Abstract
Cohesins, which mediate sister chromatin cohesion, and CTCF, which functions at chromatin boundaries, play key roles in the structural and functional organization of chromosomes. We examined the binding of these two factors on the Kaposi's sarcoma-associated herpesvirus (KSHV) episome during latent infection and found a striking colocalization within the control region of the major latency transcript responsible for expressing LANA (ORF73), vCyclin (ORF72), vFLIP (ORF71), and vmiRNAs. Deletion of the CTCF-binding site from the viral genome disrupted cohesin binding, and crippled colony formation in 293 cells. Clonal instability correlated with elevated expression of lytic cycle gene products, notably the neighbouring promoter for K14 and vGPCR (ORF74). siRNA depletion of RAD21 from latently infected cells caused an increase in K14 and ORF74, and lytic inducers caused a rapid dissociation of RAD21 from the viral genome. RAD21 and SMC1 also associate with the cellular CTCF sites at mammalian c-myc promoter and H19/Igf2 imprinting control region. We conclude that cohesin subunits associate with viral and cellular CTCF sites involved in complex gene regulation and chromatin organization.
Keywords: c-myc, cohesin, CTCF, KSHV, latency
Introduction
The establishment and maintenance of a stable gene expression pattern over multiple cellular divisions are central features of viral and cellular fate determination. Segregation of active from inactive genes is an essential component of gene expression programmes, and represents a complex challenge, especially for compact viral genomes where active and inactive genes may be in close proximity. DNA regulatory elements, such as enhancers and repressors, may exert their effects on multiple gene targets that are not contiguous in the genome, and may be separated by large distances and intervening sequences. Several chromatin-associated elements, such as enhancer blockers, insulators, promoter targeting sequences, and chromatin boundary elements, coordinate the transmission of transcription regulatory signals from enhancers and repressors to specific initiation sites for transcription (Capelson and Corces, 2004; Lin et al, 2004; Gaszner and Felsenfeld, 2006). Precisely how these chromatin regulatory elements manifest their effects is not completely understood, yet is key to our understanding of complex gene regulation and chromosome dynamics.
The gene expression programmes of latent gammaherpesviruses represent an important system for understanding chromatin architecture and boundary elements. During latency, most viral genes are repressed, but a few viral genes essential for genome maintenance and host-cell survival must be expressed. Viral genes expressed during latent infection are often associated with pathogenesis, including cancers and immune system dysfunction. Kaposi's sarcoma-associated herpesvirus (KSHV) (also referred to as HHV8) is a human gammaherpesvirus aetiologically linked to Kaposi's sarcoma (KS), primary effusion lymphoma (PEL), and multicentric Castleman's disease (Chang et al, 1994; Boshoff et al, 1995; Cesarman et al, 1995; Soulier et al, 1995; Guo et al, 2003). KSHV can be cultured as a multicopy chromatin-associated episome in latently infected PEL cells. KSHV transcription in latently infected PEL cells is restricted to a few genes, including the latency-associated nuclear antigen LANA (ORF73), vCyclin D (ORF72), vFLIP (ORF71), Kaposin (K12), and 11 microRNAs (miRNAs) (Kedes et al, 1997; Rainbow et al, 1997; Dittmer et al, 1998; Muralidhar et al, 1998; Sadler et al, 1999; Burysek and Pitha, 2001). The latency-associated viral products have growth-transforming and cell cycle-deregulating properties that are likely to contribute to KSHV-associated malignancies (Cesarman, 2002; Cotter and Robertson, 2002; Komatsu et al, 2002). Transcription regulation of these KSHV latency genes is coordinated through a primary multicistronic transcript that initiates upstream of ORF73 (Rainbow et al, 1997; Dittmer et al, 1998; Sarid et al, 1999; Talbot et al, 1999; Pearce et al, 2005). Transcripts initiating from this region also give rise to the 11 miRNAs and overlap the viral lytic origin (Pearce et al, 2005; Samols et al, 2005; Cai and Cullen, 2006). On the opposite strand adjacent to the major latency promoter is the transcription initiation site for a bicistronic message that encodes two lytic genes, K14 and vGPCR (ORF74) (Jeong et al, 2001; Liang and Ganem, 2004). vGPCR has tumorigenic and angiogenic properties, and factors that influence its gene expression are thought to be crucial for understanding KS pathogenesis (Mutlu et al, 2007). The K14/ORF74 transcript is not detected in cultured PEL cells, but low-level expression has been observed in PEL tumour biopsies and KS lesions, raising the question of whether they are expressed from a subset of lytic virus or whether these genes are leaky during latency (Nador et al, 2001). The close proximity and partial overlap of the ORF73/72/71 latency promoter with K14/ORF74 lytic promoter suggest that chromatin boundaries or other epigenetic features are required to organize this complex genetic locus.
Chromatin boundaries have been characterized in several model organisms (Gerasimova and Corces, 2001; Gerbi et al, 2002). A common molecular constituent of these boundaries is the 11-zinc finger, DNA-binding nuclear phosphoprotein CTCF that was initially discovered as a factor involved in transcriptional repression of avian, mouse, and human MYC promoters (Ohlsson et al, 2001). It was later characterized to be involved in enhancer blocking, chromatin insulation, gene activation, and imprinting on diverse genes, such as β-globin, c-myc, and the H19/Igf2 imprinting locus (Fedoriw et al, 2004; Yusufzai et al, 2004). More recent studies have implicated CTCF as a boundary factor for the latent cycle gene expression programmes of Epstein–Barr virus (EBV) (Chau and Lieberman, 2004; Chau et al, 2006; Day et al, 2007) and herpes simplex virus-1 (Amelio et al, 2006; Chen et al, 2007). CTCF uses its 11 zinc fingers to bind a wide diversity of ∼50-bp sequence elements throughout the cellular and viral genomes (Kanduri et al, 2000; Kim et al, 2007). The biochemical mechanisms underlying CTCF insulator and chromatin boundary function have not been completely elucidated. CTCF can function in transcription repression and activation, and seems to regulate interactions between DNA regulatory elements. Recent data suggest that CTCF facilitates intra-chromosomal DNA loop formation (Kurukuti et al, 2006), as well as promoting inter-chromosomal linkages (Ling et al, 2006), perhaps by tethering DNA to subnuclear structures, such as the nuclear matrix (Yusufzai et al, 2004) or through interactions with chromatin remodelling machines (Ishihara et al, 2006).
The DNA looping and scaffold attachments that coordinate gene expression programmes may be regulated by higher order chromosome structures, such as those mediated by the structural maintenance of chromosome (SMC) proteins. SMC proteins represent a class of highly conserved multiprotein complexes that include cohesins and condensin subunits (Hirano, 2006). Cohesins and condensins have a central role in chromosome assembly and segregation during mitosis, but have also been implicated in the regulation of gene expression. Cohesin subunits, in particular, have been implicated in the human developmental disorder Cornelia de Lange syndrome, suggesting that these proteins contribute to the establishment of heritable gene expression programmes (Dorsett, 2004, 2007; Gillis et al, 2004; Krantz et al, 2004). Cohesins consist of four major subunits, SMC1, SMC3, RAD21/Scc1, and Scc3, which have been shown to form a ring-like structure that is involved in sister-chromatid cohesion (Nasmyth and Haering, 2005; Losada, 2007). The sister-chromatid cohesion is essential for proper chromosome segregation, and also contributes to homologous recombination-dependent DNA damage repair and transcriptional silencing (Losada, 2007). Cohesins load on sister chromatids during DNA synthesis and then associate with centromeric regions as well as numerous other regions along the chromosome arms (Blat and Kleckner, 1999). Although several genomic mapping studies have identified cohesin attachment sites, no single mechanism has been established to account for the positioning of cohesins at specific genomic loci (Glynn et al, 2004).
In this study, we identify a cohesin-binding site in the KSHV chromosome that colocalizes with a strong CTCF site within the transcription control region of the latency transcripts. Deletion of this site causes a loss of episome stability during latency and a derepression of the K14 and ORF74 lytic transcripts. A similar colocalization of cohesins with CTCF is observed at the cellular c-myc promoter and the H19/Igf2 imprinting control region, indicating that this is not a virus-specific interaction. The significance of cohesin colocalization with CTCF-binding sites (CBSs) and the potential role of cohesins in chromatin organization and gene regulation are discussed.
Results
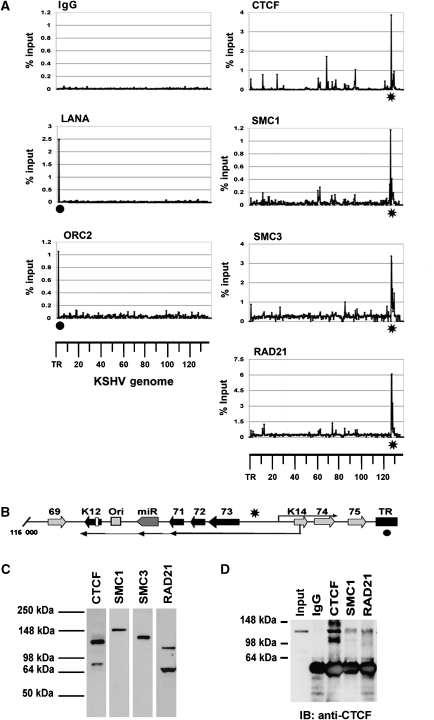
To investigate the chromatin organization of the KSHV genome during latent infection, we designed a 384-well array containing real-time PCR primer pairs that amplify ∼400-bp intervals across the ∼138 kb KSHV genome. The specificity and primer efficiency of the array were validated by several criteria (Day et al, 2007; Supplementary data). The array was used to analyse chromatin immunoprecipitation (ChIP) reactions with antibodies for several proteins that were known to be important in genome organization, including CTCF and cohesin subunits RAD21, SMC1, and SMC3 (Figure 1). We also assayed the viral-encoded LANA protein essential for stable episomal maintenance and the origin recognition complex 2 (ORC2) protein, which has been previously shown to associate with LANA-binding sites in the terminal repeat (TR) region of KSHV (Stedman et al, 2004). ChIP assays with control immunoglobulin (Ig) G showed no significant enrichments of KSHV genome sequences (Figure 1A). ChIP assays with LANA and ORC2 showed specific enrichment at the TR, with no other sites of significant interaction (Figure 1A, left panels). ChIP with CTCF revealed several sites within the KSHV genome that were significantly enriched relative to IgG controls. These major sites are listed in the table shown in Supplementary Figure S1. Among these, the most robust sites were located within genome coordinates 127 414–127 560, which lie between the divergent open reading frames (ORFs) for ORF73 and K14 (Figure 1B, indicated by asterisk). Remarkably, ChIP with three different cohesin subunits, SMC1, SMC3, and RAD21, was highly enriched at this genome location, similar to CTCF (Figure 1A, right panels). The antibodies used for ChIP recognized distinct polypeptide species in western blots (Figure 1C), indicating that the colocalization by ChIP was not a consequence of antibody cross-reaction. The colocalization of cohesin subunits with CTCF was also found in a second PEL cell line, JSC1, indicating that this is not a peculiarity of the BCBL1 cell line (Supplementary Figure S2).
Figure 1.
KSHV genome-wide ChIP analysis: colocalization of CTCF and cohesins upstream of ORF73. (A) A 384-well array of PCR primers spanning ∼400-bp intervals across the KSHV genome was used to analyse ChIP DNA isolated with antibodies specific for LANA, ORC2, CTCF, SMC1, SMC3, RAD21, or control IgG. The KSHV coordinates are indicated below each column of graphs. •LANA- and ORC2-binding sites at TR; *location of the major CTCF–cohesin-binding site upstream of ORF73. (B) Schematic of the region of KSHV genome (nt 116 000–138 000) containing the major latency transcript (leftward ORF73/72/71, vmiRNA, and K12), the rightward-oriented K14/ORF74 lytic transcript, and the TR. (C) Western blot of BCBL1 cell extracts with antibodies to CTCF, SMC1, SMC3, and RAD21 used for ChIP in (A). (D) Co-immunoprecipitation assay with antibodies to CTCF, SMC1, RAD21, or IgG control. IPs were immunoblotted with anti-CTCF.
The mechanism for cohesin loading and attachment to DNA is not completely understood (Nasmyth and Haering, 2005; Losada, 2007). To investigate the possibility that cohesin colocalization at CBSs was a result of an interaction between CTCF and cohesin subunits, we assayed the ability of these proteins to co-immunoprecipitate (co-IP) from BCBL1 cell nuclear extracts (Figure 1D and Supplementary Figure S3). IPs with antibodies to CTCF, SMC1, and RAD21 were probed by western blot with anti-CTCF. We detected CTCF protein in the IPs of these cohesin subunits, but not in control IgG (Figure 1D and Supplementary Figure S3). We also observed that SMC3 could co-IP with CTCF, but that the reverse IP with CTCF did not yield robust cohesin signals (Supplementary Figure S3A and B). To further investigate the possible interactions between CTCF and cohesins, we expressed a FLAG-CTCF construct in 293 cells and analysed these cells by co-IP (Supplementary Figure S3C and D). Using this approach, we were able to demonstrate that FLAG-CTCF could co-IP with SMC3 (Supplementary Figure S3D), but the detection of SMC1 and RAD21 was not as readily detected (data not shown). These findings suggest that the CTCF can interact with a subset of cohesin subunits, but that the interaction is either indirect, unstable, or otherwise not amenable to detection by certain co-IP conditions.
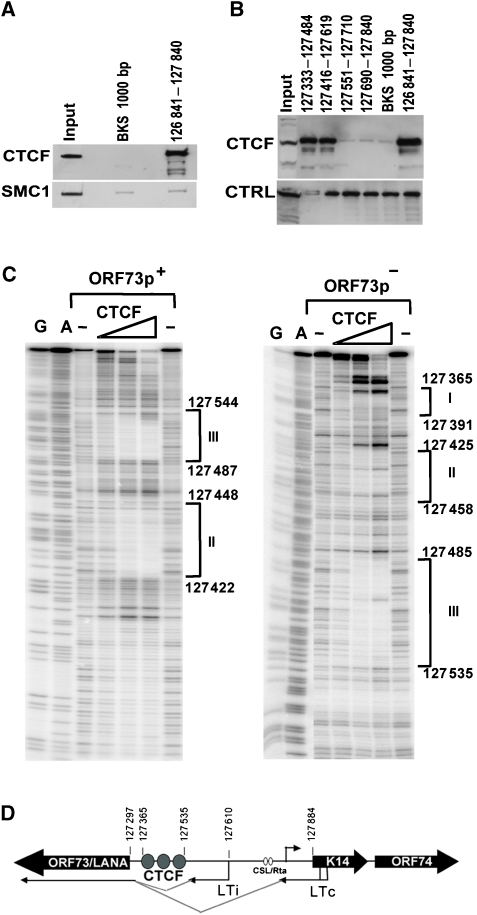
The CBSs located between ORF73 and K14 were mapped in vitro using DNA affinity purification (Figure 2A and B) and DNAse I footprinting (Figure 2C). DNA affinity was used to map the binding of CTCF from BCBL1-derived nuclear extracts (Figure 2A and B). Western blotting for CTCF revealed that CTCF bound to KSHV DNA from 126 841 to 127 841, but not to a control 1-kb DNA fragment (BKS) (Figure 2A, top panel). We also tested whether the cohesin subunit SMC1 was bound to these DNA fragments, but were unable to demonstrate any specific binding using this assay (Figure 2A, lower panel). This suggests that cohesin loading on DNA may require additional factors or processes, such as DNA replication or DNA topological constraints, that are not recapitulated in the in vitro DNA affinity assay. Nevertheless, we were able to use the DNA affinity assay to fine map the binding sites for CTCF (Figure 2B, top panel). Using smaller DNA fragments, we were able to show that CTCF bound to a region between 127 333 and 127 619, but not to regions 127 551–127 840. DNA templates were comparable in each binding reaction as demonstrated by a cross-reacting protein of faster mobility that bound nonspecifically to all DNA templates tested (Figure 2B, lower panel, CTRL).
Figure 2.
Mapping CTCF-binding sites upstream of ORF73. (A) DNA affinity purification assay with biotinylated DNA from KSHV 126 841–127 840 or a control 1000-bp fragment from pBKS. BCBL1 nuclear extract was incubated with biotinylated DNA and bound proteins were identified by western blotting with antibody specific for CTCF (top panel) or SMC1 (lower panel). (B) Smaller DNA fragments were used to fine map the CTCF-binding site upstream of ORF73 as indicated by coordinates above each lane. Bound proteins were detected by western blot with antibodies specific for CTCF (top panel). A nonspecific, cross-reacting protein is shown in lower panel as a loading control (CTRL). (C) DNase I footprinting of KSHV DNA with purified recombinant CTCF protein. KSHV coordinates are indicated to the right of each footprint boundary. G and A are chemical sequencing ladders. ORF73p+ is the sense strand (left panel) and ORF73p− is the antisense (right panel). (D) Schematic showing the location of the cluster of three CTCF-binding sites relative to ORF73 (LTi and LTc) and K14/ORF74 (rightward arrow) transcription start sites. CSL/Rta indicates the sites in K14/ORF74 promoter stimulated during lytic activation by Rta.
The nucleotide sequence of CTCF binding was further defined by DNase I footprinting using recombinant human CTCF expressed and purified from baculovirus (Figure 2C). We found that CTCF protected three major sites located at positions 127 365–127 391 (site I), 127 425–127 458 (site II), and 127 487–127 544 (site III). All three of these protected sequences contain core nucleotides that comply with the consensus CTCF recognition site (Supplementary Figure S4). These sites lie upstream of the ORF73 ATG (127 297), and within the DNA encoding the 5′ untranslated region of the major latency transcripts (Figure 2D). These sites are upstream to a splice acceptor site for the major latency transcript located at 127 314 (Sarid et al, 1999) and ∼300 bp upstream of the initiation site for the K14/ORF74 transcript located at 127 848 on the opposite strand (Jeong et al, 2001).
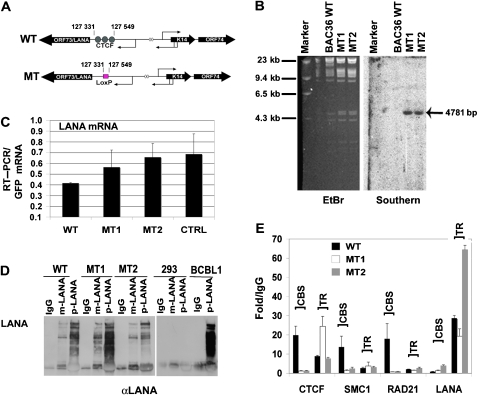
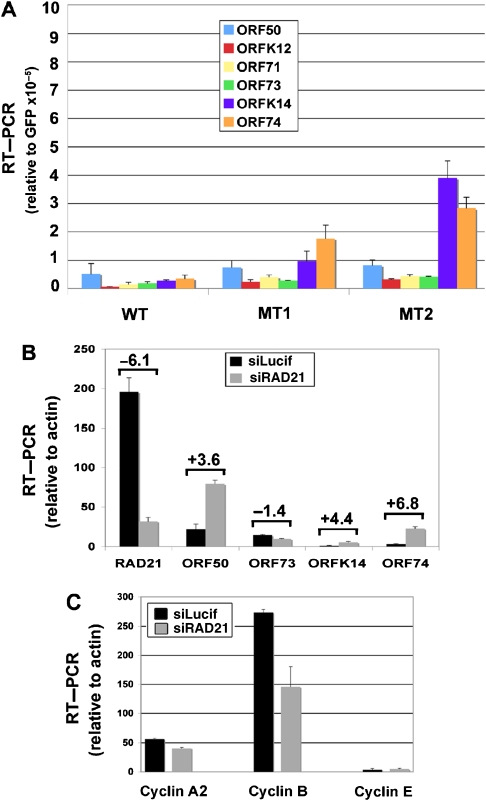
The functional significance of these CBSs was determined by site-directed deletion and substitution mutagenesis of cloned KSHV bacmid using recombination-mediated genetic engineering (Copeland et al, 2001). The CTCF sites at 127 331–127 549 were deleted and replaced by a single LoxP site (Figure 3A and Supplementary Figure S5). Two independent isolates were generated and validated using restriction enzyme digestion, PCR analysis, and direct DNA sequencing of the recombinational junctions (Chau et al, 2006) (Supplementary data). We show a representative restriction digest demonstrating the insertion of a unique EcoRI site associated with the LoxP DNA into the KSHV genome (Supplementary Figure S5A and B). A Southern blot was used to demonstrate that LoxP site was integrated into the correct target sequence, with no other detectable rearrangements (Figure 3B). Two recombinant genomes that contained the correct insertion as determined by PCR and DNA sequencing, and no other detectable rearrangements were designated MT1 and MT2, and were used for further analysis. Recombinant KSHV genomes were then purified from Escherichia coli, transfected into 293 cells, and selected for GFP expression by FACS. Similar number of GFP-positive cells were selected by FACS and then analysed for LANA mRNA by RT–PCR (Figure 3C). To control for differences in genome copy number, we used bacmid-expressed GFP as an internal control. We found that LANA mRNA was expressed at levels similar or higher than that of bacterial artificial chromosome 36 (BAC36) WT. A control deletion (CTRL) in the KSHV miRNA region showed a similar expression level for LANA. Since the MT1 and MT2 deletion was within the LANA 5′UTR, we also determined whether LANA protein levels were equally expressed in WT- and MT-transfected cells (Figure 3D). Protein extracts from identical numbers of transfected and GFP-sorted cells were analysed by western blotting of IPs with two different LANA-specific antibodies. We found that LANA protein levels were nearly identical in WT, MT1, and MT2 (Figure 3D). As expected, LANA protein was not detected in untransfected 293 cells and was expressed robustly in latently infected BCBL1 cells (Figure 3D, right panel). LANA protein was also detected in the nuclear compartment of WT, MT1, and MT2 cells by immunofluorescence assays (Supplementary Figure S6A). RT–PCR revealed that GFP mRNA levels for each bacmid were expressed to similar levels relative to cellular actin, indicating that the transfection efficiency of each bacmid was similar (Supplementary Figure S6B).
Figure 3.
CTCF sites are required for cohesin binding. (A) Schematic diagram of KSHV bacmid mutations generated to substitute the CTCF-binding site (CBS) cluster at positions 127 331–127 549 with LoxP recognition site. (B) BAC36 WT, MT1, and MT2 were analysed by ethidium bromide (EtBr) staining of agarose gels for insertion of novel EcoRI site (left) and by Southern blot of the same gel probed with LoxP-specific probe (right). (C) RT–PCR was used to measure LANA mRNA levels in WT, MT1, MT2, or a control deletion in the viral miRNAs (CTRL). bacmid-expressed GFP mRNA was used as an internal standard for genome copy number. (D) LANA protein expression was analysed by western blotting of immunoprecipitates with two different LANA-specific antibodies. Extracts were derived from identical number of cells for WT, MT1, MT2, untransfected 293, and latently infected BCBL1 cells. (E) ChIP assay of 293 cells transfected with WT, MT1, or MT2 after selection with hygromycin for 2 weeks. Antibodies for ChIP included CTCF, SMC1, RAD21, and LANA. ChIP DNA was analysed by real-time PCR with primers specific for the CBS or the terminal repeat (TR).
The effect of the deletion on CTCF and cohesin binding was analysed by ChIP assay (Figure 3E). GFP-sorted, hygromycin-resistant cell pools containing either WT, MT1, or MT2 KSHV genomes were assayed by ChIP with antibodies specific for CTCF, SMC1, RAD21, or LANA. Using primers specific for sequences adjacent to the CBS, we found that CTCF binding was completely abrogated from the CBS site in deletion mutants (MT1 and MT2), but not in BAC36 WT genomes. As control for CTCF binding, we show that CTCF binding to the TR was similar or elevated in MT1 and MT2 relative to WT. CTCF binding was higher at TR in these assays because the primer for the CBS was shifted 300 bp downstream to compensate for the deleted sequence. The effect of CBS deletion on cohesin subunits was also analysed. We found that SMC1 and RAD21 binding was eliminated from the CBS region in MT1 and MT2, but bound robustly in BAC36 WT. As controls, we show that LANA bound to the TR in WT and MT KSHV genomes with similar efficiency, indicating that LANA expression and TR binding were not disrupted in these mutants. Cohesin subunits were not detected at any region adjacent to the CTCF sites in the deletion mutants (Supplementary Figure S7). These findings indicate that deletion of the CBS from the recombinant KSHV genome eliminates CTCF binding, and more interestingly, that these CTCF sites are required for cohesin binding in vivo.
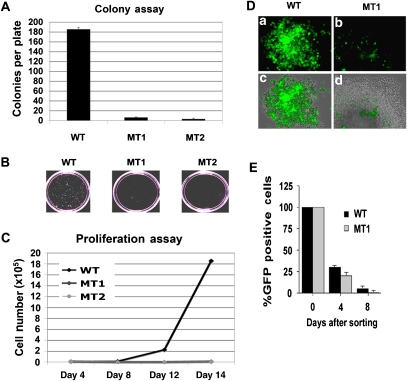
The genome stability of the MT1 and MT2 deletion mutants was compared to KSHV WT by selecting for GFP-positive, hygromycin-resistant 293 cell colonies (Figure 4). The average number of visible hygromycin-resistant colonies was ∼100-fold less for MT1 and MT2 relative to WT genomes (Figure 4A). Correspondingly, the average colony size was considerably smaller for MT genomes relative to WT (Figure 4B). The reduction in colony formation correlated with CTCF deletion, since additional deletion mutants that eliminate all or part of the CTCF sites had similar deficiency in colony formation, whereas a deletion that was immediately adjacent to the CTCF sites formed colonies similar to WT (Supplementary Figure S8). The decrease in colony formation was not due to a lack of GFP and hygromycin resistance gene expression, since all constructs expressed similar mRNA levels within the first 48 h post-transfection (Supplementary Figure S6B, and data not shown). Similarly, the decrease in colony formation could not be explained by the difference in LANA expression, since LANA mRNA and protein were detected at similar levels in WT and MT constructs (Figure 3A–D), and similar levels of LANA binding to the TR were detected by ChIP assay in all WT and MT constructs (Figure 3E). These observations suggest that a deletion of the KSHV CTCF–cohesin-binding site interferes with stable colony formation and growth in 293 cells.
Figure 4.
Host cell growth defects and plasmid instability in KSHV genomes lacking CTCF sites. (A) WT-, MT1-, and MT2-transfected 293 cells were cell sorted for GFP expression and assayed for hygromycin-resistant colony formation. After 10–15 days of selection, the plates were photographed with a 35 mm camera and the colony number was quantitated using Image-Pro Plus software. (B) Example of colony size and density for WT-, MT1-, and MT2-formed colonies. (C) WT-, MT1-, and MT2-transfected 293 cells were sorted for GFP and then cultured in hygromycin-containing media. Cells were then assayed at 4, 8, 12, and 16 days for proliferation using fluorosphere beads and FACS analysis to count cell number. (D) Plasmid stability was assayed by monitoring GFP expression of hygromycin-resistant cell colonies. Hygromycin-resistant cell colony derived with either WT (a, c) or MT1 (b, d); KSHV bacmid was assayed by fluorescence microscopy for GFP (a, b) or a merge of fluorescence with phase-contrast microscopy (c, d). (E) Plasmid stability was assayed by FACS analysis of GFP-positive cells. Identical numbers of WT- and MT1-transfected 293 cells were sorted 4 days after transfection. The percentage of cells retaining GFP-positive signals was then determined 4 or 8 days after initial sorting.
To better understand the cause of colony instability, we compared rates of cellular proliferation in WT and MT cells immediately after transfection and sorting for GFP by FACS. Similar GFP-positive cells could be obtained within 48 h post-transfection. Cells were then cultured in hygromycin-containing media for 16 days and monitored for cell number every 4 days. We found that MT1 and MT2 were significantly inhibited for cell proliferation relative to BAC36 WT-containing cells (Figure 4C). Microscopic analysis of single hygromycin-resistant colonies also revealed defects in plasmid stability (Figure 4D). Hygromycin-resistant colonies were found to have heterogeneous expression of GFP, suggesting that many cells in the colony have lost GFP expression as a result of unstable gene expression, accelerated plasmid loss, or plasmid rearrangement. To test the rate of plasmid loss, we sorted WT- or MT1-transfected cells by GFP expression, and then analysed GFP expression in culture without any hygromycin selection. We found that MT1 genomes were lost at a slightly accelerated rate relative to WT genomes, although WT genomes were also lost rapidly without selection in hygromycin (Figure 4E). No defects in the hygromycin resistance gene could be detected by restriction digest or PCR analysis of MT genomes. We conclude that deletion of the CBSs causes a significant defect in the ability to establish a stable transformation of 293 cells with KSHV bacmid genomes.
We reasoned that inhibition of 293 cell growth and plasmid instability could be caused by aberrant expression of viral gene products. To determine if the CBS deletion mutants had aberrant gene expression patterns, we compared the mRNA levels of several viral gene products by RT–PCR (Figure 5). The RNA levels were normalized to GFP to eliminate any variation in genome copy number per cell. We found that several KSHV genes were elevated relative to GFP in the CBS deletion mutants. Most notable was the increase in expression of K14 and ORF74 (Figure 5A). These data suggest that CBS deletion causes a loss of repression of the K14/ORF74 transcript typically restricted to lytic cycle gene expression.
Figure 5.
K14 and ORF74 transcription are elevated in CBS deletion mutants. (A) WT-, MT1-, and MT2-transfected 293 cells were sorted by GFP and selected in hygromycin for 4 days. RNA was isolated and analysed by RT–qPCR for primers specific for ORF50, ORFK12, ORF71, ORF73, ORFK14, ORF74, and GFP. The values were reported as fold over the internal control of GFP, which is a common component of the bacmid. (B) Latently infected BCBL1 cells were transfected with siRAD21 or control siLucif, sorted by FACS, and then assayed by RT–qPCR for RNA expression levels of RAD21, ORF50, ORF73, ORFK14, and ORF74 with normalization to cellular actin. The relative change between siRAD21 and siLucif is indicated above each pair of bars. (C) RNA samples from experiment described in (B) were evaluated for expression of Cyclin A2, Cyclin B, and Cyclin E using RT–qPCR.
To determine if cohesins contribute to the regulation of the K14/ORF74 repression during latency, we assayed the effect of RAD21 siRNA on K14 and ORF74 mRNA in latently infected BCBL1 cells (Figure 5B). siRNA depletion of RAD21 was monitored at the RNA level. We found that siRNA depletion led to a reduction of ∼80% of the RAD21 mRNA. Cells depleted for RAD21 were isolated by FACS using control fluorescent siRNA (si GLO; Dharmacon) and assayed for ORF50, ORF73, ORFK14, and ORF74 mRNA levels relative to cellular actin. We found that RAD21-depleted cells had a four to six-fold increase in K14 and ORF74 mRNA levels. RAD21 depletion had a slight increase (3.6-fold) in the lytic activator ORF50, and a slight decrease (−1.4-fold) in ORF73 mRNA levels. Control siRNA (Lucif) had no detectable effect on RAD21, K14, or ORF74 mRNA levels (Figure 5B, and data not shown). RAD21 depletion also caused a decrease in cellular Cyclin B, but had no detectable effects in Cyclin A2 or E, suggesting that it may prevent cell cycle progression into G2/M (Figure 5C). Consequently, we cannot rule out that some of the effects on KSHV gene expression are an indirect consequence of this effect on cell cycle progression. Nevertheless, these findings suggest that cohesin subunit RAD21 contributes to the stable repression of KSHV lytic genes K14 and ORF74 during KSHV latency in PEL cells through its direct physical association, and may also regulate cell cycle progression through G2/M.
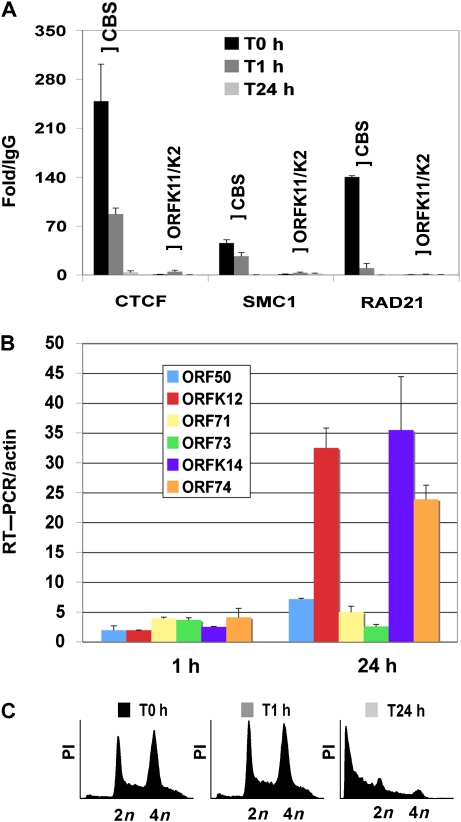
To further test if CTCF and cohesin binding correlate with transcription repression of ORF74 gene expression during KSHV latency, we assayed CTCF, SMC1, and RAD21 binding by ChIP assay in BCBL1 cells at 1 and 24 h after lytic cycle induction with sodium butyrate (Figure 6A). We found that RAD21 rapidly dissociated from the CBS after 1 h of sodium butryate treatment. Within 24 h, SMC1 and CTCF were also completely eliminated from the CBS site. CTCF, SMC1, or RAD21 was not detected at control sites located between the divergent promoters for ORF11/K2, indicating that ChIP binding was specific for the CBS. The RNA levels for KSHV genes were also measured after sodium butyrate treatment. As expected for lytic cycle genes, we found that sodium butyrate induced ORFK14, ORF74, and ORFK12 transcription ∼30-fold after 24-h treatment (Figure 6B). Propidium iodide staining indicated that sodium butyrate treatment had no significant effect on cell cycle profile after 1 h, but had a profound effect on cell viability at 24 h, as might be expected during robust lytic virus replication (Figure 6C).
Figure 6.
Dissociation of RAD21, SMC1, and CTCF upon lytic cycle induction. (A) BCBL1 cells were treated with sodium butyrate for 0, 1 or 24 h and then assayed by ChIP with antibodies to CTCF, SMC1, or RAD21. ChIP DNA was analysed by real-time PCR with primers specific for the KSHV CBS or a control region between ORFK11 and K2. ChIP data are presented as fold over IgG for each primer set and antibody. (B) RNA levels were monitored at 0, 1, or 24 h after sodium butyrate treatment and then assayed by RT–qPCR with primers specific for ORF50, ORFK12, ORF71, ORF73, ORFK14, or ORF74. RNA was quantified as fold increase over time zero relative to cellular actin. (C) FACS analysis of propidium iodide-stained BCBL1 cells at 0, 1, or 24 h after treatment with sodium butyrate (3 mM) was used for cell cycle profiling.
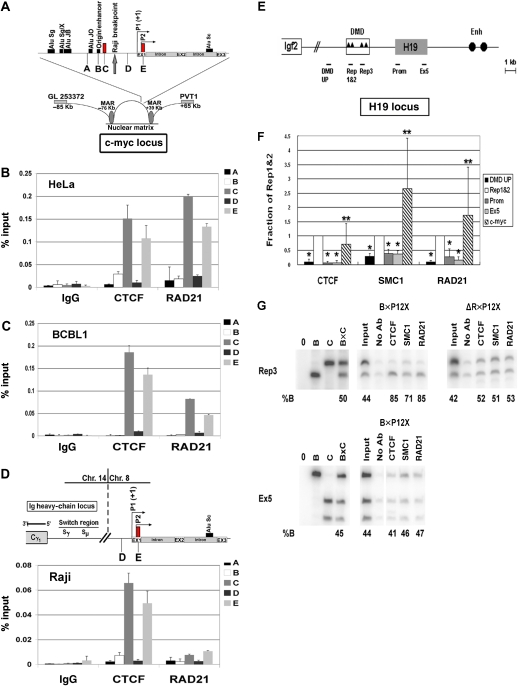
The colocalization of cohesin subunits with CTCF at the KSHV latency control region raised the question whether similar colocalizations exist at cellular CBSs. To address this, we first examined the well-characterized CBSs in the c-myc locus (Figure 7A–D). CTCF has been shown to bind to two major sites in the c-myc promoter and these sites function as enhancer blockers of c-myc transcription (Ohlsson et al, 2001; Gombert et al, 2003). We used ChIP assay to examine CTCF and RAD21 binding to c-myc in KSHV-negative HeLa cells, KSHV-positive BCBL1 cells, and also in the Burkitt lymphoma cell line Raji, where the c-myc promoter has been translocated to the Ig heavy-chain locus (Gelmann et al, 1983). Remarkably, we found that RAD21 perfectly colocalized with CTCF sites in the HeLa and BCBL1 cell lines. RAD21 did not bind to other regions of the c-myc promoter, including the origin/enhancer or Alu sequences. In contrast to these cells, we found that RAD21 did not bind to the c-myc CTCF sites in Raji cells, where c-myc promoter has been rearranged by a translocation with the Ig enhancer (Figure 7D). We also found that sodium butyrate treatment for 24 h caused the dissociation of RAD21 and CTCF from the c-myc promoter in BCLB1, similar to that observed for the KSHV CBS (Supplementary Figure S9). These findings suggest that RAD21 contributes to the transcription repression function of CTCF at the cellular c-myc locus in a manner similar to the KSHV latency control region.
Figure 7.
Colocalization of cohesin subunits at cellular CTCF sites. (A–D) Analysis of CTCF and cohesin binding at the human c-myc locus. (A) Schematic of the CTCF-binding sites and other chromatin features of the c-myc promoter. Location of primer pairs A–D are indicated. Primer pairs C and E amplify the known CTCF sites. (B–D) ChIP assays with antibodies for CTCF, RAD21, or IgG control were analysed by real- time PCR with primer pairs A–E specific for different regions of the c-myc promoter. HeLa (B), KSHV-positive BCBL1 (C), or Burkitt lymphoma Raji (D) cells were used for ChIP assays. (E, F) Analysis of CTCF and cohesin binding at the H19/Igf2-imprinted locus. (E) Schematic of the H19/Igf2 locus and primer location. Rep1&2 and Rep3 indicate the first three CTCF-binding sites in the differentially methylated domain (DMD). The H19 transcription unit is designated by a filled box and enhancers by filled circles. (F) Pattern of CTCF and cohesin binding across the H19 locus was analysed by real-time PCR relative to the input DNA. The fraction of precipitated DNA relative to input was normalized to the value obtained at Rep1&2 region, which typically had the highest level in the H19 locus. Four regions as indicated in (E) were analysed and c-myc locus was included as positive control. The graph shows the average and s.d of the normalized values. *P<0.01, CTCF, SMC1 and RAD21 binding is significantly different between Rep1&2 and other regions. **P>0.05, CTCF, SMC1, and RAD21 binding is not significantly different between Rep1&2 and c-myc regions. (G) Allele-specific ChIPs were carried out with antibodies against CTCF, SMC1, RAD21, or no antibody (no Ab). F1 hybrid B × P12X mouse embryonic fibroblast cells and neonatal liver cells from H19DMD−ΔR/+ mice isolated and examined by ChIP. DNA from control (B, C, and B × C genomic DNA alone, left-hand panels), input and precipitated samples were subjected to quantitative-PCR for the indicated region of H19 (Rep3 and Ex5) and then digested with a restriction enzyme that is polymorphic between B and C (Tsp45I for Rep3 and BglI for Ex5). The genotypes of cells and tissues are indicated at the top. In all experiments shown here, the B allele corresponds to the maternal allele, while the C allele is the paternal allele. The numbers underneath each panel indicate the percentage of B allele (maternal) relative to total. Note that ChIP assays were performed multiple times and the allele-specific assays were conducted on each ChIP sample but only one representative experiment is shown for each antibody and region.
CTCF has also been implicated in the regulation of the mammalian H19/Igf2-imprinted locus, where CTCF binds the differentially methylated maternal allele and functions as a chromatin insulator (Bell and Felsenfeld, 2000; Hark et al, 2000). We therefore asked whether cohesin subunits colocalize with the CTCF sites at the mouse H19/Igf2-imprinted locus (Figure 7E–G). CTCF binds to four sites in the differentially methylated domain with the CBSs designated as Rep1–4. CTCF binding to these sites can be detected with primers for Rep1&2 and Rep3 (Figure 7E and F) (Szabo et al, 2004; Engel et al, 2006). We found that SMC1 and RAD21 were also enriched at the Rep1&2 site relative to several other locations in mouse H19/Igf2 locus (Figure 7F). We also found RAD21 and SMC1 colocalize with CTCF at the human H19/Igf2 locus (Supplementary Figure S10). The allele-specific binding of CTCF to the Rep3-binding site was demonstrated using allele-specific PCR (Figure 7G) (Engel et al, 2006). CTCF binding was shown to be enriched on the unmethylated maternal allele (faster migrating band) as 85% of the binding was specific to that allele relative to the paternal allele (Figure 7G, left B × P12). RAD21 and SMC1 were similarly enriched (71 and 85%, respectively) at the maternal allele, and not preferentially bound at regions within the H19 exon 5 (Ex5, lower panel). Furthermore, this preferential binding to the maternal allele is lost in the liver of neonatal mice harbouring small deletions in the four CBSs (H19DMD−ΔR) (Figure 7G right panel, ΔR × P12X), indicating that cohesin binding is CTCF dependent in the imprinting control region. Real-time PCR showed that binding in these cells is background (data not shown). These data indicate that cohesins colocalize with CTCF at the H19/Igf2 imprinting control region and suggest that they play a role in CTCF-associated insulator activity.
Discussion
The structural organization of chromosomes is thought to play a central role in the regulation of gene expression. We have found that CTCF sites were co-occupied by cohesin subunits at key regulatory elements in the KSHV genome, human c-myc promoter, and mouse H19/Igf2 imprinting control region. CTCF and cohesins are known to function in chromosome structure and organization, but it has not been demonstrated previously that these proteins overlap at specific genetic loci. The colocalization of CTCF and cohesins at the KSHV major latency control region is highly significant since this region controls the transcription of a cluster of latency gene products, including LANA (ORF73), vCyclin (ORF72), vFLIP (ORF71), and the vmiRNAs. This region is also responsible for the activation of the vGPCR (ORF74) and K14 in KS lesions and during lytic replication, and their repression during latent infection. The nested arrangement of divergent latent and lytic promoters in this region of the viral genome suggests that it requires complex structure and regulation. CTCF and cohesin are likely to provide structural organization to this intricate genetic element.
CTCF is a multifunctional protein that has been implicated in gene regulation studies as a transcriptional repressor, enhancer blocker, and chromatin boundary factor (Ohlsson et al, 2001; Gaszner and Felsenfeld, 2006). Deletion of the KSHV CBS caused a derepression of several viral lytic genes (Figure 5). This was not a result of the loss of LANA mRNA or protein expression levels or DNA-binding function at TR (Figure 3C–E). The CBS is located more than 300 bp upstream of the K14 transcription initiation site, and may block activation signals emanating from ORF73/ORF72/ORF71. Earlier studies have shown that a K14- and vGPCR-activating sequence exists at ∼900 bp upstream of the transcription initiation site, and it is possible that CTCF is positioned to prevent this enhancement of K14 transcription during latency (Jeong et al, 2001). K14 and vGPCR can also be activated by the KSHV lytic activator Rta, which associates with the CSL-binding sites located in closer proximity to the K14 transcription initiation site (Liang and Ganem, 2004). Thus, Rta-mediated activity would not be blocked by CBSs, assuming they function as a physical block to upstream activation signals on linear DNA. In models of HSV1 latency, CTCF has been shown to function as an enhancer blocker, and is positioned downstream of the latency-associated transcript (LAT) promoter, similar to the orientation we have found in KSHV (Amelio et al, 2006; Chen et al, 2007). The position of CTCF downstream of the LAT promoter is thought to prevent inappropriate expression of the HSV lytic activator gene ICP0, which is further downstream from the LAT promoter than CBS. We suspect that the CTCF sites at the KSHV CBS serve a similar function, although the complexity of the nested regulatory elements upstream of ORF73 suggests that CTCF may have architectural functions distinct from a physical barrier to signal transmission along linear DNA.
Deregulation of vGPCR (ORF74) during KSHV latency is thought to be a critical initiating event in KS pathogenesis (Hayward, 2003; Sodhi et al, 2004). vGPCR has potent angiogenic activity that can induce KS-like lesions in animal models (Bais et al, 1998; Montaner et al, 2003; Mutlu et al, 2007). vGPCR is typically expressed during lytic infection, but can also be detected in KS lesions and PEL biopsies (Cesarman et al, 1996). Co-infection of KSHV with HIV increases the risk and severity of KS, and studies have shown that HIV gene products, such as TAT, and HIV-induced inflammatory cytokines, including IL-8, increase vGPCR expression and signalling (Sodhi et al, 2004). Paradoxically, we found that deregulation of KSHV lytic gene expression by deletion of the CBS caused a cytostatic effect on transfected 293 cells. The inhibition of cell growth in 293 cells may be caused by deregulation of vGPCR in the context of other lytic genes, as well as the loss of appropriate cell cycle regulation of the latency transcripts, including LANA and vCyclin. Nevertheless, our studies raise the possibility that disruption of CTCF or cohesin binding at the KSHV CBS may contribute to the deregulation of vGPCR in KS and PEL.
The most striking finding in this study is the colocalization of cohesin subunits with the CTCF sites in the KSHV latency control region (Figure 1), and at the cellular CTCF sites in the c-myc promoter and H19/Igf2 imprinting control region (Figure 7). The colocalization was specific for cohesin subunits since the related SMC5 and SMC6 proteins did not colocalize with SMC1-binding site and CBSs (Supplementary Figure S11). Cohesins are thought to have a primary function in maintaining sister-chromatid cohesion, which is necessary for both proper chromosome segregation and DNA damage repair by homologous recombination (Nasmyth and Haering, 2005; Losada, 2007). At present, it is not known whether KSHV sister chromatids adhere like cellular chromosomes, but recent studies from the related gammaherpesvirus EBV suggest that replicated sister genomes do remain associated and that these sister genomes are actively segregated similar to cellular chromosomes (Kanda et al, 2007; Nanbo et al, 2007). It is possible that cohesin binding to the ORF73 control regions does contribute to sister genome attachment, and that loss of this function contributes to the failure to maintain stable KSHV episomes in the colony formation assays shown in Figure 4. It is also likely that sister-chromatid cohesion mediated by the cohesin complex contributes to chromatin organization and transcription regulation. The ring structure formed by cohesins can be regulated at different stages of the cell cycle and may play an active role in transcriptional regulation (Nasmyth and Schleiffer, 2004; Nasmyth and Haering, 2005; Dorsett, 2007). We found that cohesin subunits contribute to the transcription repression of the lytic cycle genes K14 and ORF74, and that sodium butyrate-induced viral reactivation leads to a loss of RAD21 binding prior to the loss of other cohesin subunits and CTCF. Depletion of RAD21 from KSHV-infected BCBL1 cells caused an increase in ORF74 expression, similar to the deletion of the CBS element from KSHV genomes (Figure 5). RAD21 was rapidly depleted from the CBS sites in KSHV and c-myc within 1 h of sodium butyrate treatment (Figure 6A, Supplementary Figure S9), suggesting that it functions as a primary regulator of CTCF-mediated transcription repression, and may be sensitive to histone deacetylase inhibitors. The dynamic association of cohesins with CBSs further supports their role in regulating gene expression, which has been shown in several systems including human developmental disorders associated with Cornelia de Lange syndrome (Gillis et al, 2004; Krantz et al, 2004; Dorsett, 2007). Establishment of sister-chromatid cohesion in S phase may contribute to gene expression, and may be important for the cell cycle represssion of c-myc and LANA transcription. Additionally, sister-chromatid cohesion may provide structural features that regulate DNA looping and long-distance interactions between transcription control elements.
The association of cohesin with CBSs may help to explain some of the multifunctional properties of CTCF. At the c-myc promoter, CTCF can block the upstream enhancer activation of the proximal promoters, but these sites are also important for optimal expression of c-myc (Ohlsson et al, 2001). CTCF sites in c-myc promoter are located at boundaries of histone modification and histone variants (Gombert et al, 2003; Farris et al, 2005; Barski et al, 2007). CTCF sites may also be linked to chromatin that is elevated in H3 K4 methylation, and H3 K4 methylation has been found most enriched at the transcription initiation sites of many genes (Barski et al, 2007). We found that CTCF bound in close proximity to the initiation site of the major latency transcript, and this pattern has also been observed for the HSV LAT and for EBV latency transcripts (Amelio et al, 2006; Chen et al, 2007; Day et al, 2007). Another study indicated that CTCF can interact with the large subunit of RNA polymerase II (Chernukhin et al, 2007). In both the c-myc promoter and the KSHV major latency transcript control region, RNA polymerase pausing and RNA splicing are known to occur, suggesting that CTCF may also regulate RNA polymerase functions in elongation or splice acceptor–donor selection. At other loci, such as the β-globin gene and H19/Igf2 imprinting loci, CTCF's function as an enhancer blocker has been correlated with changes in chromatin conformation (Kurukuti et al, 2006; Splinter et al, 2006). A remarkable finding was that CTCF mediated the interactions between repressor elements on two different chromosomes (Ling et al, 2006). It is not yet known which of these CTCF functions involve cohesins, and it is likely that CTCF has cohesin-independent binding sites and activities. Nevertheless, our findings do provide a new perspective on the role of cohesins at some CTCF sites important for stable gene expression programmes.
Materials and methods
Cell lines
KSHV-positive PEL cell lines BCBL1 and JSC1 were cultured at 37°C and 5% CO2 in RPMI (Gibco-BRL) and supplemented with 10% fetal bovine serum (FBS) and penicillin–streptomycin (50 U/ml). Sodium butyrate was added to BCBL1 cells at a final concentration of 3 mM and for a period of 1 or 24 h before collection. 293 (ATCC) and HeLa cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% FBS containing penicillin–streptomycin (50 U/ml). The 293-derived cell lines were all cultured identically to 293 cells except with the addition of 200 μg/ml hygromycin B for the selection of the wild type and recombinant viral genomes.
ChIP assay and KSHV genome array
Chromatin immunoprecipitation was performed according to the protocol by Upstate Biotechnology Inc., and as previously described (Stedman et al, 2004). For KSHV genome array, the entire DNA from one IP (106 cells per IP) was mixed with Power Syber Green PCR Master Mix and aliquoted completely into one 384-well plate for a total of 12 μl per well (Day et al, 2007). Quantification of precipitated DNA was determined using real-time PCR and the Absolute Quantification program (ABI 7900HT Fast Real-Time PCR System; Applied Biosystems). Primers were designed using Primer Express, version 2.0 (Applied Biosystems) in conjunction with a batch algorithm that generated 379 real-time primer sets across the KSHV genome, yielding amplification regions spaced, on average, every 370 bp. PCR data were normalized to input values that were quantified in parallel for each experiment. IPs were performed in triplicate or quadruplicate for each antibody and the PCR reactions were repeated at least three times. The mean for each identical set of IPs was determined and values that were greater or less than the mean by more than 10% were excluded. ChIP assay methods for the H19/Igf2 imprinting locus have been described previously (Engel et al, 2006) and are described in detail in Supplementary data.
Antibodies
Chromatin immunoprecipitation assays included polyclonal antibodies to IgG (Santa Cruz Biotechnology), ORC2 (PharMingen), and CTCF (Upstate Biotechnology Inc.). A rat monoclonal antibody for ORF73 (m-LANA) was purchased from Advanced Biotechnology Inc., while a polyclonal antibody for ORF73 (p-LANA) was developed by our lab. Polyclonal antibodies for SMC1, SMC3, and RAD21 were all purchased from Bethyl Inc.
DNA affinity purification assay
The DNA affinity assay was performed as previously described (Stedman et al, 2004). Biotin-labeled DNA was generated with biotin-conjugated oligonucleotides and PCR amplification. Purified genomic KSHV DNA was used as templates for PCR to generate biotin-labeled DNA for fragments upstream of ORF73 (nucleotides 126 841–127 840; 127 333–127 484; 127 416–127 619; 127 551–127 710; and 127 690–127 840). KSHV coordinates correspond to the genome sequence submitted by Russo et al (1996). Control 1000-bp fragment was derived from BKSII DNA. Biotinylated DNA was coupled to streptavidin-coupled magnetic beads (Dynal) and incubated with BCBL1 nuclear extracts. Bound proteins were eluted from the beads with sodium dodecyl sulphate and subsequently analysed by western blotting.
Recombineering of KSHV BACs
Construction of recombinant KSHV BACs was performed as described previously (Copeland et al, 2001; Chau et al, 2006). The KSHV BAC36 clone was used for recombination-mediated genetic engineering (Zhou et al, 2002). KSHV nucleotides 127 330–127 550 were deleted and substituted with a LoxP site to construct MT1 and MT2. BAC36ΔCTCF constructs MT1 and MT2 were verified by PCR and EcoRI digestion of purified DNA, and then by direct DNA sequencing of the LoxP insertion region. The mutants were further analysed by Southern blot with a digoxygenin-labeled pL452 probe to demonstrate that the recombination was correctly targeted to a single region upstream of ORF73. MT1 and MT2 are separate but identical clonal isolates of BAC36ΔCTCF.
Colony formation, plasmid stability, and growth assay
BAC 36 WT, MT1, or MT2 DNA was purified from E. coli. and used to transfect four separate plates of 293 cells (30 μg per plate). At 48 h post-transfection, the GFP-positive cells from each condition were cell sorted using FACS. After sorting, four 10 cm plates for each condition were plated with 100 000 GFP-positive cells. After 24 h, hygromycin (200 μg/ml final concentration) was added to select for cells containing BAC constructs. The media were replaced every 3–4 days. After 10–15 days of selection, the plates were photographed with a 35 mm camera and the colony number was quantitated using Image-Pro Plus software. For growth assays, BAC DNA from WT, MT1, and MT2 was used to transfect three 6 cm plates containing 293 cells for each BAC construct. After 48 h, the cells were expanded so that there were 21 plates for each BAC construct and hygromycin (200 μg/ml final concentration) was added for selection of BAC-containing 293 cells. On day 4 post-transfection, the cells from only three plates from each BAC construct condition were collected, leaving the remaining 18 plates for each condition to grow with hygromycin selection. The collected cells were washed twice in PBS buffer and then resuspended in a final volume of 200 μl of PBS. Then, 20 μl of fluorosphere beads was added to the cells and mixed vigourously. Total cell number was calculated using FACS analysis. The procedure was repeated identically for day 8, day 12, and day 16 post-transfection. The gate used to sort the cells was reused for each condition and for each time point. Values were reported as gross number of selected cells.
Additional Materials and methods are described in Supplementary data.
Supplementary Material
Supplementary data
Acknowledgments
We thank S-J Gao for the generous gift of the BAC36 clone of recombinant KSHV and John Rux for the design of the KSHV genome array. We are grateful to the Wistar Cancer Center Core Facilities for Flow Cytometry, Genomics, Bioinformatics, and Microscopy. This study was funded by a grant from the NIH (CA117830) to PML, and GM51279 and HD42026 to MSB, and funds received from the Pennsylvania Department of Health.
References
- Amelio AL, McAnany PK, Bloom DC (2006) A chromatin insulator-like element in the herpes simplex virus type 1 latency-associated transcript region binds CCCTC-binding factor and displays enhancer-blocking and silencing activities. J Virol 80: 2358–2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bais C, Santomasso B, Coso O, Arvanitakis L, Raaka EG, Gutkind JS, Asch AS, Cesarman E, Gerhengorn MC, Mesri EA (1998) G-protein coupled receptor of Kaposi's sarcoma-associated herpesvirus is a viral oncogene and angiogenesis activator. Nature 391: 86–89 [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K (2007) High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Bell AC, Felsenfeld G (2000) Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405: 482–485 [DOI] [PubMed] [Google Scholar]
- Blat Y, Kleckner N (1999) Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell 98: 249–259 [DOI] [PubMed] [Google Scholar]
- Boshoff C, Schulz TF, Kennedy MM, Graham AK, Fisher C, Thomas A, McGee JO, Weiss RA, O'Leary JJ (1995) Kaposi's sarcoma-asociated herpesvirus infects endothelial and spindle cells. Nat Med 1: 1274–1278 [DOI] [PubMed] [Google Scholar]
- Burysek L, Pitha PM (2001) Latently expressed human herpesvirus 8-encoded interferon regulatory factor 2 inhibits double-stranded RNA-activated protein kinase. J Virol 75: 2345–2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Cullen BR (2006) Transcriptional origin of Kaposi's sarcoma-associated herpesvirus microRNAs. J Virol 80: 2234–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelson M, Corces VG (2004) Boundary elements and nuclear organization. Biol Cell 96: 617–629 [DOI] [PubMed] [Google Scholar]
- Cesarman E (2002) The role of Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) in lymphoproliferative diseases. Recent Results Cancer Res 159: 27–37 [DOI] [PubMed] [Google Scholar]
- Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM (1995) Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med 332: 1186–1191 [DOI] [PubMed] [Google Scholar]
- Cesarman E, Nador RG, Bai F, Bohenzky RA, Russo JJ, Moore PS, Chang Y, Knowles DM (1996) Kaposi's sarcoma-associated herpesvirus contains G protein-coupled receptor and cyclin D homologs which are expressed in Kaposi's sarcoma and malignant lymphoma. J Virol 70: 8218–8223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS (1994) Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266: 1865–1869 [DOI] [PubMed] [Google Scholar]
- Chau CM, Lieberman PM (2004) Dynamic chromatin boundaries delineate a latency control region of Epstein–Barr virus. J Virol 78: 12308–12319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau CM, Zhang XY, McMahon SB, Lieberman PM (2006) Regulation of Epstein–Barr virus latency type by the chromatin boundary factor CTCF. J Virol 80: 5723–5732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Lin L, Smith S, Huang J, Berger SL, Zhou J (2007) CTCF-dependent chromatin boundary element between the latency-associated transcript and ICP0 promoters in the herpes simplex virus type 1 genome. J Virol 81: 5192–5201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernukhin I, Shamsuddin S, Kang SY, Bergstrom R, Kwon YW, Yu W, Whitehead J, Mukhopadhyay R, Docquier F, Farrar D, Morrison I, Vigneron M, Wu SY, Chiang CM, Loukinov D, Lobanenkov V, Ohlsson R, Klenova E (2007) CTCF interacts with and recruits the largest subunit of RNA polymerase II to CTCF target sites genome-wide. Mol Cell Biol 27: 1631–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland NG, Jenkins NA, Court DL (2001) Recombineering: a powerful new tool for mouse functional genomics. Nat Rev Genet 2: 769–779 [DOI] [PubMed] [Google Scholar]
- Cotter MA II, Robertson ES (2002) Molecular biology of Kaposi's sarcoma-associated herpesvirus. Front Biosci 7: d358–d375 [DOI] [PubMed] [Google Scholar]
- Day L, Chau CM, Nebozhyn M, Rennenkamp AJ, Showe M, Lieberman PM (2007) Chromatin profiling of Epstein–Barr virus latency control region. J Virol 81: 6389–6401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer D, Lagunoff M, Renne R, Staskus K, Haase A, Ganem D (1998) A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J Virol 72: 8309–8315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D (2004) Adherin: key to the cohesin ring and Cornelia de Lange syndrome. Curr Biol 14: R834–R836 [DOI] [PubMed] [Google Scholar]
- Dorsett D (2007) Roles of the sister chromatid cohesion apparatus in gene expression, development, and human syndromes. Chromosoma 116: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel N, Thorvaldsen JL, Bartolomei MS (2006) CTCF binding sites promote transcription initiation and prevent DNA methylation on the maternal allele at the imprinted H19/Igf2 locus. Hum Mol Genet 15: 2945–2954 [DOI] [PubMed] [Google Scholar]
- Farris SD, Rubio ED, Moon JJ, Gombert WM, Nelson BH, Krumm A (2005) Transcription-induced chromatin remodeling at the c-myc gene involves the local exchange of histone H2A.Z. J Biol Chem 280: 25298–25303 [DOI] [PubMed] [Google Scholar]
- Fedoriw AM, Stein P, Svoboda P, Schultz RM, Bartolomei MS (2004) Transgenic RNAi reveals essential function for CTCF in H19 gene imprinting. Science 303: 238–240 [DOI] [PubMed] [Google Scholar]
- Gaszner M, Felsenfeld G (2006) Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet 7: 703–713 [DOI] [PubMed] [Google Scholar]
- Gelmann EP, Psallidopoulos MC, Papas TS, Dalla-Favera R (1983) Identification of reciprocal translocation sites within the c-myc oncogene and immunoglobulin mu locus in a Burkitt lymphoma. Nature 306: 799–803 [DOI] [PubMed] [Google Scholar]
- Gerasimova TI, Corces VG (2001) Chromatin insulators and boundaries: effects on transcription and nuclear organization. Annu Rev Genet 35: 193–208 [DOI] [PubMed] [Google Scholar]
- Gerbi SA, Strezoska Z, Waggener JM (2002) Initiation of DNA replication in multicellular eukaryotes. J Struct Biol 140: 17–30 [DOI] [PubMed] [Google Scholar]
- Gillis LA, McCallum J, Kaur M, DeScipio C, Yaeger D, Mariani A, Kline AD, Li HH, Devoto M, Jackson LG, Krantz ID (2004) NIPBL mutational analysis in 120 individuals with Cornelia de Lange syndrome and evaluation of genotype–phenotype correlations. Am J Hum Genet 75: 610–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn EF, Megee PC, Yu HG, Mistrot C, Unal E, Koshland DE, DeRisi JL, Gerton JL (2004) Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol 2: E259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gombert WM, Farris SD, Rubio ED, Morey-Rosler KM, Schubach WH, Krumm A (2003) The c-myc insulator element and matrix attachment regions define the c-myc chromosomal domain. Mol Cell Biol 23: 9338–9348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo HG, Sadowska M, Reid W, Tschachler E, Hayward G, Reitz M (2003) Kaposi's sarcoma-like tumors in a human herpesvirus 8 ORF74 transgenic mouse. J Virol 77: 2631–2639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM (2000) CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405: 486–489 [DOI] [PubMed] [Google Scholar]
- Hayward GS (2003) Initiation of angiogenic Kaposi's sarcoma lesions. Cancer Cell 3: 1–3 [DOI] [PubMed] [Google Scholar]
- Hirano T (2006) At the heart of the chromosome: SMC proteins in action. Nat Rev Mol Cell Biol 7: 311–322 [DOI] [PubMed] [Google Scholar]
- Ishihara K, Oshimura M, Nakao M (2006) CTCF-dependent chromatin insulator is linked to epigenetic remodeling. Mol Cell 23: 733–742 [DOI] [PubMed] [Google Scholar]
- Jeong J, Papin J, Dittmer D (2001) Differential regulation of the overlapping Kaposi's sarcoma-associated herpesvirus vGCR (orf74) and LANA (orf73) promoters. J Virol 75: 1798–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda T, Kamiya M, Maruo S, Iwakiri D, Takada K (2007) Symmetrical localization of extrachromosomally replicating viral genomes on sister chromatids. J Cell Sci 120: 1529–1539 [DOI] [PubMed] [Google Scholar]
- Kanduri C, Pant V, Loukinov D, Pugacheva E, Qi CF, Wolffe A, Ohlsson R, Lobanenkov VV (2000) Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr Biol 10: 853–856 [DOI] [PubMed] [Google Scholar]
- Kedes DH, Lagunoff M, Renne R, Ganem D (1997) Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi's sarcoma-associated herpesvirus. J Clin Invest 100: 2606–2610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B (2007) Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128: 1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu T, Ballestas ME, Barbera AJ, Kaye KM (2002) The KSHV latency-associated nuclear antigen: a multifunctional protein. Front Biosci 7: d726–d730 [DOI] [PubMed] [Google Scholar]
- Krantz ID, McCallum J, DeScipio C, Kaur M, Gillis LA, Yaeger D, Jukofsky L, Wasserman N, Bottani A, Morris CA, Nowaczyk MJ, Toriello H, Bamshad MJ, Carey JC, Rappaport E, Kawauchi S, Lander AD, Calof AL, Li HH, Devoto M et al. (2004) Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat Genet 36: 631–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, Lobanenkov V, Reik W, Ohlsson R (2006) CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci USA 103: 10684–10689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Ganem D (2004) RBP-J (CSL) is essential for activation of the K14/vGPCR promoter of Kaposi's sarcoma-associated herpesvirus by the lytic switch protein RTA. J Virol 78: 6818–6826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Chen Q, Lin L, Zhou J (2004) The promoter targeting sequence mediates epigenetically heritable transcription memory. Genes Dev 18: 2639–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR (2006) CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science 312: 269–272 [DOI] [PubMed] [Google Scholar]
- Losada A (2007) Cohesin regulation: fashionable ways to wear a ring. Chromosoma 116: 321–329 [DOI] [PubMed] [Google Scholar]
- Montaner S, Sodhi A, Molinolo A, Bugge TH, Sawai ET, He Y, Li Y, Ray PE, Gutkind JS (2003) Endothelial infection with KSHV genes in vivo reveals that vGPCR initiates Kaposi's sarcomagenesis and can promote the tumorigenic potential of viral latent genes. Cancer Cell 3: 23–36 [DOI] [PubMed] [Google Scholar]
- Muralidhar S, Pumfery AM, Hassani M, Sadaie MR, Kishishita M, Brady JN, Doniger J, Medveczky P, Rosenthal LJ (1998) Identification of kaposin (open reading frame K12) as a human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus) transforming gene. J Virol 72: 4980–4988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutlu AD, Cavallin LE, Vincent L, Chiozzini C, Eroles P, Duran EM, Asgari Z, Hooper AT, La Perle KM, Hilsher C, Gao SJ, Dittmer DP, Rafii S, Mesri EA (2007) In vivo restricted and reversible malignancy induced by human herpesvirus-8 KSHV: a cell and animal model of virally induced Kaposi's sarcoma. Cancer Cell 11: 245–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nador RG, Milligan LL, Flore O, Wang X, Arvanitakis L, Knowles DM, Cesarman E (2001) Expression of Kaposi's sarcoma-associated herpesvirus G protein-coupled receptor monocistronic and bicistronic transcripts in primary effusion lymphomas. Virology 287: 62–70 [DOI] [PubMed] [Google Scholar]
- Nanbo A, Sugden A, Sugden B (2007) The coupling of synthesis and partitioning of EBV's plasmid replicon is revealed in live cells. EMBO J 26: 4252–4262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH (2005) The structure and function of SMC and kleisin complexes. Annu Rev Biochem 74: 595–648 [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Schleiffer A (2004) From a single double helix to paired double helices and back. Philos Trans R Soc Lond B Biol Sci 359: 99–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson R, Renkawitz R, Lobanenkov V (2001) CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet 17: 520–527 [DOI] [PubMed] [Google Scholar]
- Pearce M, Matsumura S, Wilson AC (2005) Transcripts encoding K12, v-FLIP, v-cyclin, and the microRNA cluster of Kaposi's sarcoma-associated herpesvirus originate from a common promoter. J Virol 79: 14457–14464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainbow L, Platt GM, Simpson GR, Sarid R, Gao SJ, Stoiber H, Herrington CS, Moore PS, Schulz TF (1997) The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J Virol 71: 5915–5921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo JJ, Bohenzky RA, Chien MC, Chen J, Yan M, Maddalena D, Parry JP, Peruzzi D, Edelman IS, Chang Y, Moore PS (1996) Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc Natl Acad Sci USA 93: 14862–14867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler R, Wu L, Forghani B, Renne R, Zhong W, Herndier B, Ganem D (1999) A complex translational program generates multiple novel proteins from the latently expressed kaposin (K12) locus of Kaposi's sarcoma-associated herpesvirus. J Virol 73: 5722–5730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samols MA, Hu J, Skalsky RL, Renne R (2005) Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi's sarcoma-associated herpesvirus. J Virol 79: 9301–9305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarid R, Wiezorek JS, Moore PS, Chang Y (1999) Characterization and cell cycle regulation of the major Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) latent genes and their promoter. J Virol 73: 1438–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodhi A, Montaner S, Gutkind JS (2004) Does dysregulated expression of a deregulated viral GPCR trigger Kaposi's sarcomagenesis? FASEB J 18: 422–427 [DOI] [PubMed] [Google Scholar]
- Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d'Agay MF, Clauvel JP, Raphael M, Degos L, Sigaux F (1995) Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86: 1276–1280 [PubMed] [Google Scholar]
- Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, Galjart N, de Laat W (2006) CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev 20: 2349–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedman W, Deng Z, Lu F, Lieberman PM (2004) ORC, MCM, and histone hyperacetylation at the Kaposi's sarcoma-associated herpesvirus latent replication origin. J Virol 78: 12566–12575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo PE, Tang SH, Silva FJ, Tsark WM, Mann JR (2004) Role of CTCF binding sites in the Igf2/H19 imprinting control region. Mol Cell Biol 24: 4791–4800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot SJ, Weiss RA, Kellam P, Boshoff C (1999) Transcriptional analysis of human herpesvirus-8 open reading frames 71, 72, 73, K14, and 74 in a primary effusion lymphoma cell line. Virology 257: 84–94 [DOI] [PubMed] [Google Scholar]
- Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G (2004) CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol Cell 13: 291–298 [DOI] [PubMed] [Google Scholar]
- Zhou FC, Zhang YJ, Deng JH, Wang XP, Pan HY, Hettler E, Gao SJ (2002) Efficient infection by a recombinant Kaposi's sarcoma-associated herpesvirus cloned in a bacterial artificial chromosome: application for genetic analysis. J Virol 76: 6185–6196 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data