Abstract
A recognized feature of psoriasis and other proliferative dermatoses is accumulation in the skin of the unusual arachidonic acid metabolite, 12R-hydroxyeicosatetraenoic acid (12R-HETE). This hydroxy fatty acid is opposite in chirality to the product of the well-known 12S-lipoxygenase and heretofore in mammals is known only as a product of cytochrome P450s. Here we provide mechanistic evidence for a lipoxygenase route to 12R-HETE in human psoriatic tissue and describe a 12R-lipoxygenase that can account for the biosynthesis. Initially we demonstrated retention of the C-12 deuterium of octadeuterated arachidonic acid in its conversion to 12R-HETE in incubations of psoriatic scales, indicating the end product is not formed by isomerization from 12S-H(P)ETE via the 12-keto derivative. Secondly, analysis of product formed from [10R-3H] and [10S-3H]-labeled arachidonic acids revealed that 12R-HETE synthesis is associated with stereospecific removal of the pro-R hydrogen from the 10-carbon of arachidonate. This result is compatible with 12R-lipoxygenase-catalyzed formation of 12R-HETE and not with a P450-catalyzed route to 12R-HETE in psoriatic scales. We cloned a lipoxygenase from human keratinocytes; the cDNA and deduced amino acid sequences share ≤50% identity to other human lipoxygenases. This enzyme, when expressed in Hela cells, oxygenates arachidonic acid to 12-HPETE, >98% 12R in configuration. The 12R-lipoxygenase cDNA is detectable by PCR in psoriatic scales and as a 2.5-kilobase mRNA by Northern analysis of keratinocytes. Identification of this enzyme extends the known distribution of R-lipoxygenases to humans and presents an additional target for potential therapeutic interventions in psoriasis.
Interest in the biosynthesis of hydroxy derivatives of arachidonic acid in skin stems from the role of essential fatty acids and their derivatives in the structural integrity of normal epidermis (1–3), and from the potential involvement of arachidonate metabolites in inflammatory and proliferative skin diseases (4–6). The major products of arachidonic acid metabolism in normal human skin or keratinocytes are 12-hydroxy- and 15-hydroxyeicosatetraenoic acids (12-HETE and 15-HETE) (3–11). Biosynthesis of the 15-HETE is better understood in terms of the enzymes involved. It is formed almost exclusively as the 15S enantiomer (12, 13), and its production can be accounted for by the 15S-lipoxygenases present in skin (e.g., refs. 3, 7, 8, and 10–15). Formation of the 12-HETE in human skin is more complex, in that both 12R and 12S enantiomers are produced (9, 12, 13). The appearance of both enantiomers is not mainly attributable to autoxidation as the proportions of 12R and 12S vary considerably and, aside from the 15S-HETE, comparable amounts of the other HETE regioisomers are not formed under the usual conditions of in vitro biosynthesis. Formation of the 12S-hydroxy enantiomer can be accounted for by the platelet-type of 12S-lipoxygenase that is a constituent of normal and inflammatory human skin (5, 11, 14, 15). The enzyme or enzymes involved in the production of the 12R enantiomer have remained uncharacterized.
The first report of 12-HETE in human skin came in 1975, when Hammarström et al. (4) reported that the involved areas of epidermis in psoriasis have markedly increased concentrations of free arachidonic acid and 12-HETE. Chiral analysis of the 12-HETE in psoriasis revealed that the major enantiomer is 12R-HETE (16). It subsequently was shown that 12R-HETE is a prominent product in other nonpsoriatic proliferative dermatoses (17), and it also is formed in normal human skin as the minor 12-HETE enantiomer (9, 13). It is questioned whether the enzyme responsible for the 12R-HETE synthesis is a cytochrome P450 or a lipoxygenase. The P450-catalyzed synthesis of 12R-HETE is precedented in rat and human liver microsomes and by purified cytochromes P450 (18–20). These well-defined P450 reactions are associated, however, with the formation of many additional products that are not typically formed in incubations of skin. The alternative pathway, via a 12R-lipoxygenase, is precedented in a marine invertebrate (21, 22), but no R-lipoxygenase is known in mammals.
In this paper we present mechanistic evidence that is compatible only with a lipoxygenase pathway to 12R-HETE in human psoriatic skin. We also describe the cloning and initial characterization of a 12R-lipoxygenase from normal human keratinocytes, proving the existence of R-lipoxygenases beyond the invertebrate world.
EXPERIMENTAL PROCEDURES
Materials.
[1-14C]Arachidonic acid was purchased from NEN (DuPont). [5,6,8,9,11,12,14,15-2H8]Arachidonic acid was from a batch prepared previously (23); 2H8 (d8) was the most abundant labeled species (54%), but the sample also contained d7 (34%) and d6 (9%). As shown in Results, the deuterium content of 12R-HETE formed from this arachidonic acid was compared with that of 15-HETE prepared by reaction with soybean lipoxygenase (Sigma, type V). A detailed description of the synthesis and application of the 10-3H stereospecifically labeled arachidonic acids will be reported elsewhere. Briefly, [10R-3H]- and [10S-3H]arachidonic acids were prepared from methyl 8-ketostearate, a gift from Jin K. Cha (University of Alabama), through the following scheme: (i) reduction with NaB3H4, (ii) alkaline ester hydrolysis and preparation of the pentafluorobenzyl (PFB) ester, (iii) tosylation, (iv) resolution of the enantiomers by chiral-phase HPLC (Chiralcel OD), (v) displacement of the tosylate with LiAlH4, (vi) reoxidation at C-1 with chromic acid, (vii) coculture of the resulting [8R-3H] and [8S-3H]stearic acids mixed with [1-14C]stearic acid with the fungus Saprolegnia parasitica, and (viii) resolution of the labeled arachidonic acids essentially as described before (24).
Incubation with Deuterated Arachidonic Acid.
A sample of psoriatic scales (20–30 mg) was sonicated in 0.5 ml of Medium 199 containing 40 mM Hepes and incubated with 100 μM octadeuterated arachidonic acid for 1 h at 37°. The sample was extracted by using the Bligh and Dyer method (25), and the deuterated 12-HETE product was purified by RP-HPLC using a Beckman 5-μ ODS Ultrasphere column and a solvent of MeOH/H2O/HAc (80:20:0.01, vol/vol/vol). Care was taken to allow for the slightly more polar character (earlier elution) of the labeled product compared with unlabeled 12-HETE. The 12-HETE was further purified by straight-phase (SP)-HPLC using an Alltech 5-μ Econosil column (25 × 0.46 cm) and a solvent of hexane/isopropanol/glacial acetic acid (100:2:0.1, vol/vol/vol). It then was converted to the PFB derivative and purified again by SP-HPLC using a solvent of hexane/isopropanol (100:1, vol/vol). The resulting sample was analyzed on a Chiralcel OD HPLC column (25 × 0.46 cm) using a solvent of hexane/isopropanol (100:5, vol/vol) at a flow rate of 1.1 ml/min with UV detection at 235 nm (26).
GC-MS Analysis.
HETE PFB esters were analyzed as the trimethylsilyl ether derivatives by GC-MS in the negative ion/chemical ionization mode by using a Nermag R10–10C instrument (27). Repetitive spectra were acquired by scanning over the mass range m/z 390–404, encompassing the major M-PFB ions at m/z 391 (unlabeled HETE) and m/z 399 (d8 analogue), essentially as described previously (28).
Experiments with Stereospecifically Labeled Arachidonic Acids.
The specific activities of the two 10-3H-labeled arachidonic acids were approximately 10,000–20,000 disintegrations per min 3H per μg. The pro-S [10-3H]arachidonic acid was enriched in tritium by incubation with an 8R-lipoxygenase of Plexaura homomalla as described in principle before (29). The stereospecifically labeled arachidonic acids were admixed with [14C]arachidonic acid, which served as an internal standard for measurement of tritium retention. The final 3H/14C ratios were in the range of 1.1–2.6 in different experiments.
Incubations were conducted in a volume of 0.2 ml of 50 mM Tris, pH 7.5/100 mM NaCl, using ≈30,000 cpm 3H of stereospecifically labeled arachidonic acids (mixed with [l14C]arachidonic acid) and ≈20-mg aliquots of psoriatic scales that were known to metabolize arachidonic acid to 12-HETE (patient 1) and 15-HETE + 12-HETE (patient 2). The scales were sonicated briefly in the buffer and incubated for 90 min at 37°C. The samples were extracted with the Bligh and Dyer procedure (25), including 1 μg of triphenylphosphine to ensure reduction of any hydroperoxides. Products were purified by RP-HPLC [Beckman 5-μ ODS Ultrasphere, solvent MeOH/H2O/HAc (80:20:0.01, vol/vol/vol)], by SP-HPLC of the methyl ester [Alltech 5-μ Econosil, hexane/isopropanol (100:1, vol/vol)], and then by chiral-phase HPLC [Chiralcel OD, hexane/isopropanol (100:2, vol/vol)]. The 12R and 12S enantiomers were well resolved on the chiral column with retention times of ≈14 and 17.5 min, respectively, and >1 min of baseline separation between the peaks. Fractions of 30 sec were collected across the eluting peaks, evaporated to dryness, mixed with scintillant, and each counted for at least 60 min to define the 3H/14C ratios of the baseline and the chromatographic peaks. Recovered 12R-HETE contained ≈150–500 cpm over background in the 14C channel.
Preparation of RNA and cDNA Synthesis.
Samples of human scalp hair roots (≈30 mainly anagen follicles) or psoriatic scales (100 mg) were placed in 1 ml of TRI Reagent (Molecular Research Center, Cincinnati) and agitated in a bead beater for 20 sec using autoclaved 200-μm glass beads. Keratinocyte RNA was prepared by using 1.5 ml of TRI Reagent directly applied to a 10-cm plate of cultured cells and swirled to dissolve the RNA and protein. Total RNA then was extracted according to the manufacturer’s instructions. mRNA was prepared from total RNA by using the Oligotex mRNA Mini Kit (Qiagen). First-strand cDNA was prepared using an oligo(dT)-adapter primer. Preparation of hair follicle cDNA with adaptor primers (Marathon kit, CLONTECH) was described before (15).
PCR Cloning.
PCRs were primed with human hair follicle cDNA, keratinocyte cDNA, and in some experiments with cDNA prepared from psoriatic scales in a 50-μl reaction mixture of 10 mM Tris, pH 8.3/50 mM KCl/3 mM MgCl2 with 0.2 mM of each dNTP and 0.25 μl (1.25 units) AmpliTaq DNA polymerase (Perkin–Elmer) in a Perkin–Elmer 480 thermocycler. After addition of cDNA (1 μl from a 50-μl cDNA synthesis) at 94° (hot start), the PCR was programmed as follows: 94° for 2 min, 1 cycle; 60° for 1 min, 72° for 1 min, 94° for 1 min, 30 cycles; 72° for 10 min, 1 cycle, and then the block temperature was held at 4°C. The primers were designed based on expressed sequence tag database entry AA649213 from human tonsillar cells. The upstream primer was 5′-C-AAC-TTC-CCA-GCG-TCC-ATG-CGT-AAT-CCA-3′ versus the downstream primer 5′-TGGTGTTTTGGTCTCTGAGGTTTTTGTGTT-3′, which corresponds to the 3′ end of the ORF with the downstream primer in the untranslated region (UTR); this gave a band of 431 bp.
The 5′ rapid amplification of cDNA ends was accomplished by using the Marathon cDNA Amplication Kit (CLONTECH) using 4 μg of total RNA from beard hair follicles (15). The gene-specific downstream primers were 5′-TGGTGTTTTGGTCTCTGAGGTTTTTGTGTT-3′ and 5′-TTTTTGCTTGTTTGTTTTGTTTTGTTGAA-3′. A full-length clone was obtained by PCR using primers purified by HPLC and using a proof-reading mixture of Taq/Pwo DNA polymerases (Expand High Fidelity, Boehringer-Mannheim) as described (15). The upstream primer encoded 5′-TTGGGCCTTCGTGTGGCCCTCCA-3′, part of the 5′ UTR about 30 bp upstream of the ATG translation start site. The downstream primer encoded the C terminus of the protein: 5′-AGC-GCG-CTC-CTA-AAT-AGA-AAT-GCT-3′; After a hot start at 94°C, the reaction conditions were 94°, 2 min, 1 cycle; 60° for 1 min, 72° for 2 min, 96° 15 sec, 30 cycles; 72° 10 min, 1 cycle; hold at 4°C.
DNA Sequencing.
PCR products were subcloned into the pCR3.1 vector (Invitrogen) and sequenced by automated sequencing on an Applied Biosystems Prism 377 Genetic analyzer and fluorescence-tagged dye terminator cycle sequencing (Perkin–Elmer).
Sequence similarities were calculated by using the Jotun Hein algorithm of the Megalign program of Lasergene (DNAstar, Madison, WI).
Expression of cDNA, HPLC Analysis of Lipoxygenase Metabolism.
The PCR products corresponding to the ORF of the cDNA were ligated directly into bidirectional pCR3.1 (Invitrogen), clones with the correct orientation were selected by restriction enzyme digest, and these then were expressed by transient transfection in human Hela cells as described (30). Initially, 12 clones in pCR 3.1 were evaluated (10 expressed with equivalent activity), and subsequently an additional nine clones were expressed in pBluescript SK (four were active). After incubation with substrate (50 or 100 μM [1-14C]arachidonic acid or [1-14C]linoleic acid) for 30 min at 37°C in 50 mM Tris (pH 7.5) containing 150 mM NaCl, 0.1 mM CaCl2, the products were extracted by using the Bligh and Dyer procedure (25) and treated with triphenylphosphine to reduce any hydroperoxides to HETEs. The extracts were analyzed by RP-HPLC, normal-phase HPLC, and chiral-phase HPLC (26).
Northern Analysis.
Three nylon membranes containing mRNA from human tissues (CLONTECH) were probed by using a 32P-labeled EcoRI–NcoI 648-bp fragment of the human lipoxygenase prepared from the plasmid and labeled by Rediprime random priming (Amersham). After hybridization in ExpressHyb solution (CLONTECH) at 68°C for 1 hr, the membranes were washed finally in 0.1× SSC/0.1% SDS at 50°C for 40 min and exposed to film. The same procedure was used for Northern analysis of human keratinocyte mRNA.
Detection of the mRNA in Human Psoriatic Scales.
RNA was prepared by using Tri Reagent (Molecular Research Center). Identical aliquots of the RNA samples were used in a cDNA synthesis reaction mixture with and without reverse transcriptase (RT). PCRs were run with human keratinocyte cDNA as template, and also with psoriatic skin cDNA together with a parallel blank reaction without RT as a negative control. Two pairs of primers were used, 5′-TGCCTGCTGCACTTTGGACC-3′ with 5′-TGGTCTTCACATCCGGCAACGT-3′, giving an 852-bp product, and 5′-CAACTTCCCAGCGTCCATGCGTAATCCA-3′ with 5′-TGGTGTTTTGGTCTCTGAGGTTTTTGTGTT-3′, giving a 431-bp product. Both reactions were run using an annealing temperature of 60° in the PCR.
RESULTS
Investigation of a Potential Isomerization of 12S- to 12R-HETE.
One potential pathway to 12R-HETE is via synthesis of 12S-H(P)ETE, followed by oxidation to the 12-keto analogue and reduction back to 12R-HETE. To address the possible existence of this pathway in psoriatic scales, we measured the retention of deuterium in the biosynthesis of 12R-HETE from octadeuterated arachidonic acid. This substrate contains a deuterium label at C-12, which would be lost on formation of a keto intermediate.
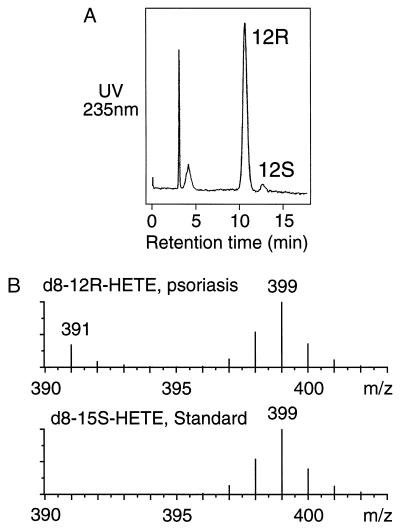
After incubation of octadeuterated arachidonic acid with psoriatic scales, the 12-HETE was isolated, the 12R and 12S enantiomers were resolved by chiral-phase HPLC (Fig. 1A), and the deuterium content of the 12R-HETE was measured by mass spectrometry (Fig. 1B). For direct comparison with a reaction involving no loss of deuterium, the 12R-HETE spectrum in Fig. 1B is compared with that of labeled 15-HETE prepared from the same batch of deuterated arachidonic acid using the soybean lipoxygenase. The deuterium content of the 12R-HETE and the 15-HETE are indistinguishable (and identical to that of d8-15-HETE formed in the psoriatic scales, not shown), indicating no loss of label in the formation of 12R-HETE and thus eliminating keto-hydroxy rearrangements as a route to 12R-HETE in psoriatic skin.
Figure 1.
Analysis of deuterated 12-HETE formed in psoriatic scales. (A) Chiral HPLC of the deuterium-labeled 12-HETE from incubation of deuterated arachidonic acid in psoriatic scales. The 12-HETE was chromatographed as the PFB ester on a Chiralcel OD column using a solvent of hexane/isopropanol (100:5, vol/vol) and detected by UV monitoring at 235 nm. (B) Partial mass spectra of the 12R-HETE from A (Upper), and 15S-HETE prepared from the same batch of deuterated arachidonic acid using the soybean lipoxygenase (Lower). The samples were analyzed by negative ion/chemical ionization GC-MS as the PFB ester trimethylsilyl ether derivatives by repetitive scanning in the range m/z 390–404. The partial mass spectra are the average of all scans collected during elution of the peaks from the GC. Unlabeled 12R-HETE also is detected in the psoriatic sample at m/z 391.
Stereospecificity of Hydrogen Abstraction in 12R-HETE Biosynthesis.
Conversion of arachidonic acid to 12-HETE requires removal of one of the two methylene hydrogens on the 10-carbon. A cytochrome P450 and 12R-lipoxygenase would show different stereoselectivity in this hydrogen abstraction. P450s tend to exhibit a suprafacial relationship between hydrogen abstraction and oxygen insertion, i.e., the two occur on the same face of the substrate (31, 32). By contrast, with lipoxygenases the two occur on opposite faces (an antarafacial relationship) (e.g., refs. 21 and 33–36). We examined this feature of 12R-HETE synthesis by conducting incubations of psoriatic scales with arachidonic acids containing a pro-R or pro-S tritium label on the 10-carbon (with [14C]arachidonic acid included to standardize the measurements of tritium retention). The 12-HETE product from each incubation was purified by HPLC, and the 12R and 12S enantiomers were resolved by chiral-phase HPLC; the 12R enantiomer accounted for 80–90% of the 12-HETE product from psoriatic skin. The tritium retention was determined from the 3H/14C ratio by liquid scintillation counting (Table 1).
Table 1.
Stereospecificity of C-10 hydrogen abstraction in 12R-HETE biosynthesis
| Sample | ProR[10-3H]20.4ω6 substrate | ProS[10-3H]20.4ω6 substrate |
|---|---|---|
| % Tritium retention in 12R-HETE
| ||
| Psoriatic scales, patient 1 | 2 | 85 |
| Psoriatic scales, patient 2 | 1 | 89 |
| 12R-lipoxygenase* | 1 | 83 |
The cDNA (Fig. 2) expressed in Hela cells.
Arachidonic acid with a pro-R 10-3H label gave rise to 12R-HETE that had lost virtually all the tritium (Table 1 ). This stereoselectivity is exactly as predicted for catalysis by a 12R-lipoxygenase (Scheme 1), and indeed it matches the result obtained with the 12R-lipoxygenase to be described below (Table 1). Using the arachidonic acid substrate with the pro-S 3H label at C-10, the 12R-HETE product retained about 85% of the tritium. This selectivity is compatible with results obtained in other lipoxygenase-catalyzed reactions in which a secondary isotope effect slightly slows the rate of reaction of the 3H-labeled molecules, resulting in less than 100% 3H retention in the product (36, 37). These results indicate no significant P450 involvement in 12R-HETE synthesis under the conditions of these experiments and directly implicate a 12R-lipoxygenase pathway. ![]()
Molecular Cloning of a Human Lipoxygenase.
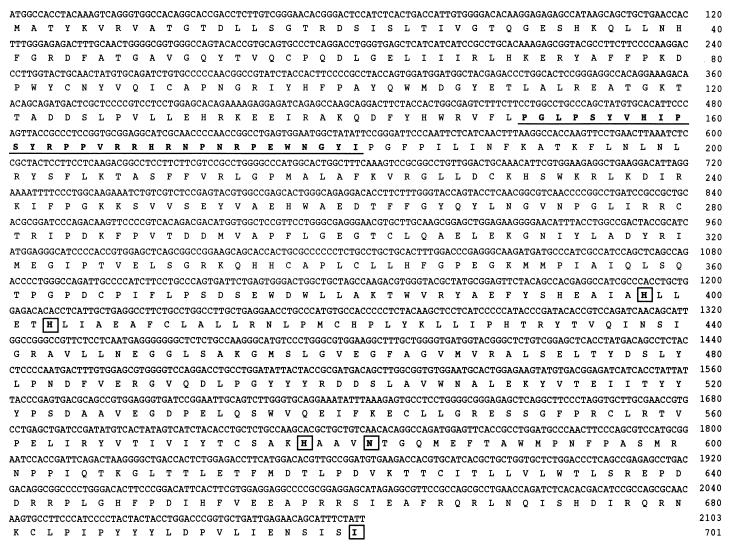
The initial clone of an additional human lipoxygenase was obtained by using hair follicle and keratinocyte cDNAs as template and primers based on sequence from a human expressed sequence tag (GenBank AA649213). The published sequence comprised approximately 500 bp encoding the 3′ end of the ORF and 150 bp of 3′ UTR. The sequence clearly encoded a previously undescribed lipoxygenase. We obtained the 5′ end of this lipoxygenase transcript by 5′ rapid amplification of cDNA ends using human hair follicle cDNA as template. The cDNA encoding the complete ORF then was prepared by PCR and subcloned into the pCR 3.1 vector. Two of the active clones described below were sequenced (Fig. 2).
Figure 2.
cDNA and deduced amino acid sequences of the lipoxygenase. Two actively expressing clones of its cDNA were sequenced and were identical. Putative iron ligands are boxed. The extra 31 amino acids referred to in Discussion are underlined. The cDNA sequence, including 5′ and 3′ UTR, is available in the GenBank/EMBL DataBank with accession no. AF038461.
This lipoxygenase cDNA has approximately 50% similarity in sequence to the second type of human 15S-lipoxygenase (15) and 40% to the human 5S-lipoxygenase. It is more distantly related to the 12S- and reticulocyte-type of 15S-lipoxygenase (38 and 35% similarity, respectively). This human sequence is closely related to a recently reported mouse lipoxygenase cDNA (≈86% identity) (38) of which it appears to be a structural homologue.
Expression of the cDNA.
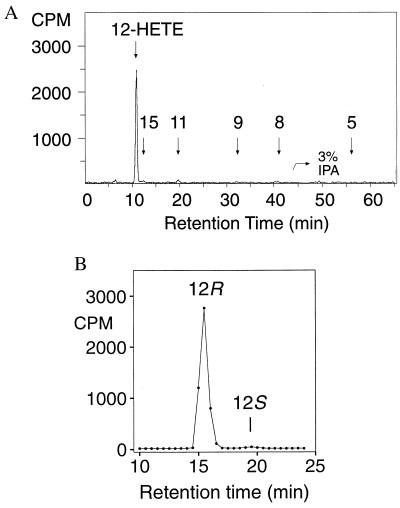
The cDNA was transfected into vaccinia-infected Hela cells, and after 20 hr the cell sonicates were evaluated for lipoxygenase activity by incubation with [14C]arachidonic acid and HPLC analysis (Experimental Procedures). RP-HPLC analysis with on-line recording of UV spectra and radioactive monitoring showed a single major product with a conjugated diene UV spectrum that cochromatographed with 12-HETE and 8-HETE (not shown). The HETEs were collected as a group from RP-HPLC and further analyzed by normal-phase HPLC as shown in Fig. 3A. The single main product was identified as 12-HETE on the basis of its cochromatography with the authentic standard and its identical UV spectrum (λmax 237 nm, indicative of the 8cis-10trans conjugation). Minor amounts of 15-HETE and 11-HETE were present. The 12-HETE product was >98% of the 12R configuration (Fig. 3B). The primary 12-lipoxygenase product (12R-HPETE) was recovered from incubations of baculovirus/insect cell-expressed enzyme, confirming that the enzyme is a 12R-lipoxygenase. Linoleic acid was a relatively poor substrate for the 12R-lipoxygenase compared with arachidonic acid (not shown).
Figure 3.
Expression in Hela cells: identification of the 12R-HETE product. (A) Product analysis by normal-phase HPLC using an Alltech 5-μ Econosil silica column (25 × 0.46 cm), a solvent system of hexane/isopropanol/glacial acetic acid (100:1:0.1, by volume, changed to the proportions 100:3:0.1 at 45 min), and a flow rate of 1.1 ml/min with on-line detection of radiolabeled products using a Packard Flo-One Radiomatic detector. Retention times of unlabeled HETE standards (coinjected with the 14C sample) are indicated on the chromatogram. (B) Chiral analysis of the methyl ester derivative of the 12-HETE using a Chiralcel OD column with a solvent of hexane/isopropanol (100:2, vol/vol) and a flow rate of 1.1 ml/min.
Tissue Expression of the 12R-Lipoxygenase.
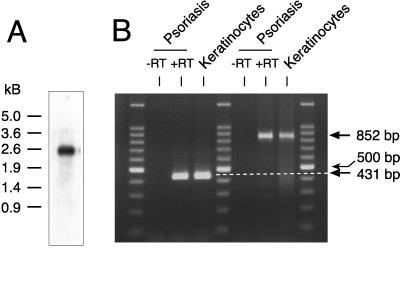
Northern analysis of human keratinocytes using a 12R-lipoxygenase-specific probe gave a single band of ≈2.5 kilobases (Fig. 4A), compatible with the predicted size of the mRNA comprising ≈260 bp of 5′ UTR, 2,103 bp ORF, and ≈150 bp 3′ UTR. No distinct hydridization was observed by Northern analysis of three human multiple tissue Northern blots comprising the following tissues from normal subjects: spleen, thymus, prostate, testis, ovary, small intestine, colon, peripheral blood leukocytes, heart, brain, placenta, lung, liver, skeletal muscle, kidney, pancreas, stomach, thyroid, spinal cord, lymph node, trachea, adrenal gland, and bone marrow. The mRNA could be detected by RT-PCR in cDNA prepared from human hair follicles, human foreskin keratinocytes, and (with uncertain prospects for recovery of mRNA) in one of two samples prepared from the scaly discarded skin of subjects with psoriasis (Fig. 4B).
Figure 4.
Expression of the 12R-lipoxygenase mRNA. (A) Northern analysis of human keratinocytes. (B) Detection of the 12R-lipoxygenase transcript in human keratinocytes and psoriatic scales by RT-PCR. Paired RNA samples prepared from psoriatic scales were run in parallel reactions with (+RT) or without (−RT) RT. PCRs then were run using two primer sets (see Experimental Procedures) including human keratinocyte cDNA as a positive control. Three lanes contain DNA size markers (100-bp ladder); the bright band in the middle is 500 bp.
DISCUSSION
The presence of 12R-HETE in psoriatic lesions is a recognized feature of the disease, yet its enzymatic origin has remained elusive. There is no convincing precedent for the specific biosynthesis of 12R-HETE in any mammalian tissue besides human skin. All of the known mammalian lipoxygenases form S configuration hydroperoxides (39), leading to a bias against a potential 12R-lipoxygenase pathway. Cytochromes P450 can convert arachidonic acid to a mixture of oxygenated derivatives that include 12R-HETE (18–20), but the distinctive aspect of 12R-HETE production in human skin is its appearance together with very few other products (mainly 12S- and 15S-HETEs). Direct biochemical characterization of the enzyme proved difficult using small amounts of human tissue. For example, NADPH-dependence is not definitively diagnostic for a P450-type of monooxygenase; lipoxygenases in tissue extracts are sensitive to the redox environment and can show changes in catalytic activity in the presence of reducing cofactors (40–42). For all of these reasons, the enzyme responsible for 12R-HETE production was uncharacterized.
In the initial series of experiments described here, we used a mechanistic approach to address the enzymatic origin of 12R-HETE in psoriatic scales. The first potential route we considered was a rearrangement from 12S-H(P)ETE through a 12-keto intermediate. Interconversion of 12R- and 12S-HETEs via the ketone is precedented in rat liver, skin, and leukocyte microsomes, although in these cases the final reduction favors formation of 12S-HETE (43, 44). Nonetheless, in principle the formation of 12R-HETE could occur in skin with the known 12S-lipoxygenase providing the initial substrate. This pathway to 12R-HETE was excluded based on the retention of a C-12 deuterium label during the biosynthesis (Fig. 1B).
The other two possibilities, a P450 type of monooxygenase or a 12R-lipoxygenase, both involve direct oxygenation into the 12R configuration. In principle, the two pathways can be distinguished by the initial formation of a 12R-hydroperoxide in the lipoxygenase-catalyzed reaction. This intermediate, however, is readily reduced in a crude tissue extract, and its detection is particularly problematic when low levels of the product are formed. We adopted an alternative method that relies on analysis of 12R-HETE, the common end product of the two potential pathways. We measured the retention of tritium in the 12R-HETE after incubation of psoriatic scales with arachidonic acid substrates containing a prochiral tritium label on the 10-carbon. Invariably, lipoxygenases catalyze a stereoselective oxygenation with removal of the prochiral hydrogen from the opposite face of the substrate (33–36). This characteristic selectivity is not observed in P450-catalyzed reactions. With P450s, the hydrogen removal and oxygenation exhibit a suprafacial relationship, often mixed with an element of stereorandom hydrogen abstraction (31, 32). Our studies in psoriatic scales provided an unequivocal result. The 12R-HETE formed from [10R-3H]arachidonic acid contained almost no tritium (Table 1). This finding indicates there is an antarafacial relationship between hydrogen abstraction and 12R oxygenation, a result compatible only with a 12R-lipoxygenase catalyzed transformation (Scheme 1). The human 12R-lipoxygenase we have cloned and expressed displays the same characteristics (Table 1).
With the emerging evidence from our mechanism-based experiments for the existence of a human 12R-lipoxygenase, we reinitiated the cloning strategy that originally had led to discovery of the second type of human 15S-lipoxygenase (15). This homology-based approach was superceded when we identified a recently released expressed sequence tag sequence (GenBank AA649213) as a clone of an additional human lipoxygenase. This partial sequence encoded the enzyme we have identified as the human 12R-lipoxygenase. Interestingly, the expressed sequence tag sequence was obtained from human tonsillar cells enriched for germinal center B cells, a facet of 12R-lipoxygenase expression that remains to be clarified.
The 12R-lipoxygenase cDNA is unusual in having ≈260 bp of 5′ UTR and a short sequence (≈150 bp) of 3′ UTR. The ORF encodes a protein with all of the typical characteristics and conserved amino acids of animal lipoxygenases. It also encodes approximately 5 kDa of extra sequence, accounted for by an insert of 31 amino acids [also represented in the murine cDNA (38)]. By reference to the crystal structure of the rabbit reticulocyte 15S-lipoxygenase (45), the extra sequence in the 12R-lipoxygenase is located after the first α-helix of the main C-terminal domain. In this position it can be accommodated on the outside of the protein without disruption of the overall tertiary structure. The 31-aa insert includes seven prolines and five arginines. Although there is not a perfect consensus sequence of, for example, a proline-rich SH3-binding domain (46), this extra sequence of the 12R-lipoxygenase could well be involved in regulatory protein–protein interactions.
We were able to establish the 12R-lipoxygenase activity of the enzyme expressed in Hela cells (and additionally in baculovirus/insect cells, not shown), yet the expressed protein has low catalytic activity. It expressed with 10-fold lower activity than the reticulocyte type of 15-lipoxygenase that we used as a positive control in each experiment. This result is similar to our observations with the murine 8S-lipoxygenase and epidermal type of 12S-lipoxygenase (30, 47), both of which also express with weak catalytic activity in vitro. In psoriatic scales, the production of 12R-HETE and 15S-HETE are often of the same order of magnitude (12). Among the possibilities to account for this finding are a major difference in the respective levels of the 12R- and 15S-lipoxygenases, the presence of an additional 12R-lipoxygenase, and/or that the activity of the 12R-lipoxygenase is increased under natural circumstances by protein modification or interactions with other component(s) of the tissue.
It was established from our cloning of 8R-lipoxygenases from coral that the R- and S-lipoxygenases are members of the same gene family (48, 49). Characterization of the 12R-lipoxygenase now extends the known occurrence of R-lipoxygenases beyond the realm of marine and freshwater invertebrates. We have detected the mRNA for the human 12R-lipoxygenase in hair roots, in primary cultures of foreskin keratinocytes, and by PCR, in psoriatic scales. With the tools made available through molecular cloning we now can approach the potential involvement of this enzyme in the cell proliferation and inflammation of psoriasis.
Acknowledgments
We thank Drs. Mitsuo Jisaka and Bih-Hwa Shieh for help and advice, and Brenda Leake for transfection of the Hela cells. Human keratinocytes were provided by the Tissue Core Laboratory of the Vanderbilt Skin Disease Research Center (supported by Grant 5P30 AR41943-03 from the National Institutes of Health/National Institute of Arthritis and Musculoskeletal and Skin Diseases). DNA sequencing used the Cancer Center Core Laboratory (Grant 1P30 CA68485). This work was supported by National Institutes of Health Grant GM-53638.
ABBREVIATIONS
- H(P)ETE
hydro(pero)xyeicosatetraenoic acid
- PFB
pentafluorobenzyl
- RT
reverse transcriptase
- UTR
untranslated region
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF038461).
References
- 1.Burr G O, Burr M M. J Biol Chem. 1929;82:345–367. [Google Scholar]
- 2.Nugteren D H, Christ-Hazelhof E, van der Beek A, Houtsmuller U M T. Biochim Biophys Acta. 1985;834:429–436. doi: 10.1016/0005-2760(85)90017-7. [DOI] [PubMed] [Google Scholar]
- 3.Nugteren D H, Kivits G A A. Biochim Biophys Acta. 1987;921:135–141. doi: 10.1016/0005-2760(87)90179-2. [DOI] [PubMed] [Google Scholar]
- 4.Hammarström S, Hamberg M, Samuelsson B, Duell E A, Stawiski M, Voorhees J J. Proc Natl Acad Sci USA. 1975;72:5130–5134. doi: 10.1073/pnas.72.12.5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hussain H, Shornick L P, Shannon V R, Wilson J D, Funk C D, Pentland A P, Holtzman M J. Am J Physiol. 1994;266:C243–C253. doi: 10.1152/ajpcell.1994.266.1.C243. [DOI] [PubMed] [Google Scholar]
- 6.Ziboh V A. Lipids. 1996;31:S249–S253. doi: 10.1007/BF02637085. [DOI] [PubMed] [Google Scholar]
- 7.Burrall B A, Cheung M, Chiu A, Goetzl E J. J Invest Dermatol. 1988;4:294–297. doi: 10.1111/1523-1747.ep12475450. [DOI] [PubMed] [Google Scholar]
- 8.Green F A. J Invest Dermatol. 1989;93:486–491. doi: 10.1111/1523-1747.ep12284046. [DOI] [PubMed] [Google Scholar]
- 9.Holtzman M J, Turk J, Pentland A. J Clin Invest. 1989;84:1446–1453. doi: 10.1172/JCI114319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henneicke-von Zepelin H-H, Schröder J-M, Smíd P, Reusch M K, Christophers E. J Invest Dermatol. 1991;97:291–297. doi: 10.1111/1523-1747.ep12480558. [DOI] [PubMed] [Google Scholar]
- 11.Takahashi Y, Reddy G R, Ueda N, Yamamoto S, Arase S. J Biol Chem. 1993;268:16443–16448. [PubMed] [Google Scholar]
- 12.Baer A N, Costello P B, Green F A. J Lipid Res. 1991;32:341–347. [PubMed] [Google Scholar]
- 13.Baer A N, Green F A. J Lipid Res. 1993;34:1505–1514. [PubMed] [Google Scholar]
- 14.Zhao H, Richards-Smith B, Baer A N, Green F A. J Lipid Res. 1995;36:24444–2449. [PubMed] [Google Scholar]
- 15.Brash A R, Boeglin W E, Chang M S. Proc Natl Acad Sci USA. 1997;94:6148–6152. doi: 10.1073/pnas.94.12.6148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woollard P M. Biochem Biophys Res Commun. 1986;136:169–175. doi: 10.1016/0006-291x(86)90891-0. [DOI] [PubMed] [Google Scholar]
- 17.Baer A N, Klaus M V, Green F A. J Invest Dermatol. 1995;104:251–255. doi: 10.1111/1523-1747.ep12612793. [DOI] [PubMed] [Google Scholar]
- 18.Capdevila J, Yadagiri P, Manna S, Falck J R. Biochem Biophys Res Commun. 1986;141:1007–1011. doi: 10.1016/s0006-291x(86)80144-9. [DOI] [PubMed] [Google Scholar]
- 19.Oliw E H. Biochim Biophys Acta. 1993;1166:258–263. doi: 10.1016/0005-2760(93)90106-j. [DOI] [PubMed] [Google Scholar]
- 20.Bylund J, Kunz T, Valmsen K, Oliw E H. J Pharmacol Exp Ther. 1998;284:51–60. [PubMed] [Google Scholar]
- 21.Hawkins D J, Brash A R. J Biol Chem. 1987;262:7629–7634. [PubMed] [Google Scholar]
- 22.Hawkins D J, Brash A R. FEBS Lett. 1989;247:9–12. doi: 10.1016/0014-5793(89)81228-1. [DOI] [PubMed] [Google Scholar]
- 23.Taber D F, Phillips M A, Hubbard W C. Methods Enzymol. 1982;86:366–369. [Google Scholar]
- 24.Maas R L, Ingram C D, Porter A T, Oates J A, Taber D F, Brash A R. J Biol Chem. 1985;260:4217–4228. [PubMed] [Google Scholar]
- 25.Bligh G H, Dyer W J. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 26.Brash A R, Hawkins D J. Methods Enzymol. 1990;187:187–192. doi: 10.1016/0076-6879(90)87024-w. [DOI] [PubMed] [Google Scholar]
- 27.Blair I A. Methods Enzymol. 1990;187:13–23. doi: 10.1016/0076-6879(90)87004-m. [DOI] [PubMed] [Google Scholar]
- 28.Song W-C, Baertschi S W, Boeglin W E, Harris T M, Brash A R. J Biol Chem. 1993;268:6293–6298. [PubMed] [Google Scholar]
- 29.Hughes M A, Brash A R. Biochim Biophys Acta. 1991;1081:347–354. doi: 10.1016/0005-2760(91)90292-p. [DOI] [PubMed] [Google Scholar]
- 30.Jisaka M, Kim R B, Boeglin W E, Nanney L B, Brash A R. J Biol Chem. 1997;272:24410–24416. doi: 10.1074/jbc.272.39.24410. [DOI] [PubMed] [Google Scholar]
- 31.White R E, Miller J P, Favreau L V, Bhattacharyya A. J Am Chem Soc. 1986;108:6024–6031. doi: 10.1021/ja00279a059. [DOI] [PubMed] [Google Scholar]
- 32.Oliw E H, Brodowsky I D, Hörnsten L, Hamberg M. Arch Biochem Biophys. 1993;300:434–439. doi: 10.1006/abbi.1993.1059. [DOI] [PubMed] [Google Scholar]
- 33.Hamberg M, Samuelsson B. J Biol Chem. 1967;242:5329–5335. [PubMed] [Google Scholar]
- 34.Egmond M R, Vliegenthart J F G, Boldingh J. Biochem Biophys Res Commun. 1972;48:1055–1060. doi: 10.1016/0006-291x(72)90815-7. [DOI] [PubMed] [Google Scholar]
- 35.Hamberg M, Hamberg G. Biochem Biophys Res Commun. 1980;95:1090–1097. doi: 10.1016/0006-291x(80)91584-3. [DOI] [PubMed] [Google Scholar]
- 36.Maas R L, Ingram C D, Taber D F, Oates J A, Brash A R. J Biol Chem. 1982;257:13525–13519. [PubMed] [Google Scholar]
- 37.Brash A R, Ingram C D, Maas R L. Biochim Biophys Acta. 1986;875:256–261. doi: 10.1016/0005-2760(86)90175-x. [DOI] [PubMed] [Google Scholar]
- 38.Krieg P, Kinzig A, Heidt M, Marks F, Fürstenberger G. Biochim Biophys Acta. 1998;1391:7–12. doi: 10.1016/s0005-2760(97)00214-2. [DOI] [PubMed] [Google Scholar]
- 39.Funk C D. Prog Nucleic Acid Res Mol Biol. 1993;45:67–98. doi: 10.1016/s0079-6603(08)60867-3. [DOI] [PubMed] [Google Scholar]
- 40.Cochran F R, Finch-Arietta M B. Biochem Biophys Res Commun. 1989;161:1327–1332. doi: 10.1016/0006-291x(89)91388-0. [DOI] [PubMed] [Google Scholar]
- 41.Riendeau D, Denis D, Choo L Y, Nathaniel D J. Biochem J. 1989;263:565–572. doi: 10.1042/bj2630565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shornick L P, Holtzman M J. J Biol Chem. 1993;268:371–376. [PubMed] [Google Scholar]
- 43.Falgueyret J P, Leblanc Y, Rokach J, Riendeau D. Biochem Biophys Res Commun. 1988;156:1083–1089. doi: 10.1016/s0006-291x(88)80743-5. [DOI] [PubMed] [Google Scholar]
- 44.Falgueyret J P, Leblanc Y, Riendeau D. FEBS Lett. 1990;262:197–200. doi: 10.1016/0014-5793(90)80188-o. [DOI] [PubMed] [Google Scholar]
- 45.Gillmor S A, Villaseñor A, Fletterick R, Sigal E, Browner M F. Nat Struct Biol. 1997;4:1003–1009. doi: 10.1038/nsb1297-1003. [DOI] [PubMed] [Google Scholar]
- 46.Lepley R A, Fitzpatrick F A. J Biol Chem. 1994;269:24163–24168. [PubMed] [Google Scholar]
- 47.Funk C D, Keeney D S, Oliw E H, Boeglin W E, Brash A R. J Biol Chem. 1996;271:23338–23344. doi: 10.1074/jbc.271.38.23338. [DOI] [PubMed] [Google Scholar]
- 48.Brash A R, Boeglin W E, Chang M S, Shieh B-H. J Biol Chem. 1996;271:20949–20957. doi: 10.1074/jbc.271.34.20949. [DOI] [PubMed] [Google Scholar]
- 49.Koljak R, Boutaud O, Shieh B-H, Samel N, Brash A R. Science. 1997;277:1994–1996. doi: 10.1126/science.277.5334.1994. [DOI] [PubMed] [Google Scholar]