Abstract
DNA ends pose specific problems in the control of genetic information quality. Ends of broken DNA need to be rejoined to avoid genome rearrangements, whereas natural DNA ends of linear chromosomes, telomeres, need to be stable and hidden from the DNA damage response. Efficient DNA end metabolism, either at induced DNA breaks or telomeres, does not result from the machine-like precision of molecular reactions, but rather from messier, more stochastic processes. The necessary molecular interactions are dynamically unstable, with constructive and destructive processes occurring in competition. In the end, quality control comes from the constant building up and tearing down of inappropriate, but also appropriate reaction steps in combination with factors that only slightly shift the equilibrium to eventually favour appropriate events. Thus, paradoxically, enzymes antagonizing DNA end metabolism help to ensure that genome maintenance becomes a robust process.
Keywords: DNA double-strand breaks, homologous recombination, non-homologous DNA end-joining, telomeres
Introduction
Maintaining genetic information, encoded in the chemical structure of DNA, is an essential biological process. This structure is, however, constantly challenged and changed in the course of normal DNA metabolism (Hoeijmakers, 2001). As DNA is essentially a very long polymer, it is susceptible to breakage. Discontinuities in this polymer are obvious threats to the integrity of genetic information. DNA ends, induced by mechanical stress, as well as endogenous and exogenous DNA-damaging agents, need to be recognized and fixed in a manner that avoids DNA sequence rearrangements (Wyman and Kanaar, 2006). In contrast, naturally occurring DNA ends at chromosome termini, telomeres, need to be maintained and shielded from the normal cellular response to DNA ends (de Lange, 2005). Conversely, telomeres should not be added to DNA ends arising internally in chromosomes.
DNA end metabolism requires quality control at several different levels. DNA breaks pose a particular problem for genetic information quality control because both strands are affected and one cannot serve as a template for the other. Thus, repairing double-stranded breaks (DSBs) can sacrifice accuracy by joining closely positioned ends without information quality control. However, although this process might be error prone at the DNA sequence level, it will avoid potentially more dangerous genome rearrangements. Alternatively, information from an intact and homologous DNA molecule can be used for recombinational repair. Recombinational repair is essentially a genome rearrangement event and therefore needs to be carefully controlled to prevent inappropriate rearrangements. Here we review recent progress in the understanding of some essential molecular mechanisms that assure quality in maintaining genetic information in the light of DNA ends, and in assuring the quality of the processes that affect their cellular processing.
DNA end metabolism by recombination
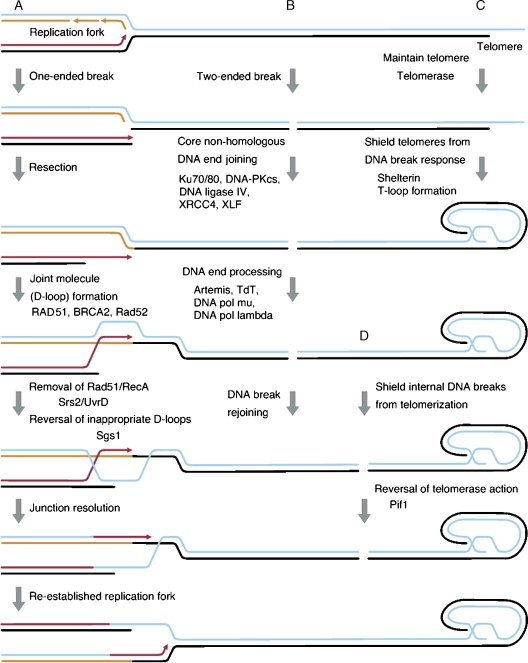
Homologous recombination can effectively and accurately repair DNA breaks using sister chromatids to recover genetic information that may have been lost at the break. Homologous recombination repair is essential during genome duplication, a particularly vulnerable event for the DNA double helix. Seemingly innocuous interruptions in one strand can be converted into DSBs and transiently exposed single strands, which can arise during lagging strand synthesis, and can break resulting in severed chromosomes (Liberi et al, 2006). This creates one-ended DNA breaks that cannot simply be fixed by joining ends (Figure 1A; Cromie et al, 2001).
Figure 1.
Metabolism at DNA ends. A schematic representation of DNA end metabolism by homologous recombination (left-hand side, under A), and non-homologous DNA end joining (middle, under B), telomerization (upper right-hand side, under C, and bottom right-hand side, under D). (A) Homologous recombination acts on one-ended DNA breaks that can arise from replication of a damaged template. Recognition and resection of the end yields single-stranded DNA with a 3′ end. Subsequently, a joint molecule is formed by D-loop formation with the identical sequence on the sister chromatid in a reaction mediated by the RAD51 nucleoprotein filament formed on the single-stranded DNA. This reaction involves numerous mediators and/or regulators, including BRCA2 in mammals and Rad52 in S. cerevisiae. The Rad51 nucleoprotein filaments in S. cerevisiae, or RecA nucleoprotein filaments in the bacterium E. coli, that form at inappropriate places and be removed by DNA helicases such as Srs2 and UvrD, respectively. Promiscuous recombination due to the formation of D-loops at incorrect locations can be avoided due to DNA helicases, such as S. cerevisiae Sgs1. Eventually, resolution of the joint between the sister chromatids results in the re-establishment of the replication fork and concludes the accurate repair of the replication-associated one-ended DNA break. (B) Two-ended DNA breaks, resulting from the direct action of DNA-damaging agents on both strands of the DNA, can be acted upon by the non-homologous DNA end-joining core components Ku70/80, DNA–Pkcs, DNA ligase IV, XRCC4, and XLF. When DNA end processing is required to create ligatable ends, endonucleases, such as Artemis, and DNA polymerases can act due to their interaction with the core components located at the ends. Rejoining of DNA breaks need not be accurate at the sequence level, but will avoid deleterious chromosomal translocations. (C) Telomeres, the natural DNA ends of linear chromosomes, are maintained during DNA replication by telomerase. They are protected form the DNA-damage response by their inclusion in a protein complex, termed shelterin. The 3′ single-strand overhang of the telomeric DNA is invaded in upstream telomeric sequences forming a D-loop-like structure, called a T-loop. (D) Conversely, DSBs need to be protected from telomerase action to prevent the formation of telomeres at inappropriate locations. In S. cerevisiae cells, the action of telomerase is antagonized by the Pif1 helicase. After Pif1's action, more appropriate DSB repair processes have another chance to act and seal the break.
The general repair pathway of homologous recombination can be described as an ordered set of DNA-processing events. These is recognition of a DNA end or single-strand gap by single-strand DNA-binding proteins, resection to reveal single-stranded DNA if necessary, recently shown to involve Sae2 in yeast and CtIP in mammalian cells (Limbo et al, 2007; Sartori et al, 2007; Takeda et al, 2007), handoff to strand exchange proteins and their assembly into filaments on the single-stranded DNA, joint molecule formation between the single strand and a homologous duplex, exchange of base-paired partners between the single strand and homologous duplex partner, dissociation of the strand exchange machinery, and in some scenarios replication and eventual resolution of the covalently connected DNA molecules (Figure 1A). Key essential proteins involved in most of these steps have been identified (Osborn et al, 2002; West, 2003; Krogh and Symington, 2004; Wyman et al, 2004; Sung, 2005). Details of dynamic interactions between the known key proteins and DNA substrates are being revealed at a rapid pace. It is becoming increasingly clear that the dynamic instability of these protein–protein and protein–DNA assemblies offers important points of control (Symington and Heyer, 2006). Indeed, since homologous recombination repair is a DNA-rearrangement event, inappropriate, incomplete, or uncoordinated activity could cause more genetic problems than those that initiated the repair process. Here we consider examples from recent work highlighting control points at key mechanistic steps in homologous recombination.
The central event of homologous recombination is a DNA strand exchange reaction catalysed by RecA-type recombinases, including Rad51 of eukaryotes and RadA of archaea (Yu et al, 2004; Wyman, 2006; Cox, 2007; Galletto and Kowalczykowski, 2007). These recombinases all form functional molecular machines assembled into filaments along single-stranded DNA. DNA strand exchange is driven by the assembly, dynamic rearrangement, and disassembly of recombinase filaments on DNA. Although the dynamic nature of recombinase nucleoprotein filaments has long been appreciated, a coherent quantitative description has recently emerged from the application of a variety of single-molecule approaches (Galletto et al, 2006; Joo et al, 2006; Miné et al, 2007; Modesti et al, 2007b; van der Heijden et al, 2007). Analysis of single complexes overcomes the complication of asynchronous reactions in bulk and provides rates for filament assembly and disassembly on single- and double-stranded DNA in defined conditions. Several key aspects of recombinase filament dynamics emerge as potential control points. For instance, Rad51, which binds well to both single- and double-stranded DNA, is now shown to disassemble 10 times slower from double-stranded DNA (van der Heijden et al, 2007). Efficient disassembly may require additional coordination in two biologically important scenarios. First, Rad51 bound inappropriately to double-stranded DNA would have to be removed to free protein for appropriate action elsewhere. Second, a Rad51 filament bound to double-stranded DNA is the end product of strand exchange, but not the end of recombinational repair. The protein filament also needs to disassemble to allow further DNA-processing events. ATP hydrolysis is coupled to recombinase filament disassembly and this transition of high-energy cofactor provides an obvious intrinsic control point. In addition, cooperating proteins such as Rad54, a DNA-translocating motor (Tan et al, 2003; Amitani et al, 2006; Heyer et al, 2006), are capable of removing Rad51 from double-stranded DNA both in vitro and in vivo (Solinger et al, 2002; Sugawara et al, 2003; Wesoly et al, 2006).
The formation of strand exchange protein filaments on DNA specifically in need of repair, and not elsewhere, requires other collaborating proteins. Such activities are described in bacteria, where specific sets of proteins initiate recombinase filaments at the different DNA structure in need of repair (Cox, 2007). Although much less well defined in mammalian cells, one likely candidate controlling this activity is BRCA2. A fungal BRCA2 homologue, and an engineered pared down version of the mammalian protein, do indeed nucleate Rad51 filaments specifically at single-strand/double-strand DNA junctions such as those created by processing of broken DNA ends or stalled replication forks (Yang et al, 2005; San Filippo et al, 2006). Although the entire mammalian BRCA2 protein thus far is refractory to purification, biochemical analysis of specific conserved parts reveals definite but contradictory effects on RAD51–DNA filaments. The BRC domains of BRCA2 bind to RAD51 and prevent filament formation in some conditions (Pellegrini et al, 2002; Shin et al, 2003) but not in others (Galkin et al, 2005). A different conserved domain of BRCA2 from its C-terminus has the opposite effect, binding across adjacent monomers in a filament and presumably stabilizing the structure (Davies and Pellegrini, 2007; Esashi et al, 2007; Petalcorin et al, 2007). This last effect of a BRCA2 C-terminal fragment stabilizing RAD51 filaments is especially intriguing because the filament stabilization may be specific for single-stranded DNA in some cases (Petalcorin et al, 2007) and is regulated by DNA damage-responsive BRCA2 phosphorylation (Esashi et al, 2007).
Recombinase–DNA filament dynamics also need to be controlled during strand exchange. This reaction, including three DNA strands bound by a recombinase filament, must be reversible at some stages. Therefore, a net strand exchange reaction likely depends on controlling the stability of the protein filament bound to these different DNA partners. Although not yet experimentally demonstrated, the same recombination partner proteins mentioned above as modulators of filament stability on single- and double-stranded DNA, also likely exercise these activities during strand exchange. In addition, the extent of strand exchange or extension of heteroduplex DNA would depend on the length of interaction between recombining DNAs. The heteroduplex length needed to repair a two-ended DNA break and generate genetic diversity, such as in meiosis, may be longer than that needed to initiate new DNA synthesis at a stalled or broken replication fork. Thus, protein partners that recognize and control filament extension and stability in the context of specific DNA structures created during strand exchange are likely to be important. At least one protein, the RAD51-associated protein RAD51AP1, has recently been described to have such activity. RAD51AP1 has the combined binding specificities needed to influence joint molecule stability; it interacts with RAD51 as well as with specific DNA structures found at the joint molecule recombination intermediate (Wiese et al, 2007; Modesti et al, 2007a).
Rad51 may also be bound to single-stranded regions that are not destined for a recombination event and the filament would have to be removed to avoid undesirable recombination events. For example, recombinase filaments may form inappropriately on single-strand DNA at stalled forks that can more easily be repaired without strand exchange, such as by translesion DNA synthesis (Eppink et al, 2006; Lehmann, 2006). Thus, proteins that disrupt recombinase filaments on single-stranded DNA before strand exchange can be initiated also control the outcome of eventual repair events. For example, bacterial UvrD and eukaryotic Srs2 can disrupt RecA or Rad51 filaments on single-stranded DNA (Krejci et al, 2003; Veaute et al, 2003, 2005). One idea is that filaments will form on single-stranded DNA exposed at stalled replication forks, but if not stabilized by other factors or proceeding into strand exchange reactions, the filaments need to be removed so that other factors, such as translesion DNA polymerases, can access the repair site. Alternatively, these enzymes act by disrupting all Rad51 filaments, including the ones destined for recombination. In this case, accessory factors (e.g., Rad52 and checkpoint proteins) that were recruited to the site remain, and these factors re-recruit Rad51 (Krogh and Symington, 2004; Paulsen and Cimprich, 2007). On the other hand, the filaments that occurred at inappropriate sites without the accessory factors do not reappear, effectively eliminating potential harmful recombination outcomes. It is this constant building up and tearing down that in the end results in a shift in the equilibrium to favour appropriate events.
One intriguing new study indicates that homologous recombination proteins may be involved in assuring genome integrity without promoting homologous recombination. Although we have focused on eukaryotic and mammalian proteins and processes, greater mechanistic detail is available for bacterial systems. Using purified bacterial components, it is possible to reconstitute sophisticated recombination and also replication scenarios in vitro. In such a defined in vitro situation, the result of stalling a leading-strand DNA polymerase has recently been determined to have two important consequences (McInerney and O'Donnell, 2007). First, the replication fork helicase continues to advance, creating a single-stranded gap on the leading-strand template. Second, the stalled DNA polymerase itself stays associated at the primer–template junction. Thus, a leading-strand block specifically results in an immobile DNA polymerase in front of a single-stranded gap, defining this structure as one needing recognition and processing. The stalled DNA polymerase has to be removed before replication restarts, which likely requires stalled nascent strand pairs with a new template or requires loading of a less accurate DNA polymerase. At least in vitro, RecA can come to the rescue by forming a filament on the single-stranded gap growing towards the DNA polymerase and displace this stalled protein (McInerney and O'Donnell, 2007). Thus, a RecA filament acts to control the quality of a stalled fork, in a manner at least in principle independent of strand exchange activity.
The DNA strand exchange reactions aimed at repairing DNA breaks create branched DNA structures with intertwined strands from different chromosomes. These are required to effect eventual repair, but can cause problems without further processing. Indeed, such DNA structures may be recognized as damage and can themselves be impediments to genome duplication and cell division. The dangers of incomplete, aberrant, or hyper recombination are evident in the absence of structure-specific helicases of the RecQ family (Myung et al, 2001b; Cheok et al, 2005). Among these helicases, the importance of BLM and WRN is especially clear, as their absence causes cancer predisposition in humans (Seki et al, 2006; Hanada and Hickson, 2007). The RecQ family helicases have been described as ‘dissolvases' or ‘disruptases' based on their presumed ability to process or reverse branched DNA structures created by strand exchange during recombination. Recently, the yeast BLM ortholog, Sgs1, has specifically been shown to suppress the formation of multi-chromatid joint molecules (Oh et al, 2007). In the same work, it was concluded that multiple and sequential strand invasion events are a consequence of the normal homologous recombination mechanism. Promiscuous recombination may not be rare at all, highlighting the need to not only control the initiation of strand exchange, but also to resolve the results of potentially deleterious unproductive strand invasion events.
DNA end metabolism by end joining
In the above sections, we discussed the homologous recombination-mediated repair of one-ended DNA breaks that mostly arise when replication becomes compromised due to irregularities in the DNA template. However, not all DNA ends are created equal and the circumstances under which DNA breaks are generated result in DNA substrates that require different repair paths. When DNA breaks are generated in the G1 phase of the cell cycle, homologous recombination will be a very ineffective repair pathway due to the lack of a sister chromatid that normally serves as the repair template. A major difference between the breaks generated in the context of replication versus those generated in a replication independent manner is that the latter will contain not one but two DNA ends. For example, two-ended DSBs can be generated directly by DNA-damaging agents such as ionizing radiation or through the improper functioning of a type 2 topoisomerases in the course of normal DNA metabolism. As opposed to one-ended breaks, repair of two-ended breaks can be accomplished by simply ligating them together. This is achieved by the non-homologous DNA end-joining pathway (Figure 1B; Lees-Miller and Meek, 2003; Lieber et al, 2003; Wilson, 2007).
The simplest breaks to be handled by non-homologous DNA end joining are those with a chemical composition that allows direct ligation. In these cases, joining requires the ‘core' components of non-homologous DNA end joining, which include the Ku70/80 heterodimer, the DNA-dependent protein kinase catalytic subunit (DNA–PKcs), XRCC4, XLF, and DNA ligase IV (Wilson, 2007). The Ku70/80 protein complex plays a central role. It provides specificity to the reaction through its ring-shaped structure that has a high affinity for DNA ends. Furthermore, it can be viewed as a ‘tool belt' because many proteins involved in the reaction interact with the Ku70/80 complex (Lieber et al, 2003). In this sense, the complex resembles proliferating cell nuclear antigen, the DNA polymerase processivity factor, which also encircles DNA and interacts with numerous proteins that provide essential DNA-processing steps during replication (Moldovan et al, 2007). Association of DNA–PKcs with the DNA end-bound Ku70/80 complex results in translocation of the complex approximately one helical turn away from the DNA ends, thereby facilitating their juxtaposition and access of other proteins to the terminal nucleotides at the break ends (Yoo and Dynan, 1999; Kysela et al, 2003). For the core reaction, this involves access of the DNA ligase IV/XRCC4/XLF complex to the ends (Lees-Miller and Meek, 2003; Meek et al, 2004; Revy et al, 2006; Sekiguchi and Ferguson, 2006; Yano et al, 2007). This complex can covalently link the ends together through the DNA-binding activity of the XRCC4 and XLF proteins and DNA ligase IV-mediated phosphodiester bond formation between 3′ hydroxyl and 5′ phosphate groups of the terminal nucleotides on either side of the break.
The core reaction described above can only work efficiently when the break arises by the precise cleavage of phosphodiester backbone of the DNA. However, the action of DNA-damaging agents on DNA will rarely results in such ‘clean' breaks. Instead, interaction of ionizing radiation and reactive oxygen species with DNA results not only in phosphodiester breakage, but also damages the structure of the bases and the deoxyribose moiety (Barlow et al, 2008). Thus, in addition to the core components of non-homologous DNA end joining, a number of other proteins will be required to process non-ligatable remnants of terminal nucleotides, remove secondary DNA structures or covalently bound modifications, such as proteins. The proteins employed to cleanse the ends are endonucleases and are template-independent as well template-directed DNA polymerases.
Artemis is one such important nuclease involved in non-homologous DNA end joining (Moshous et al, 2001; Ma et al, 2005a). This protein is a broad-specificity endonuclease (Ma et al, 2002, 2005b; Goodarzi et al, 2006) and is therefore very useful for adjusting DNA ends that can be modified in numerous and combinatorial ways. Indeed, in the absence of Artemis, a significant number of ionizing radiation-induced breaks remain unjoined (Riballo et al, 2004). Artemis physically interacts with DNA–PKcs. This interaction is not only important for the localization of Artemis to DNA ends, but also for the activation of its enzymatic activities. The activation of nucleases by other components of a pathway in which the nuclease acts, is a common theme in DNA metabolism and adds a level of quality control to the reaction (Hanada et al, 2007). Especially in the case of nucleases, their destructive action on DNA needs to be targeted to a specific location, in this case DNA ends, and must be initiated only when other components required for downstream repair steps are available. Thus, the activation of Artemis through DNA–PKcs, which in turn binds to the DNA end-marking Ku70/80 heterodimer, provides an important level of quality control to the non-homologous DNA end-joining reaction.
In addition to nucleases, the involvement of DNA polymerases in the non-homologous DNA end-joining reaction provides a great increase in the type of DNA ends that can be joined (Nick McElhinny and Ramsden, 2004). After DNA end processing results in terminal nucleotides with 3′ hydroxyl and 5′ phosphate groups, the two ends to be ligated can consist of all possible combinations of 3′ single-strand protrusions, 5′ single-strand protrusions, or flush ends. Some combinations of ends cannot be joined efficiently and others create gaps once joined. At least three DNA polymerases can play a role in sealing breaks in the context of non-homologous DNA end-joining; terminal deoxynucleotidyl transferase, which can add nucleotides to the ends of DNA in an untemplated manner, and the translesion DNA polymerases pol-mu and pol-lambda (Ma et al, 2004, 2005a; Nick McElhinny and Ramsden, 2004; Nick McElhinny et al, 2005). The translesion DNA polymerase can fill gaps that can occur when one of the two strands of each end is ligated to its partner, and they can add nucleotides to a strand at one end of the break that are templated off the strand at the other DNA end before this template strand is covalently sealed.
The plethora of activities required to seal a ‘dirty' break by non-homologous DNA end joining needs to be orchestrated. However, this may not happen in an organized linear manner in which completed reaction steps are handed down a predetermined reaction pathway. As mentioned above, a central role will be played by the Ku70/80 heterodimer because its binding specificity localizes the complex to DNA ends in need of repair. Furthermore, the complex interacts not only with other core components but also with the end-processing factors (Wilson, 2007). When repairing a clean break, the ligatable ends, held in close proximity by the Ku70/80 heterodimer complexed with DNA–PKcs, can be sealed using just the other core components of non-homologous DNA end joining. However, when dirty DNA ends are engaged by Ku70/80, accessory DNA-processing components, such as Artemis and DNA polymerases, can act if the particular DNA end is the appropriate substrate for their respective activities. A reversible interaction of the DNA end-processing factors with Ku70/80 is highly advantageous for the flexibility of the reaction in terms of the type of DNA ends that can be joined. It eliminates the need for a strict order in which processing steps need to occur, while still providing close juxtaposition of the ends throughout the different processing reactions. Furthermore, it provides an opportunity for the end-processing factors to act independently on each of the four DNA strands. Stable pre-existing holo-complexes would not provide this level of flexibility (Essers et al, 2002), which arise instead from the dynamic stochastic interaction of DNA end-processing factors with the core components of non-homologous DNA end joining. Keeping the DNA ends to be joined in close proximity during the processing steps would then be the only ‘orchestrated' step in the reaction (Kaye et al, 2004; Lisby and Rothstein, 2004; Lobachev et al, 2004).
In this sense, quality control in DSB repair through non-homologous DNA end joining is a matter of generating DNA ends that have the correct chemistry and overhang for ligation. This appears to be achieved by a level of chaos in the system rather than predetermined ordered interactions among the involved components. From the point of view of DNA sequence quality after the break has been sealed, non-homologous DNA end joining is clearly much more error prone than homologous recombination. However, the pathway is still important for genome stability, as there are situations in the cell in which homologous recombination cannot be used for DSB repair. The importance of non-homologous DNA end joining is to rejoin a broken chromosome, rather than allowing the broken chromosome to persist with the potential to eventually give rise to a chromosomal translocation (Agarwal et al, 2006; Aplan, 2006; Franco et al, 2006; Lieber et al, 2006; Povirk, 2006; Weinstock et al, 2006). In terms of the risk for cellular transformation leading to cancer, mutations at the break site are more easily tolerated in diploid cells than aneuploidy that can result from chromosomal translocations.
Telomeres and DNA end metabolism
Quality control of the natural DNA ends of linear chromosomes, telomeres, is an essential aspect of DNA metabolism. Telomeres need to be maintained in replicating cells because consecutive rounds of DNA replication will otherwise erode them (Watson, 1972). This problem is solved by telomerase, a reverse transcriptase that adds deoxynucleotides to the telomeres using an RNA template (Greider and Blackburn, 1987). Part of the normal process of telomere elongation is the recognition of the ends, a measurement of sort, to determine which ends are in need of lengthening (Chang et al, 2007). Furthermore, to avoid triggering the cellular DSB response, telomeric DNA ends are packaged in a multi-protein complex referred to as shelterin. The 3′ single-strand overhang at the telomeres is made up of repetitive sequences, which are tucked away in duplex versions of the repeats, forming T-loop structures that are structurally related to D-loops (Figure 1C; de Lange, 2005).
On the other hand, when DSBs are formed internally in chromosomes, they need to be protected from telomerization. In Saccharomyces cerevisiae, the Pif1 helicase appears to have an essential role in this process (Boule et al, 2005). The RNA–DNA hybrid intermediate in the telomerase reaction is exquisitely sensitive to the Pif1 helicase, which can efficiently unwind these hybrids. At first glance, it seems antithetical to have an efficient helicase that reverses the first step of the reaction. In fact, it is the step that will eventually lead to telomere elongatation and capping. However, it is clear from the phenotype of PIF1 deletions that this helicase activity serves an important quality-control function. First, pif1Δ cells show a dramatic increase in gross chromosomal rearrangements that are almost exclusively restricted to new telomere additions at or near a likely break point (Myung et al, 2001a). Second, the sequences that are used for the new telomere addition can be as short as 3–6 nucleotides of TG-rich telomeric repeat sequences (Kramer and Haber, 1993; Putnam et al, 2004). Although not proven, it is tempting to speculate that the one of the major roles of Pif1 is to melt incorrect DNA–RNA hybrids before they are made into natural telomeric DNA ends that will persist and result in the loss of distal information on that chromosome (Figure 1D).
The simplest interpretation is that Pif1 melts correct as well as incorrect DNA–RNA hybrids that could be used to generate telomere addition to both appropriate and inappropriate telomere addition sites. The net effect of this action is that incorrect sites are dissipated before they become permanent telomeres and break the chromosome. On the other hand, although correct sites will also be destroyed, they will reform at a much higher rate, as the precursors to telomere addition will still exist at those correct sites, increasing the rate of reformation by the telomerase complex. The net effect of these reactions is a reduction in the potential to add telomeric sequences to inappropriate internal chromosomal sites, without a subsequent reduction in the efficiency of the process at the correct sites. This kind of quality control is destined to increase the net amount of normal telomere addition.
Perspective
An important concept in quality control during DNA end metabolism is the necessary reversal of partial reactions and elimination of aberrant DNA structures. New members to the family of proteins participating in quality control at DNA ends and the different DNA structures they recognize are actively being identified. In addition, the apparent coordination of the actions needed to repair DNA breaks and maintain stable chromosome ends is not a linear pathway, but rather a path in a network of molecular interactions. The often stated ‘choices' attributed to molecules within cells are really nothing more than networks of competing pairwise interactions influenced by binding affinities, local concentrations, and reversible conformational changes. Quantitative details of interaction parameters should reveal steps where control can be effectively exercised. These details are the puzzle pieces needed to determine the interconnected interactions that result in efficient genome maintenance networks. Quality control through destruction or reversal of intermediate steps in DNA end metabolism comes at a cost. Not only will inappropriate processes be reversed, but it is inevitable that legitimate processing events will also be aborted or reversed. The appropriate reaction for specific DNA ends will eventually occur when DNA end metabolism has another go at the end. Thus, paradoxically, enzymes antagonizing DNA end metabolism ensure that genome maintenance is not only a flexible but also an extremely robust process.
Acknowledgments
Work in the CW and RK laboratories was supported by grants from the Dutch Cancer Society (KWF), the Netherlands Organization for Scientific Research (NWO), the Netherlands Genomics Initiative/NWO, the Association for International Cancer Research (AICR), and the European Commission (Integrated Projects 503259 and 512113). Work in the RR laboratory was supported by NIH grants GM50237 and GM67055. RR also thanks Beck Burgess, Roy Parker, and Ted Weinert for helpful discussions.
References
- Agarwal S, Tafel AA, Kanaar R (2006) DNA double-strand break repair and chromosome translocations. DNA Repair (Amst) 5: 1075–1081 [DOI] [PubMed] [Google Scholar]
- Amitani I, Baskin RJ, Kowalczykowski SC (2006) Visualization of Rad54, a chromatin remodeling protein, translocating on single DNA molecules. Mol Cell 23: 143–148 [DOI] [PubMed] [Google Scholar]
- Aplan PD (2006) Causes of oncogenic chromosomal translocation. Trends Genet 22: 46–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow JH, Lisby M, Rothstein R (2008) Differential regulation of the cellular response to DNA double-strand breaks in G1. Mol Cell (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boule JB, Vega LR, Zakian VA (2005) The yeast Pif1p helicase removes telomerase from telomeric DNA. Nature 438: 57–61 [DOI] [PubMed] [Google Scholar]
- Chang M, Arneric M, Lingner J (2007) Telomerase repeat addition processivity is increased at critically short telomeres in a Tel1-dependent manner in Saccharomyces cerevisiae. Genes Dev 21: 2485–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheok CF, Bachrati CZ, Chan KL, Ralf C, Wu L, Hickson ID (2005) Roles of the Bloom's syndrome helicase in the maintenance of genome stability. Biochem Soc Trans 33: 1456–1459 [DOI] [PubMed] [Google Scholar]
- Cox MM (2007) Regulation of bacterial RecA protein function. Crit Rev Biochem Mol Biol 42: 41–63 [DOI] [PubMed] [Google Scholar]
- Cromie GA, Connelly JC, Leach DR (2001) Recombination at double-strand breaks and DNA ends: conserved mechanisms from phage to humans. Mol Cell 8: 1163–1174 [DOI] [PubMed] [Google Scholar]
- Davies OR, Pellegrini L (2007) Interaction with the BRCA2 C terminus protects RAD51–DNA filaments from disassembly by BRC repeats. Nat Struct Mol Biol 14: 475–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T (2005) Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev 19: 2100–2110 [DOI] [PubMed] [Google Scholar]
- Eppink B, Wyman C, Kanaar R (2006) Multiple interlinked mechanisms to circumvent DNA replication roadblocks. Exp Cell Res 312: 2660–2665 [DOI] [PubMed] [Google Scholar]
- Esashi F, Galkin VE, Yu X, Egelman EH, West SC (2007) Stabilization of RAD51 nucleoprotein filaments by the C-terminal region of BRCA2. Nat Struct Mol Biol 14: 468–474 [DOI] [PubMed] [Google Scholar]
- Essers J, Houtsmuller AB, van Veelen L, Paulusma C, Nigg AL, Pastink A, Vermeulen W, Hoeijmakers JH, Kanaar R (2002) Nuclear dynamics of RAD52 group homologous recombination proteins in response to DNA damage. EMBO J 21: 2030–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco S, Alt FW, Manis JP (2006) Pathways that suppress programmed DNA breaks from progressing to chromosomal breaks and translocations. DNA Repair (Amst) 5: 1030–1041 [DOI] [PubMed] [Google Scholar]
- Galkin VE, Esashi F, Yu X, Yang S, West SC, Egelman EH (2005) BRCA2 BRC motifs bind RAD51–DNA filaments. Proc Natl Acad Sci USA 102: 8537–8542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletto R, Amitani I, Baskin RJ, Kowalczykowski SC (2006) Direct observation of individual RecA filaments assembling on single DNA molecules. Nature 443: 875–878 [DOI] [PubMed] [Google Scholar]
- Galletto R, Kowalczykowski SC (2007) RecA. Curr Biol 17: R395–R397 [DOI] [PubMed] [Google Scholar]
- Goodarzi AA, Yu Y, Riballo E, Douglas P, Walker SA, Ye R, Harer C, Marchetti C, Morrice N, Jeggo PA, Lees-Miller SP (2006) DNA–PK autophosphorylation facilitates Artemis endonuclease activity. EMBO J 25: 3880–3889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH (1987) The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell 51: 887–898 [DOI] [PubMed] [Google Scholar]
- Hanada K, Budzowska M, Davies SL, van Drunen E, Onizawa H, Beverloo HB, Maas A, Essers J, Hickson ID, Kanaar R (2007) The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat Struct Mol Biol 14: 1096–1104 [DOI] [PubMed] [Google Scholar]
- Hanada K, Hickson ID (2007) Molecular genetics of RecQ helicase disorders. Cell Mol Life Sci 64: 2306–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer WD, Li X, Rolfsmeier M, Zhang XP (2006) Rad54: the Swiss Army knife of homologous recombination? Nucleic Acids Res 34: 4115–4125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeijmakers JH (2001) Genome maintenance mechanisms for preventing cancer. Nature 411: 366–374 [DOI] [PubMed] [Google Scholar]
- Joo C, McKinney SA, Nakamura M, Rasnik I, Myong S, Ha T (2006) Real-time observation of RecA filament dynamics with single monomer resolution. Cell 126: 515–527 [DOI] [PubMed] [Google Scholar]
- Kaye JA, Melo JA, Cheung SK, Vaze MB, Haber JE, Toczyski DP (2004) DNA breaks promote genomic instability by impeding proper chromosome segregation. Curr Biol 14: 2096–2106 [DOI] [PubMed] [Google Scholar]
- Kramer KM, Haber JE (1993) New telomeres in yeast are initiated with a highly selected subset of TG1–3 repeats. Genes Dev 7: 2345–2356 [DOI] [PubMed] [Google Scholar]
- Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein H, Ellenberger T, Sung P (2003) DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423: 305–309 [DOI] [PubMed] [Google Scholar]
- Krogh BO, Symington LS (2004) Recombination proteins in yeast. Annu Rev Genet 38: 233–271 [DOI] [PubMed] [Google Scholar]
- Kysela B, Doherty AJ, Chovanec M, Stiff T, Ameer-Beg SM, Vojnovic B, Girard PM, Jeggo PA (2003) Ku stimulation of DNA ligase IV-dependent ligation requires inward movement along the DNA molecule. J Biol Chem 278: 22466–22474 [DOI] [PubMed] [Google Scholar]
- Lees-Miller SP, Meek K (2003) Repair of DNA double strand breaks by non-homologous end joining. Biochimie 85: 1161–1173 [DOI] [PubMed] [Google Scholar]
- Lehmann AR (2006) Translesion synthesis in mammalian cells. Exp Cell Res 312: 2673–2676 [DOI] [PubMed] [Google Scholar]
- Liberi G, Cotta-Ramusino C, Lopes M, Sogo J, Conti C, Bensimon A, Foiani M (2006) Methods to study replication fork collapse in budding yeast. Methods Enzymol 409: 442–462 [DOI] [PubMed] [Google Scholar]
- Lieber MR, Ma Y, Pannicke U, Schwarz K (2003) Mechanism and regulation of human non-homologous DNA end joining. Nat Rev Mol Cell Biol 4: 712–720 [DOI] [PubMed] [Google Scholar]
- Lieber MR, Yu K, Raghavan SC (2006) Roles of nonhomologous DNA end joining, V(D)J recombination, and class switch recombination in chromosomal translocations. DNA Repair (Amst) 5: 1234–1245 [DOI] [PubMed] [Google Scholar]
- Limbo O, Chahwan C, Yamada Y, de Bruin RA, Wittenberg C, Russell P (2007) Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol Cell 28: 134–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Rothstein R (2004) DNA repair: keeping it together. Curr Biol 14: R994–R996 [DOI] [PubMed] [Google Scholar]
- Lobachev K, Vitriol E, Stemple J, Resnick MA, Bloom K (2004) Chromosome fragmentation after induction of a double-strand break is an active process prevented by the RMX repair complex. Curr Biol 14: 2107–2112 [DOI] [PubMed] [Google Scholar]
- Ma Y, Lu H, Schwarz K, Lieber MR (2005a) Repair of double-strand DNA breaks by the human nonhomologous DNA end joining pathway: the iterative processing model. Cell Cycle 4: 1193–1200 [DOI] [PubMed] [Google Scholar]
- Ma Y, Lu H, Tippin B, Goodman MF, Shimazaki N, Koiwai O, Hsieh CL, Schwarz K, Lieber MR (2004) A biochemically defined system for mammalian nonhomologous DNA end joining. Mol Cell 16: 701–713 [DOI] [PubMed] [Google Scholar]
- Ma Y, Pannicke U, Schwarz K, Lieber MR (2002) Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell 108: 781–794 [DOI] [PubMed] [Google Scholar]
- Ma Y, Schwarz K, Lieber MR (2005b) The Artemis:DNA–PKcs endonuclease cleaves DNA loops, flaps, and gaps. DNA Repair (Amst) 4: 845–851 [DOI] [PubMed] [Google Scholar]
- McInerney P, O'Donnell M (2007) Replisome fate upon encountering a leading strand block and clearance from DNA by recombination proteins. J Biol Chem 282: 25903–25916 [DOI] [PubMed] [Google Scholar]
- Meek K, Gupta S, Ramsden DA, Lees-Miller SP (2004) The DNA-dependent protein kinase: the director at the end. Immunol Rev 200: 132–141 [DOI] [PubMed] [Google Scholar]
- Miné J, Disseau L, Takahashi M, Cappello G, Dutreix M, Viovy JL (2007) Real-time measurements of the nucleation, growth and dissociation of single Rad51–DNA nucleoprotein filaments. Nucleic Acids Res 35: 7171–7187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modesti M, Budzowska M, Baldeyron C, Demmers JA, Ghirlando R, Kanaar R (2007a) RAD51AP1 is a structure-specific DNA binding protein that stimulates joint molecule formation during RAD51-mediated homologous recombination. Mol Cell 28: 468–481 [DOI] [PubMed] [Google Scholar]
- Modesti M, Ristic D, van der Heijden T, Dekker C, van Mameren J, Peterman EJ, Wuite GJ, Kanaar R, Wyman C (2007b) Fluorescent human RAD51 reveals multiple nucleation sites and filament segments tightly associated along a single DNA molecule. Structure 15: 599–609 [DOI] [PubMed] [Google Scholar]
- Moldovan GL, Pfander B, Jentsch S (2007) PCNA, the maestro of the replication fork. Cell 129: 665–679 [DOI] [PubMed] [Google Scholar]
- Moshous D, Callebaut I, de Chasseval R, Corneo B, Cavazzana-Calvo M, Le Deist F, Tezcan I, Sanal O, Bertrand Y, Philippe N, Fischer A, de Villartay JP (2001) Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell 105: 177–186 [DOI] [PubMed] [Google Scholar]
- Myung K, Chen C, Kolodner RD (2001a) Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature 411: 1073–1076 [DOI] [PubMed] [Google Scholar]
- Myung K, Datta A, Chen C, Kolodner RD (2001b) SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homologous recombination. Nat Genet 27: 113–116 [DOI] [PubMed] [Google Scholar]
- Nick McElhinny SA, Havener JM, Garcia-Diaz M, Juarez R, Bebenek K, Kee BL, Blanco L, Kunkel TA, Ramsden DA (2005) A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Mol Cell 19: 357–366 [DOI] [PubMed] [Google Scholar]
- Nick McElhinny SA, Ramsden DA (2004) Sibling rivalry: competition between Pol X family members in V(D)J recombination and general double strand break repair. Immunol Rev 200: 156–164 [DOI] [PubMed] [Google Scholar]
- Oh SD, Lao JP, Hwang PY, Taylor AF, Smith GR, Hunter N (2007) BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell 130: 259–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn AJ, Elledge SJ, Zou L (2002) Checking on the fork: the DNA-replication stress-response pathway. Trends Cell Biol 12: 509–516 [DOI] [PubMed] [Google Scholar]
- Paulsen RD, Cimprich KA (2007) The ATR pathway: fine-tuning the fork. DNA Repair (Amst) 6: 953–966 [DOI] [PubMed] [Google Scholar]
- Pellegrini L, Yu DS, Lo T, Anand S, Lee M, Blundell TL, Venkitaraman AR (2002) Insights into DNA recombination from the structure of a RAD51–BRCA2 complex. Nature 420: 287–293 [DOI] [PubMed] [Google Scholar]
- Petalcorin MI, Galkin VE, Yu X, Egelman EH, Boulton SJ (2007) Stabilization of RAD-51–DNA filaments via an interaction domain in Caenorhabditis elegans BRCA2. Proc Natl Acad Sci USA 104: 8299–8304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povirk LF (2006) Biochemical mechanisms of chromosomal translocations resulting from DNA double-strand breaks. DNA Repair (Amst) 5: 1199–1212 [DOI] [PubMed] [Google Scholar]
- Putnam CD, Pennaneach V, Kolodner RD (2004) Chromosome healing through terminal deletions generated by de novo telomere additions in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 101: 13262–13267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revy P, Malivert L, de Villartay JP (2006) Cernunnos-XLF, a recently identified non-homologous end-joining factor required for the development of the immune system. Curr Opin Allergy Clin Immunol 6: 416–420 [DOI] [PubMed] [Google Scholar]
- Riballo E, Kuhne M, Rief N, Doherty A, Smith GC, Recio MJ, Reis C, Dahm K, Fricke A, Krempler A, Parker AR, Jackson SP, Gennery A, Jeggo PA, Lobrich M (2004) A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol Cell 16: 715–724 [DOI] [PubMed] [Google Scholar]
- San Filippo J, Chi P, Sehorn MG, Etchin J, Krejci L, Sung P (2006) Recombination mediator and Rad51 targeting activities of a human BRCA2 polypeptide. J Biol Chem 281: 11649–11657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP (2007) Human CtIP promotes DNA end resection. Nature 450: 509–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Tada S, Enomoto T (2006) Function of recQ family helicase in genome stability. Subcell Biochem 40: 49–73 [DOI] [PubMed] [Google Scholar]
- Sekiguchi JM, Ferguson DO (2006) DNA double-strand break repair: a relentless hunt uncovers new prey. Cell 124: 260–262 [DOI] [PubMed] [Google Scholar]
- Shin DS, Pellegrini L, Daniels DS, Yelent B, Craig L, Bates D, Yu DS, Shivji MK, Hitomi C, Arvai AS, Volkmann N, Tsuruta H, Blundell TL, Venkitaraman AR, Tainer JA (2003) Full-length archaeal Rad51 structure and mutants: mechanisms for RAD51 assembly and control by BRCA2. EMBO J 22: 4566–4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinger JA, Kiianitsa K, Heyer WD (2002) Rad54, a Swi2/Snf2-like recombinational repair protein, disassembles Rad51:dsDNA filaments. Mol Cell 10: 1175–1188 [DOI] [PubMed] [Google Scholar]
- Sugawara N, Wang X, Haber JE (2003) In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol Cell 12: 209–219 [DOI] [PubMed] [Google Scholar]
- Sung P (2005) Mediating repair. Nat Struct Mol Biol 12: 213–214 [DOI] [PubMed] [Google Scholar]
- Symington LS, Heyer WD (2006) Some disassembly required: role of DNA translocases in the disruption of recombination intermediates and dead-end complexes. Genes Dev 20: 2479–2486 [DOI] [PubMed] [Google Scholar]
- Takeda S, Nakamura K, Taniguchi Y, Paull TT (2007) Ctp1/CtIP and the MRN complex collaborate in the initial steps of homologous recombination. Mol Cell 28: 351–352 [DOI] [PubMed] [Google Scholar]
- Tan TL, Kanaar R, Wyman C (2003) Rad54, a Jack of all trades in homologous recombination. DNA Repair (Amst) 2: 787–794 [DOI] [PubMed] [Google Scholar]
- van der Heijden T, Seidel R, Modesti M, Kanaar R, Wyman C, Dekker C (2007) Real-time assembly and disassembly of human RAD51 filaments on individual DNA molecules. Nucleic Acids Res 35: 5646–5657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veaute X, Delmas S, Selva M, Jeusset J, Le Cam E, Matic I, Fabre F, Petit MA (2005) UvrD helicase, unlike Rep helicase, dismantles RecA nucleoprotein filaments in Escherichia coli. EMBO J 24: 180–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F (2003) The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423: 309–312 [DOI] [PubMed] [Google Scholar]
- Watson JD (1972) Origin of concatemeric T7 DNA. Nat New Biol 239: 197–201 [DOI] [PubMed] [Google Scholar]
- Weinstock DM, Richardson CA, Elliott B, Jasin M (2006) Modeling oncogenic translocations: distinct roles for double-strand break repair pathways in translocation formation in mammalian cells. DNA Repair (Amst) 5: 1065–1074 [DOI] [PubMed] [Google Scholar]
- Wesoly J, Agarwal S, Sigurdsson S, Bussen W, Van Komen S, Qin J, van Steeg H, van Benthem J, Wassenaar E, Baarends WM, Ghazvini M, Tafel AA, Heath H, Galjart N, Essers J, Grootegoed JA, Arnheim N, Bezzubova O, Buerstedde JM, Sung P et al. (2006) Differential contributions of mammalian Rad54 paralogs to recombination, DNA damage repair, and meiosis. Mol Cell Biol 26: 976–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West SC (2003) Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol 4: 435–445 [DOI] [PubMed] [Google Scholar]
- Wiese C, Dray E, Groesser T, San Filippo J, Shi I, Collins DW, Tsai MS, Williams GJ, Rydberg B, Sung P, Schild D (2007) Promotion of homologous recombination and genomic stability by RAD51AP1 via RAD51 recombinase enhancement. Mol Cell 28: 482–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson TE (2007) Nonhomologous end joining: mechanisms, conservation and relationship to illegitimate recombination. In ‘Genetic Recombination' Topics in Current Genetics, Aguilera A, Rothstein R (eds), pp 487–513. Springer [Google Scholar]
- Wyman C (2006) Monomer networking activates recombinases. Structure 14: 949–950 [DOI] [PubMed] [Google Scholar]
- Wyman C, Kanaar R (2006) DNA double-strand break repair: all's well that ends well. Annu Rev Genet 40: 363–383 [DOI] [PubMed] [Google Scholar]
- Wyman C, Ristic D, Kanaar R (2004) Homologous recombination-mediated double-strand break repair. DNA Repair (Amst) 3: 827–833 [DOI] [PubMed] [Google Scholar]
- Yang H, Li Q, Fan J, Holloman WK, Pavletich NP (2005) The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA–ssDNA junction. Nature 433: 653–657 [DOI] [PubMed] [Google Scholar]
- Yano KI, Morotomi-Yano K, Wang SY, Uematsu N, Lee KJ, Asaithamby A, Weterings E, Chen DJ (2007) Ku recruits XLF to DNA double-strand breaks. EMBO Rep 9: 91–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S, Dynan WS (1999) Geometry of a complex formed by double strand break repair proteins at a single DNA end: recruitment of DNA–PKcs induces inward translocation of Ku protein. Nucleic Acids Res 27: 4679–4686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, VanLoock MS, Yang S, Reese JT, Egelman EH (2004) What is the structure of the RecA–DNA filament? Curr Protein Pept Sci 5: 73–79 [DOI] [PubMed] [Google Scholar]