Abstract
Components of cellular stress responses can be identified by correlating changes in stress tolerance with gain or loss of function of defined genes. Previous work has shown that yeast cells deficient in Ppz1 protein phosphatase or overexpressing Hal3p, a novel regulatory protein of unknown function, exhibit increased resistance to sodium and lithium, whereas cells lacking Hal3p display increased sensitivity. These effects are largely a result of changes in expression of ENA1, encoding the major cation extrusion pump of yeast cells. Disruption or overexpression of HAL3 (also known as SIS2) has no effect on salt tolerance in the absence of PPZ1, suggesting that Hal3p might function upstream of Ppz1p in a novel signal transduction pathway. Hal3p is recovered from crude yeast homogenates by using immobilized, bacterially expressed Ppz1p fused to glutathione S-transferase, and it also copurifies with affinity-purified glutathione S-transferase-Ppz1p from yeast extracts. In both cases, the interaction is stronger when only the carboxyl-terminal catalytic phosphatase domain of Ppz1p is expressed. In vitro experiments reveal that the protein phosphatase activity of Ppz1p is inhibited by Hal3p. Overexpression of Hal3p suppresses the reduced growth rate because of the overexpression of Ppz1p and aggravates the lytic phenotype of a slt2/mpk1 mitogen-activated protein kinase mutant (thus mimicking the deletion of PPZ1). Therefore, Hal3p might modulate diverse physiological functions of the Ppz1 phosphatase, such as salt stress tolerance and cell cycle progression, by acting as a inhibitory subunit.
Because of the relatively high toxicity of sodium, the maintenance of low intracellular concentrations of this cation is a general problem for virtually every cell type. Yeast cells can actively exclude sodium (and lithium) to maintain appropriate intracellular levels of these cations when their concentrations increase in the environment. In Saccharomyces cerevisiae, the Ser/Thr protein phosphatase Ppz1 is a major determinant for salt tolerance (1). Ppz1 represents a novel type of Ser/Thr phosphatase characterized by a catalytic carboxyl-terminal half related to type 1 phosphatase and an amino-terminal half unrelated to phosphatases and rich in Ser and Thr, as well as in basic residues (2). As far as we know, the PPZ1 phosphatase gene represents the only case in which loss of function results in increased salt tolerance. Deletion of PPZ2, a very related gene (3, 4), has almost no effect, although the double mutant has a tolerance to sodium and lithium somewhat higher than the ppz1 deletant. The increased sodium tolerance of ppzΔ mutants can be attributed to an increased expression, even in the absence of salt stress, of the ENA1/PMR2A gene (1). This gene codes for a P-type ATPase (5, 6) responsible for sodium efflux and is the first unit of a tandem array of five genes (the PMR2 locus), encoding nearly identical proteins (6, 7). However, only the expression of ENA1/PMR2A is strongly induced upon exposure of the cells to sodium, lithium, or high pH, and, consequently, cells lacking ENA1/PMR2A are hypersensitive to these cations.
The PPZ phosphatases also are involved in the maintenance of cell integrity under other stress conditions. For instance, exposure of ppzΔ mutants to caffeine or high temperature leads to cell lysis. A connection between the PPZ phosphatases and the protein kinase C/mitogen-activated protein (MAP) kinase pathway has been documented (3, 8), because the deletion of PPZ1 aggravates the lytic phenotype of a slt2/mpk1 MAP kinase mutant (3). However, there is no evidence that the latter pathway might be closely connected to the sodium tolerance pathways. We have also shown that, whereas deletion of PPZ1 does not inhibit cell growth under standard conditions, strong overexpression of the phosphatase results in an intense growth defect (9).
In addition to the PPZ phosphatases, genetic approaches based on gain or loss of function have identified several yeast genes involved in sodium and lithium tolerance. Some examples are HAL1 (10), HAL2 (11, 12), HAL3/SIS2 (13), and the genes encoding casein kinase-1 (14), casein kinase-2 (15), protein phosphatase 2B (calcineurin) (16, 17), and the HOG1 MAP kinase (18). With few exceptions, the mechanisms underlying the function of these gene products and the interconnections between them are unknown. Some regulatory genes such as calcineurin (16, 17) and the HOG1 MAP kinase (18) are required for the induction of the ENA1 gene by salt stress. Others, such as HAL3 (13, 18) and PPZ1 (1), modulate both basal and salt-induced levels of ENA1 to a similar extent and, consequently, are not part of the pathways transducing the salt stress signal. They therefore could correspond to novel signal transduction pathways controlling ion homeostasis. Another similarity between HAL3 and PPZ is that changes in the expression of any of these regulatory genes influence the progression of the yeast cell cycle (9, 13). In fact, HAL3 is allelic to SIS2, which was described as a gene whose overexpression rescued the G1/S transition defect of sit4 mutants (19). These similarities prompted us to clarify the possible functional relationship between these two regulatory proteins. In this report we present evidence that Hal3p influences salt tolerance (and possibly cell cycle progression) in yeast cells by acting on the Ppz1 protein phosphatase as a negative regulatory subunit.
MATERIALS AND METHODS
Growth of Escherichia coli and Yeast Strains.
E. coli strains NM522 or DH5α were used as a host in DNA cloning and heterologous expression experiments. Bacterial cells were grown at 37°C (unless otherwise stated) in Luria–Bertani medium containing 50 μg/ml ampicillin, when needed, for plasmid selection. Yeast cells were grown at 28°C in yeast extract/peptone/dextrose (YPD) medium or, when indicated, in CM synthetic medium (20). Unless otherwise stated, all yeast strains generated in this work derive from strain JA-100 (MATa PPZ1 PPZ2 HAL3 ura 3–52 leu2–3, 112 trp1–1 his4 can-1r).
Recombinant DNA Techniques.
E. coli. cells usually were transformed by using standard calcium chloride treatment (21). Yeast cells were transformed by using a modification of described methods (22). Restriction reactions, DNA ligations, and other standard recombinant DNA techniques were carried out as described (21). Gene disruptions were performed by using the one-step technique (23). HAL3 was interrupted with the LEU2 marker as described in ref. 13. The PPZ1 disruption was as described (2). The PPZ2 gene was disrupted by replacing a 0.95-kbp HpaI-XhoI fragment containing about 100 bp of 5′ untranslated region plus most of the NH2-terminal coding region by the 0.8-kbp SalI-SmaI fragment containing the TRP1 gene, obtained from vector YDp-W (24). All disruptions were verified by PCR.
Identification of a Physical Interaction Between Hal3p and Ppz1p.
The construction of the wild-type, R451L, and NH2-terminally deleted (Δ1–344) forms of Ppz1 as GST fusions in plasmid pGEX-KT (25) for expression in E. coli has been described previously (9). A version of Ppz1p lacking residues from 17 to 193 (Δ17–193) was constructed as follows. The 1.6-kbp BamHI-HindIII fragment cloned into pBluescript SK(+) (9) was excised with HindIII and SalI and cloned into pSP72 (Promega). The construct was digested with PvuII and BamHI and cloned into the BamHI and SmaI sites of plasmid pGEX-KT. The Δ241–318 mutant was prepared similarly by using as a source plasmid pYC5Z1 (9). Expression was accomplished as in ref. 26 except that E. coli cells were induced with 0.2 mM isopropyl β-d-thiogalactoside for 3 hr at 25°C. Purification of the fusion proteins was carried out as described (26) with some modifications. Briefly, the fusion proteins were allow to bind to the glutathione-Sepharose 4B (Pharmacia) affinity matrix (typically 50–100 μl) for 1 hr at 4°C, and the beads were washed three times with buffer A (50 mM Tris⋅HCl, pH 8/150 mM NaCl/1% Triton X-100) and twice with buffer B (50 mM Tris⋅HCl, pH 8/0.1% 2-mercaptoethanol). For interaction experiments, a given volume of beads was mixed with total yeast extract for 1 hr at 4°C with gently shaking. Beads were washed three times with buffer A and twice with buffer B, boiled for 10 min in 50 μl of 2× SDS/PAGE sample buffer, electrophoresed on 8% SDS-polyacrylamide gels, and transferred to membranes. Total yeast extracts were prepared as follows. Cells were grown up to an OD660 of 3, collected by centrifugation, and disrupted with the aid of glass beads in the presence of a buffer containing 100 mM Tris⋅HCl, pH 7.5/150 mM NaCl/1 mM EDTA/1 mM DTT, plus an antiprotease mixture (0.5 mM phenylmethylsulfonyl fluoride/1 μg/ml leupeptin/0.5 mM benzamidine/1 μg/ml pepstatin). The mixture was centrifuged at 750 × g for 10 min at 4°C to remove glass beads and cellular debris, and the supernatant subsequently was used for interaction experiments.
Expression in yeast cells of GST-fused versions of wild-type and Δ1–344 forms of Ppz1p from a centromeric vector was accomplished as follows. The GST moiety was amplified from plasmid pGEX-KT (25) by using oligonucleotides E3 (5′-GCGGATCCAAAATGTCCCCTATACTAGGTTATTG-3′) and E4 (5′-CGCGTCGACACGCGGAACCAGACCACC-3′). The 0.68-kbp fragment was digested with BamHI and SalI (underlined) and cloned into these sites of the centromeric plasmid YCplac111 (27) to yield YCp-GST. Oligonucleotides E1(5′-CGGGTACCATGTTCAGATAGCTGCC-3′) and E2 (5′-GCGGATCCGAAGGAAAGATAAGCAGAG-3′) were used to amplify the promoter region of PPZ1. This 456-bp fragment was digested with KpnI/BamHI (underlined) and placed upstream the GST sequence by ligating it into these same sites of YCp-GST. This construct then was digested with SalI and HindIII and used to clone the entire PPZ1 ORF or the Δ1–344 form (obtained by SalI/HindIII digestion from plasmids pYC1Z1 and pYC2Z1, respectively, see ref. 9) to yield plasmids pYGST-C1Z1 and pYGST-C2Z1. These constructs were used to transform strain JA-101 (ppz1Δ). For affinity purification of GST-fused proteins, yeast cells were grown in 40 ml of CM medium lacking leucine, with 2% glucose, until an OD660 of 3. Cells were collected, washed in 10 mM Tris⋅HCl, pH 7.5/0.3 M sorbitol/0.1 M NaCl (buffer C) and disrupted with glass beads in the presence of 0.8 ml of buffer C supplemented with 0.1% 2-mercaptoethanol and the antiprotease cocktail. Homogenates were centrifuged at 4°C for 10 min at 750 × g and 0.5–0.6 ml of the supernatant (5 mg of proteins) mixed with 100 μl of the affinity matrix at 4°C for 75 min. After extensive washing with buffer C, the matrix was boiled for 10 min in the presence of 150 μl of 2× sample buffer and aliquots were analyzed by SDS/PAGE and transferred to membranes for immunoblotting.
In Vitro Inactivation of Ppz1p by Hal3p.
Ppz1 phosphatase activity was measured by using 32P-labeled myelin basic protein as substrate, in the presence of 25 mM MnCl2, essentially as described (26). The final concentrations of the catalytic proteins in the assays were 1.7 μg/ml (GST-Ppz1) and 3.7 μg/ml (GST-Δ1–344). The assay mixture was supplemented with different amounts of the Hal3p preparation (or buffer), to achieve final concentrations between 0–12.6 μg/ml, and the assay incubation was carried out for 10 min. Recombinant GST-Hal3p was expressed in E. coli from plasmid pGEX-KG as described (13) and purified by glutathione-Sepharose 4B chromatography. To remove the GST moiety, one volume of beads was incubated with one volume of thrombin in Tris-buffered saline (10 units/ml) for 30 min at 20°C. The sample was briefly centrifuged, and the supernatant (40–50 μg/ml of protein) was made 0.1 mM phenylmethylsulfonyl fluoride and stored at −80°C. To account for the presence of minor amounts of lower-molecular-weight polypeptides, the amount of the complete protein was evaluated in each case by running samples in SDS-polyacrylamide gels and scanning Coomassie blue-stained gels. Quantitation was made by comparison with different amounts of BSA.
Other Methods.
Salt and caffeine sensitivity assays were performed in freshly prepared YPD or CM plates containing different concentrations of the compound. Drop tests were performed as in ref. 1. SDS/PAGE was carried out as in ref. 28. For immunological detection, proteins were transferred to Immobilon P membranes (Millipore), blocked, and incubated with the appropriate polyclonal antibody. Immunoreactive proteins were identified with luminescence substrates (ECL, Amersham).
RESULTS
Hal3p Can Be Placed Upstream of the Ppz1p Phosphatase.
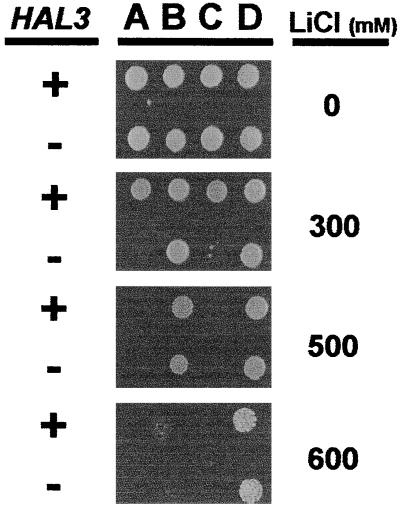
To test the possibility of a functional connection between Hal3p and Ppz1p, we constructed strains combining the hal3Δ mutation with the deletion of PPZ1, PPZ2, or both phosphatase genes and tested their growth at different concentrations of lithium chloride (Fig. 1). As it can be observed, lack of PPZ1, but not of PPZ2, results in increased salt tolerance. Also as expected, the deletion of HAL3 results in a decrease in tolerance in wild-type cells (PPZ1 PPZ2) and in the PPZ1 ppz2 background. However, in cells lacking Ppz1 (ppz1 PPZ2 and ppz1 ppz2) the effect of the HAL3 disruption is not observed, because no decrease in salt tolerance is detected when compared with a strain carrying a functional copy of HAL3. It is important to note that, despite the potency of the ppz1Δ mutation, its combination with mutations that results in salt hypersensitivity gives a clearly detectable decrease in tolerance, as it has been shown in the case of calcineurin (1). In addition, overexpression of HAL3, which has been reported to markedly increase salt tolerance in wild-type cells, does not result in an additional increase in tolerance in ppz1 or ppz1 ppz2 cells (not shown).
Figure 1.
Effect on the tolerance to lithium chloride of deletion of the HAL3 gene in wild-type, ppz1Δ, ppz2Δ, and ppz1Δ ppz2Δ backgrounds. PPZ1 PPZ2 (A), ppz1Δ PPZ2 (B), PPZ1 ppz2Δ (C), and ppz1Δ ppz2Δ (D) cells, carrying a wild-type (+) or deleted (−) HAL3 gene as indicated, were grown at 28°C on YPD medium containing the indicated concentrations of lithium chloride. Growth was scored after 3 days.
Hal3p Can Interact with Ppz1p and Inhibits Its Protein Phosphatase Activity.
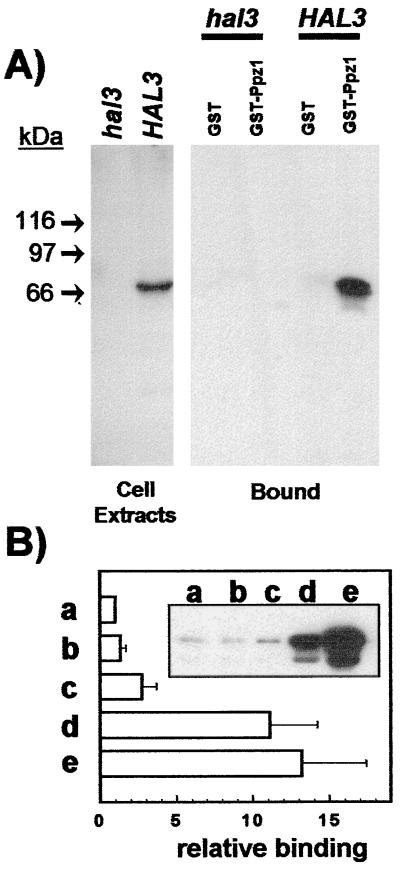
The possibility that Hal3p might actually regulate Ppz1p function on salt tolerance through direct interaction was tested initially by constructing an affinity system based in the expression of Ppz1 in bacteria as a GST-fusion protein. Recombinant GST-Ppz1p then was used to bind the Hal3p present in yeast cell extracts. As shown in Fig. 2A, Hal3p is detected specifically by immunoblot analysis of the material retained by the affinity system. Because the 692-residue Ppz1 protein has two clearly defined moieties, the carboxyl-terminal half, which corresponds to the phosphatase domain, and the Ser/Thr-rich NH2-terminal half, we then evaluated which could be the participation of both regions in the interaction with Hal3p. To this end, different versions of GST-fused Ppz1 were expressed in E. coli and used in binding experiments. Interestingly (Fig. 2B), the interaction was not modified when the noncatalytically active (R451L) version of the phosphatase was used. A relatively short deletion (Δ241–318) near the end of the NH2-terminal half modestly increased the intensity of the interaction. On the contrary, deletion of a relatively large segment at the NH2 terminus (Δ17–193) resulted in a substantial increase in binding that was enhanced further when the complete NH2-terminal half was removed (Δ1–344). These results are compatible with the notion of Hal3p interacting with the catalytic domain of the phosphatase.
Figure 2.
Physical interaction between bacterially expressed Ppz1p and cellular Hal3p. (A Left) Immunodetection of Hal3p in cell extracts. Whole yeast extracts were prepared as described from strain JA-104 (hal3Δ) or wild-type strain JA-100 (containing the HAL3 gene in a multicopy plasmid). Extracts were subjected to SDS/PAGE and immunoblotted, and the Hal3 protein was detected with polyclonal antibodies. (A Right) Cell extracts (5 mg of total protein) from the above-mentioned strains were mixed with recombinant GST or GST-Ppz1 bound to glutathione-Sepharose beads and processed as described under Materials and Methods. The presence of Hal3p bound to the affinity system was detected by using anti-Hal3p antibodies. (B) Similar binding experiments (using about 1 mg of total yeast proteins) were performed using as a bait: (a) GST-Ppz1, (b) GST-Ppz1(R451L), (c) GST-Ppz1(Δ241–318), (d) GST-Ppz1(Δ17–193), and (e) GST-Ppz1(Δ1–344). Because, in some cases, the preparations of the recombinant proteins contained small quantities of lower-molecular-weight products, the amount of the complete proteins used as a bait was estimated by Coomassie blue staining and/or immunoblot using anti-Ppz1p antibodies. After immunoblot with the anti-Hal3p antibody, the intensity of the signals was corrected, in each case, for possible differences in the amount of the bait proteins. The values correspond to the increase (-fold) with respect to the signal given by the GST-Ppz1 product and are the mean ± SEM of five independent experiments. Inset shows a typical immunoblot experiment.
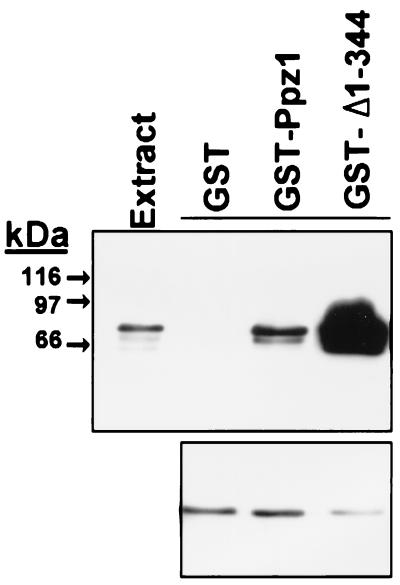
The interaction between Ppz1p and Hal3p also can be detected when both proteins are expressed in yeast cells at physiological levels. GST-fused Ppz1p was expressed from a centromeric plasmid from its own promoter to give protein levels virtually identical to those obtained from expression of the chromosomal gene (not shown). As shown in Fig. 3, affinity-purified GST-Ppz1p from yeast extracts contain bound Hal3p. Only a small fraction (10–15%) of cellular Hal3p is recovered as bound to GST-Ppz1p. Again, the amount of bound Hal3p is greatly enhanced when only the COOH-terminal moiety is expressed (about 75% of total Hal3p, as estimated by Western blot analysis, Fig. 3), thus confirming the notion that Hal3p can interact with the catalytic moiety of Ppz1p.
Figure 3.
Immunodetection of Hal3p bound to different forms of GST-Ppz1 purified from yeast extracts. The indicated constructs in plasmid YCplac111 were expressed in yeast from the PPZ1 promoter and affinity-purified from total yeast extracts through a glutathione-Sepharose matrix as described in Materials and Methods. The beads were boiled in 150 μl SDS/PAGE sample buffer, and aliquots (40 μl) were electrophoresed in 8% polyacrylamide gels. Proteins were transferred to membranes and incubated with anti-Hal3p antibodies (Upper). The material not bound to the affinity system also was subjected to SDS/PAGE (75 μg of protein) and analyzed by immunoblot for the presence of Hal3p (Lower).
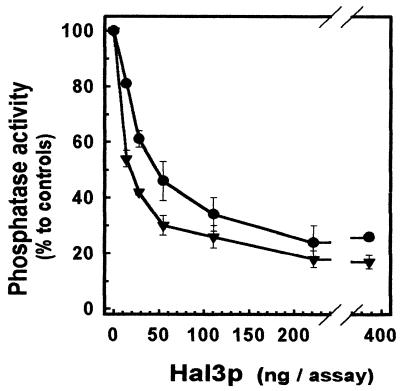
The possibility that Hal3p could actually inhibit the phosphatase activity of Ppz1 was tested by incubating GST-Ppz1p in the presence of different amounts of recombinant Hal3p and determining the phosphatase activity of the mixture. As observed in Fig. 4, recombinant Hal3p is able to produce a strong inhibition of the Ppz1 phosphatase activity (up to 25% of the initial activity) in a dose-dependent manner (calculated Ki = 30 nM). Boiling of the Hal3p preparation for 5 min completely abolished the inhibitory effect and incubation with different amounts of the noncatalytic form of Ppz1p (R451L) reduced the inhibition in a dose-dependent manner. Interestingly, the bacterially expressed COOH-terminal half of PPZ1, which has catalytic characteristics similar to those of the whole phosphatase, is inhibited even more effectively (calculated Ki = 5 nM).
Figure 4.
In vitro inactivation of recombinant Ppz1p by Hal3p. Purified GST-Ppz1 (•) or the NH2-terminally (Δ1–344) deleted version (▾) was incubated in the presence of increasing concentrations of Hal3p, and the phosphatase activity was determined as described. The assay was carried out in a final volume of 30 μl. Data are mean ± SEM of three independent experiments.
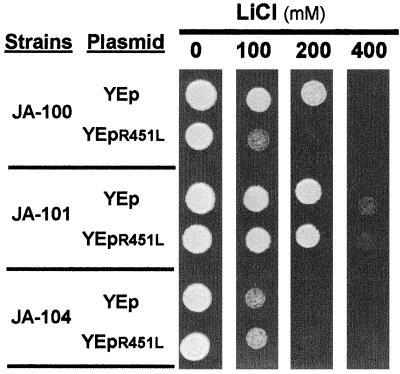
We have also observed (Fig. 5) that the overexpression of the mutated R451L form of Ppz1p on a wild-type background results in decreased salt tolerance. The intensity of the phenotype was very similar (but not additive) to that obtained by deletion of the HAL3 gene. Because the R451L mutant, although largely devoid of catalytic activity, is still able to bind Hal3p similarly to the wild-type phosphatase (Fig. 2 and data not shown), the observed phenotype can be explained on the basis that the excess of the mutated phosphatase prevents Hal3p from inhibiting the endogenous wild-type Ppz1p.
Figure 5.
Overexpression of a catalytically inactive (R451L) version of Ppz1 in wild-type yeast cells leads to lithium chloride hypersensitivity. Strains J-100 (HAL3 PPZ1), JA-101 (HAL3 ppz1Δ), and JA-104 (hal3Δ PPZ1) were transformed with the indicated plasmid and, upon selection, were plated in YPD medium containing the indicated concentrations of lithium chloride. Growth was monitored after 3 days of incubation at 28°C.
All these results support the notion that the negative effect of Hal3p on salt tolerance can be explained through a direct interaction between Hal3p and Ppz1p that results in inactivation of the phosphatase.
The Relationship Between Hal3p and Ppz1p also Extends to Other Cellular Functions.
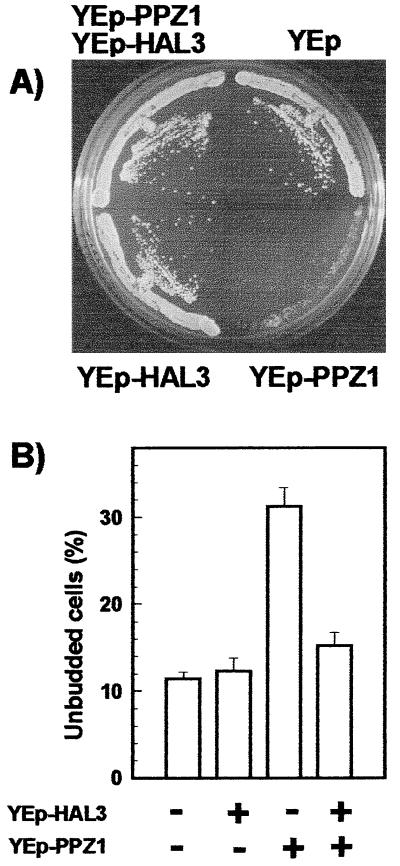
We have reported (9) that strong overexpression of PPZ1 from a GAL1 promoter causes a severe growth effect. As shown here, high-copy expression of the wild-type PPZ1 gene also results in poor growth, and this is associated to an increase in the percentage of unbudded cells (Fig. 6). We then considered the possibility that an increased expression of HAL3 might counteract the mentioned effect by blocking the excess of the Ppz1 protein. To test this hypothesis, wild-type cells were transformed with multicopy plasmids carrying PPZ1, HAL3, or both. As observed in Fig. 6, high-copy-number expression of HAL3 restores normal growth of cells overexpressing PPZ1.
Figure 6.
High-copy expression of HAL3 improves growth of cells overexpressing PPZ1. (A) Wild-type strain JA-100 was transformed with plasmid YEplac195 (27), carrying the URA3 marker (YEp), with the same plasmid bearing the PPZ1 gene (YEp-PPZ1), with plasmid YEp351 bearing the HAL3 gene (YEp-HAL3), or with both constructs, and plated in CM medium lacking uracil (to avoid loss of YEp-PPZ1). Growth was scored after 2 days. (B) The wild-type strain (JA-100) transformed with the indicated plasmids was grown in CM medium lacking uracil. The strains that did not received the YEp-PPZ1 plasmid also were transformed with plasmid YEplac195 to allow growth in selective media. Cultures were collected at optical density between 0.3 and 0.9 and the budding index was determined by microscopic examination of the samples. Data are expressed as percentage of unbudded cells (mean ± SEM from five to eight independent determinations).
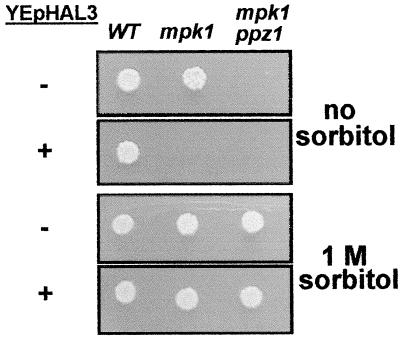
It has been reported that deletion of the PPZ1 phosphatase aggravates the lytic phenotype of a slt2/mpk1 deletion mutant, so that cells cannot grow at 30°C unless osmotically stabilized (for instance, in the presence of 1 M sorbitol). We show here (Fig. 7) that overexpression of HAL3 from a high-copy-number plasmid results in a phenotype that is reminiscent of that of the ppz1 mpk1 double mutant. This is consistent with an inhibition by Hal3p of the Ppz1p function in the MAP kinase pathway. These results strongly suggest that, in addition to the salt tolerance pathway, Hal3p can regulate other cellular functions of the Ppz1 phosphatase.
Figure 7.
High-copy-number expression of HAL3 mimics the effect of deletion of PPZ1 in a slt2/mpk1 background. Wild-type strain 1788 (WT) and its derivatives DL456 (mpk1) and DL823 (mpk1 ppz1) were transformed with plasmid YEplac181, to allow growth on synthetic medium lacking leucine (−) or with plasmid YEp351 bearing the HAL3 gene (+). Cells were grown at 30°C for 3 days on plates with or without sorbitol.
DISCUSSION
Salt homeostasis in yeast is a complex process that involves regulation of the uptake and efflux of sodium and potassium. There is evidence that the efflux of sodium through the ENA1 transporter is mediated through multiple transduction pathways (18). The study of these processes in yeast can serve to help understand the regulatory mechanisms in other organisms, such as higher plants.
In this paper we present genetic and biochemical data supporting the notion that Hal3p exerts its regulatory effect on salt tolerance by controlling the Ser/Thr protein phosphatase Ppz1. The finding that the interaction of Hal3p with the catalytic moiety of Ppz1 is more effective than the interaction with the entire protein suggests that a possible role for the NH2-terminal half of Ppz1 might be to protect the phosphatase from a complete inactivation by Hal3p. This interaction could be very specific, because it has not been observed when testing other Ser/Thr yeast phosphatases such as Glc7p or Ppg1p (unpublished results). Because our data suggest that in normal growth conditions only a small fraction of Ppz1p is bound to Hal3p, an attractive hypothesis would be that certain environmental conditions (i.e., salt stress) might result in increased binding of Hal3p to Ppz1p, which would lead to inhibition of the phosphatase and, consequently, to cellular changes (i.e., increased expression of the ENA1 gene).
The notion that Hal3p can act as a regulatory subunit of Ppz1p is reinforced by the observation that, after subcellular fractionation, both proteins are immunodetected in the same fractions (data not shown) and that, on the basis of the in vitro inhibition experiments, the affinity between Hal3p and Ppz1p is within the same range of magnitude as that described for several well defined inhibitors of the catalytic subunit of type 1 protein phosphatase (29–31). Ppz1p has been related not only to the regulation of salt tolerance (1) but also to the maintenance of cell integrity by interacting with the PKC1-activated MAP kinase pathway (3, 8). In addition, overexpression of Ppz1p negatively affects cell growth (ref. 9 and this work; see below). Our data indicate that all these phenotypes associated with the Ppz1p function are under the control of Hal3p. Therefore, this situation would be different than the one described for the catalytic subunit of yeast type 1 phosphatase (encoded by the gene GLC7). In this case, it is known that the deletion of the catalytic subunit is lethal (32, 33), whereas lack of specific regulatory components produces only specific, less dramatic phenotypes. For instance, lack of GAC1 affects glycogen accumulation but not cell viability or other cellular functions (34, 35). This suggests that the diverse cellular roles attributed to type 1 phosphatase are the result of specific interactions of the catalytic subunit with different regulatory subunits. It is remarkable that the yeast genome contains two putative genes (YOR054c and YKL088w) that code for proteins with a noticeable degree of identity with Hal3p/Sis2p. Although a possible role of these proteins as regulatory components of Ser/Thr phosphatases cannot be discarded, we are not aware of any evidence for such a function. Interestingly, a search of databanks also reveals that putative homologues of Hal3p/Sis2p can be found in other organisms, such as higher plants.
An interesting observation is that an excess of Ppz1 negatively affects cell growth. This effect is associated with an increase in the percentage of unbudded cells, a phenotype often observed in mutants delayed in the G1/S transition (36). This might indicate a role of Ppz1 in the regulation of cell growth, perhaps by affecting some aspects of the cell cycle. From our results, it is concluded that this component of the Ppz1 function is also negatively regulated by Hal3p. It is worth noting that the HAL3 gene has been isolated independently (as SIS2) and characterized on the basis of its ability to increase, when present in high-copy number, the growth rate of sit4 mutants (19). The SIT4/PPH1 gene encodes a type 2A-related Ser/Thr protein phosphatase (37) that is required in late G1 for normal G1 cyclin expression and for bud initiation (38, 39). Interestingly, overexpression of HAL3/SIS2 stimulates the rate of cyclin accumulation in sit4 mutants (19). An exciting possibility, which is being tested in our laboratory, would be that the described effect of HAL3/SIS2 on cell cycle also would be mediated by Ppz1. In this case, the phosphatase might represent a link between the existence of certain environmental conditions, such as exposure to sodium, and the decision to progress through a new cell cycle.
Acknowledgments
We thank Raquel Martínez, Mireia Zaguirre, and Anna Vilalta for skillful technical help, Dr. J. Dixon for plasmid pGEX-KT, and Dr. D. E. Levin for strains DL456 and DL823. E.d.N. is a recipient of a Fellowship from the Ministerio de Educación y Cultura, Spain. This work has been supported by Grants PB92–0585 and PB95–0663 from the Dirección General de Investigación Científica y Técnica (Ministry of Education, Spain) to J.A., and by a Project of Technological Priority of the Biotechnology Program (European Union) to R.S.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: GST, glutathione S-transferase; MAP, mitogen-activated protein; YPD, yeast extract/peptone/dextrose; CM, complete minimal.
References
- 1.Posas F, Camps M, Ariño J. J Biol Chem. 1995;270:13036–13041. doi: 10.1074/jbc.270.22.13036. [DOI] [PubMed] [Google Scholar]
- 2.Posas F, Casamayor A, Morral N, Ariño J. J Biol Chem. 1992;267:11734–11740. [PubMed] [Google Scholar]
- 3.Lee K S, Hines L K, Levin D E. Mol Cell Biol. 1993;13:5843–5853. doi: 10.1128/mcb.13.9.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hughes V, Muller A, Stark M J, Cohen P T. Eur J Biochem. 1993;216:269–279. doi: 10.1111/j.1432-1033.1993.tb18142.x. [DOI] [PubMed] [Google Scholar]
- 5.Haro R, Garciadeblás B, Rodríguez-Navarro A. FEBS Lett. 1991;291:189–191. doi: 10.1016/0014-5793(91)81280-l. [DOI] [PubMed] [Google Scholar]
- 6.Wieland J, Nitsche A M, Strayle J, Steiner H, Rudolph H K. EMBO J. 1995;14:3870–3882. doi: 10.1002/j.1460-2075.1995.tb00059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garciadeblás B, Rubio F, Quintero F J, Bañuelos M A, Haro R, Rodríguez-Navarro A. Mol Gen Genet. 1993;236:363–368. doi: 10.1007/BF00277134. [DOI] [PubMed] [Google Scholar]
- 8.Posas F, Casamayor A, Ariño J. FEBS Lett. 1993;318:282–286. doi: 10.1016/0014-5793(93)80529-4. [DOI] [PubMed] [Google Scholar]
- 9.Clotet J, Posas F, De Nadal E, Ariño J. J Biol Chem. 1996;271:26349–26355. doi: 10.1074/jbc.271.42.26349. [DOI] [PubMed] [Google Scholar]
- 10.Gaxiola R, Larrinoa I F, Villalba J M, Serrano R. EMBO J. 1992;11:3157–3164. doi: 10.1002/j.1460-2075.1992.tb05392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glaser H U, Thomas D, Gaxiola R, Montrichard F, Surdin-Kerjan Y, Serrano R. EMBO J. 1993;12:3105–3110. doi: 10.1002/j.1460-2075.1993.tb05979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murguía J R, Belles J M, Serrano R. Science. 1995;267:232–234. doi: 10.1126/science.7809627. [DOI] [PubMed] [Google Scholar]
- 13.Ferrando A, Kron S J, Rios G, Fink G R, Serrano R. Mol Cell Biol. 1995;15:5470–5481. doi: 10.1128/mcb.15.10.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson L C, Hubbard E J A, Graves P R, DePaoli-Roach A A, Roach P J, Kung C, Haas D W, Hagedorn C H, Goebl M, Culberston M R, Carlson M. Proc Natl Acad Sci USA. 1992;89:28–32. doi: 10.1073/pnas.89.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bidwai A P, Reed J C, Glover C V C. J Biol Chem. 1995;270:10395–10404. doi: 10.1074/jbc.270.18.10395. [DOI] [PubMed] [Google Scholar]
- 16.Nakamura T, Liu Y, Hirata D, Namba H, Harada S, Hirokawa T, Miyakawa T. EMBO J. 1993;12:4063–4071. doi: 10.1002/j.1460-2075.1993.tb06090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mendoza I, Rubio F, Rodríguez-Navarro A, Pardo J M. J Biol Chem. 1994;269:8792–8796. [PubMed] [Google Scholar]
- 18.Márquez J A, Serrano R. FEBS Lett. 1996;382:89–92. doi: 10.1016/0014-5793(96)00157-3. [DOI] [PubMed] [Google Scholar]
- 19.Dicomo C J, Bose R, Arndt K T. Genetics. 1995;139:95–107. doi: 10.1093/genetics/139.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sherman F, Fink G R, Hicks J B. Laboratory Course Manual for Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1986. [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 22.Schiestl R H, Gietz R D. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 23.Rothstein R J. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 24.Berben G, Dumont J, Gilliquet V, Bolle P, Hilger F. Yeast. 1991;7:475–477. doi: 10.1002/yea.320070506. [DOI] [PubMed] [Google Scholar]
- 25.Hakes D J, Dixon J E. Anal Biochem. 1992;202:293–298. doi: 10.1016/0003-2697(92)90108-j. [DOI] [PubMed] [Google Scholar]
- 26.Posas F, Bollen M, Stalmans W, Ariño J. FEBS Lett. 1995;368:39–44. doi: 10.1016/0014-5793(95)00593-x. [DOI] [PubMed] [Google Scholar]
- 27.Gietz R D, Sugino A. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 28.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 29.Bollen M, Stalmans W. Crit Rev Biochem Mol Biol. 1992;27:227–281. doi: 10.3109/10409239209082564. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Z, Bai G, Deans-Zirattu S, Browner M F, Lee E Y C. J Biol Chem. 1992;267:1484–1490. [PubMed] [Google Scholar]
- 31.Beullens M, Van Eynde A, Stalmans W, Bollen M. J Biol Chem. 1992;267:16538–16544. [PubMed] [Google Scholar]
- 32.Clotet J, Posas F, Casamayor A, Schaaf-Gerstenschläger I, Ariño J. Curr Genet. 1991;19:339–342. doi: 10.1007/BF00309593. [DOI] [PubMed] [Google Scholar]
- 33.Feng Z H, Wilson S E, Peng Z-Y, Schlender K K, Reimann E M, Trumbly R J. J Biol Chem. 1991;266:23796–23801. [PubMed] [Google Scholar]
- 34.François J M, Thompson-Jaeger S, Skroch J, Zellenka U, Spevak W, Tatchell K. EMBO J. 1992;11:87–96. doi: 10.1002/j.1460-2075.1992.tb05031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cannon J F, Clemens K E, Morcos P A, Nair B M, Pearson J L, Khalil M. Adv Prot Phosphatases. 1995;9:211–232. [Google Scholar]
- 36.Wheals A E. In: The Yeasts. 2nd Ed. Rose A H, Harrison J S, editors. London: Academic; 1987. pp. 283–390. [Google Scholar]
- 37.Arndt K T, Styles C A, Fink G R. Cell. 1989;56:527–537. doi: 10.1016/0092-8674(89)90576-x. [DOI] [PubMed] [Google Scholar]
- 38.Sutton A, Immanuel D, Arndt K T. Mol Cell Biol. 1991;11:2133–2148. doi: 10.1128/mcb.11.4.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fernandez-Sarabia M J, Sutton A, Zhong T, Arndt K T. Genes Dev. 1992;6:2417–2428. doi: 10.1101/gad.6.12a.2417. [DOI] [PubMed] [Google Scholar]