Abstract
The Tn552 transposase, a member of the DDE superfamily of transposase and retroviral integrase proteins, has been expressed in soluble form. The purified protein performs concerted strand transfer in vitro, efficiently pairing two preprocessed transposon ends and inserting them into target DNA. For maximum efficiency, both participating DNA ends must contain the two adjacent transposase-binding sites that are the normal constituents of the Tn552 termini. As is the case with transposition in vivo, the insertions recovered from the reaction in vitro are flanked by repeats of a short target sequence, most frequently 6 bp. The reaction has stringent requirements for a divalent metal ion. Concerted strand transfer is most efficient with Mg2+. Although it stimulates strand transfer overall, Mn2+ promotes uncoupled, single-ended events at the expense of concerted insertions. The simplicity and efficiency of the Tn552 transposition system make it an attractive subject for structural and biochemical studies and a potentially useful genetic tool.
The transposon Tn552 is a 6.5-kb element isolated from Staphylococcus aureus responsible for resistance to β-lactam antibiotics (1, 2). It carries genes for three proteins likely to be involved in its transposition: a transposase (TnpA, previously called p480); a protein similar to the Mu B and Tn7 TnsC proteins (TnpB, previously called p271), which, by analogy, may enhance transposition activity and play a role in target selection; and a resolvase (3). The presence of these three genes suggests that Tn552 transposes by consecutive formation and resolution of a cointegrate between donor and target DNAs; however, this remains to be demonstrated.
The Tn552 transposase belongs to the D,D(35)E superfamily of transposase proteins (3). Members of this superfamily include the integrase proteins of retroviruses and retrotransposons, the transposases of bacterial insertion sequences of the IS3 family, bacteriophage Mu A protein, and TnsB of Tn7. The defining characteristic of this family of phosphoryl-transfer proteins is a homologous catalytic core domain containing a triad of invariant acidic residues, two aspartates and a glutamate, which are essential for catalytic activity and give the superfamily its name (3, 4). Despite their homology, transposases of the D,D(35)E superfamily form a rather divergent and heterogeneous group. They range in size from the small retroviral integrases (about 290 residues) to Mu A (663 residues) and Tn7 TnsB (702 residues), and in many cases the region of homology does not appear to extend beyond the catalytic core (160–220 residues). Extensive biochemical studies of a few members of the family have provided a remarkably detailed picture of the transposition process including the substrates and proteins involved, the assembly of the transpositionally active protein–DNA complex, and the mechanism of strand transfer (5–7). However, complete understanding has been hampered by a lack of detailed structural information; despite substantial efforts, three-dimensional structures have been limited to isolated transposase domains in the absence of substrates (8–10). We have turned to Tn552 as a possible candidate for structural studies.
Of the well characterized members of the superfamily, Tn552 transposase is most closely related to Tn7 TnsB but differs significantly from it. At 480 residues, the Tn552 transposase is much smaller, with the regions N- and C-terminal to the catalytic core about 100 residues shorter. Sequence alignments outside the 160-residue central catalytic region are tentative at best, bordering on the insignificant. The Tn552 DNA substrate is also simpler. Each end of Tn552 contains two adjacent 23-bp transposase-binding sites and, as is the case with other elements of this superfamily, terminates with the dinucleotide -CAOH3′ (11). By contrast, the ends of Tn7 (and also of Mu) are complex, containing three or more transposase-binding sites spread irregularly over 90–100 bp (5).
In this paper, we present an in vitro system for Tn552 transposase-catalyzed concerted strand transfer. A previous attempt to obtain active Tn552 transposase by Rowland and colleagues was only partially successful (11). Induced expression yielded a protein that could be solubilized only under strongly denaturing conditions. The purified and renatured protein bound specifically to the ends of Tn552. However, its catalytic activity was limited to uncoupled strand transfer of single ends in a reaction that required Mn2+ as the divalent cation. The protein was also unstable under the assay conditions and appeared to aggregate irreversibly. We have overcome these problems and produced a soluble protein proficient at synapsis and concerted strand transfer with Mg2+ as the metal ion.
MATERIALS AND METHODS
Expression and Purification of Tn552 Transposase.
The expression vector pKETH-8c carrying the Tn552 transposase with an N-terminal His tag (11) and the Escherichia coli host strain BL21(DE3)[pLysS] used for the induction were kindly provided by S.-J. Rowland. Six to 10 liters of CSB1 medium (66 mM K2HPO4/33 mM NaH2PO4/15 mM (NH4)2SO4/5 mM MgCl2/5 mM sodium citrate/0.45 mM FeCl3/44 μM ZnCl2/30 μM MnCl2/25 μM CoCl2/25 μM Na2MoO4/24 μM H3BO3/20 μM CaCl2/18 μM CuCl2/0.6% glucose) supplemented with 10 μg/ml thiamin, 40 μg/ml kanamycin, and 20 μg/ml chloramphenicol were inoculated with a 1-liter overnight culture of the E. coli transposase expression strain grown in the same medium supplemented with 0.1% Casamino acids. Cells were grown at 30°C to an OD600 of 3–4, isopropyl β-d-thiogalactoside then was added to a final concentration of 0.4 mM, and growth was continued for an additional 2 hr. Cells (typically 11 g/liter) were harvested by centrifugation and the pellet was resuspended in 20 ml of 50 mM Tris⋅HCl (pH 7.5 at 4°C), 20% sucrose per liter of culture, and stored at −80°C. To lyse the cells, the suspension was thawed and brought to 1 M NaCl and passed twice through the French press at 1,300–1,500 psi. The lysate was clarified by centrifugation (130,000 × g for 60 min) and then diluted with 50 mM Tris⋅HCl (pH 7.5 at 4°C), 20% sucrose, and 3.8 M (NH4)2SO4 in 50 mM Tris·HCl (pH 7.5 at 4°C) to lower the NaCl concentration to 0.5 M and bring the final solution to 40% saturation in (NH4)2SO4. The precipitated protein was recovered by centrifugation (35,000 × g for 30 min), resuspended in about 1/6 of the original volume of 50 mM Tris⋅HCl (pH 7.5 at 4°C)/0.5 M NaCl/20% glycerol (buffer A) and dialyzed against 40 vol of the same buffer for 2–3 hr. Imidazole was added to a final concentration of 12.5 mM, and the sample was loaded onto a 10-ml Ni2+-NTA column (Qiagen) and preequilibrated in buffer A plus 12.5 mM imidazole. The column was washed with 5–10 vol of the same buffer and then 10–15 vol of buffer A plus 25 mM imidazole. Protein was eluted from the column with 10 vol of buffer A plus 200 mM imidazole and 20 mM EDTA. Fractions containing transposase were identified by SDS/PAGE, pooled, and dialyzed overnight against 50 vol of buffer A plus 750 mM (NH4)2SO4/1 mM DTT/1 mM EDTA. The dialyzed sample was loaded onto an 8-ml phenyl Superose HR10/10 column (Pharmacia) equilibrated in the same buffer. The column was washed with about 10 vol of the loading buffer followed by about 3 vol of buffer A plus 375 mM (NH4)2SO4. The column was then eluted with a 20-vol linear gradient of 375 mM–0 mM (NH4)2SO4 in buffer A plus 1 mM DTT/1 mM EDTA. Fractions containing transposase were identified by SDS/PAGE, pooled, diluted 3-fold with 50 mM Tris⋅HCl (pH 7.5 at 4°C)/20% glycerol/1 mM DTT/1 mM EDTA (buffer B) plus 200 mM NaCl and dialyzed overnight against about 70 vol of the same buffer. One half of the dialyzed sample was loaded onto a Mono S HR5/5 column (Pharmacia) equilibrated in buffer B plus 200 mM NaCl. The column was washed with 10 vol of the loading buffer and then with a 5-vol gradient of 200–350 mM NaCl in buffer B. The column was then eluted with a 40-vol gradient of 350–600 mM NaCl in buffer B. The procedure was repeated with the second half of the sample. Fractions containing transposase were pooled, the protein was precipitated by addition of 3.8 M (NH4)2SO4/50 mM Tris⋅HCl (pH 7.5 at 4°C) to 40% saturation in (NH4)2SO4, recovered by centrifugation (17,000 × g for 10 min), and resuspended to the desired final concentration in 50 mM Tris⋅HCl (pH 7.5 at 4°C)/2.0 M NaCl/50% glycerol/1 mM DTT/1 mM EDTA. Protein concentration was determined from the A280; the theoretical extinction coefficient (ɛ280 = 68,674 M−1⋅cm−1) was calculated from the amino acid sequence (12). The sample was then dialyzed to lower the NaCl concentration to 0.5 M. Final concentrations were confirmed from the A280 reading. Typical yields were around 36 mg of transposase per 100 g of cells. The transposase preparation used throughout retains the N-terminal His tag.
Preparation of DNA Substrates.
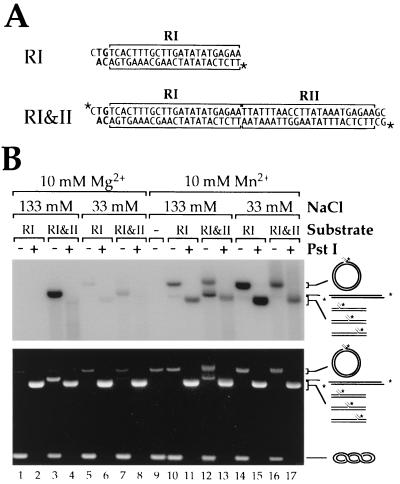
A plasmid carrying seven tandemly repeated copies of a DNA fragment containing the two binding sites for TnpA from the right end of Tn552, the CA 3′, and two additional base pairs at the “inside” end was digested with Xmn I and Tth111 I to release RI&II, a 50-bp substrate with the sequence shown in Fig. 5A. The fragment was purified on an 8% native polyacrylamide gel in TBE. RI was made by annealing AL80 (bottom strand in Fig. 5A) and AL88R (top strand in Fig. 5A), which were obtained from the Keck Biotechnology Resources Facility, Yale University Medical School. DNA concentrations were determined by measuring A260.
Figure 5.
Strand transfer reactions with oligonucleotide substrates. (A) The RI and RI&II substrates. The 23-bp transposase-binding sites are indicated with horizontal square brackets. Asterisks indicate the position of the 32P label. (B) Strand transfer reactions contained 270 ng of supercoiled pUC19 as target, about 2 pmol of a mixture of unlabeled and 32P-labeled RI or RI&II, 120 ng of TnpA, either 133 mM or 33 mM NaCl (lanes 5–8 and 14–17), and 10 mM MgCl2 or MnCl2. Reaction products were resolved by agarose gel electrophoresis and detected by both ethidium bromide staining (Lower) and autoradiography (Upper).
Various analogues of Tn552 encoding kanamycin resistance were made by PCR, by using as template a linear DNA fragment containing the Tn903 kan gene. The primers used for the left end contained LI+LII (AL25; 5′-cttggcgcgccatactagTGTTACTTTACTTGATATATGAGAATGATTTTACCTAGTAAATGAGAAcgtcaagtcagcgtaatgctc), LI (AL27; 5′-cttggcgcgccatactagTGTTACTTTACTTGATATATGAGAAcgtcaagtcagcgtaatgctc), or no repeats (AL23; 5′-cttggcgcgccatactagTGagcaatgtcctatattaaagtac). Those for the right end contained RI+RII (AL26; 5′-cttccatggactagTGTCACTTTGCTTGATATATGAGAATTATTTAACCTTATAAATGAGAAgttgtgtctcaaaatctctgatg), RI (AL28; 5′-cttccatggactagTGTCACTTTGCTTGATATATGAGAAgttgtgtctcaaaatctctgatg), or no repeats (AL24; 5′-cttccatggactagTGaaagcaacgcactttaaataag) (Tn552 sequences shown in uppercase). All primers were designed to contain the “preprocessing” SpeI site. PCR products were digested with SpeI and cloned into the SpeI site of LITMUS 28. The desired transformants were identified by restriction analysis, and the transposon-LITMUS 28 junctions—including the TnpA-binding sites—were confirmed by sequencing. The plasmid pAL101 carries the Tn552kan substrate, with the kan gene flanked by the outer 48 bp from each end of Tn552. The Tn552kan elements were obtained by digesting the appropriate plasmid with SpeI and purifying the fragments from low-melting-point agarose gels (SeaPlaque GTG, American Bioanalytical, Natick, MA) using either β-agarase (New England Biolabs) followed by ethanol precipitation and resuspension in TE or with the Geneclean II kit (Bio 101) followed by resuspension in TE.
Oligonucleotide substrates and the various Tn552kan DNAs were 5′ labeled by using [γ-32P]ATP and T4 polynucleotide kinase (New England Biolabs). Labeled fragments were separated from unincorporated nucleotides with G-25 Quick Spin columns (Boehringer Mannheim). RI&II was labeled in its double-stranded form. AL88R was hybridized to the labeled AL80 in equimolar amounts.
Strand Transfer Assays.
Typically, reaction mixtures (30 μl) contained 120 ng of transposase, 180–270 ng of pUC19; 50 fmol–3 pmol of DNA substrate; 10 mM Mg2+ (unless otherwise indicated), 133 mM NaCl, 2 mM EDTA, 50% (wt/vol) glycerol; and 50 mM Tris⋅HCl (pH 7.5 at 4°C). Reactions were incubated at 37°C for 1–3 hr and subsequently the products were precipitated with ethanol. Pellets were resuspended in 10 mM Tris⋅HCl, pH 7.5/0.1 mM EDTA, digested with PstI when appropriate, then quenched by addition of 1/4 volume of 5× Ficoll dye mix (2.5% SDS/75 mM EDTA/50 mM Tris⋅HCl, pH 7.5/25% Ficoll/bromophenol blue/xylene cyanol) and proteinase K (0.5–1.0 mg/ml final concentration). After at least 30 min at 37°C, samples were analyzed by 0.8% agarose (SeaKem GTG, American Bioanalytical) gel electrophoresis in TBE buffer, and products were detected by ethidium bromide staining and autoradiography.
Cloning and Sequencing of Transposition Products.
Competent JM109 cells (Promega) were transformed with the gel-purified concerted transposition product; alternatively, JM109 cells were electroporated directly with the outcome of a standard strand transfer reaction (resuspended in H2O). Transformants were selected with kanamycin. Plasmid DNAs were prepared by using Qiagen kits. Transposon–target junction sequences were determined using the primers 5′-GTTAATTGGTTGTAACACTGGC (AL32) and 5′-TGATGATATATTTTTATCTTGTGC (AL31) for left and right junctions, respectively. Both primers anneal within the kan gene, close to the Tn552 ends.
RESULTS
Believing that the limited activity observed earlier (11) might have resulted from the necessity to denature the initially insoluble protein, we set out to purify Tn552 transposase in a soluble form. Using the same combination of expression plasmid and host strain as Rowland et al. (11), we explored a variety of growth and induction parameters, including culture medium, growth temperature, cell density at induction, length of induction, and concentration of inducer. We found that using a defined medium and growing the cultures at 30°C were critical to our success. A pool of soluble His-tagged transposase was obtained and purified using standard methods (see Materials and Methods) to better than 95% purity.
In Vitro Concerted Strand Transfer with Tn552kan.
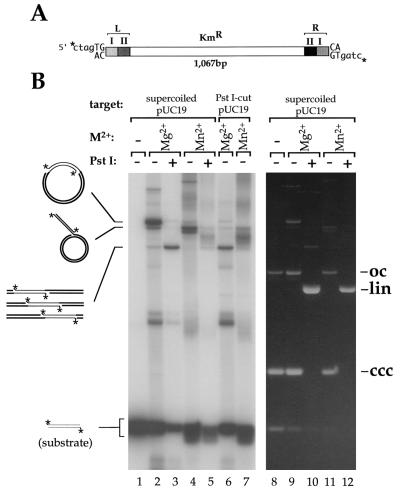
To test for activity, we incubated our soluble transposase with a labeled, preprocessed (i.e., with exposed CAOH3′ ends) substrate, Tn552kan, which consists of a kanamycin-resistance gene flanked by the terminal 48 bp from the transposon’s ends (see Fig. 1A). Concerted strand transfer of both ends of Tn552kan into target plasmid (pUC19) should result in a relaxed circle whose length will be that of pUC19 (2.7 kb) plus that of the Tn552kan (1.1 kb). Digestion with PstI (which does not cut Tn552kan) should give a 3.8-kb linear species. Insertions of a single end should produce nicked circles containing a 1.1-kb “tail.” Digestion of these products with PstI should result in Y-shaped molecules with two branches of variable length (assuming random insertion) that will not migrate as a discrete species on a gel.
Figure 1.
Strand transfer reactions with Tn552kan. (A) Tn552kan: the shaded boxes represent the 23-bp binding sites at the transposon ends. The asterisks indicate the position of the 32P label. (B) Transposition reactions (3 hr) contained 270 ng of supercoiled or PstI-linearized pUC19 as target, about 50 fmol of a mixture of unlabeled and 32P-labeled Tn552kan as the substrate, 120 ng transposase, and 10 mM MgCl2 or MnCl2 as indicated. Reactions run in lanes 3, 5, 10, and 12 were digested with PstI. Reaction products were resolved by agarose gel electrophoresis and detected by both ethidium bromide staining (Right) and autoradiography (Left). The predicted structures of the reaction products are shown; thin lines and thick lines represent the substrate and target DNAs, respectively. oc, open circle form of pUC19; lin, linear pUC19; ccc, covalently closed circular form of pUC19.
The major product of Tn552kan transposition formed in the presence of Mg2+ comigrated with a marker for the 3.8-kb nicked circle (Fig. 1B, lane 2, and data not shown) and is readily visible in the ethidium-stained gel (lane 9). The identity of this species as the product of concerted strand transfer was confirmed by its conversion, upon PstI digestion, to a 3.8-kb linear species (Fig. 1B, lane 3). Other products visible in lane 3 included (i) a band of about 2 kb that remained unchanged by PstI digestion and was also formed in reactions lacking pUC19 target DNA (data not shown)—we infer this results from concerted insertion of one molecule of Tn552kan into another; (ii) a minor band that migrated slightly faster than the 3.8-kb nicked circles and disappeared upon PstI digestion—we infer this to be the nicked form of pUC19 with a 1.1-kb tail that results from insertion of a single transposon end; (iii) a smear of radioactivity below the substrate that likely contains the products of intramolecular events; and (iv) a complex mixture of minor bands that migrated slower than the 3.8-kb nicked circles—we have not attempted to identify these but assume they result from multiple insertions into the same target and insertions into multimers of pUC19.
A divalent metal ion is essential for strand transfer activity (lane 1), and the use of alternative divalent metal ions changes the strand transfer activity of the Tn552 transposase. In the presence of Mn2+, we saw an increase in both transposition activity and product complexity (Fig. 1B, lanes 4 and 5). However, very little concerted transposition was observed; rather, the major product appeared to be the single-ended insertion. An additional band migrating just ahead of these products can be observed. Although we have not determined its identity we believe it might contain θ-shaped molecules resulting from insertion of both ends of a Tn552kan element in two independent, single-ended events. With Ca2+, transposition activity was very low and the only products detected resulted from single-end strand transfer (data not shown).
The outcome of the reactions was independent of whether the target DNA was supercoiled or linear. When we performed reactions with Mg2+ or Mn2+ using PstI-linearized pUC19 as the target (Fig. 1B, lanes 6 and 7), the patterns of products were virtually identical to those obtained after PstI digestion of the reactions with supercoiled target DNA (compare lanes 3 and 6, and 5 and 7 in Fig. 1B).
Tn552kan Insertions Are Flanked by Direct Repeats of a Target Sequence.
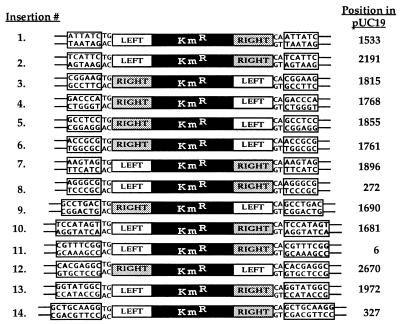
One of the hallmarks of transposition is the presence, flanking the inserted element, of copies of a short, duplicated target sequence. To determine whether insertion of Tn552kan in vitro was associated with target duplication, we performed a standard reaction with supercoiled pUC19 as target and Mg2+ as the divalent cation. Total DNA from this reaction, or DNA purified from an ethidium-stained gel slice containing the band predicted to be the concerted strand transfer product was used to transform E. coli. We analyzed plasmid DNAs from 14 colonies resistant to kanamycin. All contained single insertions of Tn552kan with the transposon ends flanked by direct repeats of pUC19 target sequences (Fig. 2). Interestingly, the size of the target duplication was variable, ranging between 6 and 9 bp (Fig. 2). The analyzed insertions did not appear to be clustered in any particular region of pUC19 (Fig. 2).
Figure 2.
Tn552kan insertion sites in pUC19. Sequences of the flanking repeats. The positions in pUC19, shown on the right, indicate the first nucleotide of the target repeat.
All Four 23-bp Repeats Are Required for Efficient Concerted Strand Transfer.
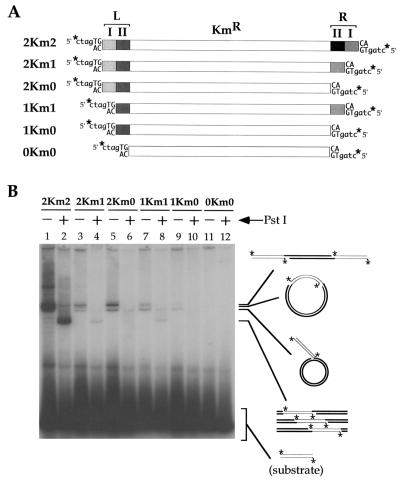
To examine the role of the two 23-bp repeats located at each Tn552 end, we constructed derivatives of Tn552kan in which these repeats were progressively removed from one or both ends (Fig. 3A). Deletion of a single (inner) 23-bp repeat from just one end severely impaired the ability of transposase to use both transposon ends in concerted strand transfer; however, such concerted strand transfer remained detectable not only with 2Km1 but also with 1Km1 (Fig. 3B; compare the yields in lanes 2, 4, and 8 of linear products obtained after digestion with PstI). For those transposons that have just one complete end (2Km1 or 2Km0), the most frequent event is coupled insertion of two different donor molecules yielding the 4.7-kb linear product (the upper band of the triplet; Fig. 3B, lanes 3 and 5). For those substrates in which the least defective end contains just a single (outer) 23-bp repeat (1Km1 or 1Km0), the preferred (though very inefficient) event is the uncoupled insertion of a single end (Fig. 3B, lanes 7 and 9).
Figure 3.
Requirement for the transposase binding sites in transposition. (A) Structure of the 2Km2–0Km0 series of Tn552kan elements (details are identical to those in Fig. 2). (B) Transposition reactions (2 hr) contained 10 mM Mg2+, 225 ng of supercoiled pUC19, and about 18 fmol of 32P-labeled substrate as indicated. Samples in even-numbered lanes were digested with PstI. Product analysis was as before.
Product Yield and Distribution Are Sensitive to Reaction Conditions.
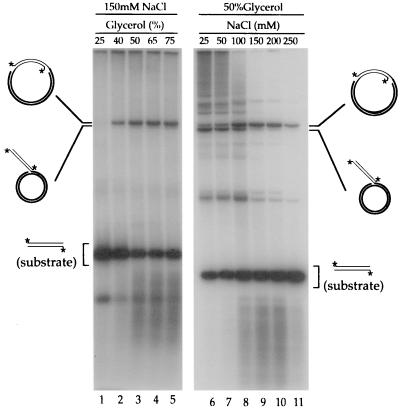
The activities of transposase are affected by glycerol and NaCl concentration (Fig. 4). Strand transfer activity under otherwise standard conditions (i.e., at 150 mM NaCl) required a high level of glycerol. Strand transfer products were undetectable at 25% glycerol or less but were obtained at high efficiency over a broad range of concentrations from 50 to 75% (Fig. 4, lanes 1–5). At a high glycerol concentration, concerted insertions were stimulated by NaCl, achieving a maximum efficiency at 100–200 mM (Fig. 4, lanes 6–11). By contrast, the level of single-ended insertions (and of most higher-molecular-weight products) decreased dramatically as NaCl concentration was increased from 25 to 150 mM.
Figure 4.
Effects of glycerol and NaCl on strand transfer activity. Reactions (1 hr) contained 180 ng (Left) or 270 ng (Right) of supercoiled pUC19 as target, about 50 fmol of 32P-labeled 2Km2 as substrate, and 10 mM Mg2+. Concentrations of NaCl and glycerol were as indicated. Product analysis was as before.
In Vitro Strand Transfer with Oligonucleotide Substrates.
A 50-bp “preprocessed” oligonucleotide substrate, RI&II, with the tandem transposase-binding sites is an efficient minimal substrate for concerted strand transfer. Coupled insertion of two oligonucleotide substrates at a single site should linearize the target plasmid. As shown in Fig. 5, a reaction with the labeled RI&II substrate in the presence of Mg2+ and high NaCl concentration gave high yields of a single product that migrates just behind linears of the pUC19 target DNA; this product also is visible in the ethidium-stained gel. We infer this to be the linear product of concerted insertion. Digestion with PstI (which cuts the target once) resulted in a heterogeneous smear of labeled DNA running ahead of linear pUC19, both confirming our conclusion and indicating an absence of any strongly preferred target sites. [Similar results were obtained using a left-end substrate (LI&II) containing both 23-bp repeats (data not shown), indicating that left and right ends are equally reactive.] The high yield of concerted insertions required an elevated NaCl concentration (compare lanes 3 and 7) and the presence of both transposase-binding sites; a 25-bp substrate, RI, with just a single transposase-binding site (and a preprocessed end) gave no products under standard conditions (lanes 1 and 2).
Replacing Mg2+ with Mn2+ had two main effects. As expected from the results with Tn552kan, it reduced the coupling of strand transfer events. Open circular products, indicative of single-end insertions, were obtained in significant amounts, even with the RI&II substrate at a high NaCl concentration (Fig. 5, compare lanes 3 and 12). Mn2+ also made the transposase permissive for the suboptimal 25-bp RI substrate, particularly at low NaCl concentration, resulting in the formation of substantial amounts of single-end insertions (Fig. 5, lanes 10, 11, 14, and 15).
DISCUSSION
We have developed a protocol for the expression of Tn552 transposase and its purification from the soluble fraction, avoiding the use of any denaturing steps. The transposase purified in this way efficiently catalyzes concerted insertions into an added target DNA, using substrate DNA with preprocessed ends, indicating that the protein is competent at both synapsis and strand transfer. The reaction requires only substrate and target DNAs, transposase protein, buffer, Mg2+, and glycerol. The substrate DNA may be a transposon analogue with two complete 48-bp Tn552 ends or a short oligonucleotide with a single 48-bp end; in either case, coupled insertions of two ends are obtained. The observed activity differs significantly from that of a previous preparation of the Tn552 transposase that was limited to single-end insertions in a Mn2+-dependent reaction (11). Similarly efficient synapsis and strand transfer reactions with short oligonucleotide substrates have been observed with Mu and Tn10 transposases (13, 14).
The concerted insertions obtained in vitro show all the hallmarks of normal transposition. When recovered from transformed E. coli cells and characterized, all insertions are accompanied by short target duplications, indicating that the target attack in vitro occurs with the expected staggered breaks. These target duplications are somewhat variable in size, ranging from 6 to 9 bp. However, variability may also be a feature of Tn552 transposition in vivo; of just three transpositions characterized, two were flanked by 6-bp repeats (1, 15) whereas the third had a 7-bp duplication (16). Within the relatively small sample analyzed, we saw no site specificity or clustering of insertion sites, suggesting that concerted strand transfer is rather nonspecific regarding target sequences. In fact, analysis of whole reactions at the molecular level has shown strand transfer to occur at virtually every position in the target DNA (unpublished data). These characteristics, coupled with the efficiency with which the reaction is catalyzed, make Tn552 transposase a potentially useful biotechnological tool.
Strand transfer activity of our transposase preparation is supported by either Mg2+ or Mn2+ but the outcome is influenced by the choice of metal. Under otherwise standard conditions, Mg2+, which is expected to be the relevant cation in vivo, leads to a much cleaner reaction profile with a higher yield of concerted strand transfer products and an overall reduction in single-ended events. In the presence of Mn2+, the total strand transfer is increased (see, for example, Fig. 5) but most products appear to result from uncoupled single-ended events and the yield of coupled events drops. Increased permissiveness in the presence of Mn2+ has been observed in several other systems, for example, retroviral integrases (17–19), Mu transposase (20), Tn10 transposase (21), and DNA polymerases (22, 23)—but the exact nature of this effect is not understood.
The concentrations of both glycerol and NaCl have a pronounced effect on strand transfer activity. Our protein requires unusually high levels of glycerol: more than 25% was necessary to detect reaction products at all, and their accumulation peaked only when the reactions were carried out in the presence of 50–75% glycerol (see Fig. 4). Glycerol stabilizes protein structure through the “solvophobic” effect; its favorable interaction with the bulk water solvent makes the contact between hydrophobic residues on the protein and water less unfavorable than those with the glycerol solution, effectively driving glycerol away from the protein surface. This preferential hydration of the protein surface is a driving force for minimizing its area (24). The extremely high concentrations of glycerol required in our reactions might be stabilizing the structure, preventing partial denaturation and aggregation, or inducing a conformational change that would normally be driven by other factor(s) in vivo.
The most interesting effect of NaCl is the selective and increasing inhibition of single-ended events and concomitant stimulation of concerted insertions as the concentration is raised from 50 to 150 mM. Possible explanations can be divided into two general classes. Full, concerted transposition normally would occur within a synaptic complex consisting of a transpososome (the two transposon ends bound and brought together by transposase—most likely four transposase protomers in the case of Tn552) plus target DNA. Within this complete complex, strand transfer events generally are concerted either because they are kinetically coupled (i.e., the rates of catalysis are faster than the rate of dissociation) or because intersubunit communication ensures that both catalytic events are primed to occur virtually simultaneously. The high incidence of single-ended events at low NaCl concentration could result either from uncoupling of the catalytic events within complete synaptic complexes or from dispensing with the requirement for a complete synaptic complex and allowing aberrant transposase-end–target complexes (lacking an end or some subunits, or incompletely engaging the target) to initiate strand transfer. High NaCl concentration may both inactivate or dissociate aberrant complexes and promote the formation of transpososomes or transpososome–target complexes.
Efficient concerted strand transfer requires both transposase-binding sites contained within the 48-bp terminal DNA segment. The reactions performed with oligonucleotide substrates (Fig. 5 and data not shown) suggest that under our standard conditions, the right and left ends are fully interchangeable. This is in contrast to what was observed in the analogous reaction with Mu transposase where two right ends proved to be far more efficient substrates (13). The requirement for a pair of binding sites at each end suggests that a Tn552 transpososome contains four protomers of the transposase (assuming each site binds one transposase monomer).
We have been unable, so far, to detect 3′ end processing activity by Tn552 transposase under our current assay conditions. This was somewhat unexpected given the efficiency with which the protein catalyzes strand transfer with preprocessed substrates. One possible explanation is that TnpB is required for this activity. Although MuB is not required for formation of the Mu transpososome and cleavage at the Mu 3′ ends (25), cleavage at the ends of Tn7 requires the TnsC protein (26). In the absence of the terminal cleavages, TnpB may be required to stabilize the transpososome or to activate the endonuclease function of the transposase.
Tn552 can now be added to a short list of transposable elements for which efficient concerted strand transfer is achievable in vitro (6, 14, 26–30). Two of the best-characterized systems are for other, distantly related members of the D,D(35)E superfamily—Mu and Tn7 (for reviews see refs. 5–7). For both of these systems the protein and substrate DNA requirements are complex. Transposition involves both the transposase (MuA or the heteromeric TnsAB of Tn7) and an ATP-dependent, nonspecific DNA-binding protein (MuB or Tn7 TnsC) that activates the transposase and plays a role in target presentation. In addition, Tn7 transposition requires one of two other proteins, either TnsD or TnsE, that determine alternative pathways of target site selection. The ends of Mu each contain three binding sites for its transposase; these are distributed irregularly over 175 bp at the left end and 80 bp at the right. In addition, Mu contains an internal sequence, the IAS, which is bound by the extreme N-terminal domain of MuA and is important for assembly of the transpososome but not for its subsequent activity. Tn7 has seven binding sites for TnsB: three, spanning 150 bp at the left end, and four, covering 90 bp at the right; again, the spacing between sites varies. Tn552 appears to provide a simpler transposition system. Not only is its transposase considerably smaller but also its natural substrate is less complex; the ends are of identical structure, containing just the two abutting transposase-binding sites, and the transposon presumably has no equivalent to the IAS.
Mizuuchi and colleagues (13, 31) have found reaction conditions that simplify dramatically the overall requirements for Mu transposition. In the presence of DMSO (as well as glycerol), dependence on MuB, the IAS, substrate superhelicity, and even the first 76 residues of MuA (the IAS-binding domain) is lost. Under the modified conditions, a DNA substrate with just the terminal 50 bp of the Mu right end and a few flanking base pairs is assembled efficiently into a transpososome, cleaved at the transposon end, and inserted into target DNA. It may well be significant that the active minimal substrate is organized similarly to the Tn552 substrate, with a pair of adjacent transposase-binding sites. This arrangement, found at only one end of the wild-type Mu and Tn7 transposons, probably provides an optimal scaffold for assembly of the Mu transpososome, which is known to contain a tetramer of the transposase (32). By adopting this arrangement at both ends, Tn552 may have obviated the necessity for the structural complexities found in Mu and Tn7. The reaction we have described here with Tn552 transposase is very similar to this simplified Mu reaction; indeed its lack of dependence on TnpB (the MuB analogue) may be attributable to the somewhat unusual reaction conditions or to the use of preprocessed transposon ends. Its overall simplicity (particularly its independence of TnpB) makes Tn552 transposase attractive for structural studies.
Acknowledgments
We are grateful to Sally J. Rowland for her generous gift of the Tn552 transposase expression strain. This work was supported by Grant GM28470 from the National Institutes of Health. A.E.L. was supported in part by a Postgraduate Fellowship B from the Natural Sciences and Engineering Research Council of Canada.
References
- 1.Rowland S J, Dyke K G. EMBO J. 1989;8:2761–2773. doi: 10.1002/j.1460-2075.1989.tb08418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murphy E, Novick R P. Mol Gen Genet. 1979;175:19–30. doi: 10.1007/BF00267851. [DOI] [PubMed] [Google Scholar]
- 3.Rowland S J, Dyke K G. Mol Microbiol. 1990;4:961–975. doi: 10.1111/j.1365-2958.1990.tb00669.x. [DOI] [PubMed] [Google Scholar]
- 4.Polard P, Chandler M. Mol Microbiol. 1995;15:13–23. doi: 10.1111/j.1365-2958.1995.tb02217.x. [DOI] [PubMed] [Google Scholar]
- 5.Craig N L. Curr Top Microbiol Immunol. 1996;204:27–48. doi: 10.1007/978-3-642-79795-8_2. [DOI] [PubMed] [Google Scholar]
- 6.Mizuuchi K. Annu Rev Biochem. 1992;61:1011–1051. doi: 10.1146/annurev.bi.61.070192.005051. [DOI] [PubMed] [Google Scholar]
- 7.Lavoie B D, Chaconas G. Curr Top Microbiol Immunol. 1996;204:83–102. doi: 10.1007/978-3-642-79795-8_4. [DOI] [PubMed] [Google Scholar]
- 8.Dyda F, Hickman A B, Jenkins T M, Engelman A, Craigie R, Davies D R. Science. 1994;266:1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- 9.Rice P, Mizuuchi K. Cell. 1995;82:209–220. doi: 10.1016/0092-8674(95)90308-9. [DOI] [PubMed] [Google Scholar]
- 10.Bujacz G, Jaskolski M, Alexandratos J, Wlodawer A, Merkel G, Katz RA, Skalka A M. J Mol Biol. 1995;253:333–346. doi: 10.1006/jmbi.1995.0556. [DOI] [PubMed] [Google Scholar]
- 11.Rowland S J, Sherratt D J, Stark W M, Boocock M R. EMBO J. 1995;14:196–205. doi: 10.1002/j.1460-2075.1995.tb06990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gill S C, von Hippel P H. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 13.Savilahti H, Rice P A, Mizuuchi K. EMBO J. 1995;14:4893–4903. doi: 10.1002/j.1460-2075.1995.tb00170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sakai J, Chalmers R M, Kleckner N. EMBO J. 1995;14:4374–4383. doi: 10.1002/j.1460-2075.1995.tb00112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rice L B, Carias L L, Marshall S H, Bonafede M E. Plasmid. 1996;35:81–90. doi: 10.1006/plas.1996.0010. [DOI] [PubMed] [Google Scholar]
- 16.Paulsen I T, Gillespie M T, Littlejohn T G, Hanvivatvong O, Rowland S J, Dyke K G, Skurray R A. Gene. 1994;141:109–114. doi: 10.1016/0378-1119(94)90136-8. [DOI] [PubMed] [Google Scholar]
- 17.Kulkosky J, Jones K S, Katz R A, Mack J P, Skalka A M. Mol Cell Biol. 1992;12:2331–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drelich M, Wilhelm R, Mous J. Virology. 1992;188:459–468. doi: 10.1016/0042-6822(92)90499-f. [DOI] [PubMed] [Google Scholar]
- 19.Katzman M, Katz R A, Skalka A M, Leis J. J Virol. 1989;63:5319–5327. doi: 10.1128/jvi.63.12.5319-5327.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim K, Namgoong S Y, Jayaram M, Harshey R M. J Biol Chem. 1995;270:1472–1479. doi: 10.1074/jbc.270.3.1472. [DOI] [PubMed] [Google Scholar]
- 21.Junop M S, Haniford D B. EMBO J. 1996;15:2547–2555. [PMC free article] [PubMed] [Google Scholar]
- 22.Beckman R A, Mildvan A S, Loeb L A. Biochemistry. 1985;24:5810–5817. doi: 10.1021/bi00342a019. [DOI] [PubMed] [Google Scholar]
- 23.Van de Sande J H, Loewen P C, Khorana H G. J Biol Chem. 1972;247:6140–6148. [PubMed] [Google Scholar]
- 24.Timasheff S N. Annu Rev Biophys Biomol Struct. 1993;22:67–97. doi: 10.1146/annurev.bb.22.060193.000435. [DOI] [PubMed] [Google Scholar]
- 25.Surette M G, Buch S J, Chaconas G. Cell. 1987;49:253–262. doi: 10.1016/0092-8674(87)90566-6. [DOI] [PubMed] [Google Scholar]
- 26.Bainton R, Gamas P, Craig N L. Cell. 1991;65:805–816. doi: 10.1016/0092-8674(91)90388-f. [DOI] [PubMed] [Google Scholar]
- 27.Chalmers R M, Kleckner N. J Biol Chem. 1994;269:8029–8035. [PubMed] [Google Scholar]
- 28.Lampe D J, Churchill M E, Robertson H M. EMBO J. 1996;15:5470–5479. [PMC free article] [PubMed] [Google Scholar]
- 29.Goryshin I Y, Reznikoff W S. J Biol Chem. 1998;273:7367–7374. doi: 10.1074/jbc.273.13.7367. [DOI] [PubMed] [Google Scholar]
- 30.Ton-Hoang B, Polard P, Chandler M. EMBO J. 1998;17:1169–1181. doi: 10.1093/emboj/17.4.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Craigie R, Mizuuchi K. Cell. 1987;51:493–501. doi: 10.1016/0092-8674(87)90645-3. [DOI] [PubMed] [Google Scholar]
- 32.Lavoie B D, Chan B S, Allison R G, Chaconas G. EMBO J. 1991;10:3051–3059. doi: 10.1002/j.1460-2075.1991.tb07856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]