Abstract
Background
The appendage domain of the γCOP subunit of the COPI vesicle coat bears a striking structural resemblance to adaptin-family appendages despite limited primary sequence homology. Both the γCOP appendage domain and an equivalent region on βCOP contain the FxxxW motif; the conservation of this motif suggested the existence of a functional appendage domain in βCOP.
Results
Sequence comparisons in combination with structural prediction tools show that the fold of the COOH-terminus of Sec26p is strongly predicted to closely mimic that of adaptin-family appendages. Deletion of the appendage domain of Sec26p results in inviability in yeast, over-expression of the deletion construct is dominant negative and mutagenesis of this region identifies residues critical for function. The ArfGAP Glo3p was identified via suppression screening as a potential downstream modulator of Sec26p in a manner that is independent of the GAP activity of Glo3p but requires the presence of the COOH-terminal ISS motifs.
Conclusion
Together, these results indicate an essential function for the predicted βCOP appendage and suggest that both COPI appendages perform a biologically active regulatory role with a structure related to adaptin-family appendage domains.
Background
The intracellular transport vesicles of eukaryotic cells are responsible for ferrying cellular cargoes between membrane-bound compartments of the secretory and endocytic systems. Many of these vesicles are formed by a protein coat which functions both to select and trap cargo and to deform the membrane as part of the budding process. COPI-coated vesicles are responsible for the retrograde transport of cargo from the cis-Golgi back to the ER and have a role in intra-Golgi transport (reviewed in [1-4]). The COPI coat consists of seven coatomer subunits which are conserved across eukaryotes [5-7]. With high salt treatment the oligomer dissociates into [8]: (i) the F-COPI sub-complex, which resembles the adaptin heterotetramers and is composed of β-, γ-, δ-, and ζCOP (Sec26p/109 kDa, Sec21p/105 kDa, Ret2p/61 kDa, and Ret3p/22 kDa in S. cerevisiae, respectively), and (ii) the B-COPI sub-complex, composed of α-, β'-, and εCOP (Sec33p/136 kDa, Sec27p/99 kDa, and Sec28p/34 kDa in S. cerevisiae, respectively) with homology to clathrin [9]. The genes encoding each of these proteins are essential for cell survival in yeast, with the exception of SEC28 (εCOP) [10].
F-COPI and adaptin tetramers such as AP2 are composed of two large subunits (α and β2 in AP2), a medium subunit (μ2 in AP2), and a small subunit (σ2 in AP2). The structure of the AP2 holocomplex reveals two large subunits composed of helical trunk domains at the NH2-terminus which form the core of the complex together with the medium and small subunits [11]. Extending from each trunk via ~100 residue unstructured linkers are the COOH-terminal appendage globular domains [12,13]; these are predicted to be capable of extending from the core to sample the environment, but to spend most of their time in a retracted position [11,14,15]. The four subunits of F-COPI are predicted to associate in a complex resembling the AP2 complex. To date, the only known structure for coatomer is of the γCOP appendage, and although the sequence identity between γCOP and the large AP2 subunits is very low, the structural homology is striking [16,17] and together with consideration of the overall architecture of the complex, hints at a similar appendage in βCOP.
The adaptin-family appendages can be divided into two subdomains, an amino-terminal β-sandwich subdomain with a fold resembling that of immunoglobulins, and a COOH-terminal "platform" subdomain. The platforms of the γCOP and AP2 appendages each contain an FxxxW motif; in AP2 this motif has been demonstrated to mediate binding to a number of ligands important in the regulation of clathrin coat formation (reviewed in [18-20]). A second ligand binding site is present in the β-sandwich subdomain of both AP2 appendages [12,14,21,22]. Other adaptins and related proteins contain similar appendages with ligand binding functions (reviewed in [4]).
COPI vesicle formation is controlled by the small GTPase Arf1p which, in turn, is regulated by guanine nucleotide exchange factors (GEFs) and GTPase activating proteins (GAPs) (reviewed in [23-28]). The ArfGEFs belong to the Sec7 family of proteins (reviewed in [29]) and catalyze the exchange of GDP for GTP by Arf1p, causing the activation and membrane recruitment of Arf1p. ArfGAPs have a complex function that is currently a source of active discussion (reviewed in [30,31]). The hydrolysis of GTP by Arf1p is a prerequisite for vesicle uncoating and requires an ArfGAP as Arf1p has no intrinsic hydrolytic activity [32]. ArfGAPs also play roles in cargo packaging and the detection of membrane curvature, in addition to being a component of the vesicle coat [33-36]. There are six identified proteins containing ArfGAP domains in yeast: Age1p, Age2p, Gcs1p, Glo3p, Gts1p, and Sps18p. Of these, the first four have been demonstrated to have ArfGAP activity on Arf1p [37-40]. There is some redundancy among the yeast ArfGAPs, with Glo3p and Gcs1p having overlapping function in retrograde trafficking and forming an essential pair [38,40].
In this study we sought to examine if βCOP contained an adaptin-family appendage and if such a domain would be critical for COPI-coated vesicle formation. Using a combination of structural modeling, genetic, biochemical, and cell biological approaches, we provide supporting evidence that βCOP does indeed have an adaptin-family appendage and demonstrate that this domain is essential for βCOP function. We also identified the ArfGAP Glo3p as a downstream effector of Sec26p. This role of Glo3p is not dependent on its catalytic GAP activity, suggesting that this protein serves distinct functions in COPI vesicle trafficking that are independent from its regulation of the nucleotide bound state of Arf1p.
Results
An adaptin-family appendage domain is predicted for Sec26p
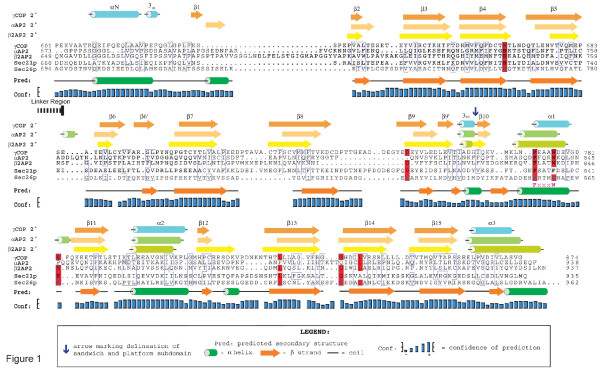
Based on the known similarities in size, sequence, and structure of members of COPI to the AP2/clathrin coat, we predicted the Sec26p COOH-terminus to contain an adaptin-family like appendage domain [16]. To evaluate this possibility, we examined the sequence of Sec26p using two protein structure prediction servers, PSIPRED and mGenTHREADER [41-45]. Using PSIPRED, we found that the NH2-terminal 730 residues are predicted to be primarily α-helical in nature with some unstructured content (data not shown). The accuracy rate (Q3) of PSIPRED is considered to be approximately 78% per residue [46,47]. From residue 730 to 840, the protein is predicted to be composed of β-strands, and from residue 840 to the end (973), there is mixed content predicted. This overall domain architecture is similar to α- and β2AP2 which are composed of an NH2-terminal helical trunk domain [11], followed by a ~100 residue flexible linker, then an appendage domain which is composed of a β-sandwich subdomain (residues 701–824 in α, 705–825 in β2) and a platform subdomain (residues 825–938 in α, 826–937 in β2) [12,13,48]. Comparison of the secondary structure prediction for Sec26p to the known structures of the large AP2 subunits appendage domains suggests that Sec26p does indeed possess a similar COOH-terminal domain. Specifically, the β-sandwich domain is formed from residues 730–840, and the platform domain from residues 841–973. The boundary in Sec26p between the trunk domain and the predicted flexible linker region is not clear based on this prediction, but we expect that the trunk terminates at residue number ~600 as with the AP2 subunits. Whether there is a flexible linker connecting the two domains or if this region forms a distinct domain in Sec26p is unclear from the prediction. The output from the PSIPRED secondary structure server is illustrated graphically in Figure 1 together with an alignment of equivalent regions of β2AP2, αAP2 and γCOP.
Figure 1.
Alignment of COPI and AP2 appendage domains. A structure-based sequence alignment of the appendage domains of α- and βAP2 with that of γCOP was generated as described in [16]. Secondary structural elements of the γCOP and the AP2 appendage domains determined previously are indicated above the sequence alignment. The blue arrow denotes the position of the boundary between the platform and the Ig-like (β-sandwich) subdomains. Highlighted in red is where four or more sequences contained a strictly conserved residue at that position. Regions of similarity, calculated using functional amino acid groupings, are boxed in blue. Residues for which no structural information is available were positioned using a primary sequence alignment using the ClustalX 1.83 program and manual inspection. The conserved FxxxW motif is indicated below the sequence alignment in pink. The predicted secondary structural elements for the appendage domain of βCOP, Sec26p are shown beneath the alignment as determined with the PSIPRED confidence level of the assignments shown in the diagram.
mGenTHREADER is a protein fold recognition server. Input of the entire Sec26p sequence returns several "Certain" hits, i.e. matches with a confidence level > 99%. Included in this list are both the AP2 and related AP1 cores, specifically the αAP2, β2AP2, and γAP1 chains. Input of the sequence for residues 700–973 (i.e., the predicted appendage region) returns a single "Certain" (p < 0.0001) hit with a net score of 0.889, namely, the αAP2 appendage, which has 11.7% primary sequence identity to the Sec26p appendage. The next closest match is the γCOP appendage with a "Medium" certainty level (p < 0.01), with a net score of 0.439 and a percent identity of 16.2%. Third on the list, with a "Guess" certainty level (p >/= 0.1), is the β2AP2 appendage with a net score of 0.317 and a sequence identity of 12.9%. The sequence relationship of the NH2-terminal trunk domain of Sec26p with the other adaptin-family trunk domains, has been previously noted [49,50]. These results emphasize the conservation of fold of the trunk domains and additionally suggest a less stringent but still significant preservation of the adaptin-appendage domain fold.
In addition to structural predictive methods, sequence alignments were also performed to predict the appendage domain in Sec26p. Alignments between βCOPs and γCOPs suggest the appendage of Sec26p to begin at approximately residue 667, whereas the alignment with αAP2s suggests a start residue of 729, closer to that predicted by the secondary structure predictions. The platform subdomain is predicted to start at residue 849 based on the alignments with γCOPs and at residue 845 based on alignments with αAP2s, both very close to the start site predicted by the secondary structural prediction models. The appendage domains include several residues which are conserved amongst appendage domains (color coded red in Figure 1) including the FxxxW motif of the platform domain.
In summary, both secondary structure and fold recognition predictors assign the Sec26p COOH-terminus to the adaptin-appendage family with a high level of confidence. The combination of predictive algorithms places the start of the platform subdomain at approximately residue 840. The start of the β-sandwich subdomain is less precisely predicted, somewhere between residue 667 and 730 in Sec26p.
The Sec26p appendage, specifically the platform subdomain, is essential in S. cerevisiae
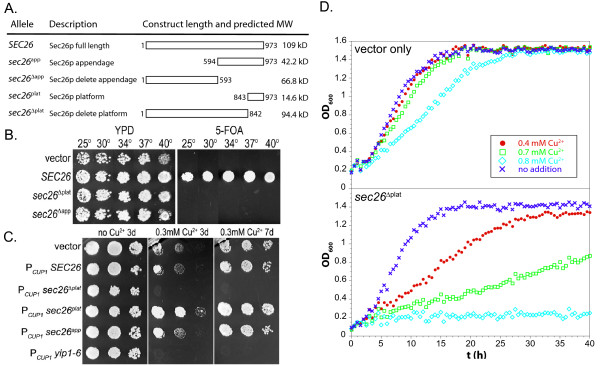
To determine if the predicted Sec26p appendage domain was a critical functional unit, a truncated construct of SEC26 (973 residues total) coding for the trunk region alone was created (sec26Δapp; pRC2482, this study, see Figure 2A for construct schematic). This plasmid was transformed into the sec26Δ tester strain and plated on 5-FOA for plasmid shuffling. The resultant strain was inviable (Figure 2B), demonstrating the essential nature of the Sec26p appendage domain. A more conservative truncation (sec26Δplat; pRC2521, this study) of the predicted platform subdomain also resulted in inviability (Figure 2B), demonstrating the critical nature of the function of the platform domain of the βCOP appendage. This is in contrast to deletion of the Sec21p appendage (sec21Δapp containing amino acids 1–676) region which is non-lethal [16].
Figure 2.
Functional importance of the βCOP appendage domain. A. Schematic illustrating the characteristics of the various Sec26p truncation constructs used for experiments. B. Plasmids coding for truncated constructs of SEC26, wild type SEC26, or vector only were transformed into the sec26Δ tester strain and spotted onto complete media (YPD) and synthetic complete media (SCD) with 5-FOA at 25, 30, 34, 37, and 40°C. C. Plasmids coding for vector alone or full length or truncated constructs under control of the copper-inducible promoter (PCUP1) were transformed into wild type yeast and replica-plated onto minimal media (SD) with and without the addition of Cu2+ (0 or 0.3 mM CuSO4). Growth was followed for three or seven days (3d and 7d respectively). The dominant negative yip1-6 construct was included as a negative control [71]. D. Transformed cells were grown in liquid minimal media with and without the exogenous CuSO4. Cultures were grown to early log phase and adjusted to an OD600 of 0.2. CuSO4 was added at the levels indicated and cells were grown at 30°C with turbidity measured every half hour for 2 days.
Over-expression of a truncated Sec26p missing the platform subdomain is dominant negative
The lethality caused by removal of the βCOP appendage or platform domain could be a result of the appendage/platform domain's critical function; however it was also possible that the truncations render the protein misfolded and subject to degradation, effectively the same as a deletion of the entire gene. Since the cell cannot function with sec26Δplat at endogenous levels as the only copy of SEC26, we wished to test if this construct could provide function when over-expressed. The truncated gene was placed under control of a copper-inducible promoter (PCUP1) for over-expression (pRC2631a, this study). The truncated constructs (see schematic in Figure 2A) were transformed into wild type cells, and screened on solid minimal media containing approximately 0.3 mM CuSO4. As depicted in Figure 2C, over-expression of sec26Δplat resulted in inviability on solid media. Growth curves in liquid media with varying amounts of CuSO4 (0, 0.4, 0.7, and 0.8 mM) reveal the dose-dependent nature of the dominant negative effect (Figure 2D). Over-expression of full length Sec26p, the appendage domain (residues 594–973; pRC2629a, this study) alone, or the platform subdomain (residues 843–973; pRC2632a, this study) alone, did not result in any detectable phenotype (Figure 2C). These results suggest that the appendage domain regulates the activity of Sec26p in the context of the holocomplex. The AP2 appendages are known to bind both unique and overlapping sets of partners, however, over-expression of the platform or appendage domain of Sec26p alone is not detrimental as might be expected if these appendage domains were to associate with binding partners and prevent their action on the holocomplex. This may be due to the fact that these interactions are of low affinity as suggested by the equivalent protein-protein interactions of the adaptin appendage domains [12-14].
Random mutagenesis of Sec26p appendage domain reveals residues critical for function
The phenotype difference between Sec26p and Sec21p appendage domain deletion highlighted the essential nature of this region for COPI function. We created point mutations of the Sec26p appendage domain to further dissect the action of this domain at a molecular level. Initially, targeted mutagenesis was performed on a subset of charged residues chosen based on sequence alignments of fungal species. Eight residues were mutated to alanine residues, singly or in pairs as listed: K667 and K669, N751, D823, E828 and D829, and D903 and D904. Silent restriction enzyme sites were incorporated to facilitate the identification of correctly mutated constructs. Constructs were transformed into the sec26Δ tester strain and plated at 25 and 40°C on 5-FOA media to assess functionality of the mutant allele. None of the mutations resulted in a detectable phenotype (data not shown). A random PCR-derived error resulted in the trunk domain mutation sec26-1 (T536A D903A D904A; pRC2653a, this study). This construct was temperature sensitive, whereas other constructs containing the D903A D904A mutations were robustly wild type, suggesting that the phenotype was attributable to the T536A mutation.
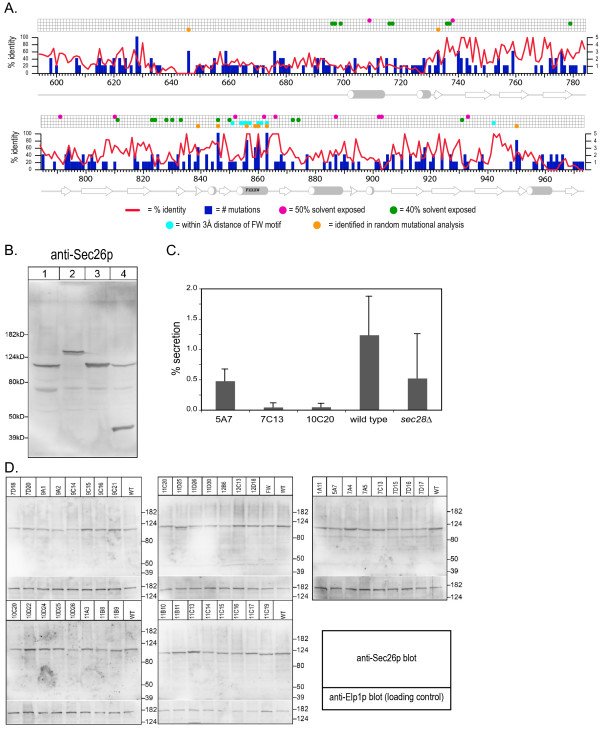
We next performed random mutagenesis on the Sec26p appendage to generate conditional lethal thermosensitive (ts) mutants. Error-prone PCR was performed on the appendage domain of Sec26p, leaving the trunk domain wild type. Tester strain transformants were screened for functionality after plasmid shuffling on 5-FOA plates incubated at 30 and 37°C. Thirty-eight unique ts mutants were generated from approximately 6000 transformants (Table 1) and contain between three and twelve point mutations each; two also contain 13-residue deletions at the COOH-terminus, and one contains an additional five residues at the COOH-terminus due to frameshift mutations. The temperature sensitivity profile of each mutant is shown in Table 1 and ranged from 34 to 40°C. In aggregate 276 mutations were identified, which occurred at 181 positions out of a total of 380 potential positions. Notably, five of these randomly generated mutants contained mutations in the FxxxW motif and there was a significant cluster of mutations identified that are close to this motif. Many of the mutated residues were represented two and occasionally three times. The number of mutations found occurring at each position is illustrated graphically in Figure 3A together with the position conservation and solvent accessibility data for residues that could be mapped onto equivalent positions in the γCOP structure.
Table 1.
Summary of sec26 mutants and genetic interactions with GLO3
| Name | Mutations | Restrictive temperature1 | GLO3 suppression? | glo3Δ synthetic lethality? |
| FW | F856A W860A | 40° | YES | YES |
| TDD | T536A D903A D904A (sec26-1) | 34° | YES | YES |
| 7A4 | K659R I681V D759V S810P C905S I938V L950S | 37° | YES | YES |
| 7D15 | S725Y K732N N772I H774R F777Y F799I K915M K964T | 37° | YES | YES |
| 9C15 | C678S F693S D759G Y823C V838A I842T Y913H | 37° | YES | YES |
| 10D22 | R672G I727T V743G K807R T877A | 37° | YES | YES |
| 10D25 | L651Q K659E H839P I846T F863I A959S | 37–40° | YES | YES |
| 11A3 | K659E K679N I733L Y744C F754S Y832N A851V F863L I893T I938L V973A | 37° | YES | YES |
| 12B6 | N680T R691W D741G F817Y M859V | 37–40° | YES | YES |
| 1A11 | F856A W860A D904A C905S | 37–40 | YES | +/- |
| 11D26 | E633D I733T D741V T769P F802L (sec26-2) | 37° | YES | +/- |
| 12D18 | Q661R S740P V806A F907Y F918S | 37° | YES | +/- |
| 11C15 | S628P I785V M859I L950S D956V H972Y | 37° | YES | +/- |
| 7D20 | S740P I785T N793S K801E F863S A951T | 37° | YES | no |
| 9A1 | I598N S728T D759V D823G I846T H855L I869L G901R | 37° | YES | no |
| 10D24 | V623A S628T P636A V757A T779S W860R K887R T895A K933E A962T | 37° | YES | no |
| 11B10 | L773Q I795T H839L N966Y | 37° | YES | no |
| 11C13 | Q607L M624V L646P H670P C678S I846T T877I N890S E920V K963E | 34° | YES | no |
| 9C16 | S626P K655R N793H T803S G818S N836D K868R E929K Q948L K963R | 37–40° | YES | no |
| 5A7 | E633G N635Y E663G K666R F690L R691W S728L V756D Q776R D921G S932T L950M | 34° | no | YES |
| 7C13 | F650L G695S H797Q I821N W860L E902G | 34° | no | YES |
| 7D17 | K655N A665T D676G Q702R Q776H G781C Y822N D844E F863Y R883S | 37° | no | YES |
| 7D18 | K614R L780H V794E A825V L885M C910R C927S A962S | 37° | no | YES |
| 9A2 | Q607R K669M I846F M859V F907L F918Y | 40° | no | YES |
| 9C14 | D697N D704H S740T I820T R831C I842V I928F S932T | 37° | no | YES |
| 10C20 | S608T D616E D676G F693L D704G S726G P735H E746K H774Y F799L K847I F856Y | 34° | no | YES |
| 10D26 | I598T R622G A718P T722I P735L K767E H839P T850S N890S D921E N966Y T971A | 37° | no | YES |
| 11B11 | I615F L646Q K715R F817S F863L K872E M891L L950S | 34–37° | no | YES |
| 11C14 | R622G K679G A694D A924V C927S V937D | 40° | no | YES |
| 11D30 | A649E C910R S917P | 37° | no | YES |
| 11C20 | D676N L730R F754L N772Y A830D I834N D852V S917T F918I | 37° | no | YES |
| 7A5 | S619F L620V D709E R957G | 40° | no | +/- |
| 11C19 | V600D F658S L712Q Q753H Q776R N836I T965A | 37° | no | +/- |
| 7D16 | S628T L712Q P789S V794E D829V I846F I961F2 | 34–37° | no | no |
| 11B8 | I615F R660G S724C S916P K964E L970H | 40° | no | no |
| 11B9 | E617V I629S K715I T889S A924V A959H3 | 34° | no | no |
| 11C16 | I662F V701G H720P V743G K784R F856I K887E | 40° | no | no |
| 11C17 | L620I M624I L646P T654S T686M I749V V794G N867S K868G | 37° | no | no |
| 11D25 | S628P A718S I733M P796T A825V Y832F3 | 37–40° | no | no |
| 12C13 | S628P G798C F802S E866G Y913C | 34° | no | no |
1 restrictive temperature on SCGalactose media
2 also contains additional residues at COOH-terminus 974WTTLS978
3 also contains deletion of COOH-terminal residues 960–973
Figure 3.
Sec26p mutant allele creation. A. Bar chart showing the relative location and number of each mutated amino acid position identified in the ts allele set created by random mutagenesis. Superimposed is the % identity of each residue derived from a sequence alignment of βCOP sequences in Clustal v1.83. For reference, a grid indicates the positions of amino acid residues that are aligned (according to Figure 1) with solvent exposed residues of γCOP, located within 3Å distance of the FxxxW motif, and identified as a minimal set of functionally important residues through GLO3 interactions. B. Expression of constructs revealed with anti-Sec26p appendage domain antibody. Whole cell lysates were prepared from (Lane 1) wild type cells expressing endogenous Sec26p (109 kDa), (Lane 2) cells expressing GFP-Sec26p as the only copy of Sec26p (136 kDa, pRC2798), (Lane 3) cells over-expressing Sec26p under control of the copper-inducible promoter (PCUP1) (42.2 kDa, pRC2630a), or (Lane 4) cells expressing both wild type levels of Sec26p plus over-expressed Sec26p appendage under control of the copper-inducible promoter (PCUP1) (pRC2629b). C. Invertase secretion in sec26ts mutant cells. Several mutant alleles were chosen and invertase secretion was measured after a one hour shift to the restrictive temperature. The graph shows % secretion relative to wild type and an alternative COPI ts mutant, the deletion of sec28 (εCOP subunit). Data plotted is the average of three experiments and confidence intervals shown with error bars. D. Sec26p allele expression levels. Cellular lysates prepared from each of the sec26ts strains after incubation at 40°C for 1 h to assess Sec26p expression levels at the restrictive temperature. Blots were subsequently probed with an anti-Elp1p (~150 kDa) antibody [70] as a loading control.
Because each mutant allele contained an average of 4.8 individual mutations that could potentially be destabilizing, we wished to determine if the temperature sensitive phenotypes of the mutant alleles of SEC26 were due to loss of expression at the restrictive temperatures. Whole cell lysates were analyzed by Western blotting using an antibody generated against a recombinant fusion protein of the Sec26p appendage domain (Figure 3B). The mutant strains were incubated at 40°C for one hour prior to lysis, a time period sufficient to observe a block in secretion measured by the accumulation of intracellular invertase (Figure 3C). Although some variation in expression is apparent in comparison to the loading control, none of the mutant strains exhibited significantly diminished protein levels or evidence of degradation after shift to the restrictive temperature, compared to wild type cells also incubated at 40°C (Figure 3D). These results suggest that the conditional temperature sensitive phenotypes are not the result of instability and altered Sec26p levels but are due to functional impairment of the Sec26p appendage domain.
The ArfGAP Glo3p genetically interacts with sec26ts mutants
One mutant allele identified through random screening, sec26-2 (11D26; pRC2948, this study), was found to contain 5 mutations restricted to the predicted β-sandwich subdomain (E633D I733T D781V T768P F802L), with no mutations in the platform subdomain. Because both of the appendages of AP2 have been identified to have two binding sites [12,14,21,22], one on the platform and one on the β-sandwich subdomain, and because related appendages without platforms (e.g., γAP1) also bind ligands [51], we selected this mutant to work with further, as a useful complement to the platform subdomain mutant, sec26FW.
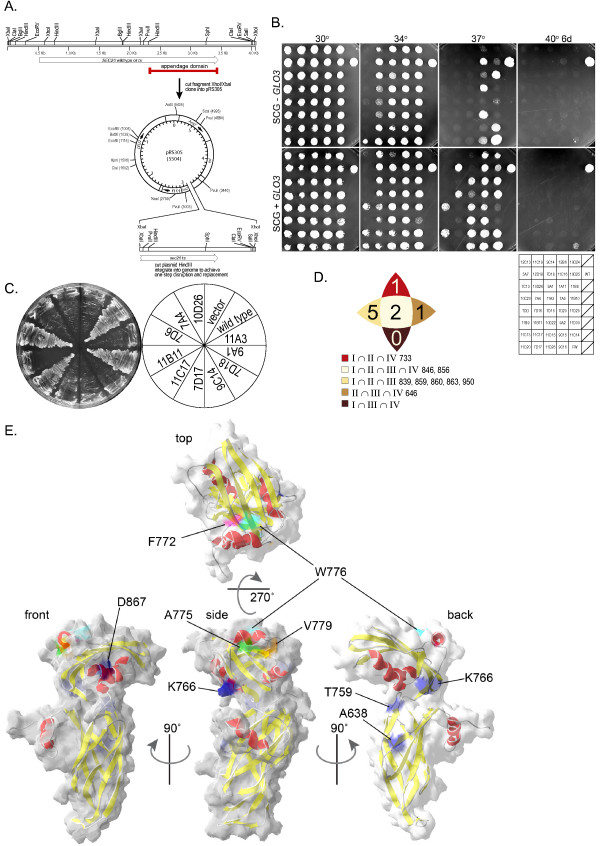
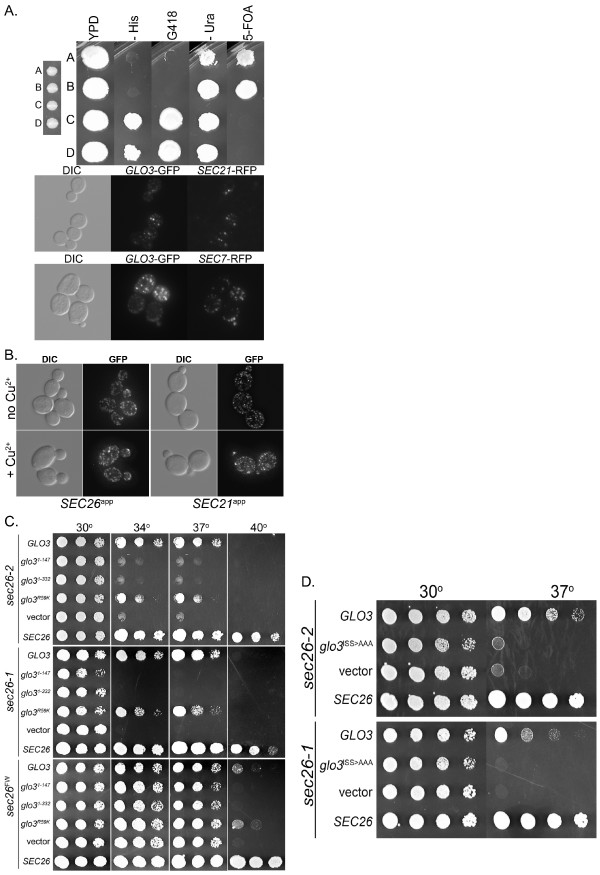
To seek candidate downstream effectors for the Sec26p appendage, we performed suppressor screens on two of the mutants to identify genetically interacting partners. The sec26FW and sec26-2 alleles were integrated into the genome as the sole copy of SEC26 for use in these screens to facilitate recovery of the suppressor plasmids (Figure 4A). These mutant strains were transformed with a galactose-inducible promoter (PGAL1/10) controlled over-expression library [52], and screened for thermoresistance at the restrictive temperature on galactose-containing media after an overnight recovery period at 30°C. Wild type SEC26 was recovered multiple times, and GLO3 was recovered twice from the sec26-2 screen. Suppression by GLO3 over-expression is partial and does not restore full wild type growth. The PGAL1/10 GLO3 plasmid (pRC2979, this study) was transformed into each of the appendage mutants and found to suppress a subset of them (Figure 4B, compare top and bottom row).
Figure 4.
Phenotype of sec26ts Mutants. A. Schematic of strategy to create appendage domain mutants for suppressor screening. ts alleles created as described in Materials and Methods were subcloned into the vector pRS305 for one-step gene disruption and replacement to create integrated genomic sec26 alleles. B. The sec26Δ tester strain was transformed with each of the sec26ts mutant plasmids and plated on synthetic complete media (SCD) with 5-FOA to counter-select for the wild type SEC26 plasmid (not shown). The resultant strains were plated at 30, 34, 37, and 40°C to demonstrate the temperature sensitive phenotype of each mutant (top row). Each strain was transformed with a GLO3 expressing plasmid under control of the galactose-inducible promoter (PGAL1/10) and plated on SCG at the above listed temperatures to evaluate the ability of GLO3 to suppress the ts phenotype (bottom row). The data is also summarized in Table 1. C. The sec26Δglo3Δ tester strain was transformed with each of the sec26ts mutant plasmids and streaked onto SCD + 5-FOA to assess growth when sec26ts was expressed as the only copy of SEC26 in the glo3Δ background. One representative plate is shown. Data for all sec26 alleles is summarized in Table 1. D. Diagram showing the numbers of residues identified at the intersection of at least three of the Groups I-IV together with the residue number in Sec26p. E. Mapping of residues identified in Figure 4D onto the known structure of γCOP appendage domain. The γCOP appendage structure is depicted as a ribbon diagram with α-helices in red and β-strands in yellow. The calculated molecular surface (transparent grey) is overlaid on the ribbon diagrams. Selected residues (K766, T759, A638, D867) were identified from the alignment shown in Figure 1 as the equivalent of the Sec26p residues chosen as described in Figure 4D and shown in blue. For clarity, the residues surrounding and including the FxxxW motif are shown in additional colors: pink (F772), light blue (W776), green (A775), and orange (V779). The appendage is shown from a "side" view (center image) with the NH2-terminal α-helix projecting toward the viewer. The image was then rotated 90° for the "front" view (left image), 90° for the "back" view (right image), and 90° towards the viewer for the "top" view (top image). Each of the selected residues has surface exposure.
Since over-expression of GLO3 suppressed several of the sec26ts mutants, we examined the effect of GLO3 deletion in combination with the sec26ts mutants. Results are shown in Figure 4C. Most of the sec26ts mutant alleles were synthetically lethal with glo3Δ, and a few exhibited impaired growth. Evaluation of the pattern of suppression and synthetic lethality for sec26ts mutants with GLO3 revealed that one set of mutants were suppressed by GLO3 over-expression and synthetically lethal with glo3Δ (Group I), another two sets showed either suppression or synthetic lethality (Group II and III), and a fourth set did not show either type of genetic interaction (Group IV). We made use of these groups as additional functional information to identify the residues most significantly contributing to function amongst the 181 mutated positions resulting from the random mutagenesis screen. Only 9 residues were shared by at least three groups (Figure 4D). Using the structure and homology based alignment of Figure 1, we mapped each of these residues onto the γCOP appendage domain structure (Figure 4E). The equivalents of these residue positions in γCOP could be identified as a surface, solvent exposed amino acid on either the "back", "side" or "top" view of the domain, suggestive of a function that engages other proteins necessary to regulate or transduce COPI function.
The relationship between Sec26p and Glo3p
Our genetic data clearly demonstrate an interaction between Sec26p and Glo3p. In order to determine if this was mediated by a direct physical interaction with the appendage, we performed in vitro co-precipitation assays with recombinant proteins. His6-tagged Sec26p appendage (His6-Sec26papp) was co-expressed with GST-tagged full-length Glo3p in E. coli however, we were unable to demonstrate the existence of a direct physical interaction between Glo3p and the Sec26p appendage in vitro (data not shown).
Since in vitro experiments did not reveal a direct physical interaction between Glo3p and the Sec26p appendage, we wished to determine if an interaction, direct or indirect, could be demonstrated in vivo. A previous immunofluorescence study in mammalian cells, showed that one of the Glo3p mammalian orthologs ARFGAP2, but not ARFGAP3, could be disturbed from its Golgi localization by over-expression of the mammalian γCOP appendage, and that this was dependent on an intact FxxxW motif [17]. To determine if Glo3p's intracellular localization was influenced by the βCOP appendage, we co-expressed Glo3p-GFP with over-expressed Sec26p appendage under control of PCUP1. This was performed in glo3Δgcs1Δ double knockout cells to eliminate any contribution from the functionally overlapping ArfGAP, Gcs1p. We first verified that Glo3p tagged with GFP at the COOH-terminus is a fully functional construct by testing its ability to act as the only retrograde Arf GAP in a double knockout of glo3Δ and gcs1Δ, a genetic background where GLO3 becomes essential for viability (Figure 5A). The appendage of Sec21p (γCOP) was also over-expressed to determine its effect on the localization of Glo3p. Results are shown in Figure 5B. We did not detect any difference in the localization of Glo3p-GFP in the presence of either appendage domain when over-expressed. Therefore, the localization of yeast Glo3p, unlike that of its ARF GAP ortholog ARFGAP2, is not dictated by the coatomer appendages and is dependent on additional, as yet unidentified factors and may be functionally be analogous to human ARFGAP3 [53].
Figure 5.
Glo3p-GFP localization in the presence of over-expressed Sec26p appendage. A. Functionality and localization of Glo3p-GFP. The functionality of the COOH-terminally tagged Glo3p-GFP construct (pRC3341A) was checked by transformation into the heterozygous diploid (MATa/α his3Δ0/his3Δ0 leu2Δ0/leu2Δ0 ura3Δ0/ura3Δ0 MET15/met15Δ0 LYS2/lys2Δ0 GLO3/glo3Δ::HIS3 GCS1/gcs1Δ::KANR). Following sporulation, haploid strains deleted for the genomic copies of GLO3 and GCS1, were identified. Viable haploids that were His+ and KANR were also positive for Glo3p-GFP and 5-FOA sensitive. Cells cotransformed with Glo3p-GFP and Sec21p-RFP or Sec7-RFP show coincidence of the green and red fluorescent signal in the punctate pattern typical of the Golgi of S. cerevisiae. B. Glo3p-GFP was expressed in a diploid glo3Δ/glo3Δ gcs1Δgcs1Δ background. This strain was transformed with a plasmid for over-expression of the Sec26p (residues E594-V973; pRC2629b) or Sec21p appendage (residues L676-Q935; pRC2628a) by a copper-inducible promoter (PCUP1). Cells were grown to early log phase, the cultures were divided, and treated with or without 0.7 mM CuSO4 for 3 hours at room temperature prior to microscopy. C. Selected sec26ts mutant strains (sec26-1, sec26-2, and sec26FW) were transformed with the glo3 truncation and point mutation constructs under control of PGAL1/10 and plated on SCG as a dilution series to evaluate the suppressive ability of the constructs. D. GLO3 suppression of sec26ts (sec26-1 and sec26-2) requires the ISS motifs located in the COOH-terminus of Glo3p. Mutant sec26 strains were transformed with the indicated constructs and tested for suppression as in 5C. The mutant ISS motif glo3 construct contained alanines at positions 388–390 and 420–422.
An additional function to the ArfGAP activity of Glo3p is suggested by genetic interactions with sec26ts mutants
The GAP domain of Glo3p is found in the first third of the protein; little is known about the function of the remaining two thirds. Each known ArfGAP, including Glo3p, contains a Zn-finger motif in the GAP domain [37,54,55] that is essential for its Arf-interaction function. A conserved arginine is also part of this domain and is also required for catalytic activity [55-58]. In order to determine what domain or function of GLO3 was responsible for the suppression activity (e.g., if suppression is dependent on catalytic activity), we created truncation and point mutants of GLO3 under control of the regulatable promoter PGAL1/10. The two Glo3p truncation mutants: glo31–147 and glo31–332 respectively includes the ArfGAP domain only (pRC3297a, this study), and the first two thirds of the protein (pRC3298a, this study). We also created a full length construct of Glo3p with the catalytic arginine mutated to lysine (glo3R59K) under control of PGAL1/10 (pRC3299b, this study). Constructs encoding GLO3 and glo3 mutants were transformed into selected sec26ts strains and plated on galactose-containing media for over-expression. Suppression required a full length version of the protein, but did not require catalytic activity (Figure 5C). This pattern was observed for each of the three mutants tested (sec26-1, sec26-2, and sec26FW). These results suggest that Glo3p has a second function that resides in its COOH-terminal domain. The COOH-terminus of Glo3p contains two repeats of the amino acid sequence ISSxxxFG (residues 388–395 ISSDQLFG and residues 420–427 ISSSSYFG) which are critical for its ability to suppress the growth defect of arf1-16 [59]. We investigated the potential contribution of these motifs to the ability of Glo3p to suppress sec26ts (Figure 5D) with a glo3 388AAA390 420AAA422 construct. Together, results show that the COOH-terminal ISS motifs in Glo3p are necessary for the ability of GLO3 to suppress sec26-1 and sec26-2 mutant cells and that this activity is independent of the ability of Glo3p to stimulate GTP hydrolysis on Arf.
Discussion
The appendage domain of the γCOP subunit of the COPI vesicle coat bears a striking structural resemblance to adaptin-family appendages despite limited primary sequence homology (12–16% identity). The adaptin appendages are integral to the regulation of clathrin-coated vesicles via FxxxW and other motifs. Both the γCOP appendage domain and equivalent region on βCOP contain the FxxxW motif; based on the conservation of this motif in βCOP, we predicted the existence of a functional appendage domain in βCOP (Sec26p in S. cerevisiae) [16]. We previously identified the thermosensitive allele sec26FW, which contains mutations of the conserved FxxxW motif (to AxxxA) in the platform subdomain [16] although mutation of the same motif in Sec21p (which is less tightly conserved as FxxxF) had no detectable effect. The 38 sec26ts appendage domain alleles identified in this study contained a total of 276 mutations distributed amongst 181 out of a possible 380 positions. Given that there were three to twelve point mutations in each allele, the set also probably contains "noise", mutations in residues that do not contribute to the phenotype, in addition to individual point mutations and groups of mutations that together contribute to the overall phenotype. To identify a core set of residues that reliably contributed to the phenotype, we considered the subset of residues that appeared to be frequently mutated and intersected in terms of a genetic interaction with GLO3. Although this is an arbitrary designation, we used this intersection of phenotypes to provide a stringent filter to select functionally important residues from the experimental set of 181 mutated positions. These 9 residues could be expected to be involved in either receiving or transmitting βCOP functionality given that these residues appeared analogous to surface exposed residues of γCOP that might be expected to engage in protein-protein interactions.
This collection of mutants represents the first set of conditional lethal sec26 alleles and will be a useful tool for dissecting Sec26p function in vivo and vitro as we gain more of a mechanistic and structural understanding of COPI function. The identification of a core set of 9 residues is undoubtedly a conservative underestimation of the mutated positions in the collection that contribute to Sec26p domain functions. With additional functional information about the appendage domain, we expect to be able to refine this analysis further and understand the mechanistic basis by which these residues contribute to appendage domain function.
In addition to shared or overlapping functions of the two appendage domains, the severity of effect in Sec26p appendage deletion or mutation suggests that its function contains at least one unique aspect that is more critical to cell physiology than for the equivalent domain in Sec21p. Similar actions have been proposed for the AP2 appendage domains suggesting a common mechanistic theme for these types of coat complexes [14,60]. Aside from the FW motif, analysis of the sequence and predicted structural alignments reveal that the residues used by adaptin domains to interact with ligands are chemically different in the case of β- and γCOP. These considerations highlight the fact that the Ig-like sandwich and the platform subdomain scaffolds provide enormous flexibility in the surfaces they offer for directed protein-protein interactions.
Does the predicted βCOP appendage have analogous function to the AP2 appendages, acting as a hub [60] to recruit protein partners essential to the regulation of COPI-coated vesicle formation? The mammalian γCOP appendage has been suggested to interact with Glo3p, an ArfGAP [17], and during the preparation of this manuscript, the βCOP appendage was demonstrated to interact with the calpain nCL-2 in stomach cells [61]. Interacting partners for the appendage domains may require the multiple low-specificity sites provided by oligomerization of the COPI coat on the two-dimensional surface of membranes, and may possibly be governed by regulatory modifications. Although we have not identified a direct binding partner for the βCOP appendage, and demonstrated that appendage domain over-expression was not dominant negative, our studies did reveal an interesting relationship between the βCOP appendage and the retrograde ArfGAP Glo3p. We demonstrate that impairment of Sec26p appendage domain function can be suppressed by Glo3p over-expression, independent of its catalytic activity, suggesting that the action of Glo3p is downstream from COPI.
Retrograde ArfGAPs such as Glo3p and mammalian ARFGAP2 and ARFGAP3 are known to bind to coatomer as demonstrated by both in vitro and in vivo assays and genetically interact with genes encoding coatomer subunits [17,58,62], however the specific region of Glo3p involved has not been clarified and the number of binding sites on coatomer also remains undefined. Mammalian ARFGAP1 has been shown to contain two coatomer binding sites, one in the catalytic domain, and one in the non-catalytic domain [63]. Additional studies to further explore the nature of the relationship between Glo3p, coatomer, membranes and cargo will be necessary to resolve these fundamental questions.
Our results do not address where coatomer and ArfGAPs interact physically in vivo, but do provide interesting insights into the multi-functional nature of Glo3p. What is most remarkable about this interaction is that glo3R59K over-expression is able to suppress the sec26ts alleles to virtually the same extent as wild type GLO3. Cells expressing glo3R59K as the only copy of GLO3 are inviable and display a variety of secretory defects including failure of vesicle budding, accumulation of ER membranes, impairment of CPY processing, and mislocalization of retrograde cargo, however the mutant protein is still able to bind coatomer in coprecipitation assays [58]. Over-expression of glo3R59K in wild type cells does not impair growth (our unpublished data) and one attractive possibility is that Glo3p combines GAP and effector interactions within the same polypeptide. Taken together with our suppression data, this strongly suggests that Glo3p has critical functions in addition to its GAP activity and that this function involves the COOH-terminus of Glo3p operating in conjunction with Sec26p and COPI. Glo3p contains two repeats of the amino acid sequence ISSxxxFG. This motif is critical for its ability to suppress the growth defect of arf1-16 [59], and, as demonstrated in this study, for the ability of GLO3 to suppress sec26ts mutants. The mechanistic underpinnings of this motif are unclear at present; a search of the yeast proteome reveals the ISSxxxFG motif in other proteins of very divergent function (Ubr2p, Vba1p, Ssm4p, Ada2p, Fur1p, Fur4p and the HO endonuclease). Further studies are required to decipher how this motif functions and if it could play a more general role. It is interesting to note that this motif is conserved in the human orthologs of Glo3p ARFGAP2 and ARFGAP3 [53,59]. Outside the GAP domain, the homology between ArfGAP proteins varies, a divergence that reflects the fact that their mechanisms of localization and specific functions are also quite disparate. An important goal will be to establish paradigms to understand how the non-catalytic regions of ArfGAPs contribute to GAP activity and respond to the signal transduction pathways that transmit to, and act downstream of Arf proteins.
Conclusion
In these studies we sought to determine if βCOP contains an adaptin-family appendage domain similar to that determined for γCOP. Sequence comparisons and structural prediction tools suggest that the fold of the COOH-terminus of Sec26p is strongly predicted to closely mimic that of adaptin-family appendages. Experimental approaches, using both deletion and mutagenic studies illustrate the critical role of the predicted Sec26p appendage domain and in particular the residues in close proximity to the FxxxW motif that are expected to form a platform-like subdomain. The essential nature of the appendage, the presence of a conserved motif (FxxxW) in the predicted platform subdomain, the weak but significant sequence homology with other appendages, and the predicted secondary structure and fold collectively support the existence of an appendage domain on βCOP that plays an important regulatory role in COPI function.
Methods
Yeast strains and media
Yeast strains were cultured using standard media and conditions [64]. Plasmid shuffling was carried out on synthetic complete media (SCD) with 2.5 mg/ml 5-fluoro-orotic acid (5-FOA). Yeast strains are listed in Table 2.
Table 2.
S. cerevisiae strains used in this study
| Strain | Genotype | Source |
| RCY269 | MATa ura3-52 leu2-3,112 | This lab |
| RCY3130 | MATα his3Δ0 leu2Δ0 ura3Δ0 lys2Δ0 SEC26ΔKANR [pRS316 SEC21 SEC26 pRC2374] | This lab |
| RCY3315 | MATa ura3-52 leu2-3,112 SEC26Δ::sec26FW (F856A W860A)LEU2 | This study |
| RCY3661 | MATa ura3-52 leu2-3,112 SEC26Δ::sec26ts 11D26 (sec26-2, E633D I733T T768P D781V F802L) LEU2 | This study |
| RCY3674 | MATα glo3Δ::KANR his3Δ0 leu2Δ0 ura3Δ0 lys2Δ0 | ResGen |
| RCY3675 | MATa/α glo3Δ::KANR his3Δ0/his3Δ0 leu2Δ0/leu2Δ0 ura3Δ0/ura3Δ0 LYS2/lys2Δ0 MET15/met15Δ0 | ResGen |
| RCY3676 | MATα gcs1Δ::KANRhis3Δ0 leu2Δ0 ura3Δ0 lys2Δ0 | ResGen |
| RCY3677 | MATa/α gcs1Δ::KANR his3Δ0/his3Δ0 leu2Δ0/leu2Δ0 ura3Δ0/ura3Δ0 LYS2/lys2Δ0 MET15/met15Δ0 | ResGen |
| RCY3782 | MATα glo3Δ::HIS3 his3Δ0 leu2Δ0 ura3Δ0 lys2Δ0 | This study |
| RCY3784 | MATa glo3Δ::KANR his3Δ0 leu2Δ0 ura3Δ0 lys2Δ0 met15Δ0 | This study |
| RCY3785 | MATa gcs1Δ::KANR his3Δ0 leu2Δ0 ura3Δ0 lys2Δ0 met15Δ0 | This study |
| RCY3786 | MATa glo3Δ::KANR his3Δ0 leu2Δ0 ura3Δ0 met15Δ0 | This study |
| RCY3787 | MATa gcs1Δ::KANR his3Δ0 leu2Δ0 ura3Δ0 met15Δ0 | This study |
| RCY3881 | MATa glo3Δ::HIS3 his3Δ0 leu2Δ0 ura3Δ0 met15Δ0 | This study |
| RCY3882 | MATa gcs1Δ::HIS3 his3Δ0 leu2Δ0 ura3Δ0 met15Δ0 | This study |
| RCY4038 | MATα his3Δ0 leu2Δ0 ura3Δ0 lys2Δ0 SEC26Δ::KANRglo3Δ::HIS3 [pRS316 SEC21 SEC26] | This study |
| RCY4233 | MATa his3Δ0 leu2Δ0 ura3Δ0 met15Δ0 gcs1Δ::KANRglo3Δ::HIS3 [pRS316 GLO3-GFP pRC3341A] | This study |
| RCY4236 | MATα his3Δ0 leu2Δ0 ura3Δ0 lys2Δ0 gcs1Δ::KANRglo3Δ::HIS3 [pRS316 GLO3-GFP] | This study |
| RCY4238 | MATa/α his3Δ0/his3Δ0 leu2Δ0/leu2Δ0 ura3Δ0/ura3Δ0 LYS2/lys2Δ0 MET15/met15Δ0 gcs1Δ::KANR/gcs1Δ::KANR glo3Δ::HIS3/glo3Δ::HIS3 [pRS316 GLO3-GFP] | This study |
Alignments and secondary structure predictions
The protein sequence for Sec26p was submitted to the PSIPRED v2.5 server [41,42,44] with "Mask Low Complexity Regions" option selected. This was repeated for GenTHREADER and mGenTHREADER [43,45]. For alignments, 17 βCOP (COPB_HUM, NP_057535.1, XP_508297.1, NP_001006467.1, NP_001002013.1, AAQ63171.1, COPB_DRO, XP_393132.1, NP_194877.2, BAC87706.1, P41810, CAH01736.1, CAB46767.1, CAF06042.1, CAB95500.1, CAD26416.1, AAT12307.1) and 20 γCOP sequences were downloaded from NCBI in FASTA format and loaded into CLUSTALX v1.83 [65]. Alignments were performed with the following penalties: for pairwise alignments, gap opening was set to 35, and gap extension 0.75; for multiple alignments, gap opening was set to 15, gap extension to 0.3. The delay was set to 30%. All βCOP sequences and all γCOP sequences were aligned separately in Multiple Alignment Mode, and the two resulting alignments were aligned in Profile Alignment Mode. The procedure was repeated for βCOP and 29 αAP2 sequences. The coordinates for the B. taurus γCOP appendage (PDB ID 1PZD) were downloaded from the RCSB Protein Data Bank [66] and loaded into DeepView/Swiss-Pdb Viewer v3.7 for image manipulation.
Mutagenesis
All SEC26 truncations and mutants were created in pRS315 [67] for evaluation of functionality in the sec26Δ tester strain (RCY3130; MATα his3Δ0 leu2Δ0 ura3Δ0 lys2Δ0 SEC26ΔKANR [pRC2374; pRS316 SEC21 SEC26] [16]) after plasmid shuffling or dominant negative activity in a wild type strain (RCY239; MATa ura3-52 leu2,3-112). Non-essential KANR single knockout strains were obtained from Research Genetics, Inc., Huntsville, AL; other knockouts were created using standard techniques.
Random mutagenic PCR was performed on the appendage domain of SEC26 using primers designed to anneal beginning at the coding sequence for residue E594 and ending 579bp downstream of the stop codon, and was carried out as described by Wilson and Keefe [68]. The mutated product was co-transformed into the sec26Δ tester strain with a linearized plasmid containing a gapped SEC26 gene (pRC2877, this study) for recombination to form the complete SEC26 ORF. Transformants were replica plated to two 5-FOA plates for plasmid shuffling and incubated at 30 and 37°C. Digital images of the plates were pseudocolored and overlaid to identify ts colonies. The plasmids were extracted and re-transformed into the tester strain to confirm thermosensitivity of the construct. Confirmed sec26ts alleles were sequenced at the Cornell Biotechnology Facility using Big Dye Terminator chemistry on an Automated 3730 DNA Analyzer (Applied Biosystems; Foster City, CA).
Suppressor screens
To facilitate suppressor plasmid recovery during the screen, the sec26ts plasmids of interest were integrated into the genome. Plasmids were digested with XbaI and XhoI to obtain a 1.9kb fragment containing the mutant appendage, which was ligated into the integrating LEU2 vector pRS305. The resulting plasmid was linearized at a unique HindIII site located immediately upstream of the appendage start and transformed into yeast for a one-step gene disruption and replacement at the SEC26 locus. The integrated sec26ts strains were transformed with a library under control of the inducible galactose promoter (PGAL1/10, kind gift of A. Bretscher) [52] and approximately 50,000 transformants screened per mutant to obtain complete coverage of the genome. Transformants were replica plated to galactose-containing synthetic drop-out media to induce gene expression and incubated at 30°C overnight before being shifted to restrictive temperature. Suppressor plasmids were isolated from thermoresistant colonies, re-transformed into the mutant strains to confirm suppressor activity, and screened by diagnostic restriction digest with HindIII to eliminate plasmids containing wild type SEC26. Novel suppressors were then sequenced as above and identified using the BLAST server at NIH.
Microscopy
GFP was fused to the COOH-terminus of GLO3 following a short linker (GGPGG) and expressed under the control of its endogenous promoter in pRS316 (pRC3441a, this study). The haploid gcs1Δ ::KANR deletion strain was crossed to a glo3Δ ::HIS3 strain transformed with the Glo3p-GFP plasmid to generate a double knockout gcs1Δglo3Δ strain containing Glo3p-GFP. Diploid double knockouts were generated by crossing complementary haploid double knockouts (RCY4238a, this study; MATa/α his3Δ0/his3Δ0 leu2Δ0/leu2Δ0 ura3Δ0/ura3Δ0 LYS2/lys2Δ0 MET15/met15Δ0 gcsΔ ::KANR/gcs1Δ ::KANR glo3Δ ::HIS3/glo3Δ ::HIS3 [pRC3341a]). Live cells were analyzed with a Nikon Eclipse E600 microscope equipped with a 100X (1.4NA) lens and 1X optivar (0.08 μm/pixel). A Sensicam EM High Performance camera (The Cooke Corporation; Romulus, MI) was used for image capture using IPLab 3.6.5 software (Scanalytics; Rockville, MD). Blind deconvolution was accomplished using AutoDeblur 9.3 software package (AutoQuant Imaging, Inc.; Watervliet, NY) for 25 ± 5 z stacks with a 0.2 μm slice size for 30 iterations.
Preparation of anti-Sec26p appendage antibodies
GST fused to Sec26p residues E594-A862 in pGEX-4T-1 (pRC2908a, this study) was expressed in E. coli and purified from inclusion bodies. Post-sonication pellets were resuspended in Buffer A (50 mM Tris pH 7.9, 0.5 mM EDTA, 50 mM NaCl, 5% glycerol) with 2% DOC and washed twice with rocking for 15 minutes at room temperature in between. The purified inclusion bodies were resuspended in equal volume 2× sample buffer (4% SDS, 120 mM Tris pH 6.8, 20% sucrose, 5% β-mercaptoethanol, 0.005% bromphenol blue) and resolved by SDS-PAGE. The band containing the fusion protein was excised, cut into slices containing ~100 μg fusion protein, and stored at -20°C. Pre-laying hens were injected intramuscularly with ~100 μg purified fusion protein suspended in PBS every two weeks for a total of three injections. The first two injections were given with 1 part by volume Freund's adjuvant and 2 parts 2% Tween 80 in sterile phosphate-buffered saline (PBS). IgY antibody was partially purified from the egg yolks once laying commenced by emulsification in 25 ml PBS per egg brought to 100 ml with chloroform, followed by centrifugation to separate the aqueous layer. 0.002% sodium azide was added to the IgY-containing aqueous extract which was stored at 4°C.
Invertase assay
2 OD600 units for each sample in duplicate were harvested from log phase cultures. One set of samples (t = 0) was washed, and resuspended in 1 ml 10 mM sodium azide and kept on ice. The other set was washed, resuspended in 1 ml YP + 0.1% D-glucose and incubated at 40°C with shaking for 1 hour (t = 1) and processed as for the t = 0 set. Each sample was processed for two measurements, external invertase was measured on whole cells and total invertase was measured after glass-bead lysis according to standard methods [69]. Percent secretion was calculated for each sample.
Western blotting
For lysate preparation, cultures were grown to early log phase, 0.7 mM CuSO4 was added to the copper-inducible strains, and all cultures were incubated for 3 hours at 30°C to mid log phase (OD600 ~0.5–0.8). 10 OD600 units of cells was collected, washed in 500 μl ice cold TAZ buffer (10 mM Tris pH 7.5, 10 mM NaN3), resuspended in 50 μl TAZ buffer plus protease inhibitors (4 mM PMSF, 10 μg/ml Pepstatin A, 2 mM benzamidine, 2 mM EDTA) and subjected to glass bead lysis for two minutes at 4°C. 70 μl 2× sample buffer was added and the samples heated to 60°C for 10 minutes prior to SDS-PAGE gel electrophoresis. The proteins were transferred to methanol pre-wetted 0.45 μm PVDF membrane (Immobilon-FL; Millipore, Billerica, MA) at 250 mA for 2 hours in transfer buffer (2 g/L Tris base, 14.4 g glycine/L) with 2% methanol. Membranes were probed with the chicken anti-Sec26p appendage antibody described above. The antibody was incubated with the membrane overnight at room temperature at 1:1000 in Tris-buffered saline plus 0.2% Tween-20 (TBST) with 5% nonfat milk. After three ten-minute washes in TBST, blots were probed with alkaline phosphatase conjugated goat anti-chicken antibody (Southern Biotechnology Inc.; Birmingham, AL) at 1:10,000 in TBST for 30 minutes, and then subjected to three additional washes in TBST. Membranes were washed in 100 mM Tris pH 9.5, 100 mM NaCl, 5 mM MgCl2 for 10 minutes and developed with phenylphosphate substituted 1,2 dioxetane (CDP-Star, Perkin-Elmer; Norwalk, CT) imaged in a Fujifilm LAS-3000 cabinet (Fuji Photo Film U.S.A., Inc.; Valhalla, NY).
To evaluate the expression levels of the Sec26p mutants, yeast expressing the sec26ts alleles as the only copy of SEC26 were grown to mid-log phase and then shifted to 40°C for 1 hour prior to lysis. The cultures were pelleted and resuspended at 10 OD600 units/ml in ice cold TAZ (10 mM Tris pH 7.5, 10 mM azide) buffer. 10 OD600 units were collected, and resuspended in 100 μl TAZ buffer plus protease inhibitors and subjected to glass bead lysis for two minutes at 4°C. An additional 150 μl lysis buffer plus protease inhibitors was added, and 300 μl 3× sample buffer (6% SDS, 180 mM Tris pH 6.8, 30% sucrose, 7.5% β-mercaptoethanol, 0.005% bromphenol blue) was added. Samples were heated at 80°C for 10 minutes and 10 μl resolved on an 8% 1.5 mm SDS-PAGE gel. Western blotting was carried out as described above, except the chicken anti-Sec26p appendage antibody was used at 1:2000. Blots were re-probed without stripping with an anti-Elp1 antibody [70] as a cell lysate loading control.
List of abbreviations
The abbreviations used are: ER, endoplasmic reticulum; COPI, Coat Protein complex I; AP2, Adaptin Protein complex 2; GST, glutathione S-transferase; GFP, green fluorescent protein; Arf1, ADP-ribosylation factor-1; GEF, guanine nucleotide exchange factor; GAP, GTPase activating protein; ts, temperature sensitive; 5-FOA, 5-fluoroorotic acid; SCD, synthetic complete media; SCG, synthetic complete media with galactose as the sole carbon source; PBS, phosphate-buffered saline.
Authors' contributions
CJD performed the experimental studies and together with RNC, was involved in the study design, analysis and interpretation of data, drafting the manuscript and figure preparation, PBR created several yeast strains and plasmids used and performed the initial experiments identifying the βCOP appendage domain as critical for viability, GRH and RAC made intellectual contributions to the conception of the study. All authors participated in the design of the study, critiqued and approved of the final manuscript.
Acknowledgments
Acknowledgements
This work was supported by NIH Grant no. 5R01GM069596 to Ruth Collins. Carol DeRegis was supported by NIH NRSA training grants (T32 RR07059 and F32 GM074463). We thank Anthony Bretscher for the kind gift of the PGAL1/10 library.
Contributor Information
Carol J DeRegis, Email: cjd37@cornell.edu.
Peter B Rahl, Email: pbr9@cornell.edu.
Gregory R Hoffman, Email: greg_hoffman@hms.harvard.edu.
Richard A Cerione, Email: rac1@cornell.edu.
Ruth N Collins, Email: rnc8@cornell.edu.
References
- Barlowe C. Traffic COPs of the early secretory pathway. Traffic. 2000;1:371–377. doi: 10.1034/j.1600-0854.2000.010501.x. [DOI] [PubMed] [Google Scholar]
- Kirchhausen T. Three ways to make a vesicle. Nat Rev Mol Cell Biol. 2000;1:187–198. doi: 10.1038/35043117. [DOI] [PubMed] [Google Scholar]
- Duden R. ER-to-Golgi transport: COP I and COP II function (Review) Mol Membr Biol. 2003;20:197–207. doi: 10.1080/0968768031000122548. [DOI] [PubMed] [Google Scholar]
- McMahon HT, Mills IG. COP and clathrin-coated vesicle budding: different pathways, common approaches. Curr Opin Cell Biol. 2004;16:379–391. doi: 10.1016/j.ceb.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Waters MG, Serafini T, Rothman JE. 'Coatomer': a cytosolic protein complex containing subunits of non-clathrin-coated Golgi transport vesicles. Nature. 1991;349:248–251. doi: 10.1038/349248a0. [DOI] [PubMed] [Google Scholar]
- Serafini T, Stenbeck G, Brecht A, Lottspeich F, Orci L, Rothman JE, Wieland FT. A coat subunit of Golgi-derived non-clathrin-coated vesicles with homology to the clathrin-coated vesicle coat protein beta-adaptin. Nature. 1991;349:215–220. doi: 10.1038/349215a0. [DOI] [PubMed] [Google Scholar]
- Hara-Kuge S, Kuge O, Orci L, Amherdt M, Ravazzola M, Wieland FT, Rothman JE. En bloc incorporation of coatomer subunits during the assembly of COP-coated vesicles. J Cell Biol. 1994;124:883–892. doi: 10.1083/jcb.124.6.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler K, Veit M, Stamnes MA, Rothman JE. Bimodal interaction of coatomer with the p24 family of putative cargo receptors. Science. 1996;273:1396–1399. doi: 10.1126/science.273.5280.1396. [DOI] [PubMed] [Google Scholar]
- Eugster A, Frigerio G, Dale M, Duden R. The alpha- and beta'-COP WD40 domains mediate cargo-selective interactions with distinct di-lysine motifs. Mol Biol Cell. 2004;15:1011–1023. doi: 10.1091/mbc.E03-10-0724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duden R, Kajikawa L, Wuestehube L, Schekman R. epsilon-COP is a structural component of coatomer that functions to stabilize alpha-COP. EMBO J. 1998;17:985–995. doi: 10.1093/emboj/17.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins BM, McCoy AJ, Kent HM, Evans PR, Owen DJ. Molecular architecture and functional model of the endocytic AP2 complex. Cell. 2002;109:523–535. doi: 10.1016/S0092-8674(02)00735-3. [DOI] [PubMed] [Google Scholar]
- Owen DJ, Vallis Y, Noble ME, Hunter JB, Dafforn TR, Evans PR, McMahon HT. A structural explanation for the binding of multiple ligands by the alpha-adaptin appendage domain. Cell. 1999;97:805–815. doi: 10.1016/S0092-8674(00)80791-6. [DOI] [PubMed] [Google Scholar]
- Owen DJ, Vallis Y, Pearse BM, McMahon HT, Evans PR. The structure and function of the beta 2-adaptin appendage domain. EMBO J. 2000;19:4216–4227. doi: 10.1093/emboj/19.16.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praefcke GJ, Ford MG, Schmid EM, Olesen LE, Gallop JL, Peak-Chew SY, Vallis Y, Babu MM, Mills IG, McMahon HT. Evolving nature of the AP2 alpha-appendage hub during clathrin-coated vesicle endocytosis. Embo J. 2004;23:4371–4383. doi: 10.1038/sj.emboj.7600445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser JE, Keen J. Deep-etch visualization of proteins involved in clathrin assembly. J Cell Biol. 1988;107:877–886. doi: 10.1083/jcb.107.3.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman GR, Rahl PB, Collins RN, Cerione RA. Conserved structural motifs in intracellular trafficking pathways: structure of the gammaCOP appendage domain. Mol Cell. 2003;12:615–625. doi: 10.1016/j.molcel.2003.08.002. [DOI] [PubMed] [Google Scholar]
- Watson PJ, Frigerio G, Collins BM, Duden R, Owen DJ. Gamma-COP appendage domain – structure and function. Traffic. 2004;5:79–88. doi: 10.1111/j.1600-0854.2004.00158.x. [DOI] [PubMed] [Google Scholar]
- Slepnev VI, De Camilli P. Accessory factors in clathrin-dependent synaptic vesicle endocytosis. Nat Rev Neurosci. 2000;1:161–172. doi: 10.1038/35044540. [DOI] [PubMed] [Google Scholar]
- Lafer EM. Clathrin-protein interactions. Traffic. 2002;3:513–520. doi: 10.1034/j.1600-0854.2002.30801.x. [DOI] [PubMed] [Google Scholar]
- Dafforn TR, Smith CJ. Natively unfolded domains in endocytosis: hooks, lines and linkers. EMBO Rep. 2004;5:1046–1052. doi: 10.1038/sj.embor.7400276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett TJ, Traub LM, Fremont DH. Accessory protein recruitment motifs in clathrin-mediated endocytosis. Structure (Camb) 2002;10:797–809. doi: 10.1016/S0969-2126(02)00784-0. [DOI] [PubMed] [Google Scholar]
- Edeling MA, Mishra SK, Keyel PA, Steinhauser AL, Collins BM, Roth R, Heuser JE, Owen DJ, Traub LM. Molecular switches involving the AP-2 beta2 appendage regulate endocytic cargo selection and clathrin coat assembly. Dev Cell. 2006;10:329–342. doi: 10.1016/j.devcel.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Nie Z, Hirsch DS, Randazzo PA. Arf and its many interactors. Curr Opin Cell Biol. 2003;15:396–404. doi: 10.1016/S0955-0674(03)00071-1. [DOI] [PubMed] [Google Scholar]
- Spang A. ARF1 regulatory factors and COPI vesicle formation. Curr Opin Cell Biol. 2002;14:423–427. doi: 10.1016/S0955-0674(02)00346-0. [DOI] [PubMed] [Google Scholar]
- Kahn RA, Volpicelli-Daley L, Bowzard B, Shrivastava-Ranjan P, Li Y, Zhou C, Cunningham L. Arf family GTPases: roles in membrane traffic and microtubule dynamics. Biochem Soc Trans. 2005;33:1269–1272. doi: 10.1042/BST0331269. [DOI] [PubMed] [Google Scholar]
- Donaldson JG, Honda A, Weigert R. Multiple activities for Arf1 at the Golgi complex. Biochim Biophys Acta. 2005;1744:364–373. doi: 10.1016/j.bbamcr.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Donaldson JG, Honda A. Localization and function of Arf family GTPases. Biochem Soc Trans. 2005;33:639–642. doi: 10.1042/BST0330639. [DOI] [PubMed] [Google Scholar]
- D'Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- Jackson CL, Casanova JE. Turning on ARF: the Sec7 family of guanine-nucleotide-exchange factors. Trends Cell Biol. 2000;10:60–67. doi: 10.1016/S0962-8924(99)01699-2. [DOI] [PubMed] [Google Scholar]
- Randazzo PA, Hirsch DS. Arf GAPs: multifunctional proteins that regulate membrane traffic and actin remodelling. Cell Signal. 2004;16:401–413. doi: 10.1016/j.cellsig.2003.09.012. [DOI] [PubMed] [Google Scholar]
- Nie Z, Randazzo PA. Arf GAPs and membrane traffic. J Cell Sci. 2006;119:1203–1211. doi: 10.1242/jcs.02924. [DOI] [PubMed] [Google Scholar]
- Kahn RA, Kern FG, Clark J, Gelmann EP, Rulka C. Human ADP-ribosylation factors. A functionally conserved family of GTP-binding proteins. J Biol Chem. 1991;266:2606–2614. [PubMed] [Google Scholar]
- Bigay J, Gounon P, Robineau S, Antonny B. Lipid packing sensed by ArfGAP1 couples COPI coat disassembly to membrane bilayer curvature. Nature. 2003;426:563–566. doi: 10.1038/nature02108. [DOI] [PubMed] [Google Scholar]
- Lanoix J, Ouwendijk J, Stark A, Szafer E, Cassel D, Dejgaard K, Weiss M, Nilsson T. Sorting of Golgi resident proteins into different subpopulations of COPI vesicles: a role for ArfGAP1. J Cell Biol. 2001;155:1199–1212. doi: 10.1083/jcb.200108017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malsam J, Gommel D, Wieland FT, Nickel W. A role for ADP ribosylation factor in the control of cargo uptake during COPI-coated vesicle biogenesis. FEBS Lett. 1999;462:267–272. doi: 10.1016/S0014-5793(99)01543-4. [DOI] [PubMed] [Google Scholar]
- Bigay J, Casella JF, Drin G, Mesmin B, Antonny B. ArfGAP1 responds to membrane curvature through the folding of a lipid packing sensor motif. EMBO J. 2005;24:2244–2253. doi: 10.1038/sj.emboj.7600714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon PP, Wang X, Rotman M, Huber I, Cukierman E, Cassel D, Singer RA, Johnston GC. Saccharomyces cerevisiae Gcs1 is an ADP-ribosylation factor GTPase-activating protein. Proc Natl Acad Sci USA. 1996;93:10074–10077. doi: 10.1073/pnas.93.19.10074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon PP, Cassel D, Spang A, Rotman M, Pick E, Singer RA, Johnston GC. Retrograde transport from the yeast Golgi is mediated by two ARF GAP proteins with overlapping function. EMBO J. 1999;18:555–564. doi: 10.1093/emboj/18.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon PP, Nothwehr SF, Singer RA, Johnston GC. The Gcs1 and Age2 ArfGAP proteins provide overlapping essential function for transport from the yeast trans-Golgi network. J Cell Biol. 2001;155:1239–1250. doi: 10.1083/jcb.200108075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CJ, Bowzard JB, Anido A, Kahn RA. Four ARF GAPs in Saccharomyces cerevisiae have both overlapping and distinct functions. Yeast. 2003;20:315–330. doi: 10.1002/yea.966. [DOI] [PubMed] [Google Scholar]
- Bryson K, McGuffin LJ, Marsden RL, Ward JJ, Sodhi JS, Jones DT. Protein structure prediction servers at University College London. Nucleic Acids Res. 2005:W36–38. doi: 10.1093/nar/gki410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- Jones DT. GenTHREADER: an efficient and reliable protein fold recognition method for genomic sequences. J Mol Biol. 1999;287:797–815. doi: 10.1006/jmbi.1999.2583. [DOI] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- McGuffin LJ, Jones DT. Improvement of the GenTHREADER method for genomic fold recognition. Bioinformatics. 2003;19:874–881. doi: 10.1093/bioinformatics/btg097. [DOI] [PubMed] [Google Scholar]
- Koh IY, Eyrich VA, Marti-Renom MA, Przybylski D, Madhusudhan MS, Eswar N, Grana O, Pazos F, Valencia A, Sali A, et al. EVA: Evaluation of protein structure prediction servers. Nucleic Acids Res. 2003;31:3311–3315. doi: 10.1093/nar/gkg619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B, Eyrich VA. EVA: large-scale analysis of secondary structure prediction. Proteins. 2001:192–199. doi: 10.1002/prot.10051. [DOI] [PubMed] [Google Scholar]
- Traub LM, Downs MA, Westrich JL, Fremont DH. Crystal structure of the alpha appendage of AP-2 reveals a recruitment platform for clathrin-coat assembly. Proc Natl Acad Sci USA. 1999;96:8907–8912. doi: 10.1073/pnas.96.16.8907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm M, Bonifacino JS. Adaptins: the final recount. Mol Biol Cell. 2001;12:2907–2920. doi: 10.1091/mbc.12.10.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schledzewski K, Brinkmann H, Mendel RR. Phylogenetic analysis of components of the eukaryotic vesicle transport system reveals a common origin of adaptor protein complexes 1, 2, and 3 and the F subcomplex of the coatomer COPI. J Mol Evol. 1999;48:770–778. doi: 10.1007/PL00006521. [DOI] [PubMed] [Google Scholar]
- Kent HM, McMahon HT, Evans PR, Benmerah A, Owen DJ. Gamma-adaptin appendage domain: structure and binding site for Eps15 and gamma-synergin. Structure (Camb) 2002;10:1139–1148. doi: 10.1016/S0969-2126(02)00801-8. [DOI] [PubMed] [Google Scholar]
- Liu H, Krizek J, Bretscher A. Construction of a GAL1-regulated yeast cDNA expression library and its application to the identification of genes whose overexpression causes lethality in yeast. Genetics. 1992;132:665–673. doi: 10.1093/genetics/132.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigerio G, Grimsey N, Dale M, Majoul I, Duden R. Two human ARFGAPs associated with COPI-coated vesicles. Traffic. 2007;8:1644–1655. doi: 10.1111/j.1600-0854.2007.00631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CJ, Cavenagh MM, Kahn RA. A family of Arf effectors defined as suppressors of the loss of Arf function in the yeast Saccharomyces cerevisiae. J Biol Chem. 1998;273:19792–19796. doi: 10.1074/jbc.273.31.19792. [DOI] [PubMed] [Google Scholar]
- Cukierman E, Huber I, Rotman M, Cassel D. The ARF1 GTPase-activating protein: zinc finger motif and Golgi complex localization. Science. 1995;270:1999–2002. doi: 10.1126/science.270.5244.1999. [DOI] [PubMed] [Google Scholar]
- Yanagisawa LL, Marchena J, Xie Z, Li X, Poon PP, Singer RA, Johnston GC, Randazzo PA, Bankaitis VA. Activity of specific lipid-regulated ADP ribosylation factor-GTPase-activating proteins is required for Sec14p-dependent Golgi secretory function in yeast. Mol Biol Cell. 2002;13:2193–2206. doi: 10.1091/mbc.01-11-0563.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg J. Structural and functional analysis of the ARF1-ARFGAP complex reveals a role for coatomer in GTP hydrolysis. Cell. 1999;96:893–902. doi: 10.1016/S0092-8674(00)80598-X. [DOI] [PubMed] [Google Scholar]
- Lewis SM, Poon PP, Singer RA, Johnston GC, Spang A. The ArfGAP Glo3 is required for the generation of COPI vesicles. Mol Biol Cell. 2004;15:4064–4072. doi: 10.1091/mbc.E04-04-0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahara N, Sato K, Nakano A. The Arf1p GTPase-activating protein Glo3p executes its regulatory function through a conserved repeat motif at its C-terminus. J Cell Sci. 2006;119:2604–2612. doi: 10.1242/jcs.02997. [DOI] [PubMed] [Google Scholar]
- Schmid EM, Ford MG, Burtey A, Praefcke GJ, Peak-Chew SY, Mills IG, Benmerah A, McMahon HT. Role of the AP2 beta-appendage hub in recruiting partners for clathrin-coated vesicle assembly. PLoS Biol. 2006;4:e262. doi: 10.1371/journal.pbio.0040262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata S, Koyama S, Kawahara H, Doi N, Maeda T, Toyama-Sorimachi N, Abe K, Suzuki K, Sorimachi H. Stomach-specific Calpain, nCL-2, Localizes in Mucus Cells and Proteolyzes the beta-Subunit of Coatomer Complex, beta-COP. J Biol Chem. 2006;281:11214–11224. doi: 10.1074/jbc.M509244200. [DOI] [PubMed] [Google Scholar]
- Eugster A, Frigerio G, Dale M, Duden R. COP I domains required for coatomer integrity, and novel interactions with ARF and ARF-GAP. EMBO J. 2000;19:3905–3917. doi: 10.1093/emboj/19.15.3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SY, Yang JS, Hong W, Premont RT, Hsu VW. ARFGAP1 plays a central role in coupling COPI cargo sorting with vesicle formation. J Cell Biol. 2005;168:281–290. doi: 10.1083/jcb.200404008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 2002;350:3–41. doi: 10.1016/s0076-6879(02)50954-x. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An Information Portal to Biological Macromolecular Structures http://www.rcsb.org/pdb/home/home.do
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DS, Keefe AD. Random Mutagenesis by PCR. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editor. Current Protocols in Molecular Biology. Vol. 1. John Wiley & Sons; 2000. pp. 1–8. [DOI] [PubMed] [Google Scholar]
- Nair J, Muller H, Peterson M, Novick P. Sec2 protein contains a coiled-coil domain essential for vesicular transport and a dispensable carboxy terminal domain. J Cell Biol. 1990;110:1897–1909. doi: 10.1083/jcb.110.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Chen CZ, Collins RN. Elp1p, the yeast homolog of the FD disease syndrome protein, negatively regulates exocytosis independently of transcriptional elongation. Mol Cell. 2005;17:841–853. doi: 10.1016/j.molcel.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Chen CZ, Calero M, DeRegis CJ, Heidtman M, Barlowe C, Collins RN. Genetic analysis of yeast Yip1p function reveals a requirement for Golgi-localized rab proteins and rab-Guanine nucleotide dissociation inhibitor. Genetics. 2004;168:1827–1841. doi: 10.1534/genetics.104.032888. [DOI] [PMC free article] [PubMed] [Google Scholar]