Abstract
Nitric oxide (NO) is known to have various biologic and pathophysiologic effects on organisms. The molecular mechanisms by which NO exerts harmful effects are unknown, although various O2 radicals and ions that result from reactivity of NO are presumed to be involved. Here we report that adaptive cellular response controlled by the transcription factor hypoxia-inducible factor 1 (HIF-1) in hypoxia is suppressed by NO. Induction of erythropoietin and glycolytic aldolase A mRNAs in hypoxically cultured Hep3B cells, a human hepatoma cell line, was completely and partially inhibited, respectively, by the addition of sodium nitroprusside (SNP), which spontaneously releases NO. A reporter plasmid carrying four hypoxia-response element sequences connected to the luciferase structural gene was constructed and transfected into Hep3B cells. Inducibly expressed luciferase activity in hypoxia was inhibited by the addition of SNP and two other structurally different NO donors, S-nitroso-l-glutathione and 3-morpholinosydnonimine, giving IC50 values of 7.8, 211, and 490 μM, respectively. Inhibition by SNP was also observed in Neuro 2A and HeLa cells, indicating that the inhibition was not cell-type-specific. The vascular endothelial growth factor promoter activity that is controlled by HIF-1 was also inhibited by SNP (IC50 = 6.6 μM). Induction generated by the addition of cobalt ion (this treatment mimics hypoxia) was also inhibited by SNP (IC50 = 2.5 μM). Increased luciferase activity expressed by cotransfection of effector plasmids for HIF-1α or HIF-1α-like factor in hypoxia was also inhibited by the NO donor. We also showed that the inhibition was performed by blocking an activation step of HIF-1α to a DNA-binding form.
Nitric oxide (NO) mediates a variety of biological effects including relaxation of blood vessels (1), cytotoxicity of activated macrophages (2), and formation of cGMP by activation of glutamate receptors of neurons (3). NO has also been implicated for such pathophysiological conditions as destruction of tumor cells by macrophages (2), rheumatoid arthritis (4), and focal brain ischemia (5). Some of these effects of NO are associated with hypoxic conditions. Although mechanisms of the toxic effects exhibited by NO are unclear, various types of proposal have been reported, including monoADP ribosylation (6–8), S-nitrosylation of glyceraldehyde-3-phosphate dehydrogenase (6–8), inhibition of mitochondrial enzymes such as cis-aconitase (9), inhibition of mitochondrial electron transport chain (2), inhibition of ribonucleotide reductase (10, 11), DNA damage (12, 13), and activation of poly(ADP-ribose) synthetase (14–17).
Hypoxic stress causes a set of adaptive responses by changing metabolism and gene expression. When O2 is limited, expression of genes encoding components of electron transport chain is repressed, while transcription of genes encoding enzymes of the glycolytic pathway is activated (18). Expression levels of erythropoietin (EPO), the primary growth factor for erythroid progenitor cells and, therefore, a major physiologic regulator of O2 supply in mammals, increases several hundredfold in rodent liver and kidney in response to hypoxia or anemia (19). In addition to these genes, a number of genes, including genes for vascular endothelial growth factor (VEGF) and inducible nitric oxide synthase, are inducibly expressed in hypoxia (20).
To date, the relationship between NO action in hypoxia and hypoxic responses has not been well understood. In addition to the direct action of NO such as peroxidation of lipids, nitrosylation of proteins, and production of O2 radicals, we hypothesized that some indirect effect of NO, such as modulation of hypoxic responses, is possibly involved in the cytotoxicity of NO in hypoxic cells. Here we report that NO donors inhibit transcription of genes that result in adaptive cellular responses in hypoxia. This inhibition resulted from blocking of activation of a transcription factor, hypoxia-inducible factor 1 (HIF-1), which plays a key role in adaptive cellular response under stressful conditions (21, 22).
MATERIALS AND METHODS
Materials.
Sodium nitroprusside (SNP), S-nitroso-l-glutathione, and 3-morpholinosydnonimine were obtained from Wako Pure Chemicals (Osaka). Restriction enzymes and T4 DNA ligase were from Takara Shuzo (Kyoto). [γ-32P]ATP and [α-32P]dCTP were obtained from Dupont/NEN. pGL3-control vector containing simian virus 40 promoter and enhancer was obtained from Promega. All other chemicals were of reagent grade, and used without further purification.
Cell Culture and DNA Transfection.
Hep3B and Neuro 2A cells were maintained in DMEM supplemented with 10% fetal bovine serum. HeLa cells were grown in Eagle’s minimal essential medium with 10% fetal bovine serum. DNA transfection was carried out by the calcium-phosphate coprecipitation method as described (23). Four hours after the transfection medium was changed, and NO donors were added to the medium, and the cells were then incubated in a humidified 1% O2, 5% CO2 atmosphere in a hypoxic condition for an appropriate time at 37°C. Luciferase reporter assay was performed by using extracts from cells incubated for 15 hr after transfection with the use of β-galactosidase to measure transfection efficiency. Data are presented as the relative luciferase activity by calculating the average of at least three separate experiments. Reporter and effector plasmids were constructed as described (24).
Northern Blot Analysis.
Hep3B cells were treated with 100 μM SNP for 10 hr, and total RNA was extracted by the guanidine thiocyanate/CsCl method (25). A 20-μg portion of the total RNA was applied to an 0.8% agarose gel containing 2.2 M formaldehyde. Filter hybridization was carried out by using full-length cDNAs for human EPO, aldolase A, glyceraldehyde-3-phosphate dehydrogenase, and CYP1A1 as probes.
Gel Shift Assay.
Nuclear extracts of Hep3B cells incubated in the normoxic (20% O2) or hypoxic (1% O2) condition for 4 hr in the presence or absence of SNP were prepared as described (26). Gel shift assays were performed as described using hypoxia-response element (HRE) (W18; ref. 27) in the EPO gene as the probe.
RESULTS AND DISCUSSION
Effect of NO Donors on EPO and Aldolase A mRNA Expression in Hypoxic Cells.
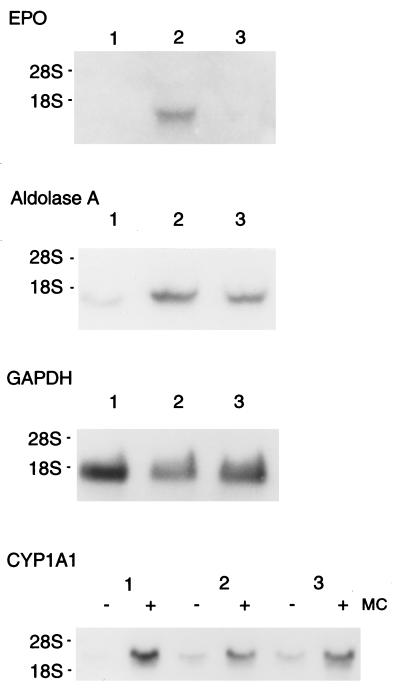
The effect of NO donors on the EPO and aldolase A mRNA expression was examined using a human hepatoma cell line Hep3B that hypoxically induces EPO and glycolytic enzyme mRNAs in a physiologic manner. The inducible gene expression occurs mainly as a result of activation of a transcription factor, HIF-1 (21). As shown in Fig. 1, expression of EPO mRNA was strongly induced in hypoxia (1% O2) whereas no signals in normoxia (20% O2) was observed as reported (28). However, addition of 100 μM SNP, which spontaneously releases NO, completely inhibited the induced expression of the EPO mRNA in hypoxia (Fig. 1 Upper, compare lanes 2 and 3). Expression of glycolytic aldolase A mRNA was induced ≈4-fold and partially inhibited by SNP (40% inhibition, an average of two separate experiments). This partial inhibition agreed with previous observation that inducible expression of aldolase A in hypoxic cells is, to some extent, performed in a HIF-1 independent manner (29). Expression of GADPH and CYP1A1 mRNAs that were not hypoxically induced (29) was unchanged by the addition of SNP in hypoxic cells. Taken together, these results demonstrate that hypoxic induction of the genes by HIF-1 is blocked by NO.
Figure 1.
Repression of induction of EPO and aldolase mRNAs in hypoxic Hep3B cells by the addition of SNP. Total RNA was extracted from Hep3B cells cultured in a normoxic or hypoxic condition with or without 100 μM SNP, electrophoresed on an 0.8% agarose gel containing formaldehyde, and transferred to a nylon membrane. Filter hybridization was carried out using human full-length EPO, aldolase A, glyceraldehyde-3-phosphate dehydrogenase, or CYP1A1 cDNA as hybridization probes. Lane 1, RNA from normoxic cells; lane 2, RNA from hypoxic cells; lane 3, RNA from hypoxic cells treated with 100 μM SNP. MC, 3-methylcholanthrene (final concentration, 1 μM).
Inhibition of HIF-1 Activity by NO Donors in Hypoxia.
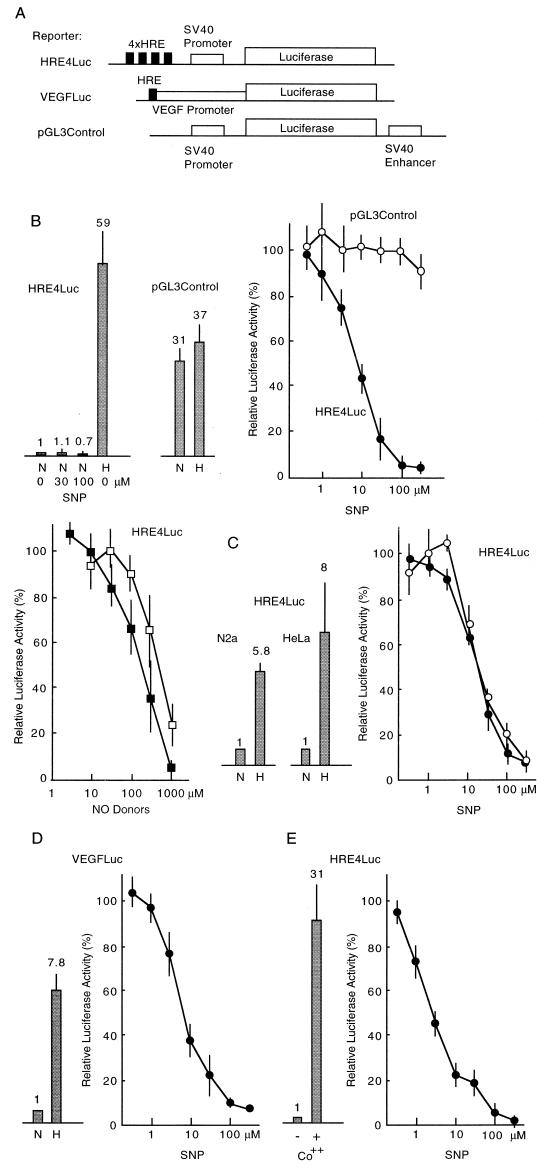
To examine HIF-1 activity in hypoxia, we introduced a reporter plasmid (Fig. 2A, see ref. 24) containing four tandemly repeated sequences of HRE that was found in the 3′ enhancer region of the EPO gene, into Hep3B cells, and the cells were incubated at a condition of 1% O2 for 15 hr. Fig. 2B shows that the luciferase activity increased ≈60-fold in hypoxia. To investigate the effect of NO donors on the expression of luciferase activity driven by HREs, we added SNP (0.3–300 μM) to the medium. As shown in Fig. 2B, hypoxic expression of luciferase decreased dose-dependently (IC50 = 7.8 μM), while the addition of SNP (up to 100 μM) to normoxic cells showed only weak inhibition on expressed luciferase activity; the luciferase activity in normoxic cells was relatively low, but was measurable quantitatively. This low IC50 value in hypoxic cells strongly suggests that the concentration of NO that effectively inhibits luciferase activity is in a range similar to that produced in physiologic and pathophysiologic conditions in the organism, when taking into account that the half life of NO released from SNP is several seconds in the medium (30). It has also been reported that the concentration of NO is ≈20 nM in a buffer containing 1 mM SNP (31). In brain ischemia, concentration of NO increases from 10 nM to 2.2 μM after middle cerebral artery occlusion (32). Pretreatment of Hep3B cells for 4 hr with 100 μM SNP in a normoxic condition showed no effect on the luciferase activity of cells successively incubated in the hypoxic condition (data not shown). Inhibition was also observed by two other structurally different NO donors, S-nitroso-l-glutathione (IC50 = 211 μM) and 3-morpholinosydnonimine (IC50 = 490 μM) as shown in Fig. 2B, confirming that NO is the direct inhibitor to the luciferase expression, although the possibility cannot be excluded that immediate products derived from reaction of NO inhibit the luciferase activity. Addition of 8-bromo-cGMP (0.1 mM) showed no effect (data not shown), suggesting that this inhibition was not mediated by the soluble guanylate cyclase-dependent pathway (3). Luciferase activity driven by simian virus 40 early promoter and enhancer carrying no HRE sequences was only modestly induced hypoxically, and was not repressed by the addition of SNP in hypoxic cells (Fig. 2B). To examine the effect of NO on neural cells, we tested the inhibition of the luciferase activity by SNP in a human neuroblastoma cell line, Neuro 2A cells. As shown in Fig. 2C, inhibition was observed with a similar dose-dependency (IC50 = 18 μM) to that of Hep3B cells. Furthermore, similar inhibition was also observed in HeLa cells (IC50 = 18 μM), suggesting that the inhibitory effect of NO on the hypoxic transcription is not cell-type specific, and possesses a common mechanism. We next examined the inhibition of luciferase activity generated from a plasmid containing an upstream sequence of the VEGF gene, which contains one HRE (Fig. 2 A and D), and results showed the luciferase activity was inhibited in a similar manner. The IC50 value (6.6 μM) was quite similar to that of the synthetic reporter containing four HREs, although the extent of induction in hypoxia was much lower because it held only one HRE sequence. It is reported that VEGF gene expression was inhibited by NO through repression of activator protein 1 activity (33). The result shown here suggests that in addition to the repression of activator protein 1 activity by NO, inhibition of HIF-1 activity by NO also contributes to repression of the VEGF gene expression in hypoxic conditions.
Figure 2.
Inhibition of reporter activity containing HRE by NO donors. (A) Structure of reporter plasmids used in the experiments. (B) Induction ratio of luciferase activity of HRE4Luc and pGL3Control reporters from normoxic to hypoxic Hep3B cells is shown (bars with SD). Dose-dependent inhibition curves of luciferase activity in hypoxic cells by NO donors are shown at right. •, Luciferase activity of HRE4Luc treated with SNP; ○, pGL3Control with SNP; ▪, HRE4Luc with S-nitroso-l-glutathione; □, HRE4Luc with SIN-1. (C) Induction ratios in Neuro 2A and HeLa cells are shown (bars with SD). Dose-dependent inhibition curves by SNP are shown at right. • and ○ for Neuro 2A and HeLa cells, respectively. (D) Induction ratio of luciferase activity from the VEGF-luciferase expression plasmid is shown (bars with SD). Dose-dependent inhibition curves by SNP are shown at right. (E) Induction ratios by CoCl2 are shown (bars with SD). Dose-dependent inhibition curves of CoCl2-induced luciferase expression by SNP are shown at right. A mixture of a reporter plasmid (2 μg) and an internal control plasmid of lac Z (3 μg) was used for transfection of Hep3B cells in 60-mm dishes. N, normoxia; H, hypoxia.
It is known that the addition of particular transition metals such as cobalt and nickel can mimic hypoxic conditions (34). Thus, we examined inhibition of HIF-1 activity by SNP in Hep3B cells in the presence of 100 μM cobalt chloride. The luciferase activity increased 31-fold in the presence of cobalt, a somewhat lower induction than that in the hypoxic induction (Fig. 2E) as reported (34). The increased luciferase activity was inhibited by the addition of SNP with a more sensitive dose-dependence curve than that in hypoxia (IC50 = 2.5 μM).
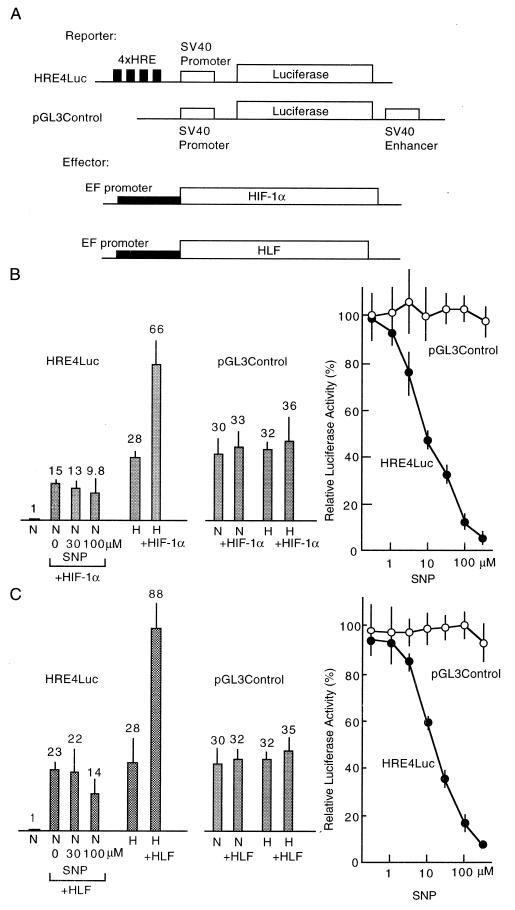
HIF-1 is composed of two subunits, HIF-1α and HIF-1β (also named Arnt), and the latter is a common partner of a group of transcription factors containing bHLH and PAS domains like Ah receptor and Sim (21, 35, 36). Thus, HIF-1α is a specific factor responsible for hypoxic responses. Recently, HIF-1α-like factor (HLF, also designated EPAS1), which shows sequence similarity to HIF-1α, has been cloned and shown to have functions similar to HIF-1 (24, 37). The two factors possess identical DNA-binding specificity. Therefore, we introduced a HIF-1α or HLF expression plasmid into Hep3B cells simultaneously with the HRE4Luc reporter plasmid. As shown in Fig. 3, overexpression of HIF-1α and HLF increased the luciferase activity in hypoxic cells 2.4- and 3.1-fold, respectively, when compared with activity without the effector plasmid. These increased luciferase activities were almost completely inhibited by the addition of SNP (IC50 = 9.0 and 14.9 μM, respectively) with dose-dependency roughly the same as that without overexpression of HIF-1α or HLF. As shown in Fig. 3, elevated basal levels of luciferase activity driven by the overexpression of HIF-1α or HLF in a normoxic condition were only weakly inhibited even at higher concentrations of SNP. This result suggests that inhibition by NO may arise from blocking of the activation process of HIF-1α and HLF by a hypoxic signal. Luciferase activity driven by simian virus 40 early promoter and enhancer was not activated by coexpression of HIF-1α or HLF as shown in Fig. 3 B and C, and was not inhibited by the addition of SNP in hypoxic cells. We constructed and introduced into Hep3B cells an expression plasmid encoding a green fluorescent protein-HIF-1α chimeric protein (38) to examine the subcellular localization of HIF-1α in the presence or absence of SNP. The chimeric protein remained in the nucleus regardless of presence or absence of cobalt (data not shown). This result suggests that diffusion of HIF-1α from nucleus to cytosol by the addition of SNP can be excluded, and that decrease in the concentration of HIF-1α in the nucleus by increased proteolytic cleavage can also be excluded as a cause of the inhibition by SNP.
Figure 3.
Inhibition of HIF-1α and HLF transcriptional activity by SNP. (A) Structure of two reporter and two effector plasmids are shown. (B and C) Increase of basal activities of HRE4Luc and pGL3Control reporters by overexpression of HIF-1α and HLF, and their hypoxic induction are shown (bars with SD). Inhibition of basal activity of HRE4Luc produced by overexpression of HIF-1α or HLF by SNP is also shown (bars with SD). Dose-dependent inhibition curves of luciferase activity in hypoxic cells overexpressed with HIF-1α or HLF by SNP are shown at right. A mixture of the reporter plasmid (2 μg), an effector plasmid (2 μg), and the internal control plasmid of lac Z (1 μg) was used for transfection of Hep3B cells in 60-mm dishes. N, normoxia; H, hypoxia.
Inhibition of DNA Binding Activity of HIF-1 by SNP.
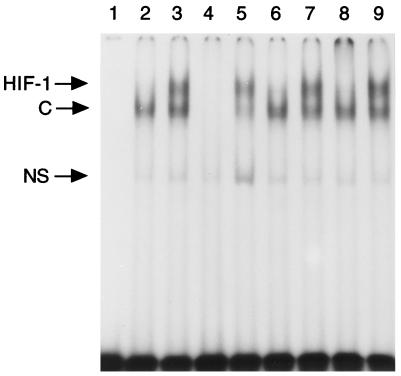
We examined whether or not the activation of HIF-1α to a DNA-binding form by a low oxygen signal was affected by the addition of NO donors by using the gel mobility shift assay. As shown in Fig. 4, nuclear extracts from hypoxic Hep3B cells gave rise to doublet bands, which were specific for activated HIF-1 and reacted on anti-Arnt antibody (lane 8) as reported (27). However, these bands disappeared when the nuclear extracts from hypoxic cells treated with 100 μM SNP were used, demonstrating that NO prevents activation of HIF-1 to a DNA-binding form.
Figure 4.
Inhibition of DNA binding activity of HIF-1 by SNP. Nuclear extracts of Hep3B cells incubated in hypoxia (1% O2/5% CO2) in the presence or absence of 100 μM SNP for 4 hr were used for the gel mobility shift assay using synthetic HRE (W18, ref. 27) as the probe. Lane 1, no protein; lane 2, nuclear extracts of normoxic Hep3B cells; lanes 3–5 and 7–9, nuclear extracts of hypoxic Hep3B cells; unlabeled oligonucleotides (300-fold) of W18 and mutated W18 (M18, ref. 27) were used as competitors in lanes 4 and 5, respectively. Lane 6, nuclear extracts from SNP-treated hypoxic Hep3B cells. Antiserum to Arnt (ref. 35) or control serum was added to the reaction mixture before binding reaction (lanes 8 and 9). C, constitutive binding activity; NS, nonspecific binding activity.
Although the molecular mechanism that activates HIF-1 to a DNA binding form is unclear, the hypothetic oxygen sensor of a heme protein that localizes in the cell membrane is implicated in the oxygen-sensing mechanism (34). In the model, a ligand-dependent conformational change of the heme protein accounts for the mechanism. Because it is known that NO binds a heme with affinity higher than O2 (39, 40), the binding of NO would fix the conformation of the sensor protein to an “oxy form” even at hypoxic conditions that shut down signal transduction. Our data presented here, therefore, satisfy this hypothesis.
Biologic and pathophysiologic effects of NO were frequently associated with hypoxia and anemia, including acute and chronic inflammation, rejection of transplanted organs, and necrosis of neoplastic lesions (41–44). Furthermore, in hypoxic-ischemic brain injury, neural cells—especially hypocampal excitatory glutaminergic neurons at CA1—are markedly weak for ischemia. It is reported that the toxic effect of hypoxically produced glutamate is mediated by NO (45). For this neural damage, Huang et al. demonstrated that neuronal, but not endothelial, NO synthase activity exacerbates acute ischemic injury using mice lacking neuronal NO synthase (46), although investigations about beneficial effects of NO synthase inhibitors in in vivo ischemia models are controversial (47, 48). In rheumatoid arthritis where O2 is limited, NO concentration increased significantly for its adverse effect (4). It is also reported that production of EPO was not adequately elevated as expected for the degree of anemia in syndromes such as anemia of rheumatoid arthritis and anemia of solid tumors (49), where the serum NO level had increased significantly. In these cases, we propose that suppression of the hypoxic adaptive response by NO through inhibition of HIF-1 activity is at least partly a cause of the toxic effects of NO; suppression of glycolytic enzymes may lead cells to energy depression.
Recently, it has been reported that the gene for inducible NO synthase of macrophages is activated in hypoxia (50) through a HRE sequence present in the promoter region, demonstrating that response to hypoxia is important for macrophages that can be exposed in a broad range of O2 tension from aerobic lung to hypoxic inflammatory tissues. This finding and the data presented here clearly show the presence of a cross-talk between the NO synthesis system and hypoxia response system at the transcriptional level.
Acknowledgments
We thank Dr. Ken-ichi Tsutsumi for his generosity in supplying us with human aldolase A cDNA. This work was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sport and Science of Japan.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: EPO, erythropoietin; HIF-1, hypoxia-inducible factor 1; HRE, hypoxia-response element; SNP, sodium nitroprusside; VEGF, vascular endothelial growth factor; HLF, HIF-1α-like factor.
References
- 1.Moncada S, Palmer R M J, Higgs E A. Pharmacol Rev. 1991;43:109–142. [PubMed] [Google Scholar]
- 2.Nathan C F, Hibbs J B., Jr Curr Opin Immunol. 1991;3:65–70. doi: 10.1016/0952-7915(91)90079-g. [DOI] [PubMed] [Google Scholar]
- 3.Bredt D S, Snyder S H. Neuron. 1992;8:3–11. doi: 10.1016/0896-6273(92)90104-l. [DOI] [PubMed] [Google Scholar]
- 4.Farrell A J, Blake D R, Palmer R M, Moncada S. Ann Rheum Dis. 1992;51:1219–1222. doi: 10.1136/ard.51.11.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dawson V L, Dawson T M, Batley D A, Uhl G R, Snyder S H. J Neurosci. 1993;13:2651–2661. doi: 10.1523/JNEUROSCI.13-06-02651.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang J, Snyder S H. Proc Natl Acad Sci USA. 1992;89:9382–9385. doi: 10.1073/pnas.89.20.9382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dimmeler S, Lottspeich F, Brune B. J Biol Chem. 1992;267:16771–16774. [PubMed] [Google Scholar]
- 8.Vedia L M, McDonald B, Reep B, Brune B, Silvio M D, Billiar T R, Lapetina E G. J Biol Chem. 1992;267:24929–24932. [PubMed] [Google Scholar]
- 9.Drapier J-C, Hibbs J B., Jr J Clin Invest. 1986;78:790–797. doi: 10.1172/JCI112642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lepoivre V, Chenais B, Yapo A, Lemaire G, Thelander L, Tenu J-P. J Biol Chem. 1990;265:14143–14149. [PubMed] [Google Scholar]
- 11.Kwon N S, Stuehr D J, Nathans C F. J Exp Med. 1991;174:761–767. doi: 10.1084/jem.174.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wink D A, Kasprzak K S, Maragos C M, Elespuru R K, Misra M, Dunam T M, Cebula T A, Koch W H, Andrews A W, Allen J S, et al. Science. 1991;254:1001–1003. doi: 10.1126/science.1948068. [DOI] [PubMed] [Google Scholar]
- 13.Nguyen T, Brunson D, Crespi C L, Penman B W, Wishnok J S, Tannenbaum S R. Proc Natl Acad Sci USA. 1992;89:3030–3034. doi: 10.1073/pnas.89.7.3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Mucia G, Menissier-de Mucia J, Schreiber V. BioEssays. 1991;13:455–462. doi: 10.1002/bies.950130905. [DOI] [PubMed] [Google Scholar]
- 15.Cleaver J E, Morgan W F. Mutat Res. 1991;257:1–18. doi: 10.1016/0165-1110(91)90016-o. [DOI] [PubMed] [Google Scholar]
- 16.Gaal J C, Smith K R, Pearson C K. Trends Biol Sci. 1987;12:129–130. [Google Scholar]
- 17.Berger N A. Radiat Res. 1985;101:4–15. [PubMed] [Google Scholar]
- 18.Webster K A, Gunning P, Hardeman E, Wallace D C, Kedes L. J Cell Physiol. 1990;142:566–572. doi: 10.1002/jcp.1041420316. [DOI] [PubMed] [Google Scholar]
- 19.Bondurant M C, Koury M J. Mol Cell Biol. 1986;6:2731–2733. doi: 10.1128/mcb.6.7.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wenger R H, Gassmann M. Biol Chem. 1997;378:609–616. [PubMed] [Google Scholar]
- 21.Wang G L, Jiang B-H, Rue E A, Semenza G L. Proc Natl Acad Sci USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bunn H F, Poyton R O. Physiol Rev. 1996;76:839–885. doi: 10.1152/physrev.1996.76.3.839. [DOI] [PubMed] [Google Scholar]
- 23.Fujisawa-Sehara A, Sogawa K, Nishi C, Fujii-Kuriyama Y. Nucleic Acids Res. 1986;14:1465–1477. doi: 10.1093/nar/14.3.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ema M, Taya S, Yokotani N, Sogawa K, Matsuda Y, Fujii-Kuriyama Y. Proc Natl Acad Sci USA. 1997;94:4273–4278. doi: 10.1073/pnas.94.9.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chirgwin J M, Przybyla A E, MacDonald R J, Rutter W J. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 26.Schreiber E, Matthias P, Mullar M M, Schaffner W. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Semenza G L, Wang G L. Mol Cell Biol. 1992;12:5447–5454. doi: 10.1128/mcb.12.12.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goldberg M A, Glass G A, Cunningham J M, Bunn H F. Proc Natl Acad Sci USA. 1987;84:7972–7976. doi: 10.1073/pnas.84.22.7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Semenza G L, Roth P H, Fang H-M, Wang G L. J Biol Chem. 1994;269:23757–23763. [PubMed] [Google Scholar]
- 30.Kikuchi K, Nagano T, Hayakawa H, Hirata Y, Hirobe M. J Biol Chem. 1993;268:23106–23111. [PubMed] [Google Scholar]
- 31.Matthews J R, Botting C H, Panico M, Morris H R, Hay R T. Nucleic Acids Res. 1996;24:2236–2242. doi: 10.1093/nar/24.12.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malinski T, Bailey F, Zhang Z G, Chopp M. J Cereb Blood Flow Metab. 1993;13:355–358. doi: 10.1038/jcbfm.1993.48. [DOI] [PubMed] [Google Scholar]
- 33.Tsurumi Y, Murohara T, Krasinski K, Chen D, Witzenbichler B, Kearney M, Couffinhal T, Isner J M. Nat Med. 1997;3:879–886. doi: 10.1038/nm0897-879. [DOI] [PubMed] [Google Scholar]
- 34.Goldberg M A, Dunning S P, Bunn H F. Science. 1987;242:1412–1415. doi: 10.1126/science.2849206. [DOI] [PubMed] [Google Scholar]
- 35.Matsushita N, Sogawa K, Ema M, Yoshida A, Fujii-Kuriyama Y. J Biol Chem. 1993;268:21002–21006. [PubMed] [Google Scholar]
- 36.Wharton K A, Jr, Franks R G, Kasai Y, Crews S T. Development (Cambridge, UK) 1994;120:3563–3569. doi: 10.1242/dev.120.12.3563. [DOI] [PubMed] [Google Scholar]
- 37.Tian H, McNight S L, Russell D W. Genes Dev. 1997;11:72–82. doi: 10.1101/gad.11.1.72. [DOI] [PubMed] [Google Scholar]
- 38.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 39.Rose E J, Hoffman B M. J Am Chem Soc. 1983;105:2866–2873. [Google Scholar]
- 40.Moore E G, Gibson Q H. J Biol Chem. 1976;251:2788–2794. [PubMed] [Google Scholar]
- 41.Moncada S, Higgs A. N Engl J Med. 1993;329:2002–2012. doi: 10.1056/NEJM199312303292706. [DOI] [PubMed] [Google Scholar]
- 42.Mulligan M S, Hevel J M, Marletta M A, Ward P A. Proc Natl Acad Sci USA. 1991;88:6338–6342. doi: 10.1073/pnas.88.14.6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang X, Chowdhury N, Cai B, Brett J, Marboe C, Sciacca R R, Michler R E, Cannon P J. J Clin Invest. 1994;94:714–721. doi: 10.1172/JCI117390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cobbs C S, Brenman J E, Aldape K D, Bredt D S, Israel M A. Cancer Res. 1995;55:727–730. [PubMed] [Google Scholar]
- 45.Dawson V L, Dawson T M, London E D, Bredt D S, Snyder S H. Proc Natl Acad Sci USA. 1991;88:6368–6371. doi: 10.1073/pnas.88.14.6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang Z, Huang P L, Panahian N, Dalkara T, Fishman M C, Moskowitz M A. Science. 1994;265:1883–1885. doi: 10.1126/science.7522345. [DOI] [PubMed] [Google Scholar]
- 47.Dalkara T, Moskowitz M A. Brain Pathol. 1994;4:49–57. doi: 10.1111/j.1750-3639.1994.tb00810.x. [DOI] [PubMed] [Google Scholar]
- 48.Iadecola C, Pelligrino D A, Moskowitz M A, Lassen N A. J Cereb Blood Flow Metab. 1994;14:175–192. doi: 10.1038/jcbfm.1994.25. [DOI] [PubMed] [Google Scholar]
- 49.Barosi G. Ann Hematol. 1994;68:215–223. doi: 10.1007/BF01737420. [DOI] [PubMed] [Google Scholar]
- 50.Melillo G, Musso T, Sica A, Taylor L S, Cox G W, Varesio L. J Exp Med. 1995;182:1683–1693. doi: 10.1084/jem.182.6.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]