Abstract
Background
Idiopathic pulmonary arterial hypertension (IPAH) is a progressive and still incurable disease. Research of IPAH-pathogenesis is complicated by the lack of a direct access to the involved tissue, the human pulmonary vasculature. Various auto-antibodies have been described in the blood of patients with IPAH. The purpose of the present work was therefore to comparatively analyze peripheral blood B lymphocyte RNA expression characteristics in IPAH and healthy controls.
Methods
Patients were diagnosed having IPAH according to WHO (mean pulmonary arterial pressure ≥ 25 mmHg, pulmonary capillary occlusion pressure ≤ 15 mmHg, absence of another explaining disease). Peripheral blood B-lymphocytes of patients and controls were immediately separated by density gradient centrifugation and magnetic beads for CD19. RNA was thereafter extracted and analyzed by the use of a high sensitivity gene chip (Affymetrix HG-U133-Plus2) able to analyze 47000 transcripts and variants of human genes. The array data were analyzed by two different softwares, and up-and down-regulations were defined as at least 1.3 fold with standard deviations smaller than fold-changes.
Results
Highly purified B-cells of 5 patients with IPAH (mean pulmonary artery pressure 51 ± 13 mmHg) and 5 controls were analyzed. Using the two different analyzing methods we found 225 respectively 128 transcripts which were up-regulated (1.3–30.7 fold) in IPAH compared with healthy controls. Combining both methods, there were 33 overlapping up-regulated transcripts and no down-regulated B-cell transcripts.
Conclusion
Patients with IPAH have a distinct RNA expression profile of their peripheral blood B-lymphocytes compared to healthy controls with some clearly up-regulated genes. Our finding suggests that in IPAH patients B cells are activated.
Introduction
Idiopathic pulmonary arterial hypertension (IPAH) is histopathologically characterized by endothelial cell proliferation and formation of plexiform lesions. Plexiform lesions are often found to be surrounded by immune cells, which have been identified as B- and T-lymphocytes, macrophages and mast cells [1,2]. The possible pathogenetic role of the immune system in IPAH is further supported by the close association between various immune disorders and pulmonary arterial hypertension and the frequent finding of autoimmune antibodies and altered cytokine status in serum of IPAH patients [3,4]. B-lymphocytes (B-cells) are fundamental for the humoral immune response due to their potential to differentiate into antibody-producing plasma B-cells. But B-cells also play a crucial role in cell-mediated immune regulation through antigen presentation, production of various cytokines, differentiation of T effector cells and collaboration with antigen-presenting dendritic cells and lymphoid organogenesis. Antibodies directed against pulmonary endothelial cells have been found in IPAH[5]. The generation of these autoantibodies from mature B-cells might be explained by a different RNA expression profile between B-cells of patients with IPAH and healthy control cells and thus might present a different translational functionality. Moreover, the differential RNA-pattern of B-cells in IPAH might provide helpful information to elucidate the pathogenetic role of the immune system in IPAH and might be of diagnostic value in the early detection of the disease.
Methods
Subjects
Patients were diagnosed with IPAH according to WHO if the mean pulmonary artery pressure was ≥ 25 mmHg by right heart catheterization and an extensive clinical work-up did not reveal other conditions responsible for pulmonary arterial hypertension[6]. Patients and healthy volunteers gave their written informed consent and the study was approved by the local institutional review board (Colorado Multiple Institution Review Board [COMIRB]).
Blood collection and B cell separation and RNA-extraction
10 ml of peripheral blood was collected in tubes containing ethylenediamineteraacetic acid and samples were processed within 30–60' after blood drawn under careful and frequent decontamination of the working space and all materials needed (RNAseZAP, Ambion, TX, US, Cat #9790). The blood was diluted in three volumes of phosphate-buffered saline + 2-mM ethylenediamineteraacetic acid + 0.5% bovine serum albumin. The peripheral blood mononuclear cell (PBMC) layer was isolated via density gradient centrifugation (Histopaque 1077, Sigma-Aldrich, St. Louis, USA), at 1200 rpm for 30 min. B cells were magnetically separated from PBMC's using MACS anti-CD19 micro beads (Miltenyi Biotec, Bergisch-Gladbach, Germany). Purity of the B-cell separation was assessed by flow-cytometry after staining 100'000 cells with fluorescently labelled monoclonal antibody against CD 19 (anti-CD19-APC respectively FACS Calibur, BD Biosciences, NY, USA). B-cell pellets were dissolved in 1 ml of TRI reagent (Ambion, Tx, US, Cat) and stored at -80°C until RNA extraction.
All frozen B cells samples were simultaneously defrosted on ice. After thawing, the RNA was extracted using RiboPureTM-Kit (Ambion, Tx, US Cat # 1924) according to the manufacture's instruction and stored at -20°C until RNA microarray was performed.
Microarray data generation
RNA quality assessment, sample preparation, reverse transcription, labeling, high-density oligonucleotide array hybridization, scanning and data analysis were performed according to standard practice [7-10]. Samples were analyzed by the use of the Affymetrix HG-U133-Plus2 gene chip, which is able to analyze 47,000 transcripts and variants, including 38,500 well-characterized human genes due to its high resolution (Affymetrix, CA, US). Fluorescence intensities were quantified using the affymetrix Microarray Analysis Suite 5.0 (MAS5) and Robust Multichip Analysis (RMA) statistical algorithm with default parameters for the array type used in this study (Affymetrix HG-U133-Plus2, CA, US).
Data analysis and statistics
Detailed protocols for data analysis of Affymetrix microarrays and extensive documentation of the sensitivity and quantitative aspects of the method have been described[11]. In brief, the array data were analyzed by GeneChip® Operating Software (GCOS, Affymetrix, CA, US) and genesprings software (GSS, Agilent Technologies, CA, US). Both softwares are able to statistically analyze quantitative signal expression levels retrieved from the Affymetrix microarray with GCOS mainly used for comparison of expression profiles between single patients across groups and GSS used to compare differential expression profiles between groups (e.g. healthy vs diseased). The raw data from array scans were averaged across all gene probes on each array by MAS5 and RMA, two different mathematical algorithms to process, background-correct and normalize raw data from microarray gene chips, thereafter, a scaling factor was applied to bring the average intensity for all probes on the array to 500. For further normalization of the raw data all signal values were log transformed (log base 2), values below 0.01 were set to 0.01, each measurement was divided by the 50.0th percentile of all measurements in that sample and each gene was divided by the median of its measurements in all samples. To define up- and down-regulated genes in IPAH versus healthy controls, all genes whose flags were present or marginal in all 5 IPAH samples (for up-regulated genes) or not-present (for down-regulated genes) in comparison with controls were selected as first filter technique by GSS. As second filter, up- and down regulated genes were selected if they were present or marginal in 4 of the 5 IPAH versus control samples and if a statistical difference was present between IPAH and control values (Student's t-test, p-value cut-off 0.05). By using GSS, all genes with normalized data values >1.3 fold higher respectively lower than in the other groups were selected. By our second software, the GCOS, we select all genes which were up- or down-regulated in IPAH vs. controls in at least 15 of the possible 25 comparisons with a fold change (FC) greater than the calculated standard deviation for each gene.
Results
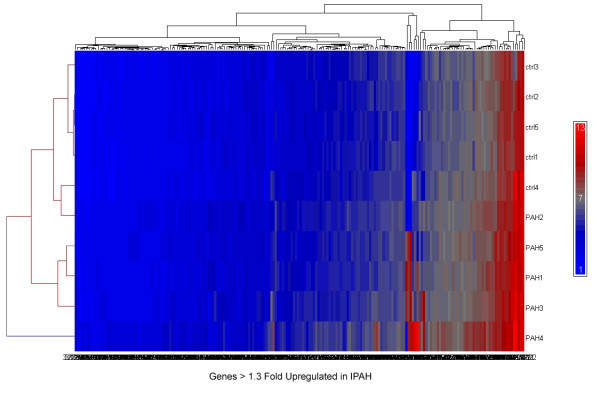
B-cells from five Caucasian patients with severe IPAH (mean age 46 ± 8.1 years, 3 females, mean pulmonary artery pressure 51 ± 13 mmHg, mean cardiac index 2.2 ± 0.2 ml/min/m-2) and five healthy Caucasian controls (mean age 47 ± 8.7 years, 3 females) were analyzed. Four of the patients were on intravenous epoprostenol therapy. The B-cell purity checked by flow cytometry detecting fluorescently labeled CD-19 was > 97%. B-cell RNA and raw data quality was good as per the specialist in the GeneChip processing core facility of the university of Colorado Health Science Centre (UCHSC). Using GCOS we found 225 genes which were at least 1.3 fold up-regulated in IPAH vs. controls (1.3 – 30.7 fold, SD < FC). Using GSS we found 128 up-regulated genes (1.3–178 fold). Combining analysis by both methods, we found overlap of 33 up-regulated genes (table 1, figure 1). Of interest, many of the up-regulated genes belong to biological processes involved in inflammation and immune responses, suggesting the activation of B cells in patients with IPAH In contrast, we found no down-regulated genes.
Table 1.
Up-regulated genes in IPAH vs. controls: at least 1.3 fold up-regulated in GCOS and GSS
| Gene Name | Fold Change | Description | Gene Symbol | GO Biological Process Description |
| 205033_s_at | 30.76 | defensin, alpha 1, myeloid-related sequence | DEFA1 /// DEFA3 | xenobiotic metabolism /// response to virus /// defense response to bacteria /// defense response to fungi /// defense response /// response to pest, pathogen or parasite |
| 204351_at | 15.05 | S100 calcium binding protein P | S100P | endothelial cell migration |
| 232629_at | 10.03 | prokineticin 2 | PROK2 | activation of MAPK activity /// angiogenesis /// anti-apoptosis /// chemotaxis /// inflammatory response /// elevation of cytosolic calcium ion concentration /// neuropeptide signaling pathway /// spermatogenesis /// cell proliferation /// sensory percept |
| 202917_s_at | 5.678 | S100 calcium binding protein A8 (calgranulin A) | S100A8 | inflammatory response |
| 211654_x_at | 5.479 | major histocompatibility complex, class II, DQ beta 1 | HLA-DQB1 | immune response /// immune response /// antigen presentation, exogenous antigen /// antigen processing, exogenous antigen via MHC class II |
| 209773_s_at | 5.435 | ribonucleotide reductase M2 polypeptide | RRM2 | DNA replication /// deoxyribonucleoside diphosphate metabolism /// DNA replication |
| 220005_at | 5.189 | G protein-coupled receptor 86 | P2RY13 | signal transduction /// G-protein coupled receptor protein signaling pathway |
| 225987_at | 4.787 | likely ortholog of mouse tumor necrosis-alpha-induced adipose-related protein | STEAP4 | fat cell differentiation /// electron transport |
| 209773_s_at | 5.435 | ribonucleotide reductase M2 polypeptide | RRM2 | DNA replication /// deoxyribonucleoside diphosphate metabolism /// DNA replication |
| 220005_at | 5.189 | G protein-coupled receptor 86 | P2RY13 | signal transduction /// G-protein coupled receptor protein signaling pathway |
| 225987_at | 4.787 | likely ortholog of mouse tumor necrosis-alpha-induced adipose-related protein | STEAP4 | fat cell differentiation /// electron transport |
| 212999_x_at | 4.322 | major histocompatibility complex, class II, DQ beta 1 | HLA-DQB1 | immune response /// immune response /// antigen presentation, exogenous antigen /// antigen processing, exogenous antigen via MHC class II |
| 220167_s_at | 3.448 | TP53TG3 protein | TP53TG3 | --- |
| 223204_at | 3.429 | hypothetical protein DKFZp434L142 | DKFZp434L142 | --- |
| 213975_s_at | 2.807 | lysozyme (renal amyloidosis) | LYZ /// LILRB1 | carbohydrate metabolism /// cell wall catabolism /// cytolysis /// defense response to bacteria /// immune response /// response to virus /// tRNA aminoacylation for protein translation |
| 209514_s_at | 2.779 | RAB27A, member RAS oncogene family | RAB27A | intracellular protein transport /// small GTPase mediated signal transduction /// protein transport |
| 228898_s_at | 2.13 | similar to putative NADH oxidoreductase complex I subunit homolog. | SMARCB1 | chromatin remodeling /// transcription /// regulation of transcription from RNA polymerase II promoter /// cell cycle /// DNA integration /// negative regulation of progression through cell cycle /// retroviral genome replication /// regulation of transcription |
| 201310_s_at | 2.039 | chromosome 5 open reading frame 13 | C5orf13 | --- |
| 227724_at | 1.956 | hypothetical gene supported by AK091744 | LOC439987 | --- |
| 208704_x_at | 1.92 | amyloid beta (A4) precursor-like protein 2 | APLP2 | G-protein coupled receptor protein signaling pathway |
| 222688_at | 1.916 | yf40c04.s1 Soares fetal liver spleen 1NFLS Homo sapiens cDNA clone IMAGE:129318 3', mRNA sequence. | PHCA | protein biosynthesis /// ceramide metabolism |
| 214864_s_at | 1.852 | glyoxylate reductase/hydroxypyruvate reductase | GRHPR | L-serine biosynthesis /// excretion /// metabolism /// metabolism |
| 230126_s_at | 1.774 | KIAA0876 protein | JMJD2B | regulation of transcription, DNA-dependent |
| 207426_s_at | 1.774 | tumor necrosis factor (ligand) superfamily, member 4 (tax-transcriptionally activated glycoprotein 1, 34kDa) | TNFSF4 | immune response /// signal transduction /// cell-cell signaling /// positive regulation of cell proliferation |
| 212998_x_at | 1.759 | major histocompatibility complex, class II, DQ beta 2 | HLA-DQB2 | immune response /// immune response /// antigen presentation, exogenous antigen /// antigen processing, exogenous antigen via MHC class II |
| 212495_at | 1.745 | KIAA0876 protein | JMJD2B | regulation of transcription, DNA-dependent |
| 221581_s_at | 1.676 | Williams-Beuren syndrome chromosome region 5 | LAT2 | intracellular signaling cascade /// calcium-mediated signaling /// immune response /// mast cell degranulation /// B cell activation |
| 208248_x_at | 1.672 | amyloid beta (A4) precursor-like protein 2 | APLP2 | G-protein coupled receptor protein signaling pathway |
| 226456_at | 1.66 | hypothetical protein MGC24665 | MGC24665 | --- |
| 208703_s_at | 1.651 | amyloid beta (A4) precursor-like protein 2 | APLP2 | G-protein coupled receptor protein signaling pathway |
| 212496_s_at | 1.625 | KIAA0876 protein | JMJD2B | regulation of transcription, DNA-dependent |
| 223445_at | 1.588 | dystrobrevin binding protein 1 | DTNBP1 | organelle organization and biogenesis /// sensory perception /// visual perception /// response to stimulus |
| 225593_at | 1.578 | U7 snRNP-specific Sm-like protein LSM10 | LSM10 | nuclear mRNA splicing, via spliceosome /// mRNA processing |
| 223361_at | 1.537 | similar to HSPC280 | C6orf115 | --- |
| 224948_at | 1.477 | mitochondrial ribosomal protein S24 | MRPS24 | --- |
Figure 1.
Cluster dendrogram of up-regulated genes in 5 patients with idiopathic pulmonary arterial hypertension (lower rows, PAH-number) and 5 healthy controls (upper rows, ctrl-number). The color-scale goes from blue (not up-regulated) to red (highly up-regulated) as indicated (scale on the right).
Discussion
In the present study we comparatively investigated B-cell RNA expression profiles in patients with IPAH and healthy controls. We hereby found that IPAH patients slightly differed from healthy controls with some clearly up-regulated genes consistently found by two different analysis methods.
IPAH is a devastating and progressive condition of unknown etiology affecting the pulmonary circulation with a dismal prognosis [6]. Research on the pathobiology of IPAH on the molecular level is limited by a lack of a direct and early access to the site of pathology, the human lung. One research strategy therefore lays in the analysis of easily obtainable peripheral blood samples from IPAH patients in order to retrieve both information on possible underlying disease mechanisms and potential diagnostic tools. Recently, a strategy of assessing RNA-expression profiles of peripheral blood mononuclear cells by microarrays was introduced and shown to be able to differentiate variously classified pulmonary arterial hypertension patients and healthy controls[12]. In this work we focus this strategy based on microarray technology towards peripheral blood B-cells, based on the hypothesis to find specifically differential RNA expression profiles in a disease where various auto-antibodies have been found in the peripheral blood[4,5,13]. Indeed we found a slightly distinct RNA-expression profile with some up-regulated genes on the transcript level. At this point however, we can only speculate about the biological significance of these up-regulated genes and will in the following discuss some of them with potential value in respect to the pathogenesis of IPAH (table 1). Strikingly, many of the up-regulated transcripts are involved in inflammatory mechanisms, host defense or endothelial function. Human defensins are small cationic peptides involved in various biological processes associated primarily with defensive and regulatory responses to infections by pathological agents but they also have immunoregulatory properties, associated with their ability to bind and activate the G(i)-protein-coupled seven-transmembrane receptors and are chemoattractants for dendritic cells and memory T cells[14]. Increased airway epithelial defensin concentrations were found in association with various pulmonary infections[15] and plasma alpha-defensin concentrations were found increased in pulmonary sarcoidosis, a disease often associated with an until now unidentified infectious agent [16]. It is increasingly recognized that a deregulated immune system plays a pathogenetic role in IPAH[17,18], although a clear idea about an initial trigger and potentially involved pathways is still lacking. The herein found clear up-regulation of the B-cell RNA encoding for defensin alpha 1 in IPAH may indicate that a hitherto unknown infectious trigger may be pathogenetically involved. Other herein-found up-regulated transcripts associated with inflammatory mechanisms are sequences encoding for the major histocompatibility complex class II (HLA_DQB1 and 2), ribonucleotide reductase M2 polypeptides (which confer resistance to hydroxyurea in lymphoblastic and other tumor cell lines [19] and members of the tumor necrosis factor superfamily.
Other herein found up-regulated transcripts in B-cells of IPAH patients are involved in vessel biology, vasomotor regulation, angiogenesis or cell proliferation. Tumor-like proliferating endothelial and smooth muscle cell accumulating in the so called plexiform lesions are the cornerstone of histologic finding in IPAH. The S-100 calcium binding protein is not only a marker of tumor cell lines (e.g. melanoma or neurogenous tumors), it is also involved in cell proliferation and vasoconstriction[20,21]. Prokineticins, herein found 10-fold up-regulated in IPAH patient B-cells vs. healthy controls, are multifunctional secreted proteins able to activate distinct endogenous G-protein coupled pathways, thereby stimulating Ca2+ mobilization and cAMP accumulation[22]. Prokineticins also play a role in circadian rhythms [23], they also seem to have different pathophysiological roles in various endothelial cell systems[24,25]. Another potentially interesting herein found up-regulated transcript encodes for the purinergic receptor P2Y (or G-protein coupled receptor 86), which has not only been described playing a role in some leukemias and cancers[26,27], but has recently been implicated in the risk of atherothrombosis, namely ischemic stroke, myocardial infarction and venous thromboembolism[28]. This protein deserves being evaluated by future research in IPAH, the intrinsic illness of the pulmonary vasculature where microthrombosis is one of the key pathologic features. Interestingly, also transcripts encoding for different types of amyloid beta precursor-like proteins are herein consistently found up-regulated in IPAH. Amyloid beta proteins are important initiating molecules in the pathogenesis of Alzheimer's disease[29]. But the beta amyloid precursor protein is also highly expressed in the endothelium on neoforming vessels suggesting that it may play a role during angiogenesis[30]. An association between pulmonary arterial hypertension and Alzheimer's disease has not been described so far; however, autopsy studies reveal that venous thrombosis and atherosclerotic cardiovascular diseases are highly common comorbidities in Alzheimer patients, so the question of a potential association with pulmonary vascular disease as well may merit evaluation in light of our findings[31].
Our study has several limitations: our sample size investigating 5 patients and controls each is rather small, however, it included a broad and costly gene chip in order to retrieve the highest amount of possibly involved genes. Another limitation of our study is that we do not know if changes in the peripheral blood B-cell RNA expression profiles found are related to, cause or consequence of the pressure elevation found in the pulmonary vasculature. Furthermore, therapy might influence gene expression profiles in general. Preliminary data of the present study however could not observe such effect between B-cells from patients with and without epoprostenol treatment. The study of peripheral blood B-cell RNA expression profiles has further intrinsic limitation, as we do not know whether similar up-regulated transcripts would be found in the pulmonary vasculature itself. Finally, the biological significance of the genes detected has not been investigated by functional analyses. These issues will be addressed by subsequent studies. Despite these limitations, our studies suggest that B cells are activated in patient with IPAH. We strongly believe that the results present herein contribute significantly to our understanding of the pathogenesis of IPHA and thus might help to find new treatment strategies for this still incurable, devastating disease.
Conclusion
We found that patients with IPAH express a distinct RNA expression profile in their peripheral blood B-lymphocytes that clearly suggests activation of B cells when compared with healthy controls The up-regulated transcripts herein described may help to direct future research on the pathogenesis of pulmonary arterial hypertension.
Abbreviations
B-cells = B lymphocytes, IPAH = idiopathic pulmonary arterial hypertension, PBMC = peripheral blood mononuclear cells, RNA = ribonucleic acid
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
SU, LTS and NV have made substantial contributions to conception and design, acquisition, analysis and interpretation of data. SU wrote the manuscript. LCH and RS have been involved in drafting the manuscript and revised it critically for important intellectual content, LTS and NV have given final approval of the version to be published. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
The authors are grateful to Dr. Andrew Fontenot at the University of Colorado Denver and Health Sciences Center and the people in his laboratory for their most valuable technical assistance with B cell purification and characterization and to Ted Shade in the microarray core facility for assistance with data analysis.
Contributor Information
Silvia Ulrich, Email: ulris@bluewin.ch.
Laima Taraseviciene-Stewart, Email: laima.taraseviciene@uchsc.edu.
Lars C Huber, Email: lars.huber@usz.ch.
Rudolf Speich, Email: rudolf.speich@usz.ch.
Norbert Voelkel, Email: nvoelkel@mcvh-vcu.edu.
References
- Voelkel NF, Cool C. Pathology of pulmonary hypertension. Cardiol Clin. 2004;22(3):343–351. doi: 10.1016/j.ccl.2004.04.010. [DOI] [PubMed] [Google Scholar]
- Voelkel NF, Cool C, Lee SD, Wright L, Geraci MW, Tuder RM. Primary pulmonary hypertension between inflammation and cancer. Chest. 1998;114(3 Suppl):225S–230S. doi: 10.1378/chest.114.3_Supplement.225S. [DOI] [PubMed] [Google Scholar]
- Dorfmuller P, Perros F, Balabanian K, Humbert M. Inflammation in pulmonary arterial hypertension. Eur Respir J. 2003;22(2):358–363. doi: 10.1183/09031936.03.00038903. [DOI] [PubMed] [Google Scholar]
- Yanai-Landau H, Amital H, Bar-Dayan Y, Levy Y, Gur H, Lin HC, Alosachie IJ, Peter JB, Shoenfeld Y. Autoimmune aspects of primary pulmonary hypertension. Pathobiology. 1995;63(2):71–75. doi: 10.1159/000163936. [DOI] [PubMed] [Google Scholar]
- Tamby MC, Chanseaud Y, Humbert M, Fermanian J, Guilpain P, Garcia-de-la-Pena-Lefebvre P, Brunet S, Servettaz A, Weill B, Simonneau G, Guillevin L, Boissier MC, Mouthon L. Anti-endothelial cell antibodies in idiopathic and systemic sclerosis associated pulmonary arterial hypertension. Thorax. 2005;60(9):765–772. doi: 10.1136/thx.2004.029082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonneau G, Galie N, Rubin LJ, Langleben D, Seeger W, Domenighetti G, Gibbs S, Lebrec D, Speich R, Beghetti M, Rich S, Fishman A. Clinical classification of pulmonary hypertension. J Am Coll Cardiol. 2004;43(12 Suppl S):5S–12S. doi: 10.1016/j.jacc.2004.02.037. [DOI] [PubMed] [Google Scholar]
- Geraci MW, Moore M, Gesell T, Yeager ME, Alger L, Golpon H, Gao B, Loyd JE, Tuder RM, Voelkel NF. Gene expression patterns in the lungs of patients with primary pulmonary hypertension: a gene microarray analysis. Circ Res. 2001;88(6):555–562. doi: 10.1161/01.res.88.6.555. [DOI] [PubMed] [Google Scholar]
- Lockhart DJ, Dong H, Byrne MC, Follettie MT, Gallo MV, Chee MS, Mittmann M, Wang C, Kobayashi M, Horton H, Brown EL. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996;14(13):1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- Lee CK, Klopp RG, Weindruch R, Prolla TA. Gene expression profile of aging and its retardation by caloric restriction. Science. 1999;285(5432):1390–1393. doi: 10.1126/science.285.5432.1390. [DOI] [PubMed] [Google Scholar]
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, Bloomfield CD, Lander ES. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286(5439):531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- Massague J, Blain SW, Lo RS. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103(2):295–309. doi: 10.1016/S0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- Bull TM, Coldren CD, Geraci MW, Voelkel NF. Gene expression profiling in pulmonary hypertension. Proc Am Thorac Soc. 2007;4(1):117–120. doi: 10.1513/pats.200605-128JG. [DOI] [PubMed] [Google Scholar]
- Tamby MC, Humbert M, Guilpain P, Servettaz A, Dupin N, Christner JJ, Simonneau G, Fermanian J, Weill B, Guillevin L, Mouthon L. Antibodies to fibroblasts in idiopathic and scleroderma-associated pulmonary hypertension. Eur Respir J. 2006;28(4):799–807. doi: 10.1183/09031936.06.00152705. [DOI] [PubMed] [Google Scholar]
- Wiehler S, Proud D. Interleukin-17A modulates human airway epithelial responses to human rhinovirus infection. Am J Physiol Lung Cell Mol Physiol. 2007;293(2):L505–15. doi: 10.1152/ajplung.00066.2007. [DOI] [PubMed] [Google Scholar]
- Donnarumma G, Paoletti I, Buommino E, Iovene MR, Tudisco L, Cozza V, Tufano MA. Anti-inflammatory effects of moxifloxacin and human beta-defensin 2 association in human lung epithelial cell line (A549) stimulated with lipopolysaccharide. Peptides. 2007 doi: 10.1016/j.peptides.2007.09.009. [DOI] [PubMed] [Google Scholar]
- Ashitani J, Matsumoto N, Nakazato M. Elevated alpha-defensin levels in plasma of patients with pulmonary sarcoidosis. Respirology. 2007;12(3):339–345. doi: 10.1111/j.1440-1843.2007.01061.x. [DOI] [PubMed] [Google Scholar]
- Humbert M, Morrell NW, Archer SL, Stenmark KR, MacLean MR, Lang IM, Christman BW, Weir EK, Eickelberg O, Voelkel NF, Rabinovitch M. Cellular and molecular pathobiology of pulmonary arterial hypertension. J Am Coll Cardiol. 2004;43(12 Suppl S):13S–24S. doi: 10.1016/j.jacc.2004.02.029. [DOI] [PubMed] [Google Scholar]
- Nicolls MR, Taraseviciene-Stewart L, Rai PR, Badesch DB, Voelkel NF. Autoimmunity and pulmonary hypertension: a perspective. Eur Respir J. 2005;26(6):1110–1118. doi: 10.1183/09031936.05.00045705. [DOI] [PubMed] [Google Scholar]
- Efferth T, Verdorfer I, Miyachi H, Sauerbrey A, Drexler HG, Chitambar CR, Haber M, Gebhart E. Genomic imbalances in drug-resistant T-cell acute lymphoblastic CEM leukemia cell lines. Blood Cells Mol Dis. 2002;29(1):1–13. doi: 10.1006/bcmd.2002.0530. [DOI] [PubMed] [Google Scholar]
- Calabretta B, Battini R, Kaczmarek L, de Riel JK, Baserga R. Molecular cloning of the cDNA for a growth factor-inducible gene with strong homology to S-100, a calcium-binding protein. J Biol Chem. 1986;261(27):12628–12632. [PubMed] [Google Scholar]
- Lefranc F, Golzarian J, Chevalier C, DeWitte O, Pochet R, Heizman C, Decaestecker C, Brotchi J, Salmon I, Kiss R. Expression of members of the calcium-binding S-100 protein family in a rat model of cerebral basilar artery vasospasm. J Neurosurg. 2002;97(2):408–415. doi: 10.3171/jns.2002.97.2.0408. [DOI] [PubMed] [Google Scholar]
- Buchi S, Brandli O, Klingler K, Klaghofer R, Buddeberg C. [Inpatient rehabilitation in inpatients with chronic obstructive lung diseases (COPD): effect on physical capacity for work, psychological wellbeing and quality of life] SchweizMedWochenschr. 2000;130(5):135–142. [PubMed] [Google Scholar]
- Prosser HM, Bradley A, Chesham JE, Ebling FJ, Hastings MH, Maywood ES. Prokineticin receptor 2 (Prokr2) is essential for the regulation of circadian behavior by the suprachiasmatic nuclei. Proc Natl Acad Sci U S A. 2007;104(2):648–653. doi: 10.1073/pnas.0606884104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podlovni H, Ovadia O, Kisliouk T, Klipper E, Zhou QY, Friedman A, Alfaidy N, Meidan R. Differential expression of prokineticin receptors by endothelial cells derived from different vascular beds: a physiological basis for distinct endothelial function. Cell Physiol Biochem. 2006;18(6):315–326. doi: 10.1159/000097609. [DOI] [PubMed] [Google Scholar]
- Pasquali D, Rossi V, Staibano S, De Rosa G, Chieffi P, Prezioso D, Mirone V, Mascolo M, Tramontano D, Bellastella A, Sinisi AA. The endocrine-gland-derived vascular endothelial growth factor (EG-VEGF)/prokineticin 1 and 2 and receptor expression in human prostate: Up-regulation of EG-VEGF/prokineticin 1 with malignancy. Endocrinology. 2006;147(9):4245–4251. doi: 10.1210/en.2006-0614. [DOI] [PubMed] [Google Scholar]
- Fujiwara S, Yamashita Y, Choi YL, Watanabe H, Kurashina K, Soda M, Enomoto M, Hatanaka H, Takada S, Ozawa K, Mano H. Transforming activity of purinergic receptor P2Y, G protein coupled, 8 revealed by retroviral expression screening. Leuk Lymphoma. 2007;48(5):978–986. doi: 10.1080/10428190701225882. [DOI] [PubMed] [Google Scholar]
- Hatanaka H, Takada S, Choi YL, Fujiwara S, Soda M, Enomoto M, Kurashina K, Watanabe H, Yamashita Y, Sugano K, Mano H. Transforming activity of purinergic receptor P2Y, G-protein coupled, 2 revealed by retroviral expression screening. Biochem Biophys Res Commun. 2007;356(3):723–726. doi: 10.1016/j.bbrc.2007.03.048. [DOI] [PubMed] [Google Scholar]
- Zee RY, Michaud SE, Diehl KA, Chasman DI, Emmerich J, Gaussem P, Aiach M, Ridker PM. Purinergic receptor P2Y, G-protein coupled, 12 gene variants and risk of incident ischemic stroke, myocardial infarction, and venous thromboembolism. Atherosclerosis. 2007 doi: 10.1016/j.atherosclerosis.2007.07.001. [DOI] [PubMed] [Google Scholar]
- Chen JX, Yan SD. Pathogenic role of mitochondral amyloid-beta peptide. Expert Rev Neurother. 2007;7(11):1517–1525. doi: 10.1586/14737175.7.11.1517. [DOI] [PubMed] [Google Scholar]
- Paris D, Quadros A, Patel N, DelleDonne A, Humphrey J, Mullan M. Inhibition of angiogenesis and tumor growth by beta and gamma-secretase inhibitors. Eur J Pharmacol. 2005;514(1):1–15. doi: 10.1016/j.ejphar.2005.02.050. [DOI] [PubMed] [Google Scholar]
- Fu C, Chute DJ, Farag ES, Garakian J, Cummings JL, Vinters HV. Comorbidity in dementia: an autopsy study. Arch Pathol Lab Med. 2004;128(1):32–38. doi: 10.5858/2004-128-32-CID. [DOI] [PubMed] [Google Scholar]