Abstract
The most abundant base-substitution mutation resulting from oxidative damage to DNA is the GC to AT transition mutation. 5-hydroxyuracil (5-OHU), produced by the oxidative deamination of cystosine, has been established as the major chemical precursor for this most abundant transition mutation. Results from NMR spectroscopy and UV melting experiments show that 5-OHU would form the most stable pair with G, and the least stable pair with C. The hydroxyl group in the 5th position of the 5-OHU residue may play a role in increasing the stability of the 5-OHU:G pair over the normal Watson-Crick pair, the 5-OHU:A. The 5-OHU:C base pair would be least stable, and would destabilize the base-stacking in the duplex. Our results explain why certain DNA polymerases preferentially incorporate G opposite to 5-OHU over A and why C doesn’t get incorporated against 5-OHU during DNA replication in vivo.
Keywords: DNA repair, oxidative DNA damage, base-pair mismatch, hydroxy uracil
Introduction
Oxidative DNA damage results from exposure to both endogenous and exogenous reactive oxygen species (ROS) and has been implicated in a variety of diseases, including cancer, rheumatoid arthritis, and aging [1,2]. ROS are formed endogenously as a by-product pf oxidative metabolism and exogenously by various environmental factors including chemicals and ionizing radiations. Oxidative DNA damage occurs when ROS interacts and covalently damage the heterocyclic bases in DNA duplexes and in cellular dNTP pools. The most frequent base substitution mutation observed in aerobic organisms and the most abundant genetic change resulting from oxidative DNA damage is the GC to AT transition mutation [3]. Many point mutations in oncogenes and tumor suppressor genes have been identified in cancers, among which GC to AT transitions are the most common mutations [4]. Several recent studies have established that the oxidized cytosine product 5-hydroxy uracil (5-OHU) as the major chemical precursor to the GC to AT transition mutations [5].
Oxidation of pyrimidine residues frequently modifies the 5th position of the base. 5-OHU is generated by the oxidative deamination of cytosine. Oxidation of cytosine gives rise to Cytosine glycol (Cg), an unstable DNA base that can either dehydrate, deaminate, or undergo both reactions to form 5-hydroxy cytosine, (5-OHC), Uracil glycol, (Ug), and 5-hydroxyuracil (5-OHU), respectively [6,7] (Fig.1). Among the oxidized cytosine products, 5-OHC and 5-OHU have been reported to be the major stable products [7]. The steady state levels of 5-OHC and 5-OHU have been observed in significant quantities in untreated DNA and the levels increase substantially when the DNA is exposed to ionizing radiation or oxidizing agents. Among the oxidized cytosine products, Ug and 5-OHU showed the highest mutation frequencies [7].
Figure 1.
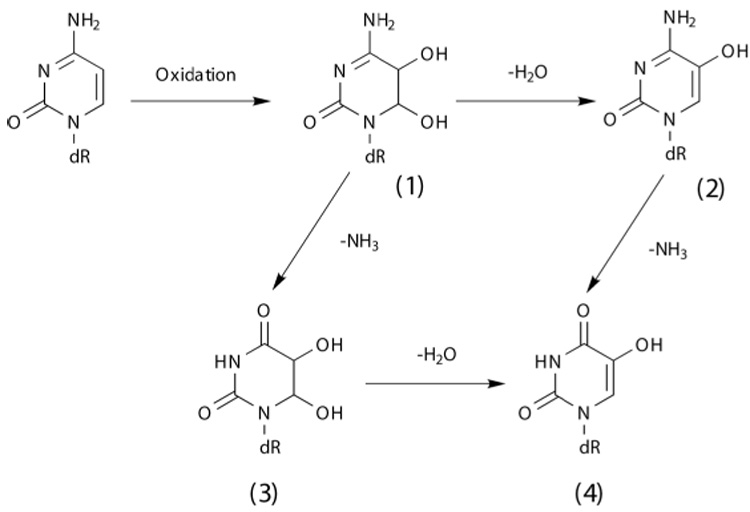
Scheme showing the formation of 5-hydroxyuracil (5-OHU) through the oxidative deamination of cytosine.
The chemical structure of 5-OHU shows that it can undergo keto-enol tautomerism through a 1,3 hydrogen shift mechanism [8]. However, the enol form (at C-5 position) is favored because the enolic double bond is in conjugation, and this form is capable of forming an intra molecular hydrogen bond. UV absorption studies also showed that 5-OHC and 5-OHU retain the enol rather than the keto configuration, at the 5th position [9]. The chemical structure of 5-OHU, shows that it would form a normal Watson-Crick-like base pair with adenine. However, base pairing with other bases is possible.
During DNA replication, different DNA polymerases incorporate A, G and T opposite 5-OHU. Both Ug and 5-OHU are readily bypassed by E.coli DNA polymerase I Klenow fragment during DNA replication in vitro [10]. It was further shown that 5-OHU triphosphate is a more efficient substrate than the 8-OG triphosphate for the E.coli Klenow DNA polymerase [11]. When 5-OHU, derived from C, pair with A during DNA replication, it would result in GC to AT transition mutation. The mammalian bypass polymerase Poli preferentially incorporates T (and to a lesser extent G) opposite to 5-OHU, over the Watson-Crick base, A [12]. The 5-OHU:T pair would result in a GC to TA transversion mutation that is also observed frequently in vivo [13]. However, the major source for the GC to TA transversions is believed to be 8-oxoguanine. While the 5-OHU:T mispairs were extended poorly, the 5-OHU:G mispairs were extended with equal or greater efficiency than the correctly paired (5-OHU:A) primer termini by Poli.
Previously, we reported results from theoretical studies based on ab-initio calculations on base pairs formed between 5-OHU and the four DNA bases (A, C, G or T) [14]. In this paper, we report results from experimental methods using NMR, CD and UV spectroscopies, on the stabilities of the base pairs formed between 5-OHU and other DNA bases. The DNA duplexes were made from the self-complementary 12-mer sequence, d(CGCXAATTOH-UGCG, where X=A, C, G or T)
Materials and methods
Oligonucleotides
The 12-mer oligonucleotides, d(CGCXAATTOH-UGCG, where X=A, C, G or T) were synthesized by the standard phosphoramidite method on an ABI Expedite model 8900 DNA synthesizer using 5-hydroxy-dU-CE phosphoramidite (Glen Research) with off-line coupling of the reagent. The 5’-dimethoxytrityl(DMT)-oligonucleotides were cleaved from the solid support and the base protecting groups were removed using concentrated NH4OH containing 0.25M 2-mercaptoethanol (16 hours at 55°C). The full-length 5’-DMT-protected oligonucleotides were separated from failure sequences using a C-18 Sep-Pak column (Waters Corp.) followed by removal of the 5’DMTgroup with 1% aqueous TFA and elution of the product with 20% acetonitrile. The oligonucleotides were further purified by reverse-phase HPLC using a Waters Delta-Pak C-18 column and gradient elution (0–20% acetonitrile, 20 minutes in 0.1M TEAA buffer pH 7.2). The products were then dialyzed against 10 mM NaCl and once again in distilled water before final lyophilization. Purities of the oligonucleotides were tested by gel electrophoresis and the molecular weights were confirmed by mass spectrometry.
NMR Spectroscopy
Samples for NMR experiments were made by dissolving the oligonucleotides in buffer containing 20 mM sodium phosphate (pH 7.2), 0.02 mM EDTA, and 100 mM NaCl. The final concentrations of DNA in the samples, calculated from UV absorbance measurements, were 1.8 to 2.6 mM. All proton NMR experiments were done at 750 MHz on a Varian UnityPlus instrument equipped with pulsed field gradients. One-dimensional proton NMR spectra were acquired as a function of temperature, using a 15000 Hz spectral width. Data sets were collected with 128 scans using 4096 data points. Water suppression was achieved by a combination of a selective 270°-composite pulse and gradient pulses [15]. Proton signals were referenced to DSS at 0.0 ppm.
UV melting experiments
Melting curves were recorded at 260 nm using a Cary 1-Bio spectrophotometer (Varian Associates) equipped with a temperature controller. Samples for the melting experiments were prepared in a buffer containing 100 mM NaCl, 10 mM sodium phosphate (pH 7.2) and 0.02 mM EDTA. The concentrations of the duplexes were ~1 µM. Absorbance at 260 nm was monitored every 0.5°C, from 20°C to 95°C. Heating rate was set at 1°C per minute. Samples were allowed to equilibrate at the starting temperature until the absorbance remained constant for at least 10 minutes. Quartz cuvettes were sealed to prevent sample evaporation. Three scans were performed for each duplex. Melting temperatures were calculated from the sigmoid melting curves using a two-state model that assumes a simple equilibrium between the duplex and single strand forms [16]. Percentage hyperchromicity is defined as (Ass-Ads)/Ass, where Ass is the absorbance of the single-strand state and Ads is the absorbance of the double-stranded state [17].
CD spectroscopy
Circular dichroism spectra were recorded on an AVIV model 215 spectropolarimeter equipped with a temperature controller. Spectra were recorded from 220 nm to 300 nm at a rate of 50 nm/min. Data points were collected every 2 nm with a response time of 2s. Three scans were collected for each sample. Duplex DNA samples (400 µl) were prepared in the same buffer used for NMR experiments, to yield concentrations of ~50 µM. A 1mm path length quartz cell was used and the sample temperature was maintained at 25°C.
Ab initio calculations
As described previously [14] all ab-initio calculations were performed at the DFT (B3LYP) level on a 12-node Sun64-SVR-4 computer with Gaussian98, using the 6-311G (d,p) basis set. Calculations were performed using trimmed nucleotides where the N1 (for pyrimidines) or the N9 (for purines) nitrogen atoms were capped with a methyl group in place of the C1’ carbon atom. Our results showed that a constant interaction energy change of −0.2 kcal mol-1 between the full nucleotides and trimmed nucleotides. Therefore, calculations were performed on trimmed nucleotides that significantly lowered the need for computational resources.
Results
NMR spectroscopy of DNA duplexes containing 5-OHU
Proton NMR spectra of the DNA duplexes in H2O showed sharp, downfield shifted signals corresponding to the imino protons of the base residues, for all four DNA duplexes. The imino proton region of the duplexes, obtained at 5°C intervals, are shown in Figure 2. Although UV-absorbance monitored thermal melting curves provide valuable information about the stability of the duplex DNA, NMR spectra of imino regions give information about the melting of individual base pairs. In the 5-OHU:C duplex, the 5-OHU imino proton signal appeared as a low intensity peak only at 10°C. All imino signals disappeared at 30°C. For the 5-OHU:G and 5-OHU:T duplexes, the imino protons resonated at a wider spectral range than those in the 5-OHU:A and 5-OHU:C duplexes. In all four duplexes, the signal corresponding to the G12 imino proton was not observed, even at 10°C, possibly due to end-fraying. Two-dimensional NOESY spectra collected with 250 ms mixing time, showed the sequential connectivity of the imino protons through the center of the DNA duplex (data not shown). The imino-imino and imino-amino sequential NOE cross peaks showed that all the bases are involved in hydrogen bonding and are paired in all four duplexes. NOE data also showed that the bases are stacked upon each other throughout the duplex in all four duplexes.
Figure 2.
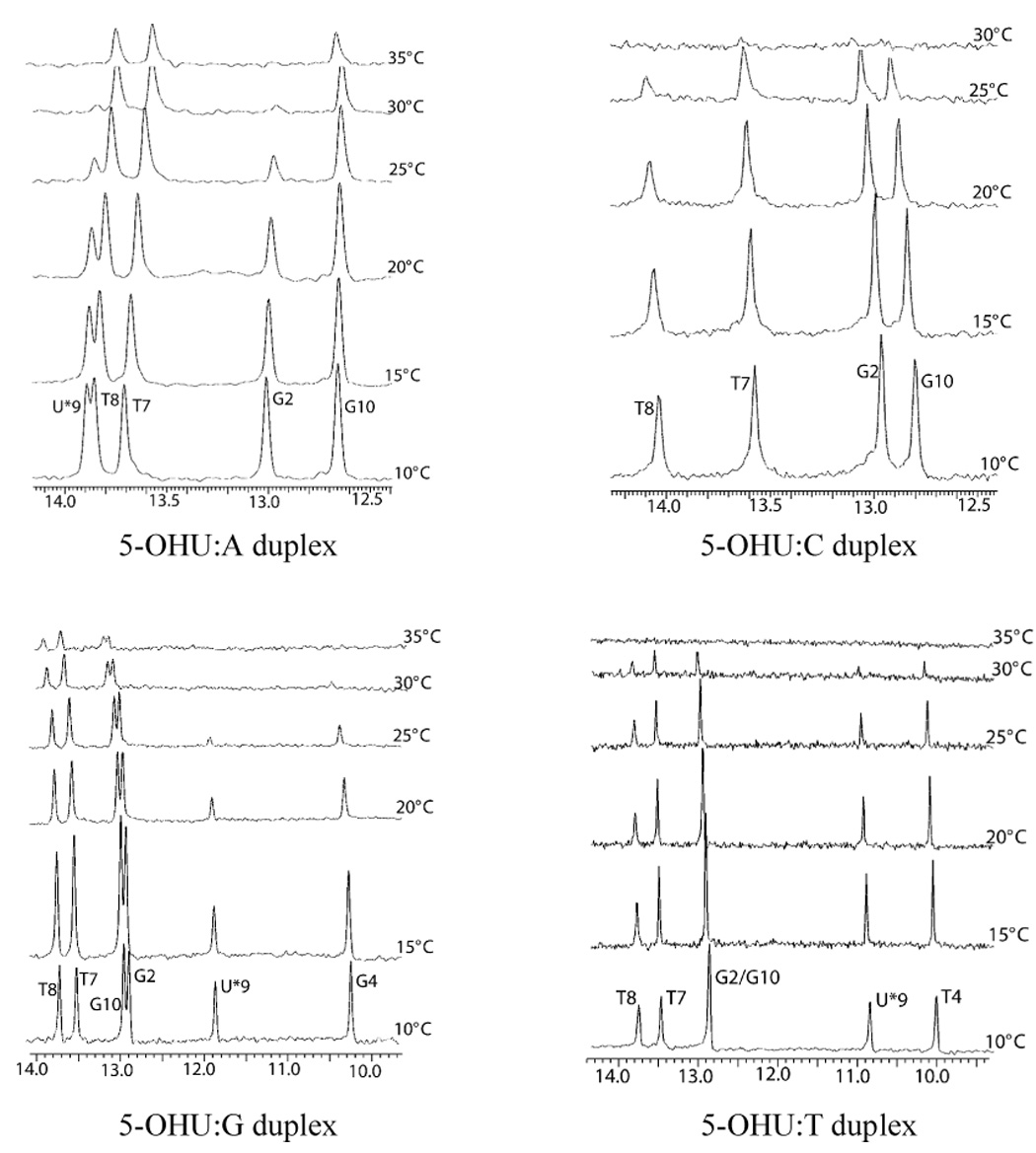
The imino proton region of the NMR spectra of the four duplexes, as a function of temperature. The imino proton signal corresponding to the 5-OHU is marked with U*9. (A) 5-OHU:A duplex (B) 5-OHU:C duplex (C) 5-OHU:G duplex and (D) 5-OHU:T duplex.
Melting temperatures derived from UV absorbance curves
Melting temperatures of the DNA duplexes containing 5-OHU:X (where X= A, C, G, or T), determined from the UV-monitored thermal melting curves obtained under identical conditions, are listed in Table 1. Hyperchromicity, the increase in absorbance during the denaturation of the duplex, results from the nearest neighbor interactions and is a measure of base-stacking in the duplex. The melting profiles (Supplementary Figure 1) for the four DNA duplexes showed that the %hyperchromicity differ significantly among the four duplexes. The 5-OHU:A and 5-OHU:G duplexes showed higher %hyperchromicity, 14 and 16%, respectively, while the 5-OHU:T duplex showed a 9% increase. In general, pyrimidines have lower hyperchromicity than purines. The 5-OHU:C duplex showed only a 2% hyperchromicity, indicating that the bases are stacked poorly in the 5-OHU:C duplex compared to the other three duplexes. The low melting temperature and the poor %hyperchromicity observed for the 5-OHU:C duplex showed that the 5-OHU forms the least stable pair with the C residue and that the 5-OHU:C pair stacks poorly in the duplex. Both 5-OHU:A and 5-OHU:G melting profiles followed a sigmoidal curve, consistent with the two-state model. The 5-OHU:C melting curve was linear.
Table 1.
Melting temperatures and hyperchromicity determined by UV absorbance for dodecamer DNA duplexes containing 5-OHU:X base pairs (where X= A, C, G, or T)
| Duplex | tm(°C) | %hyperchromicity |
|---|---|---|
| 5-OHU:A | 50.3 | 16 |
| 5-OHU:C | 40.7 | 2 |
| 5-OHU:G | 53.7 | 14 |
| 5-OHU:T | 46.6 | 9 |
Circular Dichroism
The CD spectra of the oligonucleotides, showing the molar ellipticity (Θ) as a function of wavelength, are shown in Figure 3. Despite the quantitative differences, the CD spectra of all four DNA duplexes showed characteristic patterns of B-form geometry [18]. The 2D NOESY spectra confirmed the overall B-DNA structures for all 4 duplexes. Since the CD spectrum of a DNA duplex is a function of its nucleotide composition, sequence and conformation [18,19], the very similar nature of the four CD spectra show that the four duplexes adopt very similar geometries. Both the dip and the raise in molar ellipticities is significantly lower for the 5-OHU:T duplex. Two different hydrogen bonding patterns are possible for the 5-OHU:T base-pair, as shown in Figure 4. As the energy difference between these two conformations is only 0.7–0.9 kcal/mol [14], both conformations will be present in significant amounts in the sample.
Figure 3.
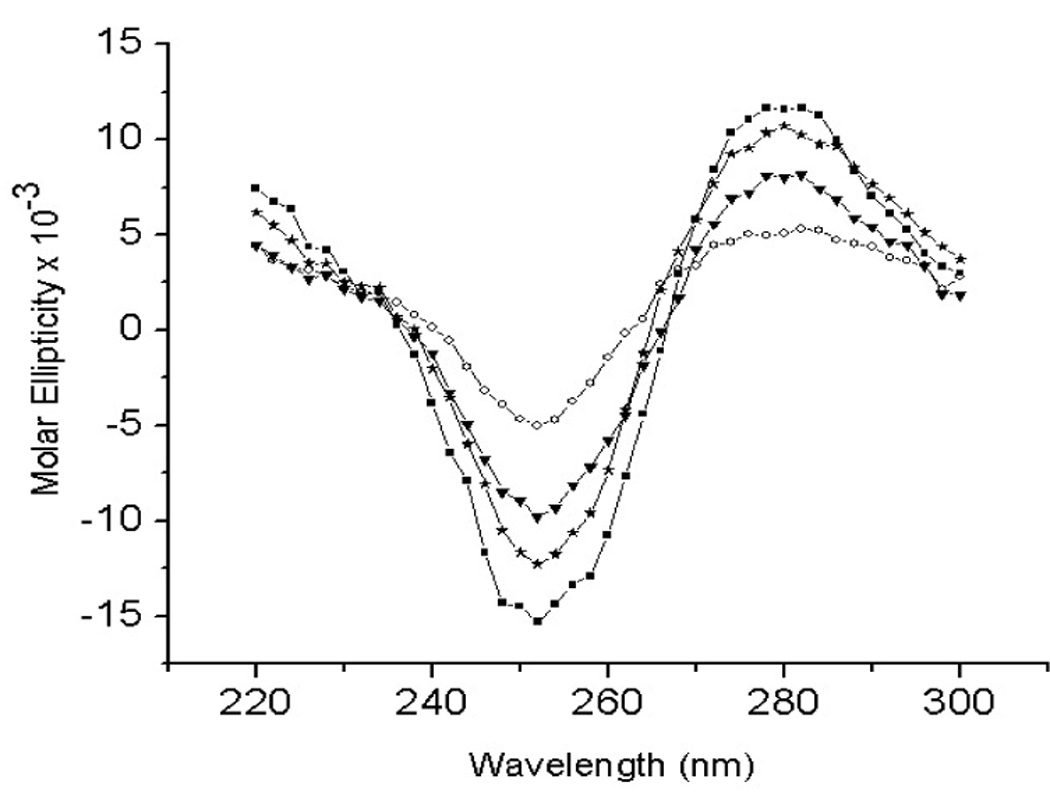
CD Spectra of the 5-OHU:X duplexes. (ν -: 5-OHU:A duplex; h- 5-OHU:C duplex; τ- 5-OHU:G duplex; o- 5-OHU:T duplex)
Figure 4.
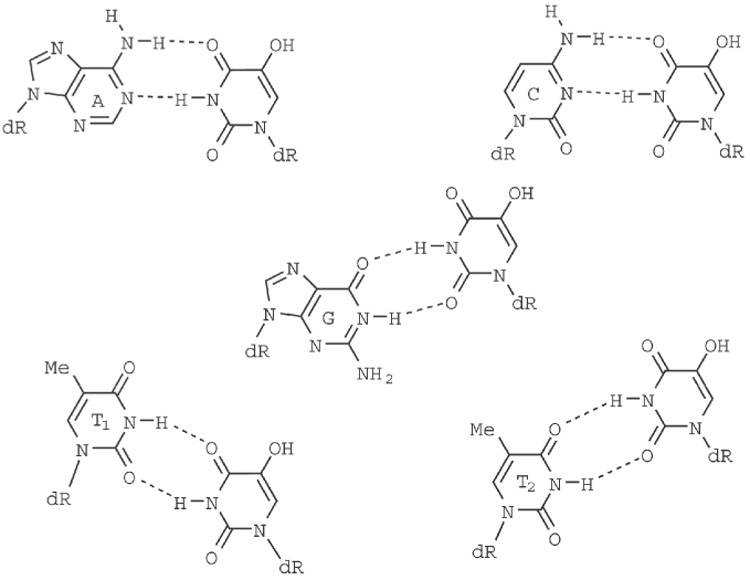
Schematic diagram showing the hydrogen bonding pattern formed in the base-pairs between 5-OHU and A, C, G and T. T can form two different types of base-pairing with 5-OHU.
Discussion
5-OHU has been established as the major chemical precursor for the GC to AT transition mutations, the most frequent base substitution mutation observed in aerobic organisms. Although 5-OHU can undergo keto-enol tatomerism, the enol form (at the 5th position) would be more stable. At the 2nd and 4th positions, the keto form is more stable and would be the predominant form, as the electron donating –OH group would make the imino hydrogen (at the N3 position) less acidic. The pKa of the hydroxyl group at the 5th position is determined to be 7.68 [20], and therefore, under physiological conditions, the ionized form of 5-OHU would also be present in cells.
At least four human enzymes are able to excise 5-OHU lesions from DNA, indicating that 5-OHU is a significant damage, both qualitatively and quantitatively, that it requires multiple enzymes for its repair. Often times, the DNA repair enzymes recognize the damaged bases by the local perturbations caused by the damaged bases. The efficiency of the DNA repair enzymes to bind and excise the damaged residue would depend on the extent of structural perturbations caused by the presence of the damage residue, which in turn would depend on the instability of the base-pair. Kino et al., [21] showed that the excision activity and the substrate binding affinity of the DNA repair enzyme XPC-HR23, involved in the repair of 5-formyl uracil lesions, increased with the increase in the instability of the base pair.
The geometry of the base pair and the energetics of the base-pair formation may play significant roles in determining which reside gets incorporated opposite to the 5-OHU residue, during DNA replication. Both G and A would form very stable base-pairs with 5-OHU without causing significant perturbation in the geometry of the DNA duplex. T would also form stable base-pair with 5-OHU. However, as T is a pyrimidine base, a T:5-OHU base-pair would perturb the DNA duplex. Pyrimidine-pyrimidine base-pairs are less frequently observed in double-stranded nucleic acids (e.g., double-stranded RNA regions). All our results presented here support that C is least likely to get incorporated against 5-OHU.
Both the ab-initio calculations performed on the isolated base pairs and the UV-monitored thermal melting curves obtained for the DNA duplexes, indicate that, among the four normal DNA bases, G forms the most stable pair with 5-OHU. This is biologically significant. Since 5-OHU is produced from the oxidative damage of cytosines, when 5-OHU is paired with G during DNA replication, the mutation will not be passed on to the subsequent generation. However, when 5-OHU is paired with A, it would produce a transition mutation in the next generation. Vaisman & Woodgate showed that when bypassing 5-OHU, DNA polymerase ι (pol ι) inserted G with a 26-fold preference over A [12]. Since G:5-OHU mispairs were extended with equal or greater efficiency than the A:5-OHU pairs, pol ι provides a mechanism to reduce the mutagenic potential of 5-OHU.
Experimental and theoretical studies show that 5-OHU:G pair is energetically more favorable than the 5-OHU:A pair. Although the 5-OHU:G base pair is 1.6 kcal/mol more stable than the 5-OHU:A base pair, the U:G base pair is only 0.5 kcal/mol more stable than the U:A base pair [14]. Thus, hydroxylating the uracil in a U:G base pair makes base pair formation 1.1 kcal/mol more stable. So what could account for the greater difference in the affinities for the 5-hydroxyuracil base pairs? The hydroxyl group in the 5th position may play a significant role in determining the stability of the base pair. In the 5-OHU:A pair, the carbonyl oxygen on the 4th position of 5-OHU residue is involved in hydrogen bonding with the amino hydrogen of the A. In the 5-OHU:G pair, the carbonyl oxygen in the 2nd position of 5-OHU is involved in hydrogen bonding with the imino hydrogen of G (Fig. 4). In 5-OHU, an intra-molecular hydrogen bond can be formed between the 4th carbonyl oxygen and the hydroxyl hydrogen of the – OH group in the 5th position, making the carbonyl oxygen at the 4th position a weaker hydrogen bond acceptor than the carbonyl oxygen at the 6th position. Therefore the hydrogen bond in the 5-OHU:A pair would be weaker than the hydrogen bond in the 5-OHU:G pair.
Presence of 5-OHU would challenge the integrity of the genome in two different ways. During DNA replication 5-OHU could be incorporated into the DNA from the cellular dNTP pool. 5-OHdUTP is an efficient substrate for Klenow polymerase [12]. Different DNA polymerases are known to incorporate G, A, and T residues opposite to the 5-OHU residue. However, C doesn’t get incorporated opposite to 5-OHU. Our results here show that C forms the least stable pair with 5-OHU. Ab-initio calculations showed that in the 5-OHU:C pair the bases are twisted by ± 40 degrees relative to each other in the 5-OHU:C pair. The twist in the base pair is expected, in order to reduce the electrostatic repulsion between the O2 atoms of the C and 5-OHU. In the duplex, this higher propeller twist would also affect the neighboring base-pairs, disrupting the stacking of bases in the duplex, resulting in decreased %hyperchromicity. In the 5-OHU:C duplex, disappearance of T8 imino signal at a lower temperature than for the T7 signal, further supports the theoretical calculations that the 5-OHU:C pair would disrupt neighboring bases in the DNA duplex. For all the other three duplexes, T8 and T7 imino signals disappeared at about the same temperature.
Supplementary Material
Acknowledgments
This research was supported by grants from NIAID (U01 AI054827), NIEHS (ES06676) and the Welch Foundation (H-1296). The authors thank Dr Richard Hodge and the NIEHS synthetic chemistry core facility at UTMB for the synthesis of oligonucleotides and Dr. Andrew Russo for assistance with the CD measurements.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ames BN, Shigenaga MK, Hagen TM. Oxidants, antioxidants, and the degenerative diseases of aging. Proc Natl. Acad. Sci. USA. 1993;90:7915–7922. doi: 10.1073/pnas.90.17.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ward JF. The complexity of DNA damage: relevance to biological consequences. Int. J Radiat. Biol. 1994;66:427–432. doi: 10.1080/09553009414551401. [DOI] [PubMed] [Google Scholar]
- 3.Schaaper RM, Danforth BN, Glickman BW. Mechanisms of spontaneous mutagenesis: an analysis of the spectrum of spontaneous mutation in the Escherichia coli lacI gene. J. Mol. Biol. 1986;189:273–284. doi: 10.1016/0022-2836(86)90509-7. [DOI] [PubMed] [Google Scholar]
- 4.Moraes EC, Keyse SM, Tyrrell RM. Mutagenesis by hydrogen peroxide treatment of mammalian cells: a molecular analysis. Carcinogenesis. 1990;11:283–293. doi: 10.1093/carcin/11.2.283. [DOI] [PubMed] [Google Scholar]
- 5.Feig DI, Sowers LC, Loeb LCLA. Reverse chemical mutagenesis: identification of the mutagenic lesions resulting from reactive oxygen species-mediated damage to DNA. Proc Natl. Acad. Sci. USA. 1994;91:6609–6613. doi: 10.1073/pnas.91.14.6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dizdaroglu M, Holwitt E, Hagan MP, Blakely WF. Formation of cytosine glycol and 5,6-dihydroxycytosine in deoxyribonucleic acid on treatment with osmium tetroxide. Biochem J. 1986;235:531–536. doi: 10.1042/bj2350531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kreutzer DA, Essigman JM. Oxidized, deaminated cytosines are a source of C→T transitions in vivo. Proc. Natl. Acad. Sci. USA. 1998;95:3578–3582. doi: 10.1073/pnas.95.7.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rappoport Z. The chemistry of functional groups. Wiley: Chichester; 1990. The chemistry of enols. [Google Scholar]
- 9.Moschel RC, Behrman EJ. Oxidation of nucleic acid bases by potassium peroxodisulfate in alkaline aqueous solution. J. Org. Chem. 1974;39:1983–1989. doi: 10.1021/jo00928a001. [DOI] [PubMed] [Google Scholar]
- 10.Purmal AA, Lampman GW, Bond JP, Hatahet Z, Wallace SS. Enzymatic processing of uracil, a major oxidative product of DNA cytosine. J. Biol. Chem. 1998;273:10026–10035. doi: 10.1074/jbc.273.16.10026. [DOI] [PubMed] [Google Scholar]
- 11.Prumal AA, Kow YW, Wallace SS. 5-Hydroxypyrimidine deoxynucleoside triphosphates are more efficiently incorporated into DNA by exonuclease-free Klenow fragment than 8-oxopurine deoxynucleoside triphosphates. Nucleic Acids Res. 1994;22:3930–3935. doi: 10.1093/nar/22.19.3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vaisman A, Woodgate R. Unique misinsertion specificity of poliota may decrease the mutagenic potential of deaminated cytosines. EMBO J. 2001;20:6520–6529. doi: 10.1093/emboj/20.22.6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng KC, Cahill DS, Kasai H, Nishimura S, Loeb LA. 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G→T and A→C substitutions. J Biol. Chem. 1992;267:166–172. [PubMed] [Google Scholar]
- 14.Volk DE, Thiviyanathan V, Somasunderam A, Gorenstein DG. Ab initio base-pairing energies of uracil and 5-hydroxyuracil with standard DNA bases at the BSSE-free DFT and MP2 theory levels. Org. Biomol. Chem. 2006;4:1741–1745. doi: 10.1039/b602263d. [DOI] [PubMed] [Google Scholar]
- 15.Zhang S, Yang X, Gorenstein DG. Enhanced suppression of residual water in a "270" WET sequence. J. Magn. Reson. 2000;143:382–386. doi: 10.1006/jmre.1999.1987. [DOI] [PubMed] [Google Scholar]
- 16.Canton CR, Schimmel PR. Biophysical Chemistry, Part II. New York: W.H. Freeman; 1980. Techniques for the Study of Biological Structure and Function. [Google Scholar]
- 17.Devoe H, Tinoco I. The hypochromism of helical polynucleotides. J. Mol. Biol. 1962;4:518–527. doi: 10.1016/s0022-2836(62)80106-5. [DOI] [PubMed] [Google Scholar]
- 18.Gray DM, Hung SH, Johnson KH. Absorption and circular dichroism spectroscopy of nucleic acid duplexes and triplexes. Methods Enzymol. 1995;246:19–34. doi: 10.1016/0076-6879(95)46005-5. [DOI] [PubMed] [Google Scholar]
- 19.Gary DM, Ratliff RL, Vaughan MR. Circular dichroism spectroscopy of DNA. Methods Enzymol. 1992;211:389–406. doi: 10.1016/0076-6879(92)11021-a. [DOI] [PubMed] [Google Scholar]
- 20.LaFrancois CJ, Jang YH, Cagin T, Goddard WA, Sowers LC. Conformarion and proton configuration of pyrimidine deoxynucleoside oxidation damage products in water. Chem. Res. Toxicol. 2000;13:462–470. doi: 10.1021/tx990209u. [DOI] [PubMed] [Google Scholar]
- 21.Kino K, Shimizu Y, Sugasawa K, Sugiyama H, Hanaoka F. Nucleotide excision repair of 5-formyluracil in vivo is enhanced by the presence of mismatched bases. Biochemistry. 2004;43:2682–2687. doi: 10.1021/bi0361416. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


