Abstract
The polymerase (PB2) and nucleocapsid (NP) genes encoded by the genome of influenza virus are essential for replication of the virus. When synthetic genes that express RNAs for external guide sequences targeted to the mRNAs of the PB2 and NP genes are stably incorporated into mouse cells in tissue culture, infection of these cells with influenza virus is nonproductive. Endogenous RNase P cleaves the targeted influenza virus mRNAs when they are in a complex with the external guide sequences. Targeting two different mRNAs simultaneously inhibits viral particle production more efficiently than does targeting only one mRNA.
A variety of agents have been used as inhibitors of viral replication in cultured cells and in mammals. These include small molecules, some as selective inhibitors of nucleic acid replication or protease function, and higher molecular weight agents, such as antisense molecules, the hairpin and other ribozymes, and vaccines (1–15). External guide sequences (EGSs) are oligoribonucleotides (16) that have also been used in conjunction with endogenous RNase P to diminish both the expression of herpes virus thymidine kinase in tissue culture and of several genes in Escherichia coli (4, 5, 17–21). This method takes advantage of RNase P to cleave a targeted mRNA or viral RNA when it is in a complex with the appropriate EGS (16). When more than one site in a particular mRNA is targeted, the method is particularly effective (20). In this report, we demonstrate inhibition of influenza (flu) virus production in cells in tissue culture by the simultaneous targeting of two flu virus mRNAs. We presumed that the mechanisms of action of two EGSs in the same cell, each directed at a different gene, would be independent of each other, thereby increasing the efficiency of inhibition of flu virus replication in that single cell. The mRNAs we targeted are those encoded by the PB2 (polymerase subunit 2) and NP (nucleocapsid) genes.
Influenza virus is a negative strand RNA virus (22, 23); i.e., each of the eight (negative) RNA strands is copied into a complementary (positive) strand mRNA for translation of a flu protein. The 5′ termini of the mRNAs consist of capped segments of 15 bases acquired during their synthesis from host cell mRNAs (22). The early gene product, PB2, is essential for this process (24, 25). Replication and packaging of flu genomic RNA requires the product of the NP gene (26, 27). The NP protein may also be important in mRNA synthesis (28). Thus, inhibition of the expression of these genes should prevent flu virus replication (29–32). Since we utilized a nonpermissive mouse cell line (C127) which produces noninfectious flu virus in our experiments, we have measured the inhibition of flu virus particle production.
MATERIALS AND METHODS
Details of general bacteriological methods and plasmid and vector constructions are available from the authors and are described in ref. 19 as indicated below.
Plasmids.
pGEM1-PB2, a plasmid carrying the gene for the influenza A polymerase subunit PB2 under control of the T7 promoter, and pGEM1-NP, a plasmid carrying the gene form the influenza A nucleoprotein, were generous gifts from Robert Krug (Rutgers University, New Brunswick, NJ). pmU6(-315/1), a plasmid carrying the mouse U6 promoter (33) in pBluescript, was a gift from Yan Yuan (University of Pennsylvania, Philadelphia). LXSN, a retroviral vector based on Moloney murine leukemia virus and carrying a neomycin resistance gene (originally constructed by A. D. Miller; ref. 34), and RVY, a retroviral vector similar to LXSN but carrying a hygromycin resistance gene instead of the neomycin resistance gene (35), were both generous gifts from Daniel DiMaio (Yale University).
Enzymes and Chemicals.
T4 DNA ligase, DNA polymerase I Klenow fragment, and restriction endonucleases were purchased from New England Biolabs; T7 RNA polymerase and RNasin (ribonuclease inhibitor) were obtained from Promega; RNase-free DNase I was purchased from Worthington; T4 RNA ligase and nucleoside triphosphates from Pharmacia; alkaline phosphatase (CIP) from Boehringer Mannheim, and Sequenase from United States Biochemical. Partially purified RNase P from HeLa cells was prepared according to refs. 36 and 37. Radiolabeled chemicals were purchased from Amersham; Sephadex G-50 spin columns were obtained from Boehringer Mannheim; ampicillin from Sigma. The sequencing kit was obtained from United States Biochemical. G418 and Fungizone were purchased from GIBCO. Hygromycin was obtained from Calbiochem. DMEM was purchased from GIBCO; fetal bovine serum from Mediatech Laboratories (Cody, NY). DNA oligonucleotides were made by an automated synthesizer. For a complete list of oligonucleotides used in this work, please consult the authors or ref. 19.
PCR and Construction of EGS-mU6 Plasmids.
All PCRs were carried out according to the manufacturer’s (Perkin–Elmer) protocol. The EGSs directed against PB2 and NP mRNAs were generated by PCR for transcription in vitro by T7 RNA polymerase. The details of the construction of EGSs and EGS-mU6 plasmids bearing the genes encoding them can be obtained from the authors or ref. 19.
Construction of Retroviral Vectors Containing EGS Sequences.
To clone the EGSs into the retroviral vector LXSN, EGS-mU6 plasmids were used as templates for PCR to acquire the correct restriction enzyme sites (EcoRI and XhoI). Details can be obtained from the authors.
RNA Manipulations.
RNA was transcribed in vitro by T7 RNA polymerase as described (38). RNA was prepared for 5′ end labeling as generally described in ref. 39. Alkaline hydrolysis and partial RNase T1 digestion of 5′ end-labeled RNA were then also carried out as generally described in ref. 39.
Assay of RNase P Activity with Complexes of EGSs and PB2 or NP mRNA as Substrates.
Two thousand counts per minute of a 5′ end-labeled fragment of PB2 mRNA (798 nt, generated from T7 transcription of pGEM-PB2 cut with HindIII) was mixed with 2.5 pmol of EGS. Assays were carried out with 1 μl of partially purified human RNase P at 37°C for 30 min in PA buffer [50 mM Tris·HCl (pH 7.5), 10 mM MgCl2, 100 mM NH4Cl] in the presence of 0.5 μl of RNasin. The reactions were stopped by addition of 1 μl of phenol and 10 M urea dye that contained 0.02% bromphenol blue and xylene cyanol. The samples were electrophoresed on an 8% polyacrylamide/7 M urea gel (19).
Similar reactions were also carried out with a shorter PB2 mRNA fragment of 483 nt, generated from T7 transcription of pGEM-PB2 cut with BglII and with 2000 cpm of a 5′ end-labeled fragment of NP mRNA (480 nt, generated using T7 RNA polymerase transcription in vitro of pGEM-NP cut with BamHI) mixed with 12.5 pmol of EGS. Cleavage was measured by analysis with a Bio-Image Analyzer BAS2000 (Fuji).
Packaging of LXSN Retroviral Vectors and Generation of Cell Lines.
The following procedure was adapted from ref. 40 by David Riese in Daniel DiMaio’s laboratory (Yale University). Briefly, PA317 amphotropic packaging cells (41) were transfected with LXSN retroviral vector DNA. The low titer amphotropic retrovirus produced by these cells was harvested and used to infect Ψ-2 ecotropic packaging cells (42). Colonies that grew under selection with 600 μg/ml G418 were pooled. These cells produced high-titer ecotropic retrovirus that was used to infect C127 mouse cells. Colonies were selected again with G418. Individual colonies were isolated and propagated and cell lines were stored in liquid nitrogen.
Packaging of RVY Retroviral Vectors and Generation of Cell Lines Expressing Two EGSs.
PA317 amphotropic packaging cells were transfected with RVY retroviral DNA. The low-titer amphotropic retrovirus produced by these cells was harvested and used to infect Ψ-2 ecotropic packaging cells. Colonies were selected with 300 units/ml hygromycin and pooled. These cells produced high-titer ecotropic retrovirus that was used to infect EF1-2 cells. Colonies were selected with hygromycin as above. Individual colonies were selected and propagated and cell lines were stored in liquid nitrogen.
Manipulations with Influenza Virus.
Growth and titration of viral stocks and infection of cells in flasks or multiwell plates were carried out as described previously (43).
Labeling of Flu Virus Protein.
For labeling of cells in 24-well plates, the cells were split 3 or more days before infection to have 100,000 cells per well in 1 ml of DME-10 that contained 10% fetal bovine serum. On the day of infection, the medium was changed to DME that contained 0.2% BSA (DMEB buffer; Sigma). After one wash, the virus was added at the desired multiplicity of infection (MOI) in 0.2 ml of DMEB. One hour later, the medium was aspirated and 1 ml of DMEB was added back. Five hours after infection, the medium was aspirated and 0.2 ml of DMEB (without cysteine/methionine) containing 20 mCi of [35S]cysteine/methionine was added to the cells. After 1 h, the medium was aspirated and the cells were lysed in 0.2 ml of dissociation buffer (10 mM Tris·HCl, pH 8/250 mM NaCl/0.5% Triton X-100/0.5% sodium deoxycholate/0.1% SDS) per well. The unlabeled cells from a duplicate plate were also lysed and the protein was used for Western blotting. The plates were placed on ice for 5 min, after which the lysates were collected and frozen at −20°C for later use. For infection of cells in flasks, this protocol was scaled up accordingly. Twenty-four hours after infection, the supernatants from triplicate flasks were collected for hemagglutination (HA) assays and frozen at −70°C.
Radioactive cell lysates were thawed on ice and sheared with a 28-gauge needle. Ten microliters of lysate was mixed with 10 μl of 2X protein loading buffer (19). Ten microliters of each sample was run on a polyacrylamide/SDS gel of the desired percentage, and Rainbow markers (Amersham) were run as size markers.
Western Blot.
Unlabeled cell lysates were thawed on ice and sheared with a 28-gauge needle. Ten microliters of lysate was mixed with 10 μl 2X protein loading buffer. Ten microliters of each sample was run on a polyacrylamide/SDS gel of the desired percentage. Rainbow markers (Amersham) were run as size markers. The proteins were transferred to nitrocellulose and blots carried out as described in ref. 37.
HA Assays.
Twofold serial dilutions in PBS were made of each sample of viral particles in a volume of 100 μl in each well of a 96-well microtiter plate, on ice. To each well was added 100 μl of a 0.5% suspension of red blood cells (Ortho Diagnostics) in PBS. The plates were kept at 4°C until HA was observed. Influenza virus stock of a known titer (determined by plaque assay) was used as a standard, to which samples of unknown titer were compared.
RESULTS
Design and Testing of EGSs in vitro.
Transcripts that contained the 5′ third of the PB2 and NP genes were synthesized in vitro and treated with RNase T1 under mild conditions of digestion. The digestion buffer (see Materials and Methods) contained 10 mM Mg2+ to ensure that the RNA transcripts maintained a folded structure. Three sites in the PB2 transcript (Fig. 1) and two in the NP transcript that were accessible to digestion with RNase T1 were chosen for further analysis. The particular sequences near the sites of RNase T1 cleavage were selected to provide a G at the 3′ side of the putative cleavage site by RNase P in the target mRNA:EGS complex and a uracil 8 nts downstream, as is found in all tRNAs.
Figure 1.
Sequences of target mRNAs. (A) 5′ proximal sequence of the plus strand of the PB2 gene of the influenza strain A/PR/8/34 (see ref. 45). The start codon is enlarged and in bold type. The sites in the gene that were chosen for targeting by EGSs are underlined. The first underlined nucleotide at each site is a guanosine that was shown to be accessible to cleavage by RNase T1. The sites were chosen to have a uracil residue in position 8 after the guanosine. (B) 5′ proximal sequence of the plus strand of the NP gene of the influenza strain A/PR/8/34 (see ref. 46).
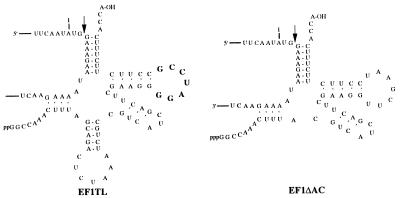
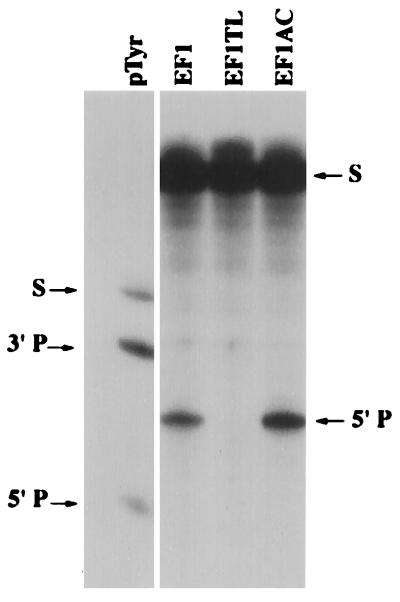
EGSs that simulated three-quarters of a tRNA were designed to each of the five sites chosen for analysis (19, 21). The EGSs were synthesized and tested for the efficacy in vitro in promoting cleavage of target mRNA transcripts by RNase P (EF1, EF2, EF3; NP1, NP2). In each case, efficient cleavage of the target mRNAs was observed at EGS:mRNA ratios of about 100 (data not shown). In addition to these EGSs, one was designed as a negative control because it contained an altered T-loop analogue sequence (Fig. 2, EF1-TL; ref. 44) that prevented binding to, and therefore cleavage, by RNase P (19). Another was designed for further comparison of RNase P cleavage efficiencies in vitro versus those in tissue culture cells. The latter EGS (Fig. 2, EF1-ΔAC; refs. 19, 44) suffered a deletion of the anticodon stem and loop analogue of its “tRNA” domain: it was about twice as efficient in promoting RNase P cleavage of a target mRNA as was the intact EGS EF1 (Fig. 3).
Figure 2.
Proposed secondary structures of the EF1-TL and EF1-ΔAC EGSs. EF1-TL has an altered sequence in the T-loop analogue (bold type) of the tRNA domain (the wild-type sequence is UUCGAAU) and ΔAC is missing the anticodon stem and loop analogue of the tRNA domain.
Figure 3.
Cleavage in vitro of PB2 mRNA by human RNase P and the EGSs EF1, EF1-TL, and EF1-ΔAC. Reaction mixtures contained 2.5 pmol of EGS RNA and 2000 cpm of 5′ end-labeled PB2 mRNA substrate (corresponding to about 0.031 pmol of RNA), yielding an EGS to target ratio of 80. Uniformly labeled pTyr (precursor to E. coli tRNA Tyr) was used as a positive control for cleavage by human RNase P. The arrows on the left indicate the positions of the full-length pTyr substrate (S), the 3′ product (3′P), and the 5′ product (5′P). The arrows on the right indicate the positions of the 5′ end-labeled PB2 mRNA substrate (S) and the 5′ products of human RNase P cleavage directed by the EGSs EF1 and EF1-ΔAC (5′P).
Selection and Testing of Clones That Contained Genes Coding for a Single EGS.
Of the five EGSs tested in vitro, only EF3 was not chosen for further study. Sequences coding for the remaining four EGSs were cloned into the LXSN vector under control of the U6 pol III promoter (see Materials and Methods). The vectors bearing the genes coding for the EGSs were stably integrated into mouse C127 cells. Several clones were selected and tested for their ability to inhibit flu virus protein and particle production (see below). An examination of the cell morphology and generation time of the selected clones showed no differences from cells with no vector or no EGSs. The MOI used in these experiments was 3–20 (see below). In natural flu infections, the MOI is thought to be <1, a factor that would balance the use of EGSs in favor of greater efficiency of inhibition. Nevertheless, for experimental purposes, a higher MOI was used to ensure infection of every cell in culture.
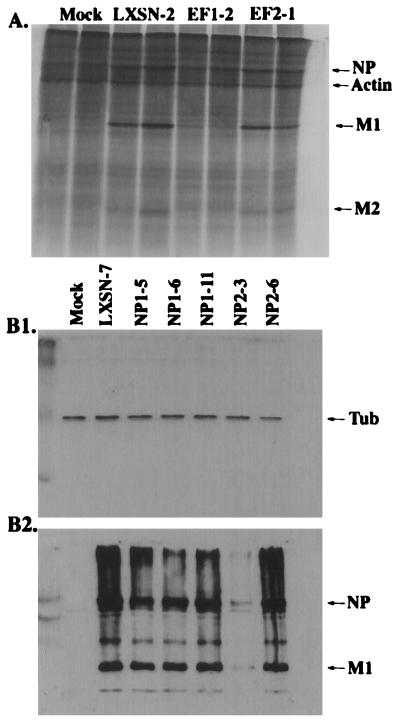
Six or more clones that served as controls (no EGS), and a similar number that contained genes coding for EGSs against either PB2 or NP mRNAs were picked and tested. None of the randomly picked negative control clones inhibited flu protein or virus production. The degree of inhibition of flu protein and virus production in the clones that contained genes coding for EGS was variable. A representative experiment is shown in Fig. 4, in which the results with a sampling of clones are shown. The production of flu virus M1 protein (a major viral protein) was measured by labeling infected cells with 35S and by Western blotting of cell extracts as shown (Fig. 4A and and Tables 1 and 2). The production of viral NP protein was also measured by Western blotting (Fig. 4B2; Fig. 4B1 serves as an internal control for Fig. 4B2) and viral particle production was measured by a HA assay (Tables 1 and 2). Some EGS-containing clones inhibited flu virus protein production by as much or more than 80%. The experiments in which flu proteins were labeled (Fig. 4A) were carried out at a MOI of 3 to maximize the efficiency of the EGS by reducing the infectious genome copy number to the lowest number that would still guarantee that every cell was infected. Under these conditions, about 90% inhibition was observed with the most efficient clone, EF1-2. The experiments from which the Western blot (Fig. 4B) and HA assays (Tables 1 and 2) were derived were carried out at a MOI equal to 20 to determine whether the efficiency of inhibition was less effective under these conditions. The HA assay is a crude measure of particle production but, in most cases, the results with that assay agreed with the Western blot, which shows, as expected, different amounts of inhibition in the different clones. Clone NP2-3 is consistently the most effective. We note that in all cases the negative controls, i.e., mock infection and the TL mutant EGS, behaved as expected, as did the positive control vector with no EGSs. The variation in efficiency of inhibition of flu replication noted with different clones is a result, presumably, of variations in the production of EGS RNA (see Discussion). The general results with EF and NP clones were sufficiently promising to warrant an attempt to target both genes simultaneously (see below).
Figure 4.
A. Polyacrylamide gel analysis of proteins labeled with [35S]cysteine/methionine after infection of cells with influenza virus at a MOI of 3. The samples were run in duplicate. The cells contain a retroviral vector (LXSN) expressing no EGS, EF1, or EF2 under the control of the mouse U6 pol III promoter. As a negative control, LXSN-2 cells (lacking an EGS) were mock infected (first two lanes). The arrows on the right indicate the position of actin and the positions of various viral proteins. B. Western blot of protein lysates from cells collected 6 h after infection with influenza at a MOI of 20. Control (first lane) as for A. Infected LXSN-7 (second lane) expressing no EGS. Other lanes, infected cell lines that express NP EGSs as noted. (B1) Western blot using anti-β-tubulin antibodies, carried out as an internal control. The position of the tubulin band (Tub) is indicated at the right.(B2) Western blot using IBO antibodies which recognize influenza proteins. The positions of the NP and M1 bands are indicated at the right. For experimental details, see Materials and Methods.
Table 1.
Results of assays of clones with genes for a single EGS (EF)
Average of five experiments. See Figure 4A (MOI = 3) and text for details.
For each individual experment, this measurement was the average of the end point of twofold serial dilutions of two samples. The average result of two individual experiments in each case is shown here.
Table 2.
Results of HA of clones expressing a single EGS (NP)
| Cell line | HA, units/ml | % inhibition (HA) |
|---|---|---|
| LXSN-7 | 240 | 0 |
| NP1-5 | 180 | 25 |
| NP1-6 | 180 | 25 |
| NP1-11 | 90 | 62 |
| NP2-3 | 45 | 81 |
| NP2-6 | 45 | 81 |
See Fig. 4B (MOI = 20) and text for details.
Construction and Testing of Clones That Contained Genes Coding for EGSs for both the PB2 and NP Genes.
EF1, directed against the PB2 gene, was the EGS with highest efficiency of inhibition of flu replication. NP1 and NP2, each directed against the NP gene, were both reasonably efficient. The genes for these latter two EGSs were cloned separately into the retroviral vector, RVY, which has an hygromycin-selectable marker, packaged (see Materials and Methods) into the retrovirus and used to superinfect the clone EF1-2. Several clones that contained both the hygromycin and neomycin (for LXSN that contained EF1) markers were selected and propagated for further testing. These cells did grow more slowly than the appropriate parent clones (that harbored only one drug marker) in medium that contained one or the other antibiotic.
The efficiency of inhibition of flu virus protein and particle production was assayed in the clones that contained two EGSs in the same fashion as the experiments performed with clones that contained one EGS. Positive controls contained the retroviral vectors with no EGSs. Negative controls were mock infected. The EF1-TL mutant was used as a control for potential antisense effects of the EF1 EGS. All of the experiments were performed with a MOI of 10 to effectively extend the range of inhibition measurements compared with those obtained at MOI = 3. Separate experiments were performed to determine that the various cell lines used in these experiments were efficiently infected by flu particles.
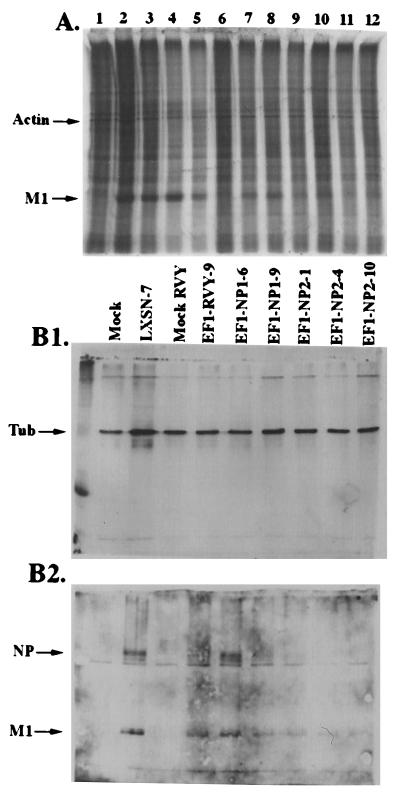
Representative analyses of protein production are shown in Fig. 5. The results of these assays and the HA assay are summarized in Table 3. At least two clones that contained EGSs against both the PB2 and NP genes, EF1-NP2-1 and EF1-NP2-10, inhibited protein production and particle production between 90 and 100% (Fig. 5 and Table 3). The success in targeting two genes in these experiments is emphasized by the fact that inhibition by a single EGS, EF1-2, yielded an inhibition of about 50% compared with an average of about 90% at a lower MOI. The expression of the NP2 EGSs, determined by RNase protection assays, was roughly equivalent in the clones that coded for EF1-NP2-1 and EF1-NP-10 as normalized to the endogenous levels of the RNA subunit of RNase P (data not shown). The HA assay is intrinsically of low resolution (Table 3). Nevertheless, it is clear in this experiment that the use of two EGSs in one cell, each targeted to a different mRNA, is more efficient than one, EF1-2, alone. Furthermore, the negative control with EF1-TL showed no inhibitory effect. EF1-ΔAC was more efficient than its corresponding parent EF1. These results with the “mutant” EGSs correlate well with those obtained from enzymatic assays in vitro (Fig. 3).
Figure 5.
Inhibition of flu protein production in cells with two EGSs. (A) Polyacrylamide gel analysis of proteins labeled with [35S]cysteine/methionine 6 h after infection of the cells with influenza virus at a MOI of 10. Cells expressing two EGSs simultaneously contain LXSN to express EF1 and RVY to express NP1 or NP2 (for details, see text and Materials and Methods). Lanes: 1, mock infection, LXSN-7; 2, LXSN-7; 3, EF1-2; 4, EF1TL-6; 5, EF1ΔAC-1; 6, mock infection, EF1-RVY-9; 7, EF1-RVY-9; 8, EF1-NP1-6; 9, EF1-NP1-9; 10, EF1-NP2-1; 11, EF1-NP2-4; and 12, EF1-NP2-10. (B) Western blot of proteins collected 6 h after infection of the cells with influenza virus at a MOI of 10. Cell lines as described for A. As a control, LXSN-7 cells (lacking an EGS) were mock infected (first lane, labeled Mock). EF1-RVY-9 cells were also mock infected, as a control for the use of the RVY vector (labeled as Mock RVY). (B1) Western blot using anti-β-tubulin antibodies, carried out as an internal control. The position of the tubulin band (Tub) is indicated at the right. (B2) Western blot using IBO antibodies (see Materials and Methods) which recognize influenza proteins on an aliquot of the same sample that was used in B1. The positions of the NP and M1 bands are indicated on the right.
Table 3.
Results of assays of clones that contain genes for two EGSs
| Cell line | % inhibition as measured by
|
|
|---|---|---|
| Synthesis of M1 protein | HA units | |
| LXSN-7 | 0 | 0 |
| EF1-2 | 42 | 50 |
| EF1TL-6 | 0 | 0 |
| EF1ΔAC-1 | 57 | 50 |
| EF1-RVY-9 | 71 | 50 |
| EF1-NP1-6 | 76 | 50 |
| EF1-NP1-9 | 83 | 50 |
| EF1-NP2-1 | 89 | 100 |
| EF1-NP2-4 | 70 | 50 |
| EF1-NP2-10 | 95 | 100 |
See Fig. 5A (NOI =10) and text for details. Error in measurements from scanned gels is ≈10%.
DISCUSSION
The combined functions of EGSs and RNase P have been used previously to reduce the expression of single gene targets in both bacteria and tissue culture cells. When two sites were targeted in a single gene responsible for chloramphenicol resistance in E. coli, the host strain was phenotypically converted to chloramphenicol sensitivity (20). Since flu virus genomes are subject to high rates of variation because of recombination and spontaneous mutagenesis, we targeted two essential genes that contain sequences highly conserved among flu strains. All of the sites chosen for targeting were found to have identical sequences in the two flu strains used in this study: A/PR/8/34 (45, 46), used for studies in vitro, and A/WSN/33 (47, 48), used for infection of cells in tissue culture. Less desirable targets than PB2 and NP would have been the HA or NA genes. Variants of the latter may be preferentially selected in nature to evade host immune systems.
We have shown that the targeting of two separate flu virus genes did, indeed, result in virtually complete inhibition of flu virus replication in cells in tissue culture within the constraints of the resolution of the methods we have used. The results with two EGSs were clearly better than those with a single EGS targeting one flu virus gene in the cell construct used for the experiments shown in Fig. 5. This is a further demonstration of the validity of a multiple targeting strategy for all antisense and ribozyme methodologies for the reduction of expression of targeted genes (6). The general methodology we describe here is quite effective.
The target genes chosen for study in this report, PB2 and NP, were previously shown to be indispensable for flu virus replication through a variety of methods, including the use of conditional lethal mutants and interferon. Although we did not assay the entire population of flu-encoded proteins during the infectious process, the synthesis of major proteins such as NP and M1 were followed in addition to the production of mature flu particles. The results concerning flu virus biology obtained here with the EGS technology are in agreement with those obtained by the other methods cited above: both the products of the PB2 gene and the NP gene are required for synthesis of other flu proteins. We note also that the EGS methodology is relatively insensitive to mutational variation in the target flu genes since a single nucleotide change in the EGSs for either target gene will not change the stability of the EGS:mRNA complex sufficiently to alter cleavage rates by RNase P (20). Three or four mutations, at a minimum, would be needed to affect the stability of the two complexes, an unlikely scenario considering the actual sequences targeted in this report (see above).
The results with the EF1-TL EGS provide strong evidence that the inhibitory effects on flu replication that we observed are EGS mediated. The only functional difference between this latter EGS, which was unable to exert an inhibitory effect on flu replication, and its “wild-type” counterpart is the inability of RNase P to bind to the complex it forms with its target RNA. The experiment with EF1-TL also shows that the inhibitory effect with other EGSs is not the result of an antisense phenomenon since EF1-TL forms a specific, hydrogen-bonded complex with the target mRNA. Although we did not quantitate relative levels of target RNA and EGSs in the clones we used, we infer that the reduction of flu virus production is a consequence of a lowering of the amounts of the target mRNAs, as has been shown previously for EGS-mediated effects with other genes (5, 17, 20). Furthermore, since none of the control clones (no EGS) we picked exhibited an unexpected or unexplained ability to inhibit flu replication, other explanations for our results are unlikely. We also determined that the majority of EGS transcripts originated from the upstream long terminal repeat promoter in the retroviral vectors and not from the U6 promoter directly in front of the EGS genes (data not shown).
The goal of the experiments described in this report was to demonstrate the general utility of the EGS technology for reduction of gene expression in an eukaryotic viral system. In the specific instance of flu virus, one may ask whether the methodology may lend itself to the production of an antiviral therapy. Such a therapy may come about through the permanent integration of EGS genes into the pulmonary system via a nonpathogenic delivery vehicle or through the transient delivery of such genes during the flu virus infectious cycle. In both cases, the ability to target a particular aspect of mammalian physiology through a directed (inhalation) therapy is advantageous. However, the efficiency of the delivery process, through biological or chemical packaging vectors, is still a major problem to be investigated and solved.
Acknowledgments
We thank Drs. Robert Krug, Ari Helenius, and Dan DiMaio and members of their laboratories for their generous gifts of materials and advice and several colleagues for comments on the manuscript. We thank members of our laboratory for many helpful discussions and encouragement. This work was supported by U.S. Public Health Service Grant 19422 (to S.A).
ABBREVIATIONS
- EGS
external guide sequence
- flu
influenza
- MOI
multiplicity of infection
- HA
hemagglutinin
References
- 1.Carrat F, Valleron A-J. Rev Mal Respir. 1994;11:239–255. [PubMed] [Google Scholar]
- 2.Couture L A, Stinchcomb D T. Trends Genet. 1996;12:510–515. doi: 10.1016/s0168-9525(97)81398-4. [DOI] [PubMed] [Google Scholar]
- 3.Hatta T, Takai K, Nakada S, Yokota T, Takaku H. Biochem Biophys Res Commun. 1997;232:545–549. doi: 10.1006/bbrc.1997.6185. [DOI] [PubMed] [Google Scholar]
- 4.Li Y, Altman S. Nucleic Acids Res. 1996;24:835–842. doi: 10.1093/nar/24.5.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu F, Altman S. Genes Dev. 1995;9:471–480. doi: 10.1101/gad.9.4.471. [DOI] [PubMed] [Google Scholar]
- 6.Ohkawa J, Yuyama N, Takebe Y, Nishikawa S, Taira K. Proc Natl Acad Sci USA. 1993;90:11302–11306. doi: 10.1073/pnas.90.23.11302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun L Q, Ely J A, Gerlach W, Symonds G. Mol Biotechnol. 1997;7:241–251. doi: 10.1007/BF02740815. [DOI] [PubMed] [Google Scholar]
- 8.Ulmer J B, Donnelly J J, Parker S E, Rhodes G H, Felgner P L, Dwarki V J, Gromkowski S H, Deck R R, DeWitt C M, Friedman A, et al. Science. 1993;259:1745–1749. doi: 10.1126/science.8456302. [DOI] [PubMed] [Google Scholar]
- 9.von Itzstein M, Wu W-Y, Kok G B, Pegg M S, Dyason J C, Jin B, Phan T V, Smythe M L, White H F, Oliver S W, et al. Nature (London) 1993;363:418–423. doi: 10.1038/363418a0. [DOI] [PubMed] [Google Scholar]
- 10.Yu M, Ojwang J, Yamada O, Hampel A, Rapapport J, Looney D, Wong-Staal F. Proc Natl Acad Sci USA. 1993;90:6340–6344. doi: 10.1073/pnas.90.13.6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zakharchuk A N, Lazarev V N, Naroditskii B S, Kaverin N V. Mol Biol. 1996;30:857–860. [PubMed] [Google Scholar]
- 12.Altman S. Proc Natl Acad Sci USA. 1993;90:10898–10900. doi: 10.1073/pnas.90.23.10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rittner K, Sczakiel G. Nucleic Acids Res. 1991;19:1421–1426. doi: 10.1093/nar/19.7.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sczakiel G, Pawlita M. J Virol. 1991;65:468–472. doi: 10.1128/jvi.65.1.468-472.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang X, Hobom G, Luo D. J Med Virol. 1994;42:385–395. doi: 10.1002/jmv.1890420411. [DOI] [PubMed] [Google Scholar]
- 16.Forster A C, Altman S. Science. 1990;249:783–786. doi: 10.1126/science.1697102. [DOI] [PubMed] [Google Scholar]
- 17.Guerrier-Takada C, Li Y, Altman S. Proc Natl Acad Sci USA. 1995;92:11115–11119. doi: 10.1073/pnas.92.24.11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, Guerrier-Takada C, Altman S. Proc Natl Acad Sci USA. 1992;89:3185–3189. doi: 10.1073/pnas.89.8.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Plehn-Dujowich D. Ph.D. thesis. New Haven, CT: Yale University; 1998. [Google Scholar]
- 20.Guerrier-Takada C, Salavati R, Altman S. Proc Natl Acad Sci USA. 1997;94:8468–8472. doi: 10.1073/pnas.94.16.8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan Y, Altman S. Science. 1994;263:1269–1273. doi: 10.1126/science.8122108. [DOI] [PubMed] [Google Scholar]
- 22.Krug R M, Alonso-Caplen F V, Julkunnen I, Katze M G. In: The Influenza Viruses. Krug R M, editor. New York: Plenum; 1989. pp. 89–152. [Google Scholar]
- 23.Lamb R A. In: The Influenza Viruses. Krug R, Krug M, editors. New York: Plenum; 1989. pp. 1–87. [Google Scholar]
- 24.Ulmanen I, Broni B, Krug R M. Proc Natl Acad Sci USA. 1981;78:7355–7359. doi: 10.1073/pnas.78.12.7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ulmanen I, Broni B, Krug R M. J Virol. 1983;45:27–35. doi: 10.1128/jvi.45.1.27-35.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beaton A R, Krug R M. Proc Natl Acad Sci USA. 1986;83:6282–6286. doi: 10.1073/pnas.83.17.6282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shapiro A I, Krug R M. J Virol. 1988;62:2285–2290. doi: 10.1128/jvi.62.7.2285-2290.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barcena J, Ochoa M, De Le Luna S, Melero J A, Nieto A, Ortin J, Portela A. J Virol. 1994;68:6900–6909. doi: 10.1128/jvi.68.11.6900-6909.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang T S, Palese P, Krystal M. J Virol. 1990;64:5669–5673. doi: 10.1128/jvi.64.11.5669-5673.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klumpp K, Ruigrok R W H, Baudin F. EMBO J. 1997;16:1248–1257. doi: 10.1093/emboj/16.6.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skorko R, Summers D F, Galarza J M. Virology. 1991;180:668–677. doi: 10.1016/0042-6822(91)90080-u. [DOI] [PubMed] [Google Scholar]
- 32.Stranden A M, Staeheli P, Pavlovic J. Virology. 1993;197:642–651. doi: 10.1006/viro.1993.1639. [DOI] [PubMed] [Google Scholar]
- 33.Das G, Henning D, Wright D, Reddy R. EMBO J. 1988;7:503–512. doi: 10.1002/j.1460-2075.1988.tb02838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller A D, Rosman G J. Biotechniques. 1989;7:980–990. [PMC free article] [PubMed] [Google Scholar]
- 35.Riese D J, II, DiMaio D. Oncogene. 1995;10:1431–1439. [PubMed] [Google Scholar]
- 36.Yuan Y, Hwang E-S, Altman S. Proc Natl Acad Sci USA. 1992;89:8006–8010. doi: 10.1073/pnas.89.17.8006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eder P S, Kekuda R, Stolc V, Altman S. Proc Natl Acad Sci USA. 1997;94:1101–1106. doi: 10.1073/pnas.94.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vioque A, Arnez J, Altman S. J Mol Biol. 1988;202:835–848. doi: 10.1016/0022-2836(88)90562-1. [DOI] [PubMed] [Google Scholar]
- 39.Guerrier-Takada C, Altman S. Biochemistry. 1984;23:6327–6334. doi: 10.1021/bi00321a006. [DOI] [PubMed] [Google Scholar]
- 40.Leptak C, Ramon y Cajal S, Kulke R, Horwitz B H, Riese D J, II, Dotto G P, DiMaio D. J Virol. 1991;65:7078–7083. doi: 10.1128/jvi.65.12.7078-7083.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller A D, Buttimore C. Mol Cell Biol. 1986;6:2895–2902. doi: 10.1128/mcb.6.8.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mann R, Mulligan R C, Baltimore D. Cell. 1983;33:153–159. doi: 10.1016/0092-8674(83)90344-6. [DOI] [PubMed] [Google Scholar]
- 43.Shapiro G I, Gurney T, Jr, Krug R M. J Virol. 1987;61:764–773. doi: 10.1128/jvi.61.3.764-773.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yuan Y, Altman S. EMBO J. 1995;14:159–168. doi: 10.1002/j.1460-2075.1995.tb06986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fields S, Winter G. Cell. 1982;28:303–313. doi: 10.1016/0092-8674(82)90348-8. [DOI] [PubMed] [Google Scholar]
- 46.Van Rompuy L, Min Jou W, Huylebroeck D, Devos R, Fiers W. Eur J Biochem. 1981;116:347–353. doi: 10.1111/j.1432-1033.1981.tb05341.x. [DOI] [PubMed] [Google Scholar]
- 47.Kaptein J S, Nayak D P. J Virol. 1982;42:55–63. doi: 10.1128/jvi.42.1.55-63.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li R, Palese P, Krystal M. Virus Res. 1989;12:97–112. doi: 10.1016/0168-1702(89)90057-9. [DOI] [PubMed] [Google Scholar]