Abstract
Abf2p is a high mobility group (HMG) protein found in yeast mitochondria that is required for the maintenance of wild-type (ρ+) mtDNA in cells grown on fermentable carbon sources, and for efficient recombination of mtDNA markers in crosses. Here, we show by two-dimensional gel electrophoresis that Abf2p promotes or stabilizes Holliday recombination junction intermediates in ρ+ mtDNA in vivo but does not influence the high levels of recombination intermediates readily detected in the mtDNA of petite mutants (ρ−). mtDNA recombination junctions are not observed in ρ+ mtDNA of wild-type cells but are elevated to detectable levels in cells with a null allele of the MGT1 gene (Δmgt1), which codes for a mitochondrial cruciform-cutting endonuclease. The level of recombination intermediates in ρ+ mtDNA of Δmgt1 cells is decreased about 10-fold if those cells contain a null allele of the ABF2 gene. Overproduction of Abf2p by ≥ 10-fold in wild-type ρ+ cells, which leads to mtDNA instability, results in a dramatic increase in mtDNA recombination intermediates. Specific mutations in the two Abf2p HMG boxes required for DNA binding diminishes these responses. We conclude that Abf2p functions in the recombination of ρ+ mtDNA.
High mobility group (HMG) proteins are ubiquitous eukaryotic DNA binding proteins that participate in a variety of DNA transactions including packaging, replication, and transcription (1). Recent in vitro findings also suggest that some HMG proteins may have a role in recombination: they bind preferentially to DNA structures containing sharp bends, including four-way junction DNA (2, 3), the equivalent of Holliday recombination intermediates, and they have been shown to stimulate the DNA double-strand cleavage step in V(D)J recombination (4) and the integration of HIV-1 cDNA (5). In prokaryotes, proteins such as HU and integration host factor, which, like HMG proteins, can also bend DNA, are known to play direct roles in recombination processes (6).
Several HMG proteins have been identified in Saccharomyces cerevisiae. One of these, Abf2p, is an abundant 20-kDa nuclear-encoded mitochondrial protein containing two HMG boxes separated by six amino acids (7). Unlike vertebrate homologs of this mitochondrial HMG protein, which appear to have a direct role in transcription (8, 9), Abf2p lacks a C-terminal extension believed to interact with the transcription machinery (10). Yeast cells lacking Abf2p (Δabf2) are able to maintain and express their wild-type (ρ+) mitochondrial genomes as long as they are provided with a nonfermentable carbon source; however, ρ+ mtDNA is lost from such cells during growth on rich medium containing a fermentable carbon source (7). Although Abf2p is not essential for mtDNA replication, recent studies indicate that it may play some role in that process, because the copy number of mtDNA is altered by changes in the level of Abf2p (11).
The mechanism of yeast mtDNA replication is poorly understood. However, recent data point to the possibility that replication may be driven by recombination processes. As an alternative to RNA-primed DNA replication, several organisms, including T-even phage and Escherichia coli, are able to initiate DNA replication by recombination via priming from the 3′ ends of single strands that have invaded duplex DNA (12, 13). That some mitochondrial genomes may replicate by such a recombination-driven process is suggested from electron microscope analyses of mtDNA from yeast (14, 15) and Plasmodium falciparum (16), which reveal the presence of single-strand lariats, large single-strand regions, and complex aggregates. In addition, gel electrophoretic analysis of S. cerevisiae mtDNA shows the presence of a significant fraction of molecules much larger than unit length (17).
Thus, the effects of Abf2p on mtDNA stability and copy number could be caused by, at least in part, a role of this protein in mtDNA recombination. The most direct evidence to date for this hypothesis was our finding that the frequency of recombination between mtDNA markers in ρ+ by ρ+ crosses is severely depressed when both parental strains lack Abf2p (11). Those data, combined with recent in vitro findings that HMG proteins and functionally similar proteins from prokaryotes stimulate recombinogenic processes (4–6), led us to seek more direct evidence for a role of Abf2p in mtDNA recombination. Here, we show that Abf2p directly influences the level of mtDNA recombination intermediates. These data provide in vivo evidence for the participation of a HMG protein in recombination.
MATERIALS AND METHODS
Strains and Growth Conditions.
All strains used in this study were isogenic derivatives of wild-type 14WW (MATa ade2 trp1 leu2 ura3–52 cit1∷LEU2). The abf2∷TRP1 derivative of 14WW (1–14WW) has been described (18). The MGT1 gene (YKL011C), including 261 bp of 5′ and 35 bp of 3′ flanking sequences, was amplified by PCR from DNA of strain 14WW and ligated into the BamHI and XhoI sites of pGEM7f (Promega), creating pGEM-MGT1. The MGT1 coding sequence between the SalI and EcoRI sites was replaced with the URA3 gene to yield pGEM-mgt1∷URA3. A strain carrying the Δmgt1 (null) allele, 14WW Δmgt1, was constructed by transforming strain 14WW with a linearized SstI–XhoI fragment from pGEM-mgt1∷URA3, and transformants were verified by Southern blotting. The double-null mutant strain, 14WW Δabf2 Δmgt1, was isolated from random spores generated from the diploid formed in a cross between a MAT α derivative of 14WW Δabf2 and (MAT a) 14WW Δmgt1. The expression vectors pGal-Abf2 and pGal-Abf21−2− encoding, respectively, wild-type and mutant forms of Abf2p under control of the GAL1–10 promoter are described elsewhere (11). Cells were grown in YP (1% yeast extract, 2% bactopeptone) or YNB + cas (0.67% yeast nitrogen base, 1% casamino acids) medium containing either 2% dextrose (YPD, YNBD), 3% glycerol (YPG, YNBG), or 2% galactose (YPGal, YNBGal). All strains were grown at 30°C unless specified otherwise.
mtDNA Stability.
The stability of ρ+ mtDNA was assayed by using an experimental protocol developed earlier (11). Cells were grown overnight at 30°C in YPG medium, and then transferred to YPD medium. At intervals, aliquots of the cells were plated on YPD medium, grown for 3 days at 30°C, and the colonies were scored for the presence of ρ+ cells by the 2,3,5-triphenyl tetrazolium chloride-overlay procedure (19).
DNA Isolation and Enrichment for Replication Intermediates.
Cells were grown to a density of 1 × 107 cells/ml in 500 ml of YPG or YPD medium. Total DNA was isolated by using modifications to a previously reported protocol (20). Briefly, cells were harvested, washed, and resuspended in 1 M sorbitol containing 100 mM EDTA, pH 7.4. Spheroplasts were prepared by treatment with 100 units/ml of yeast lytic enzyme (ICN) for 15 min. The spheroplasts were harvested by centrifugation and resuspended in 10 mM Tris-Cl, pH 9.5. An equal volume of 10 mM Tris-Cl, 0.5M EDTA, pH 9.5, and 2% SDS was added to lyse the cells. Immediately after lysis, Proteinase K was added to a final concentration of 2 mg/ml, and the samples were incubated for 4 hr at 37°C. An equal volume of TE (10 mM Tris-Cl, 1 mM EDTA, pH 8.0) was used to dilute the samples before extraction with phenol/chloroform and chloroform. The DNA was precipitated by the addition of absolute ethanol at room temperature. Replication and recombination intermediates were enriched by batch absorption to BND-cellulose, which enriches for DNA with single-stranded character (21).
Electrophoresis and DNA Blots.
Two-dimensional (2D) gel electrophoresis was carried out by the method of Brewer and Fangman (22). The DNA was transferred to S&S Nytran Plus by capillary action and subsequently hybridized with cloned mtDNA sequences (a 2.6-kb EcoRI–BamHI fragment from the COXI gene and a 1-kb HpaII fragment from the VAR1 gene) radiolabeled by random priming with [α-32P]dATP. Hybridizations were carried out at 65°C in 0.5 M NaPO4, pH 7.0, containing 7% SDS, 1× Denhardts, 1 mM EDTA, and 0.2 mg/ml sheared and denatured salmon sperm DNA (Sigma). Stringent washes were performed at 65°C in 0.1 M NaPO4, pH 7.0 containing 2% SDS.
Western Immunoblotting.
Whole-cell extracts were fractionated on 15% polyacrylamide gels, blotted to S&S Protran nitrocellulose paper, and probed with a polyclonal antiserum raised against Abf2p, as described previously (11).
RESULTS
Recombination Intermediates Are Reduced in mtDNA from ρ+ Δabf2 Cells.
To determine whether Abf2p directly functions in mtDNA recombination, we have used 2D gel electrophoresis (22, 23) to detect levels of Holliday structures. The diagnostic pattern of X-shaped DNA molecules indicative of Holliday junctions, as well as other DNA structures resolved by the 2D procedure, are diagramatically illustrated in Fig. 1A. mtDNA was isolated from midlog phase glycerol medium (YPG) cultures of ρ+ strains ABF2 14WW and Δabf2 14WW and digested with EcoRI. Blots of 2D gels of the digested mtDNA were hybridized with a probe specific for a 5-kb EcoRI fragment of the mitochondrial COXI gene. As shown in Fig. 1B, a distinct Y-arc from the 1N to the 2N position is readily detected in the mtDNA from both strains; this Y-arc is characteristic of a replication fork that initiated outside of the region probed. Recombination intermediates, which would be found as a vertical spike arising from the 2N position, were not evident in the wild-type or mutant samples shown, respectively, in Fig. 1B-a and b.
Figure 1.
(A) Schematic of 2D gel electrophoresis (22). Restriction fragments of total DNA enriched for single-stranded character (21) are separated by agarose gel electrophoresis in the first dimension according to their mass. The second-dimension agarose gel resolves the restriction fragments by both mass and shape. The DNA then is transferred to a membrane and hybridized with a specific probe. The majority of the DNA will be present as linear molecules at the 1N spot. Replication intermediates will have a characteristic pattern depending on whether an origin fires inside (giving rise to a bubble arc) or outside (giving rise to a Y-arc) of the fragment of interest. Recombination intermediates are detected as a vertical spike (X-arc) rising from the 2N position. (B) Abf2p influences the levels of Holliday junctions in ρ+ mtDNAs. Replication and recombination intermediates in ρ+ mtDNAs of 14WW strains ABF2 MGT1 (a), Δabf2 MGT1 (b), ABF2 Δmgt1 (c), and Δabf2 Δmgt1 (d) were resolved by 2D gel electrophoresis and hybridized with a probe specific for a 5-kb EcoRI fragment from the COX1 gene. Replication and recombination intermediates are indicated in a and c, respectively. Positions of linear 5-kb (1N) and 10-kb (2N) fragments are indicated in a. DNA was prepared from log-phase cells cultured in YPG medium.
The failure to see recombination structures in the blots shown in Fig. 1B-a and b, is most likely because the steady-state levels of recombination intermediates in those DNAs were below a detectable threshold. Lockshon et al. (24) reported that disruption of the MGT1 gene, which encodes a mitochondrial endonuclease (CCE1) that specifically cleaves Holliday junctions (25), causes ρ+ cells to have elevated levels of recombination structures, which are readily detectable by 2D gel electrophoresis. Therefore, to increase sensitivity, we analyzed ρ+ mtDNA from ABF2 and Δabf2 strains, both of which contain a null allele of the MGT1 gene. These Δmgt1 strains were grown on YPG medium and extracted DNA was analyzed by the same 2D gel procedure. Thus, any differences that might exist in the level of recombination structures in the mtDNA of ABF2 and Δabf2 cells should be revealed in the Δmgt1 background. Fig. 1B-c shows the presence of a strong recombination spike (X-arc) extending from the 2N position in the mtDNA from ABF2 Δmgt1 cells. In contrast, the level of X-arc in ρ+ mtDNA from the Δmgt1 Δabf2 double mutant (Fig. 1B-d) was dramatically reduced. From these experiments, we conclude that Abf2p influences the level of recombination intermediates in ρ+ mtDNA.
The Level of Recombination Intermediates in ρ− mtDNA Is Unaffected by Abf2p.
Mitochondrial genomes of petite (ρ−) strains have sustained large deletions of ρ+ mtDNA sequences, and the remaining DNA sequences are stably maintained as an array of tandem repeats. Whereas Abf2p is essential for the maintenance of ρ+ mtDNA in cells grown on rich dextrose medium (7), ρ− mtDNAs are stable in Δabf2 cells grown under the same conditions (11). Because of the repeated sequences, ρ− mtDNAs have a high potential for recombination, and, therefore, we would expect there to be detectable levels of recombination intermediates in ρ− ABF2 MGT1 strains.
To determine whether Abf2p influences the level of recombination intermediates in ρ− mtDNA, we analyzed by 2D gel electrophoresis the mtDNA from ABF2 and Δabf2 strains harboring the 2–33 petite genome that consists of a 2-kb repeat of sequences from the VAR1 region of mtDNA (26). Total cellular DNA was isolated from YPD cultures of these strains and first digested with BamHI (which does not cut in the mtDNA repeat) to fragment nuclear DNA. After BND-cellulose chromatography, the DNA was digested with HincII, which cleaves once in the ρ− repeat unit, yielding 2-kb fragments. This material was fractionated by 2D gel electrophoresis to reveal replication and recombination intermediates. As shown in Fig. 2A, replication intermediates indicative of a fork moving through the fragment were detected in both DNA samples, and their levels appeared to be similar. In addition, a prominent X-arc of recombination intermediates was detected in both samples. We also detected a trailing signal extending along the path of the Y-arc and continuing through the recombination spike to higher molecular weights species (see arrows, Fig. 2A). Others have interpreted such a trailing pattern as indicative of single-stranded tailing (27, 28); thus, its presence may reflect intermediates of rolling circle-type replication. Such trailing is much less evident in samples of ρ+ mtDNA.
Figure 2.
The levels of Holliday junctions in ρ− mtDNAs are independent of Abf2p. (A) 2D gel electrophoresis of replication and recombination intermediates from the 2-kb VAR1 petite, 2–33, in ABF2 (Upper) and Δabf2 (Lower) 14WW cells. The blots were hybridized with a probe specific for petite mtDNA. The arrows indicate high molecular weight species, which may be representative of single-strand tailing resulting from rolling circle replication. (B) One-dimensional gel electrophoresis of ρ− 2–33 mtDNA linearized with increasing concentrations of HincII (0.25, 0.5, and 1 unit/μl) from ABF2 and Δabf2 14WW cells. Molecular size markers and the expected migration positions of linear double-stranded and Holliday junction DNAs are indicated.
Because recombination structures were detectable in both samples, we decided to use one-dimensional (1% agarose) gels to quantify the amounts of these intermediates. In this system, linear duplex DNAs are expected to migrate as 2-kb fragments, whereas molecules linked by a recombination junction should migrate as 4-kb molecules. To ensure that any material present at the 4-kb position represents recombination intermediates and not an artifact of partial digestion, the DNA was digested with several concentrations of HincII, from 0.25 units/μl to 1 unit/μl after BND chromatography. Fig. 2B shows that discrete bands were detected by using a VAR1 probe at both the 2-kb and 4-kb positions with DNAs from both ABF2 and Δabf2 cells, and that the amount of these fragments was not changed by increasing the HincII concentration. These data suggest that the 4-kb fragment is a recombination intermediate and that, relative to total mtDNA, the amount of this fragment (≈1%) is the same in ABF2 and Δabf2 cells.
Overproduction of Abf2p Increases the Level of Recombination Junctions in ρ+.
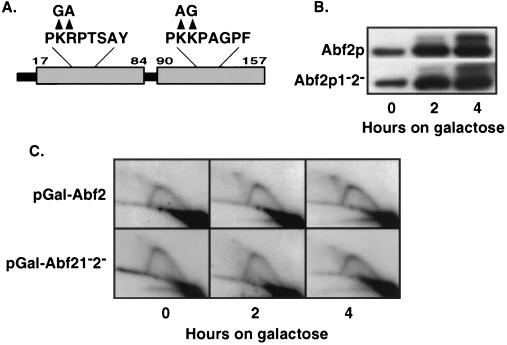
mtDNA. Lockshon et al. (24) showed that there is a mitotic instability of mtDNA, and particularly of some ρ− mtDNAs, in cells lacking CCE1. They suggested that mtDNA containing high levels of recombination intermediates in Δmgt1 cells is trapped as large DNA networks, and thus is unable to segregate efficiently to daughter cells. We have observed that in cells induced to overexpress Abf2p (≥ 10-fold), there is a marked instability both of ρ+ and ρ− mtDNAs, such that most cells are converted to ρ0 petites after as little as three generations of growth after induction (11). This mtDNA instability was not the result of some secondary effect of overproduction of Abf2p: a mutant protein (Abf2p1−2−) containing mutations in conserved basic residues within each of the HMG boxes of Abf2p (Fig. 3A) that inhibit its binding to DNA in vitro and to mtDNA in vivo is much less effective in inducing petites when overexpressed to the same level as the wild-type protein (11). One possible explanation for the instability of mtDNA in cells overproducing wild-type Abf2p is that there is an increased level of recombination intermediates that interferes with efficient transmission of mtDNA. If so, then the ineffectiveness of the mutant protein, Abf2p1−2−, to induce ρ0 petites could be explained if that protein did not cause a similar increase in recombination intermediates when overexpressed.
Figure 3.
Overproduction of Abf2p results in an increase in Holliday junctions in ρ+ mtDNA. (A) Schematic of Abf2p with the HMG-box domains represented by the gray boxes. Mutations were made in conserved basic residues, as indicated, from both HMG-box domains resulting in mutant protein Abf2p1−2−. (B) Overproduction of Abf2p and Abf2p1−2− in 14WW ρ+ cells after the transition from YNBG to YNBGal medium for the indicated time points. Abf2p and Abf21−2− were overexpressed from transformants containing the CEN plasmid GAL1–10 promoter constructs pGal-Abf2 and pGal-Abf21−2−, respectively. Protein levels were detected by Western blotting with polyclonal antiserum raised against Abf2p (18). (C) 2D gel electrophoresis of ρ+ mtDNA replication and recombination intermediates from 14WW cells containing either pGal-Abf2 or pGal-Abf21−2− after the transition from YNBG to YNBGal medium. The blots were hybridized with a probe specific for a 5-kb EcoRI fragment from the COX1 gene.
To examine this possibility, we compared the effects of overproduction of Abf2p and Abf2p1−2− on mtDNA stability and on the level of recombination intermediates of ρ+ mtDNA in strain 14WW. In these experiments, wild-type Abf2p and the mutant protein, Abf2p1−2−, were overexpressed from the GAL1–10 promoter by transferring glycerol-grown cells to YNBGal + cas medium. As shown in Fig. 3B, both the wild-type and mutant proteins were induced with similar kinetics over a 4-hr period of induction. As shown in Fig. 3C, overproducing wild-type Abf2p resulted in an increase in X-arc recombination intermediates to detectable levels by 2 hr of induction, whereas there was no discernible X-arc signal at this time in the mtDNA from cells overexpressing Abf2p1−2−. By 4 hr of induction, there was a further increase of intensity of the X-arc signal in the mtDNA from cells overexpressing Abf2p, and a faint X-arc signal in the mtDNA from the cells overexpressing Abf2p1−2−. Taken together, these data provide direct evidence that increasing the level of Abf2p increases the level of mtDNA recombination intermediates, and that this response requires the integrity of key residues of the HMG boxes involved in DNA binding.
mtDNA Stability and Levels of Recombination Intermediates.
Previously, we determined the kinetics of petite induction in experiments where Δabf2 cells were shifted from glycerol to glucose medium (11) and found that petite induction was significantly slower than in cells in which Abf2p was overexpressed, as in the experiment described in the preceding section. To determine whether there is any relation between the level of mtDNA recombination intermediates and the stability of ρ+ mtDNA, we compared the kinetics of petite formation in cultures of ABF2 Δmgt1, Δabf2 MGT1, and Δabf2 Δmgt1 ρ+ cells with that of the wild-type ABF2 MGT1 parent after a shift from glycerol to dextrose medium (Fig. 4). In ABF2 Δmgt1 cells, whose mtDNA contains a high level of recombination intermediates (Fig. 1C), there was only a slight increase in the formation of petites during growth on YPD medium. As expected, ρ+ mtDNA was unstable in Δabf2 MGT1 cells: after six generations of growth on dextrose, ≈60% of the cells gave rise to petite colonies. However, in Δabf2 Δmgt1 cells, which have significantly higher levels of mtDNA recombination intermediates than do either ABF2 MGT1 or Δabf2 MGT1 cells, the kinetics of petite induction were essentially the same as observed with Δabf2 MGT1 cells after six generations of growth on dextrose medium. Thus, although Abf2p can profoundly influence the level of recombination intermediates in ρ+ mtDNA, that property is not likely to be a major factor in accounting for mtDNA instability in cells that either lack Abf2p or have a much greater than wild-type amount of the protein.
Figure 4.
Effect of ABF2 and MGT1 alleles on the stability of ρ+ mtDNA. The kinetics of petite induction were determined for ABF2 MGT1 (○), Δabf2 MGT1 (□), ABF2 Δmgt1 (▴), and Δabf2 Δmgt1 (⧫) 14WW cells. Cells were shifted from YPG to YPD medium, and aliquots were plated at various intervals and on YPD medium. The fraction of ρ+ colonies was scored by overlaying with 2,3,5-triphenyl tetrazolium chloride soft agar as described in Materials and Methods.
DISCUSSION
In this paper we show that the steady-state level of Holliday junction intermediates in ρ+ mtDNA is dependent on the HMG protein, Abf2p. This finding provides in vivo evidence for a direct role of an HMG protein in recombination. To demonstrate the effect of an abf2 null allele on the level of mtDNA recombination structures, we took advantage of the observation of Lockshon et al. (24) that the low steady-state level of recombination structures in ρ+ mtDNA of wild-type cells could be increased to detectable levels in mtDNA from cells lacking the mitochondrial Holliday junction-cleaving enzyme, CCE1, encoded by the MGT1 gene. We confirmed the observations of Lockshon et al. (24) that there is a high density of recombination structures in samples of ρ+ mtDNA from Δmgt1 cells, and found that the amount of these structures was decreased by about 10-fold in cells that also lacked Abf2p. This finding is consistent with the observations that recombination between markers on yeast mtDNA is severely depressed in crosses between Δabf2 cells (11) and that HMG proteins can stimulate recombination processes in vitro (4, 5).
Abf2p could function in recombination by promoting a DNA conformation that is particularly favorable for strand exchange. HMG proteins are known to induce bending (29) as well as negative supercoiling of DNA (30). Such structures could facilitate the interaction of proteins of the recombination apparatus, as has been suggested for the in vitro stimulation by HMG-1 of V(D)J cleavage by RAG1 and RAG2 (4). Alternatively, if bound preferentially to recombination junctions, Abf2p could stabilize recombination structures by reducing their accessibility to cleavage by CCE1. In that case, however, the lower steady-state amount of joint molecules observed in ρ+ mtDNA from Δabf2 Δmgt1 cells would have to have been the result of cleavage by one or more, presumably nonspecific, endonucleases capable of resolving a Holliday junction.
In contrast to the results for ρ+ mtDNA, the level of recombination intermediates in ρ− mtDNA was found to be independent of Abf2p. Possibly, the high density of repeated sequences (and thus the high potential for recombination) in ρ− mtDNA may be sufficient to allow the formation of stable recombination junctions in the absence of Abf2p. This distinction between the effects of Abf2p on ρ+ versus ρ− mtDNAs is reminiscent of the observation that Abf2p is required for the maintenance of ρ+ mtDNA in cells grown on fermentable carbon sources, but is not required for the maintenance of ρ− mtDNAs in petite cells grown under the same conditions (11). These effects could be related by indications that yeast mtDNA replication may be a recombination-dependent process (11, 17, 24, 31). The control exerted by Abf2p over mtDNA recombination also could account for the direct relation between the dosage of Abf2p and ρ+ mtDNA copy number (11). Although hyperexpression of Abf2p leads to a dramatic increase in the level of recombination intermediates of mtDNA, that DNA is not propagated under any growth conditions tested, most likely because the binding of such large amounts of Abf2p to mtDNA interferes with the replication process. Indeed, hyperexpression of the mutant protein, Abf2p1−2−, which has reduced ability to bind DNA, does not lead to the same extent of recombination intermediates or mtDNA instability seen for the wild-type protein.
Is there a relation between mtDNA stability and recombination? Although the data of Lockshon et al. (24) show that the high level of recombination intermediates in ρ− mtDNA that was generated by the mgt1 null mutation clearly interferes with the transmission of the petite mtDNA, ρ+ mtDNA transmission was only marginally affected by the mgt1 mutation. In our experiments, the instability of ρ+ mtDNA in Δabf2 cells as measured by the frequency of petite formation was found to be independent of the MGT1 allele despite the fact that some recombination intermediates could be detected in Δabf2 Δmgt1 cells as compared with undetectable levels in the Δabf2 MGT1 strain. Thus, the overriding factor in determining ρ+ mtDNA stability was the presence or absence of Abf2p. Moreover, the rapid loss of mtDNA during the overproduction of Abf2p cannot be solely attributed to increased levels of recombination intermediates, because mtDNA from Δmgt1 cells contains similar levels of recombination junctions, yet are relatively stable. Thus, although maintenance of ρ+ mtDNA does not appear to be affected to any appreciable extent by the steady-state levels of recombination intermediates, it remains possible that mtDNA copy number depends, at least in part, on the formation of recombination structures, which may serve to prime replication.
Acknowledgments
We thank S. Newman for some of the constructs used in this study and for helpful discussion and G. Kapler (Texas A & M) for advice on the 2D gel electrophoresis system. This work was supported by Grant GM33510 from the National Institutes of Health and Grant I-0642 from the Robert A. Welch Foundation.
ABBREVIATIONS
- HMG
high mobility group
- YP
yeast extract/peptone
- YNB
yeast nitrogen base
- YPD
yeast extract/peptone/dextrose
- YPG
yeast extract/peptone/glycerol
- 2D
two-dimensional
References
- 1.Landsman D, Bustin M. BioEssays. 1993;15:1–8. doi: 10.1002/bies.950150807. [DOI] [PubMed] [Google Scholar]
- 2.Varga-Weisz P, Zlatanova J, Leuba S H, Schroth G P, van Holde K. Proc Natl Acad Sci USA. 1994;91:3525–3529. doi: 10.1073/pnas.91.9.3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teo S H, Grasser K D, Hardman C H, Broadhurst R W, Laue E D, Thomas J O. EMBO J. 1995;14:3844–3853. doi: 10.1002/j.1460-2075.1995.tb00054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Gent D C, Hiom K, Pauli T T, Gellert M. EMBO J. 1997;16:2665–2670. doi: 10.1093/emboj/16.10.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farnet C M, Bushman F D. Cell. 1997;88:483–492. doi: 10.1016/s0092-8674(00)81888-7. [DOI] [PubMed] [Google Scholar]
- 6.Nash H A. In: The Hu and IHF Proteins: Accessory Factors for Complex Protein-DNA Assemblies. Lin E C C, Lynch A S, editors. Austin, TX: Landes; 1996. pp. 149–179. [Google Scholar]
- 7.Diffley J F, Stillman B. Proc Natl Acad Sci USA. 1991;88:7864–7868. doi: 10.1073/pnas.88.17.7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Antoshechkin I, Bogenhagen D F. Mol Cell Biol. 1995;15:7032–7042. doi: 10.1128/mcb.15.12.7032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parisi M A, Clayton D A. Science. 1991;252:965–969. doi: 10.1126/science.2035027. [DOI] [PubMed] [Google Scholar]
- 10.Dairaghi D, Shadel G, Clayton D. J Mol Biol. 1995;249:11–28. doi: 10.1006/jmbi.1995.9889. [DOI] [PubMed] [Google Scholar]
- 11.Zelenaya-Troitskaya O, Newman S M, Okamoto K, Perlman P S, Butow R A. Genetics. 1998;148:1763–1776. doi: 10.1093/genetics/148.4.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luder A, Mosig G. Proc Natl Acad Sci USA. 1982;78:1101–1105. doi: 10.1073/pnas.79.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asai T, Sommer S, Bailone A, Kogoma T. EMBO J. 1993;12:3287–3295. doi: 10.1002/j.1460-2075.1993.tb05998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sena E P, Revet B, Moustacchi E. Mol Gen Genet. 1986;202:421–428. doi: 10.1007/BF00333272. [DOI] [PubMed] [Google Scholar]
- 15.Maleszka M R, Skelly P J, Clark-Walker G D. EMBO J. 1991;10:3923–3929. doi: 10.1002/j.1460-2075.1991.tb04962.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Preiser P R, Wilson R J M, Moore P W, McCready S M, Hajibagheri M, Blight A N, Strath K J, Williamson D H. EMBO J. 1996;15:684–693. [PMC free article] [PubMed] [Google Scholar]
- 17.Bendich A J. J Mol Biol. 1996;255:564–588. doi: 10.1006/jmbi.1996.0048. [DOI] [PubMed] [Google Scholar]
- 18.Newman S M, Zelenaya-Troitskaya O, Perlman P S, Butow R A. Nucleic Acids Res. 1996;24:386–393. doi: 10.1093/nar/24.2.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ogur M, St. John R, Nagai S. Science. 1957;125:928–929. doi: 10.1126/science.125.3254.928. [DOI] [PubMed] [Google Scholar]
- 20.MacAlpine D M, Zhang Z, Kapler G M. Mol Cell Biol. 1997;17:4517–4525. doi: 10.1128/mcb.17.8.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huberman J A, Spotila L D, Nawotka K A, Sufian A M, Davis L R. Cell. 1987;51:473–481. doi: 10.1016/0092-8674(87)90643-x. [DOI] [PubMed] [Google Scholar]
- 22.Brewer B J, Fangman W L. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- 23.Zou H, Rothstein R. Cell. 1997;90:87–96. doi: 10.1016/s0092-8674(00)80316-5. [DOI] [PubMed] [Google Scholar]
- 24.Lockshon D, Zweifel S G, Freeman-Cook L L, Lorimer H E, Brewer B J, Fangman W L. Cell. 1995;81:947–955. doi: 10.1016/0092-8674(95)90014-4. [DOI] [PubMed] [Google Scholar]
- 25.Kleff S, Kemper B, Sternglanz R. EMBO J. 1992;11:699–704. doi: 10.1002/j.1460-2075.1992.tb05102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopez I C, Farelly F, Butow R A. J Biol Chem. 1981;256:6496–6501. [PubMed] [Google Scholar]
- 27.Belanger K G, Mirzayan C, Kreuzer H E, Alberts B M. Nucleic Acids Res. 1996;24:2166–2175. doi: 10.1093/nar/24.11.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhiyi H, Stachow C. Chromosoma. 1994;103:162–170. doi: 10.1007/BF00368008. [DOI] [PubMed] [Google Scholar]
- 29.Paull T T, Johnson R C. J Biol Chem. 1995;270:8744–8754. doi: 10.1074/jbc.270.15.8744. [DOI] [PubMed] [Google Scholar]
- 30.Javaherian K, Liu L F, Wang J C. Science. 1978;199:1345–1346. doi: 10.1126/science.628842. [DOI] [PubMed] [Google Scholar]
- 31.Lorimer H E, Brewer B J, Fangman W L. Mol Cell Biol. 1995;15:4803–4809. doi: 10.1128/mcb.15.9.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]