct the protein to sites in heterochromatin, but it is not known whether either domain can direct the protein to the limited sites in euchromatin that are visualized by polytene chromosome staining (13). Like Polycomb, HP1 does not bind DNA directly (14). Proteins bearing both a CD and a CSD have been found in a wide array of organisms (15).
Previous work has shown that so-called “nuclear bodies” (NBs), found in nuclei of mammalian cells, contain the proteins SP100A (16), PML (17), and PIC1 (18). SP100A is an autoantigen recognized by antibodies from patients suffering from primary biliary cirrhosis, (16) and PML has been implicated to play a role in acute promyelocytic leukemia (17). Both SP100A and PML are up-regulated by interferons (19, 20), and overexpression of PML results in slow growth (21). It has been suggested that the NBs might play a role in cellular antiviral defense mechanisms (22).
In this report, we show that DSP1 and its mammalian homolog hHMG2 interact with SP100B, a splice variant of SP100A (23), and that SP100B, in turn, binds to mammalian homologs of the Drosophila heterochromatin protein 1, HP1. We further show that each of these proteins behaves as a transcriptional repressor when tethered to DNA. Our results therefore suggest the existence of an HMG/SP100B/HP1-repressing complex that could be recruited to DNA by interaction with various members of the rel family of transcriptional activators such as Dorsal. The accompanying paper by Seeler et al. (41) provides evidence for the existence of such a complex in NBs of mammalian cells.
MATERIALS AND METHODS
Chloramphenicol Acetyltransferase (CAT) Assays.
HeLa cells were grown and transiently transfected as described (2). The GAL4(1–147) fusions were made by using M1, a mammalian expression vector containing the DNA-binding domain of GAL4 under the control of the early SV40 enhancer (24). Ten micrograms of plasmids encoding the GAL4 fusions were transfected, except for the experiments of Fig. 4, in which only 1 μg was used. The TK reporter was made by cloning five GAL4 sites into the polylinker of BLCAT2 (25). The SV40 reporter was a gift from P. Broad (Zeneca Pharmaceuticals, Macclesfield, Cheshire, U.K.). Then, 0.3 μg of the TK reporter was transfected in the experiments shown in Figs. 1 and 2, and 0.1 μg of the SV40 reporter was transfected in the experiments as shown in Fig. 3. One microgram of the SP100CAT reporter, 0.1 μg of the SV40CAT reporter, and 1 μg of the TKCAT reporter were used for the experiments shown in Fig. 5 together with 3 μg of cDNA3 plasmid (Invitrogen) containing the depicted effector protein. Each transfection was done in the presence of 1 μg of pAdVAntage (Promega), and CAT activity was normalized for transfection efficiency using 1 μg of CH110 (26) except for the experiments shown in Fig. 5, in which 0.5 μg of CMV-lacZ (Promega) was used. All presented normalized CAT activities were calculated the same way: CAT activity = acetylated chloramphenicol/total chloramphenicol; normalized CAT activity = CAT activity/β-galactosidase activity. Each number represents the average of at least three independent experiments with a SD of <20%.
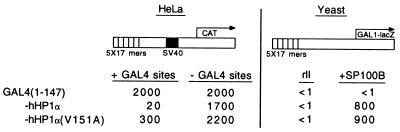
Figure 4.
A mutant of the hHP1α chomo shadow domain that is specifically deficient in the repressing function. (Left) As in Fig. 3C, except that one of the fusions bears the V151A mutation (line 3) and that 1 μg (instead of 10 μg) of plasmid expressing the GAL4(1–147) derivative was used. (Right) As in Fig. 3A (Bottom).
Figure 1.
GAL4-DSP1 and GAL4-hHMG2 repress transcription in human cells but not in yeast. (Left) CAT activities in HeLa cells transiently transfected with the diagrammed reporter and with DNA encoding the indicated effector protein. The reporter was activated by endogenous SP1 and a C/EBP homolog. (Right) β-Galactosidase activities in yeast bearing the diagrammed reporter and transformed with a plasmid encoding GAL4(1–147) or GAL4(1–147) fused to the indicated molecule. The reporter was activated by endogenous GCN4.
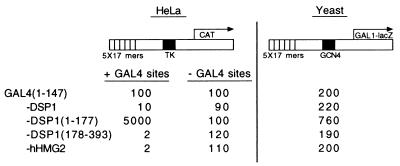
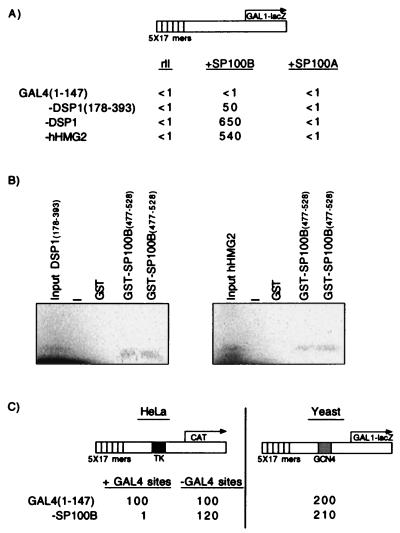
Figure 2.
SP100B interacts with DSP1 and with hHMG2 in vivo (A) and in vitro (B), and GAL4-SP100B represses transcription in human cells but not in yeast (C). (A) β-Galactosidase activities in yeast bearing the diagrammed reporter and two additional plasmids. One of the plasmids expressed either rII alone (Left) or rII fused to SP100B (Center) or rII fused to SP100A (Right). The second plasmid expressed GAL4(1–147) or GAL4(1–147) fused to the indicated molecule. (B) In vitro translated DSP1(178–393) interacts with GST-SP100B(477–528), and in vitro translated hHMG2 interacts with GST-SP100B(477–528). A “−” stands for a free lane. Presented are two independent experiments. (C Left) CAT activities in cells transiently transfected with the diagrammed reporter and with DNA encoding GAL4(1–147) or GAL4(1–147) fused to SP100B. The reporter was activated by endogenous activators that recognize sequences taken from the TK promoter. (Right) β-Galactosidase activities in yeast cells bearing the diagrammed reporter and a plasmid encoding the depicted effector. The reporter was activated by endogenous GCN4.
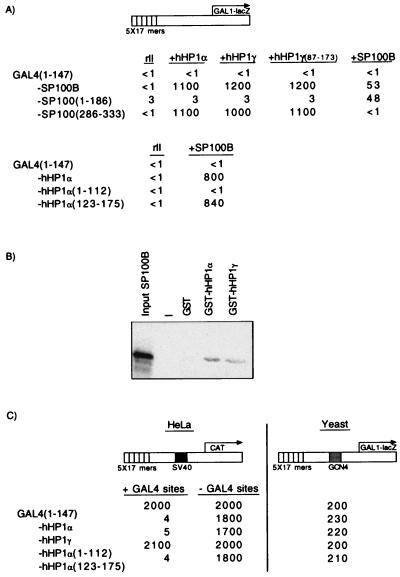
Figure 3.
hHP1α and hHP1γ interact with SP100B in vivo (A) and in vitro (B), and GAL4-hHP1α and GAL4-hHP1γ repress transcription in human cells but not in yeast (C). (A) β-Galactosidase activities in yeast bearing the diagrammed reporter and two additional plasmids. One of the plasmids expressed either rII alone or rII fused to the indicated molecule. The second plasmid expressed GAL4(1–147) or GAL4(1–147) fused to the indicated molecule. (B) In vitro translated SP100B(5–528) interacts with GST-hHP1α and GST-hHP1γ but not with GST. A “−” stands for a free lane. (C) As in Fig. 2C, except that in this case the repressing molecule was hHP1α, hHP1γ, or deletion derivatives of hHP1α fused to GAL4(1–147), and the SV40 enhancer was used instead of the TK promoter.
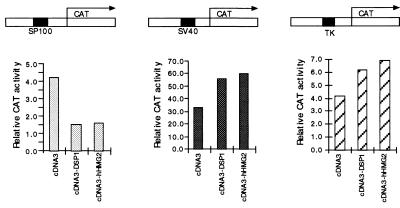
Figure 5.
DSP1 and hHMG2 inhibit the SP100 promoter but not the SV40 enhancer or the TK promoter in transient transfection assays. CAT activities in HeLa cells transiently transfected with the diagrammed reporter and with DNA encoding the indicated effector protein. Presented are relative CAT activities normalized to CMV-LacZ and compared with the empty expression vector cDNA3.
β-Galactosidase Assays.
Yeast cells were grown and transformed as described (27). The GAL4(1–147) fusions were made by using Y1, a TRP1-marked yeast expression vector containing the DNA-binding domain of GAL4 under the control of the ADH1 promoter (24). The fusions to the activation domain of GAL4 were made by using pACT (CLONTECH). The reporter bearing five GAL4 sites upstream of a single GCN4 site has been described (28), as well as the reporter with just five GAL4 sites (27). Both reporters were integrated into the yeast chromosome of NLY2 (28). β-Galactosidase assays were performed, and β-galactosidase activity was calculated as described (28). Each number represents the average of at least three independent measurements with a SD of <20%.
Two-Hybrid Screens.
The two-hybrid screens were performed by using the yeast strain HF7c (CLONTECH) and a cDNA library derived from human B cells fused to the activation domain of GAL4 (CLONTECH), following the protocol given from the manufacturer. Each time 106 transformants were screened.
Glutathione S-transferase (GST)-Pulldown Assay.
The GST-pulldown assay was carried out according to ref. 29.
RESULTS
Fig. 1 shows that DSP1, fused to a GAL4 DNA-binding domain and bound to DNA at GAL4 sites, functions as a transcriptional repressor in HeLa cells. The reporter used in this experiment bore five GAL4 sites upstream of DNA sequences, taken from the TK promoter, that bind the activators C/EBP and SP1. The figure shows that a GAL4 fusion bearing the complete DSP1 repressed some tenfold in mammalian cells (line 2). A fusion containing the carboxyl half of DSP1, which includes both HMG boxes, repressed even more efficiently than did the full length fusion (line 4), whereas the amino half of DSP1 increased activation, a result of unknown significance (line 3). Intact human HMG2 fused to GAL4 also worked as an efficient repressor in mammalian cells (line 5). In all cases, repression was abolished by deleting the GAL4-binding sites. As Fig. 1 (Right) shows, in no case did we observe repression in yeast, suggesting that the repression we see in higher eukaryotes requires proteins absent from yeast.
To identify such proteins, we performed a yeast two-hybrid screen by using GAL4-DSP1(178–393) as bait and challenging with a human cDNA library derived from B cells (CLONTECH). The cDNAs were fused to DNA encoding rII, an activating region derived from GAL4 (30). We found one interacting candidate, a previously sequenced protein called SP100B (23), a splice variant of the NB protein SP100A (16). SP100A is 479 amino acids long and SP100B contains 688 amino acids. Both proteins are identical in their first 476 amino acids. Our cDNA clone represents amino acids 5–528 of SP100B, referred to as SP100B here. The two-hybrid reconstruction experiment of Fig. 2A shows that SP100B, but not SP100A, interacted with the carboxyl half of DSP1, (lines 2 and 3), as well as with human HMG2 (line 4). We obtained a stronger signal with GAL4-DSP1(178–393) than with GAL4-DSP1 (cf. lines 2 and 3). However, Western blot analysis revealed that full-length DSP1 was expressed at lower levels than the C-terminal half in S. cerevisiae, which might explain this difference (data not shown). Fig. 2B shows that the B-specific domain of SP100B (amino acids 477–528) was sufficient for the interaction with both DSP1(178–393) and hHMG2 in vitro. Fig. 2C shows that the fusion protein GAL4-SP100B, like GAL4-DSP1 and GAL4-hHMG2, worked as a repressor in mammalian cells but not in yeast. The repression depended on the presence of the GAL4 DNA-binding sites in the reporter.
We performed a second two-hybrid screen, similar to the first, except that in this case the bait was GAL4-SPl00B. Two strongly interacting candidates were recovered, each of which contained both a CD and a CSD. Both proteins, hHPlα (31) and hHP1γ (32), are homologs of the Drosophila protein HPl (also called Su(var)205 as described above). Fig. 3A shows that, in a yeast two-hybrid assay system, hHPlα and hHP1γ interacted with SP100B. The interaction could be seen regardless of which protein was fused to the DNA-binding domain of GAL4 (Fig. 3A, cf. Top and Bottom). The figure further shows that the interaction required the CSD of hHP1 and not the CD (hHP1α: Bottom, cf. lines 3 and 4; hHP1γ: Top, cf. columns 3 and 4; see Fig. 6 for the location of the chromo and shadow domains of hHP1α and hHP1γ). Fig 3A also shows that for SP100B, the interaction domain is located between amino acids 286 and 333, a region identical in both SP100A and SP100B. SP100B also interacts with itself and this self-association determinant is probably located in the N terminus of the protein (Fig. 3A). Fig. 3B shows that SP100B interacts with both hHP1 proteins in vitro. Fig. 3C shows that both GAL4-hHP1 fusion proteins worked as repressors when bound to GAL4 sites in mammalian cells. In all of the cases, repression was abolished by deleting the GAL4-binding sites. Like the interaction with SP100B, repression requires the chromo shadow and not the CD (Fig 3C, cf. lines 4 and 5). As before, no repression was observed in yeast. GAL4-hHPl repressed approximately tenfold more efficiently than did GAL4-DSP1(178–393) and fivefold more efficiently than did GAL4-SP100B (cf. Figs. 1–3).
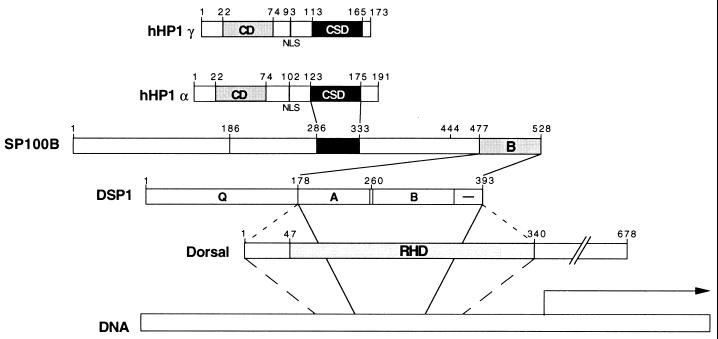
Figure 6.
Interaction scheme for the putative HMG/SP100B/HP1 repressing complex. Indicated are the regions of the proteins that mediate the interactions. The HMG domain of e.g., DSP1 interacts with the rel homology domain (RHD) of, e.g., Dorsal, and both proteins bind cooperatively to DNA. Thus, the repressing complex would be recruited to specific sites on DNA. (The interactions represented by the dashed lines and the cooperative binding of HMG-RHD to DNA are unpublished results from J. Brickman and M.P.).
The experiments shown in Fig. 3 show that the CSD of hHP1α, comprising 53 amino acids, bears two functions: it confers the repression function when fused to GAL4, and it interacts with SP100B. Further deletions into this domain from either end eliminated both functions (data not shown). To identify residues required for one or the other function, we performed an alanine scan experiment. Seven glycine and alanine residues are found in this 53-residue CSD. We separately replaced each of the remaining 46 residues with alanine and tested each variant for repression activity and for interaction with SP100B in experiments similar to those of Fig. 3. Thirty-four variants were unaffected for either function, and 11 were defective in both (data not shown). One, V151A, interacted like wild type with SP100B as assayed in yeast but was markedly deficient in its repression function as assayed in mammalian cells (Fig. 4). Western blot analysis failed to reveal hHP1, either wild type or mutant, in these transfected mammalian cells, and so we cannot exclude the possibility that the mutant was preferentially degraded.
The experiment of Fig. 5 suggests that the SP100 promoter itself can be inhibited by a complex that includes hHMG2/DSP1. The figure shows that the SP100 promoter (33), cloned upstream of a CAT reporter gene, was inhibited some threefold by transient overexpression of DSP1 or hHMG2 (Left). Two other promoters with their enhancers, the strong SV40 promoter and the weak TK promoter, were not inhibited by overexpression of DSP1/hHMG2 (Center and Right).
DISCUSSION
Our experiments suggest that a complex comprising at least four proteins—a human homolog of the Drosophila rel protein Dorsal, a human homolog of the Drosophila HMG protein DSP1, SP100B, and hHP1—works as a repressor in mammalian cells, and the corresponding complex works similarly in Drosophila (Fig. 6). Thus, yeast two-hybrid experiments, as well as experiments performed in vitro, showed interactions between a rel protein (e.g., Dorsal) and DSP1 (J. Brickman and M.P., unpublished results), between DSP1 and SP100B, and between SP100B and hHP1. Each of these components, with the exception of the rel protein, functioned as a repressor in mammalian cells when tethered to DNA as a GAL4-fusion protein. As assayed from GAL4-binding sites positioned upstream of an activator, the extent of repression mediated by these molecules was GAL4-hHP1 > GAL4-SP100B > GAL4-DSP1. These findings suggest that the sole role of each component (with the possible exception of hHP1, see below) is to recruit to DNA an additional member of the complex. According to this idea, the rel protein, at certain promoters, recruits DSP1 (or its human homolog) (J. Brickman and M.P., unpublished results). DNA-tethered DSP1 then recruits SP100B, and SP100B in turn recruits hHP1. We have no direct evidence for the existence of such a complex. As described in the accompanying paper by Seeler et al., however, hHP1 and SP100, as well as an SP100-HMG fusion protein, are found in NBs, suggesting that the interactions we have described may indeed produce the complex suggested here.
Our experiments do not explain the promoter-specific effect of Dorsal, that is, its ability to repress from certain promoters and to activate from others. As reported elsewhere, we have failed to identify sequences outside the Dorsal-binding sites in the zen ventral repression element that affect cooperative binding to DNA of Dorsal and DSP1. In particular, the so-called NRE (negative regulatory element), a sequence found in the zen VRE, is irrelevant for that effect (J. Brickman and M.P., unpublished results). That result, taken with others (J. Brickman and M.P., unpublished results) suggests that the NRE is also irrelevant for repression in vivo. Other proteins (e.g., Groucho, ref. 34) and DNA sequences (35) have been implicated in specifically determining the negative effect of Dorsal.
Our finding that GAL4-DSP1 and GAL4-hHMG2 work as transcriptional repressors in mammalian cells but not in S. cerevisiae (at least at the promoter tested) could be explained if these repression effects required proteins absent from S. cerevisiae. The complete sequence of the S. cerevisiae genome is now available (36) and indeed there is no S. cerevisiae SP100B homolog. There are S. cerevisiae proteins with CDs, but not with CSDs, and we have shown that both repressing and interacting functions are comprised within the CSD. Finally, there is at least one S. cerevisiae protein with an HMG box that functions as a transcriptional repressor (ROX1), but this repression is mediated by the SSN6/TUP1 pathway (37).
Our analysis of hHP1α and hHP1γ showed that the CSDs of these proteins also bear two functions: the separated CSD interacted with SP100B and mediated repression as efficiently as did the intact molecule. In contrast, no other portion of hHP1α, including the CD, manifested either function. It is possible that the specificity determinants on the CSD that direct hHP1–SP100B interaction are similar or identical to those that direct HP1 to its specific sites on the Drosophila chromosome. Our experiments delineated the CSD of hHP1α as comprising residues 123–175. For HP1, the corresponding residues would be 149–201. Thus, our result is consistent with the finding of Powers and Eissenberg (38) that HP1(95–206), but not HP1(152–206), localizes to heterochromatin in Drosophila. Others (39, 40) have found that Polycomb represses transcription as a GAL4 fusion but that its CD is neither necessary nor sufficient for this function.
Acknowledgments
We thank Josh Brickman, Jacob Seeler, and Anne Dejean for communicating results before publication, Ivonne Seffen and Sandra Kröber for excellent technical help, and Dale Dorsett and all of the members of the Ptashne and Lehming laboratories for critical comments on the manuscript. This work was supported by a fellowship from the American Cancer Society, by grants from the Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie and the Max-Planck-Gesellschaft to N.L., by scolarships from the Max-Planck-Gesellschaft to A.L.S. and J.S., and by a grant from the National Institutes of Health to M.P.
ABBREVIATIONS
- CD
chromo domain
- CSD
chromo shadow domain
- HMG
high mobility group
- NB
nuclear body
- rII
GAL4 activating region II (768–881)
- CAT
chloramphenicol acetyltransferase
References
- 1.Doyle H J, Harding K, Hoey T, Levine M. Nature (London) 1986;323:76–79. doi: 10.1038/323076a0. [DOI] [PubMed] [Google Scholar]
- 2.Lehming N, Thanos D, Brickman J M, Ma J, Maniatis T, Ptashne M. Nature (London) 1994;371:175–179. doi: 10.1038/371175a0. [DOI] [PubMed] [Google Scholar]
- 3.Gasser S M, Paro R, Stewart F, Aasland R. Cell Mol Life Sci. 1998;54:1–5. doi: 10.1007/s000180050120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jürgens G. Nature (London) 1985;316:153–155. [Google Scholar]
- 5.Struhl G. Nature (London) 1981;293:36–41. doi: 10.1038/293036a0. [DOI] [PubMed] [Google Scholar]
- 6.Paro R, Hogness D S. Proc Natl Acad Sci USA. 1991;88:263–267. doi: 10.1073/pnas.88.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zink B, Engstrom Y, Gehring W J, Paro R. EMBO J. 1991;10:153–162. doi: 10.1002/j.1460-2075.1991.tb07931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiang A, O’Connor M B, Paro R, Simon J, Bender W. Development (Cambridge) 1995;121:1681–1689. doi: 10.1242/dev.121.6.1681. [DOI] [PubMed] [Google Scholar]
- 9.Messmer S, Franke A, Paro R. Genes Dev. 1992;6:1241–1254. doi: 10.1101/gad.6.7.1241. [DOI] [PubMed] [Google Scholar]
- 10.Eissenberg J C, James T C, Foster-Hartnett D M, Hartnett T, Ngan V, Elgin S C. Proc Natl Acad Sci USA. 1990;87:9923–9927. doi: 10.1073/pnas.87.24.9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muller H J. J Genet. 1930;22:299–334. [Google Scholar]
- 12.James T C, Eissenberg J C, Craig C, Dietrich V, Hobson A, Elgin S C. Eur J Cell Biol. 1989;50:170–180. [PubMed] [Google Scholar]
- 13.Platero J S, Hartnett T, Eissenberg J C. EMBO J. 1995;14:3977–3986. doi: 10.1002/j.1460-2075.1995.tb00069.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh P B, Miller J R, Pearce J, Kothary R, Burton R D, Paro R, James T C, Gaunt S J. Nucleic Acids Res. 1991;19:789–794. doi: 10.1093/nar/19.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aasland R, Stewart A F. Nucleic Acids Res. 1995;23:3163–3173. doi: 10.1093/nar/23.16.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szostecki C, Guldner H H, Netter H J, Will H. J Immunol. 1990;145:4338–4347. [PubMed] [Google Scholar]
- 17.Dyck J A, Maul G G, Miller W H, Jr, Chen J D, Kakizuka A, Evans R M. Cell. 1994;76:333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- 18.Boddy M N, Howe K, Etkin L D, Solomon E, Freemont P S. Oncogene. 1996;13:971–982. [PubMed] [Google Scholar]
- 19.Guldner H H, Szostecki C, Grotzinger T, Will H. J Immunol. 1992;149:4067–4073. [PubMed] [Google Scholar]
- 20.Lavau C, Marchio A, Fagioli M, Jansen J, Falini B, Lebon P, Grosveld F, Pandolfi P P, Pelicci P G, Dejean A. Oncogene. 1995;11:871–876. [PubMed] [Google Scholar]
- 21.Mu Z M, Chin K V, Liu J H, Lozano G, Chang K S. Mol Cell Biol. 1994;14:6858–6867. doi: 10.1128/mcb.14.10.6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carvalho T, Seeler J S, Ohman K, Jordan P, Pettersson U, Akusjarvi G, Carmo-Fonseca M, Dejean A. J Cell Biol. 1995;131:45–56. doi: 10.1083/jcb.131.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dent A L, Yewdell J, Puvion-Dutilleul F, Koken M H, de The H, Staudt L M. Blood. 1996;88:1423–1426. [PubMed] [Google Scholar]
- 24.Sadowski I, Bell B, Broad P, Hollis M. Gene. 1992;118:137–141. doi: 10.1016/0378-1119(92)90261-m. [DOI] [PubMed] [Google Scholar]
- 25.Luckow B, Schutz G. Nucleic Acids Res. 1987;15:5490. doi: 10.1093/nar/15.13.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hall C V, Jacob P E, Ringold G M, Lee F. J Mol Appl Genet. 1983;2:101–109. [PubMed] [Google Scholar]
- 27.Lehming N, McGuire S, Brickman J M, Ptashne M. Proc Natl Acad Sci USA. 1995;92:10242–10246. doi: 10.1073/pnas.92.22.10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saha S, Brickman J M, Lehming N, Ptashne M. Nature (London) 1993;363:648–652. doi: 10.1038/363648a0. [DOI] [PubMed] [Google Scholar]
- 29.Ausubel F, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1987. [Google Scholar]
- 30.Ma J, Ptashne M. Cell. 1987;48:847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- 31.Saunders W S, Chue C, Goebl M, Craig C, Clark R F, Powers J A, Eissenberg J C, Elgin S C, Rothfield N F, Earnshaw W C. J Cell Sci. 1993;104:573–582. doi: 10.1242/jcs.104.2.573. [DOI] [PubMed] [Google Scholar]
- 32.Ye Q, Worman H J. J Biol Chem. 1996;271:14653–14656. doi: 10.1074/jbc.271.25.14653. [DOI] [PubMed] [Google Scholar]
- 33.Grotzinger T, Jensen K, Will H. J Biol Chem. 1996;271:25253–25260. doi: 10.1074/jbc.271.41.25253. [DOI] [PubMed] [Google Scholar]
- 34.Dubnicoff T, Valentine S A, Chen G, Shi T, Lengyel J A, Paroush Z, Courey A J. Genes Dev. 1997;11:2952–2957. doi: 10.1101/gad.11.22.2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang J, Rushlow C A, Zhou Q, Small S, Levine M. EMBO J. 1992;11:3147–3154. doi: 10.1002/j.1460-2075.1992.tb05387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cherry J M, Adler C, Ball C, Chervitz S A, Dwight S S, Hester E T, Jia Y, Juvik G, Roe T, Schroeder M, Weng S, Botstein D. Nucleic Acids Res. 1998;26:73–79. doi: 10.1093/nar/26.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balasubramanian B, Lowry C V, Zitomer R S. Mol Cell Biol. 1993;13:6071–6078. doi: 10.1128/mcb.13.10.6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Powers J A, Eissenberg J C. J Cell Biol. 1993;120:291–299. doi: 10.1083/jcb.120.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Müller J. EMBO J. 1995;14:1209–1220. doi: 10.1002/j.1460-2075.1995.tb07104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bunker C A, Kingston R E. Mol Cell Biol. 1994;14:1721–1732. doi: 10.1128/mcb.14.3.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seeler J S, Marchio A, Sitterlin D, Transy C, Dejean A. Proc Natl Acad Sci USA. 1998;95:7316–7321. doi: 10.1073/pnas.95.13.7316. [DOI] [PMC free article] [PubMed] [Google Scholar]