Abstract
Background
The Yersinia enterocolitica flagellar master regulator FlhD/FlhC affects the expression levels of non-flagellar genes, including 21 genes that are involved in central metabolism. The sigma factor of the flagellar system, FliA, has a negative effect on the expression levels of seven plasmid-encoded virulence genes in addition to its positive effect on the expression levels of eight of the flagellar operons. This study investigates the phenotypes of flhD and fliA mutants that result from the complex gene regulation.
Results
Phenotypes relating to central metabolism were investigated with Phenotype MicroArrays. Compared to the wild-type strain, isogenic flhD and fliA mutants exhibited increased growth on purines and reduced growth on N-acetyl-D-glucosamine and D-mannose, when used as a sole carbon source. Both mutants grew more poorly on pyrimidines and L-histidine as sole nitrogen source. Several intermediates of the tricarboxylic acid and the urea cycle, as well as several dipeptides, provided differential growth conditions for the two mutants. Gene expression was determined for selected genes and correlated with the observed phenotypes. Phenotypes relating to virulence were determined with the chicken embryo lethality assay. The assay that was previously established for Escherichia coli strains was modified for Y. enterocolitica. The flhD mutant caused reduced chicken embryo lethality when compared to wild-type bacteria. In contrast, the fliA mutant caused wild-type lethality. This indicates that the virulence phenotype of the flhD mutant might be due to genes that are regulated by FlhD/FlhC but not FliA, such as those that encode the flagellar type III secretion system.
Conclusion
Phenotypes of flhD and fliA mutants are related to central metabolism and virulence and correlate with gene regulation.
Background
The flagellar hierarchy in Yersinia enterocolitica is believed to be similar to E. coli with FlhD/FlhC constituting the master regulator [1] and FliA the sigma factor [2]. FlhD/FlhC is expressed from the flhD operon, that consists of the flhD and flhC genes [3]. The two proteins form a heterohexameric complex [4] and are referred to as class I in the three-tiered regulatory hierarchy of flagellar expression [for reviews, please, see [5-8]]. The FlhD/FlhC complex binds to upstream sequences of class II genes, including the fliA gene [9]. FliA is the sigma factor of the flagellar system [10] and is required for the expression of the class III flagellar genes [11].
A more detailed investigation of the flagellar hierarchy of Y. enterocolitica demonstrated FliA regulation of eight of the 15 flagellar operons [12]. The expression levels of the remaining seven operons were affected by FlhD/FlhC in a FliA independent manner. In accordance with the E. coli nomenclature, genes that were regulated by FliA were classified as class III [12]. It is noteworthy that the two operons that contain the chemotaxis genes, motA and tar, are both regulated by FliA, while the three operons that contain genes of the flagellar type III secretion system, flhB, fliF and fliL, are independent from FliA. Regulation of the three Y. enterocolitica flagellin genes, fleA, fleB, and fleC, by FliA had been previously shown [13,14].
In addition to their function as flagellar regulators, FlhD/FlhC and FliA are both global regulators that affect the expression of a large number of non-flagellar genes [for a review, please, see [15]]. FlhD/FlhC affects the expression levels of many metabolic genes [16], while FliA affects the expression levels of several plasmid-encoded virulence genes [12]. Genes with a large difference in expression between the wild-type cells and the flhD mutants were genes involved in carbamoylphosphate synthesis and degradation, a pathway that leads into the biosynthesis of pyrimidines and hut genes that are involved in the utilization of histidine [16]. It was concluded that FlhD/FlhC might be involved in the flux of nitrogen, using carbamoylphosphate as a checkpoint. Regulation of the plasmid-encoded virulence genes by FliA was most likely through inhibition of their transcriptional activator VirF [12].
In this study, we extend our phenotypic analysis of Y. enterocolitica flhD and fliA mutants. In agreement with our previous microarray analysis [12], wild-type cells grew better on pyrimidines as nitrogen sources. Interestingly, both mutants grew better on purines as carbon sources. In agreement with their higher expression of the hut genes, wild-type cells grew better on histidine.
Since FliA appeared to be a regulator of plasmid-encoded virulence genes [12], we needed an animal model to study the effect of flagellar proteins on virulence of Y. enterocolitica. A chicken embryo lethality assay (ELA) was previously developed to determine the pathogenic potential of E. coli strains [17]. This assay has been exhaustively studied [17-21]. The typical variability and proper statistical analysis for dichotomous data has been established using the logistic regression analysis and resulting odds ratios. This analysis compares the likelihood of death of any given embryo with a specific bacterial isolate. Although the ELA has been used primarily as an assay for comparing the pathogenic potential of avian E. coli isolates, its design was also intended to permit comparative virulence studies for isogenic mutants of the Enterobacteriacea [17]. For this work, the ELA was modified for Y. enterocolitica strains.
Dose response curves for the virulent 8081v isolate and the 8081c isolate that had been cured of the pYV virulence plasmid were obtained. Lethality of the 8081v isolate, as well as its isogenic flhD and fliA mutants were determined. Lethality caused by the flhD mutant was reduced relative to the 8081v strain, while the fliA mutation did not affect virulence in this model. The possibility of the reduced lethality of the flhD mutant being the consequence of a lacking flagellar type III secretion system is discussed.
Results and discussion
Mutants in flhD and fliA have pleiotropic phenotypes on carbon and nitrogen sources
Phenotypes of the 8081c Y. enterocolitica wild-type strain and its isogenic flhD and fliA mutants were investigated with Phenotype MicroArrays (PM; Biolog, Hayward, CA). PM1 plates contain carbon sources and PM3 plates contain nitrogen sources. To correlate the data with the previously described gene regulation [16], the experiments were performed at 25°C, 37°C, and 4°C. Growth patterns were compared between wild-type 8081c bacteria and their isogenic flhD or fliA mutant strains.
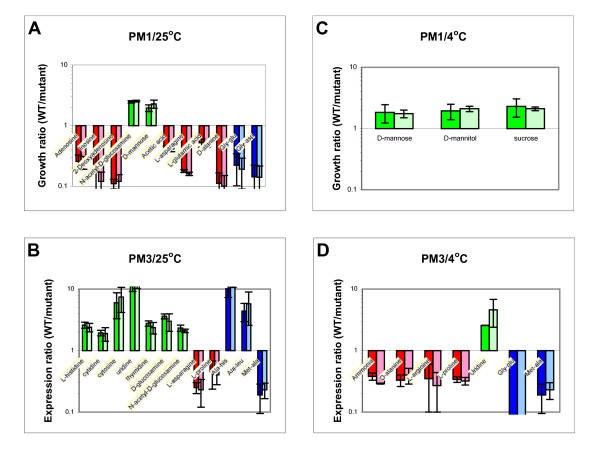
Fig. 1 shows the determined phenotypes of the strains on carbon and nitrogen sources. At 25°C (Panel A), flhD mutants grew better on the purines adenosine, inosine, and 2-deoxyadenosine as carbon sources, as well as acetic acid, L-asparagine, L-glutamate, and D-alanine. Wild-type cells grew better on N-acetyl-D-glucosamine and D-mannose. Wild-type cells also grew better on L-histidine, the pyrimidines cytodine, cytosine, uridine, and thymidine, in addition to D-glucosamine, and N-acetyl-D-glucosamine as nitrogen sources (Panel B). The mutants grew better on L-asparagine and L-proline. In addition, several dipeptides provided differential growth conditions for wild-type cells and flhD mutants. No difference was observed between flhD and fliA mutants.
Figure 1.
Phenotype MicroArrays, nutrients. Growth on sole carbon and nitrogen sources was determined with Phenotype MicroArrays (Biolog, Hayward, CA) for wild-type 8081c cells and their isogenic flhD and fliA mutants. Bacterial solutions in IF-0 were pipetted onto PM1 (carbon sources, Panels A and C) and PM3 (nitrogen sources, Panels B and D) plates. Plates were incubated at 25°C (Panels A and B) or 4°C (Panels C and D). Growth on all plates was determined as OD630. The background OD630 (obtained from well A1) was subtracted from the OD630 for each nutrient on each plate. Growth ratios are expressed as OD630 of the wild-type cells divided by OD630 of each of the mutant strains (darker bars, flhD mutant; lighter bars, fliA mutant) for each nutrient. Nutrients that provided better growth conditions for the wild-type cells are printed in green, nutrients that provided better growth conditions for the mutants are printed in red. Dipeptides are printed in blue. The experiment was performed three to four times and the mean of the population is presented. Error bars indicate one standard deviation (1 s).
At 37°C, bacteria did not grow on the PM1 and PM3 plates. At 4°C, only three carbon sources provided better growth conditions for the wild-type cells (Panel C). These were D-mannose, D-mannitol, and sucrose. Several intermediates of the urea cycle provided better growth conditions for the mutants when used as single nitrogen sources (Panel D). These were ammonia, D-alanine, L-arginine, and L-proline. Uridine was the only one among the pyrimidines that still provided better growth conditions for wild-type cells. The dipeptides met-ala and gly-glu provided better growth conditions for the mutants. Again, there was no difference between the phenotypes of the flhD and the fliA mutants.
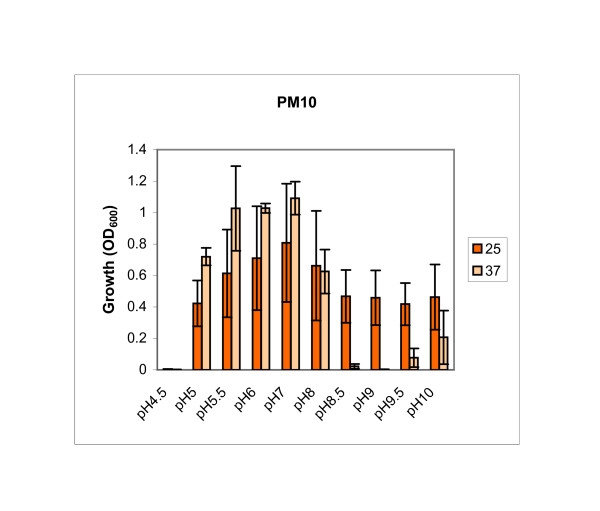
In addition to the PM1 and PM3 plates, growth profiles of wild-type cells and flhD and fliA mutants were determined on PM10 plates. PM10 plates contain media of different pH, supplemented at low pH (4.5) with substrates of amino acid decarboxylases and at high pH (9.0) with substrates of amino acid deaminases. No difference was observed between wild-type cells and both mutants (data not shown). However, it is noteworthy that Y. enterocolitica can grow between a pH of 5 and 10 at 25°C (Fig. 2). At 37°C, all tested strains grew best between a pH of 5 and 8, but growth at pH 5 is better than at 25°C. This might be an adaptation to the human or animal host, where growth conditions are 37°C and the pH is low.
Figure 2.
Phenotype MicroArrays, pH. Growth at different pH levels was determined with Phenotype MicroArrays (Biolog) for wild-type 8081c cells. Bacterial solutions in IF-10 were pipetted onto PM10 plates. Plates were incubated at 25°C (dark orange bars) or 37°C (light orange bars), growth was determined and is expressed as OD630. The experiment was performed three times, the mean of the population is presented. Error bars indicate 1 s.
In summary, flhD and fliA mutants grew better on purines as carbon sources and wild-type cells grew better on pyrimidines as nitrogen sources and L-histidine. Carbon sources that are being degraded by glycolysis provided better growth conditions for wild-type cells. In contrast, carbon sources that are being degraded through the tricarboxylic (TCA) acid cycle provided better growth conditions for the mutants. Mutants also grew better than wild-type cells on intermediates of the urea cycle as nitrogen sources. The mutants did not have a phenotype on the pH plates that was different from the wild-type.
Gene regulation correlates with the observed phenotypes
To correlate gene regulation with the expression of phenotypes, the levels of mRNA were determined for genes involved in the above metabolic pathways and possible transport systems (Table 1). These are the hutC and hutU operons for histidine utilization, the carA and pyrB operons for carbamoylphosphate synthesis and degradation, the deoC operon for purine metabolism, and the acrE operon for the pyruvate dehydrogenase complex. Remarkably, Y. enterocolitica has four operons for the transport of peptides [22]. Of particular interest are the two oligopeptide transport systems. The first one, opp1, contains a hybrid of oppD and oppF (the OppD/OppF hybrid protein contains two ABC_tran domains, but is lacking a C-terminal oligo_HPY domain [23]). The second one, opp2, contains a duplication of the oppA gene. Quantitative PCR (qPCR) was performed for selected genes (Table 2) with RNA isolated from cultures of wild-type 8081c cells and flhD and fliA mutants grown in LB at 25°C until an OD600 of 0.75.
Table 1.
Annotation information for selected metabolic genes and operons
| Operon | Genes in operon | Function of the operon |
| fleC | fleC | Flagellin |
| hutC | hutC, hutI, hutG | Histidine utilization |
| hutU | hutU, hutH, hutP | Histidine utilization |
| carA | carA, carB | Carbamoyl phosphate synthetase |
| pyrB | pyrB, pyrI | Aspartate carbamoyl transferase |
| deoC | deoC, deoA, deoB, deoD | Purine metabolism |
| acre | acrE, lpdA, aceF | Pyruvate dehydrogenase |
| opp1 | oppA1, oppB1, oppC1, oppD/F | Oligopeptide transport |
| opp2 | oppA2, oppA2, oppB2, oppC2, oppD2, oppF2 | Oligopeptide transport |
| dpp1 | dppA1, dppB1, dppC1, dppD1, dpp1F | Dipeptide transport |
| dpp2 | dppA2, dppB2, dppC2, dppD2, dppF2 | Dipeptide transport |
Column 1 indicates the name of the operon that was analyzed.
Column 2 contains all genes in that operon.
Column 3 indicates the function of the operon in metabolism and/or transport. Annotation information was obtained from Sanger [22].
Table 2.
Primers for qPCR
| Gene | Forward primer | Reverse primer |
| dnaE | 5'-AATCACACTCTGCCGCTTATG-3' | 5'-CCATCCGCCAGCACTCAT-3' |
| polA | 5'-GCTGGCTTGCGGATGTAGAT-3' | 5'-AGCACGGCGGTCACTTCA-3' |
| gapA | 5'-CCATCCGTGTTACCGCAGAG-3' | 5'-TCTTAGCACCAGCAGCAATGT-3' |
| fleC | 5'-GTAGTAGCGGTTTTCAGTGCTGT-3 | 5'-CCGGTGGTATTTCAGGTGAT-3' |
| hutI | 5'-ACTCGCAGGTGTCACTTG-3' | 5'-GAGCCAGTAAGCCAATTCAG-3' |
| hutU | 5'-CGATATTCTTCCCAGCTCCA-3' | 5'-GCAACAGATTTTGATGATGCG-3' |
| carA | 5'-AAGGCAGTTGGACTCTGGAA-3' | 5'-CCGTTGGATAAGAAAATGCC-3' |
| carB | 5'-GTTAATTGGCGGCATTATGG-3' | 5'-GACTTCAATCAGGTAAACTTC-3' |
| pyrB | 5'-CCGATTGTCGTTTGAAACCT-3' | 5'-ATTGACCACCGGCACATTGCC-3' |
| deoB | 5'-GCTGACTGAAGGCGGTTACAAT-3' | 5'-GGTGCTGGCGGTTCAACTG-3' |
| acrE | 5'-AGATTCTGGCTAACGACTACG-3' | 5'-ATCACCTGAGCGATGTATGG-3' |
| oppA1 | 5'-TCAAGAACAGCCAGAAGCCCTA-3' | 5'-AAGCGAGCGTGGTTGGAATAC-3' |
| oppB1 | 5'-TGGCGGTGATAGTGGATT-3' | 5'-TGATCTCGGCAATAACTTCC-3' |
| oppC1 | 5'-ACCAAGATCCGTATGTCATT-3' | 5'-TACCGAGCCAGTTAGTGT-3' |
| oppDF | 5'-TGAGATTATCCAGTGCCTTGA-3' | 5'-TAGTCAGAACCGCCATAGC-3' |
| oppB2 | 5'-GCATTCCTACTGGCGGTGATT-3' | 5'-AGGACTAACAGAGGAGCAACGA-3' |
| dppB1 | 5'-CCTTACTGACAGTGATTGC-3' | 5'-ATAATGGCGGTGGTATCG-3' |
| dppB2 | 5'-ACCATTCCTCTCGCCGTTAT-3' | 5'-GACCACAATCACTCGCATCC-3' |
Column 1 presents the genes that were analyzed.
Columns 2 and 3 indicate the primers that were used for the qPCR reaction. Sequence information was obtained from Sanger [22].
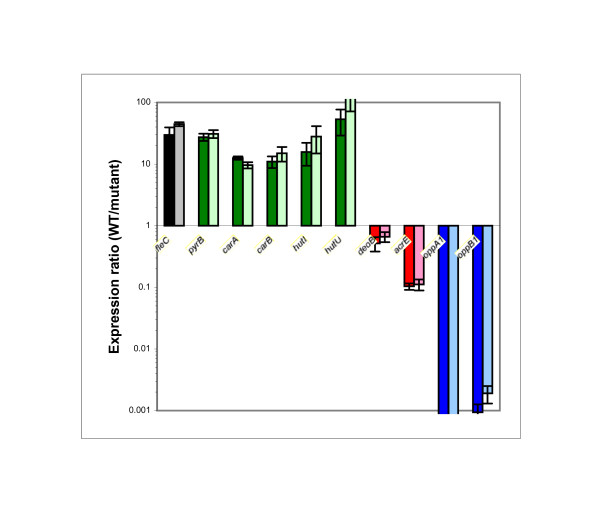
The fleC gene (encoding flagellin) was used as a control for all qPCR experiments and exhibited an expression ratio of approximately 30 fold between the wild-type strain and the mutants (Fig. 3). This is consistent with previous studies [12,16]. The pyrB, carA, carB, hutI, and hutU genes were expressed at least ten times higher in the wild- type cells than in each of the mutants. In contrast, deoB, acrE, oppA1 and oppB1 were expressed higher in the mutants than in the wild-type cells. For the oppA1 and oppB1 genes, the expression ratio was more than 1000 fold. The oppC1 and oppD/F genes appeared unregulated by FlhD/FlhC and FliA, this was also seen in the remaining peptide transport systems (oppB2, dppB1, and dppB2) (data not shown).
Figure 3.
Quantitative PCR. Gene expression was determined for selected genes. Total RNA was isolated from wild-type 8081c cells and their isogenic flhD and fliA mutants. mRNA was reverse transcribed to cDNA and qPCR was performed. Data were normalized with dnaE, polA, and gapA (housekeeping genes). Expression ratios compare the expression levels between wild-type cells and flhD (darker bars) and fliA (lighter bars) mutants. Genes that are expressed higher in wild-type cells are printed in green, genes that are expressed higher in the mutants are printed in red, peptide transport genes are printed in blue. The fleC gene constitutes our internal control and is printed in black. The experiment was performed three times with each of three independently grown bacterial cultures and the mean of the population is presented. Error bars indicate 1 s.
In a previous study [12], FliA did not have an effect on the mRNA levels of the pyrB, carA, carB, hutI, and hutU genes. These data were obtained with RNA from cultures grown to an OD600 of 0.5. It appears that the growth phase of the culture has an impact on regulation. As one example, the class I operon flhD is expressed earlier in growth than the class II gene fliA [24-26]. The pyrB, carA, carB, hutI, and hutU genes seem affected by FlhD/FlhC in a FliA independent manner early in growth. Later in growth, FlhD/FlhC regulation of these genes did require FliA.
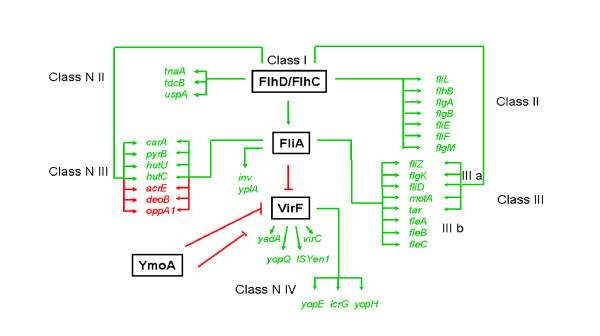
Fig. 4 summarizes our current knowledge of gene regulation in Y. enterocolitica by FlhD/FlhC and FliA. As reported previously, the class I flagellar master regulator FlhD/FlhC regulates class II flagellar genes in a FliA independent manner and class III flagellar genes through FliA [12]. While class IIIb genes are regulated by FlhD/FlhC through FliA alone, class IIIa genes are also affected by FlhD/FlhC in a FliA independent manner. Regulation of metabolic genes is similar to the flagellar genes and a new classification for FlhD/FlhC regulated non-flagellar genes is proposed (Fig. 4). The tnaA, tdcB, and uspA operons (class N II) are regulated by FlhD/FlhC without any current evidence of FliA regulation [12,16]. The carA, pyrB, hutU, hutC, acrE, deoC, and opp1 operons (class N III) are regulated by FliA in late exponential growth (this study), possibly in addition to a FliA independent FlhD/FlhC dependent mechanism in mid-exponential growth [12,16]. In addition (class N IV), FliA represses a number of plasmid-encoded virulence genes [12], most likely through repression of their activator VirF [27]. Regulation of many plasmid-encoded virulence genes is an interplay between VirF [28-30] and the chromatin structure [31,32] that is believed to dislodge the histone-like protein YmoA [33]. The invasion gene inv and the phospholipase gene yplA are regulated by FliA [34,35].
Figure 4.
Summary of the FlhD/FlhC and FliA regulated genes. The cartoon demonstrates our current knowledge of Y. enterocolitica FlhD/FlhC and FliA dependent gene regulation [this study and [12,16,22,27,33-35]]. Regulators are printed in black and are surrounded by boxes. Regulated genes are printed in color. Genes that are positively regulated are printed in green (includes green arrow) and genes that are negatively regulated are printed in red (includes red arrow). The four classes of flagellar and non-flagellar genes are indicated.
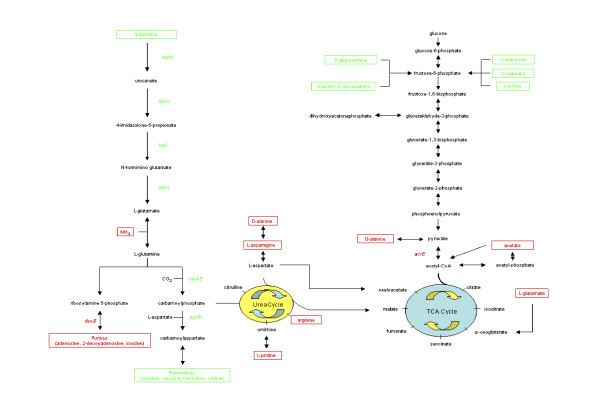
Fig. 5 summarizes our current knowledge of FlhD/FlhC and FliA regulated pathways. L-histidine was one of the nitrogen sources, on which flhD and fliA mutants had exhibited a growth deficiency (Fig. 1). In agreement with this, several hut genes demonstrated higher expression in wild-type cells than in both mutants (Fig. 3). The hut genes serve histidine degradation in numerous bacteria, such as Salmonella typhimurium [36], Klebsiella aerogenes [37], and Bacillus subtilis [38]. Regulation of the hut genes by FlhD/FlhC and FliA (Fig. 3) likely explains the phenotype of reduced growth on L-histidine with the flhD and fliA mutants (Fig. 1).
Figure 5.
Summary of the FlhD/FlhC and FliA regulated pathways. The cartoon demonstrates our knowledge of Y. enterocolitica FlhD/FlhC and FliA affected pathways [this study and [16]]. Metabolites of major pathways are printed in black. Metabolites that feed into these pathways and provide better growth conditions for wild-type cells are printed in green. Metabolites that provide better growth conditions for the mutants are printed in red. Regulated genes are color coded accordingly. Data from the 25°C and 4°C experiments are combined and dipeptides are omitted.
The numerous differential phenotypes on dipeptides can most likely be explained with high regulaton of the Opp1 oligopeptide transport system. Other metabolic phenotypes are more difficult to understand. In addition to the flux of nitrogen through carbamoylphosphate that was postulated previously [16], FlhD/FlhC seems to affect the flux of carbon through pyruvate (Fig. 5). This hypothesis is supported by the observations that FlhD/FlhC and FliA support growth on carbon sources that are degraded through glycolysis, mainly fructose-6-phosphate [39], whereas carbon sources that are being degraded via the TCA cycle provide better growth conditions for the flhD and fliA mutants. In addition, the acrE gene, encoding one of the components of pyruvate dehydrogenase, was more highly expressed in the mutants than in the wild-type cells.
Modification of the ELA for Y. enterocolitica strains
The group of genes that are included in Fig. 4 as regulated by FlhD/FlhC and have not been addressed yet are the virulence genes. This includes early virulence genes, such as inv and yplA, and late virulence genes, such as the yop genes. In an attempt to determine virulence of the Y. enterocolitica flhD and fliA mutants, we searched for a suitable animal model. The ELA is a virulence assay that was developed in an effort to determine the pathogenicity of E. coli strains that were isolated from healthy and diseased poultry. The assay correlated well with a number of phenotypic and genotypic traits that are associated with avian E. coli virulence [17-21]. In related studies, the ELA was compared to an intravenous chicken challenge method [18] and a subcutaneous challenge model [19]. A total of 20 strains yielded nearly identical results for the intraveneous challenge and the ELA [18]. So far, the ELA has been used solely to determine the virulence potential of E. coli isolates. In this study, we modified the well established assay for the use of Y. enterocolitica strains.
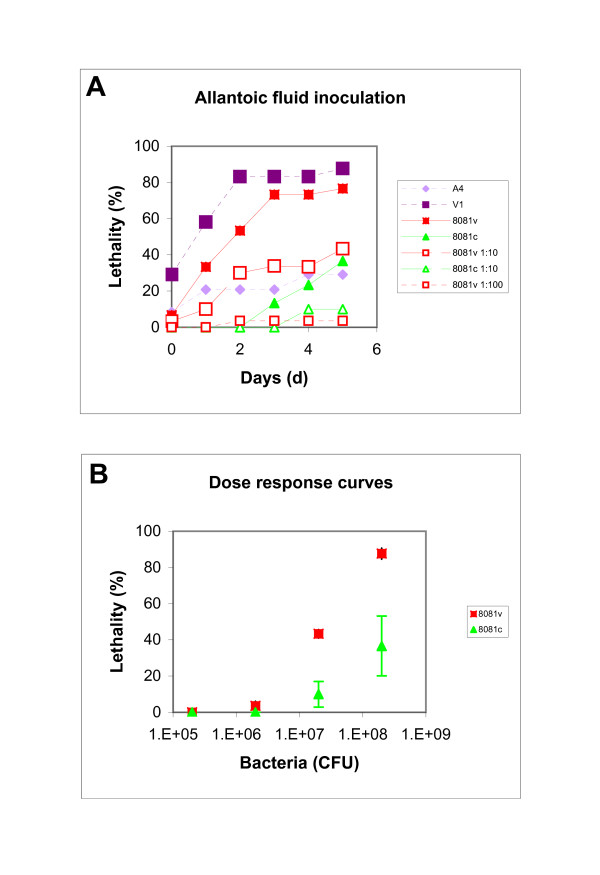
In the first experiment, we determined dose response curves for the Y. enterocolitica 8081v and 8081c strains after allantoic fluid inoculation. Previously described E. coli strains [19] were used as positive and negative controls. Inoculation into the allantoic fluid yielded good lethality by the V1 (88%) and 8081v (77%) strains (Fig. 6A). Lethality caused by the A4 (29%) and 8081c (37%) strains was considerably lower. According to the dose response curve (Fig. 6B), an inoculum of 108 colony forming units (CFU) was chosen for Y. enterocolitica strains.
Figure 6.
Dose response curve for the ELA. The ELA was performed with Y. enterocolitica 8081v and 8081c strains and E. coli A4 and V1 strains as controls. Undiluted Y. enterocolitica cultures contained 2 × 109 CFU/ml and serial dilutions were performed in steps of 1:10. Bacterial cultures (100 μl) were administered to the chicken embryos into the allantoic fluid and lethality of the embryos was determined for five consecutive days (Panel A). The experiment was done twice, the mean of the average is presented. Standard deviations varied between 5 and 30%. Panel C is the dose response curve for the two Y. enterocolitica strains between inocula of 1 × 105 and 1 × 109 CFU after five days. Data are expressed as the mean over the two experiments and error bars indicate 1 s.
While the ELA has previously been shown to correlate with numerous virulence traits in E. coli [17-21], the common animal model that is being used for Y. enterocolitica is the mouse model [2,40]. The first target after orogastric infection of mice are the Peyer's patches of the small intestine [41,42]. Systemic infection involves the spleen, liver, and lungs, a progression that is believed to be similar to humans [43]. To compare the course of infection in chicken embryos with that in mice, we analyzed infected embryo organs and determined their bacterial load. A total of nine embryos (infected with 8081v) from two experiments were opened and the colonization of their internal organs was determined. Organs were colonized at 3.3 × 105 CFU/g for livers, 1.3 × 105 CFU/g for hearts, and 4.6 × 104 CFU/g for spleens. As a comparison, spleens of infected mice are colonized at approximately 104 CFU/g of organ after infection with an inoculum that is identical to ours at 108 CFU [40].
Mutants in flhD, but not fliA, have reduced lethality in the chicken embryo lethality assay
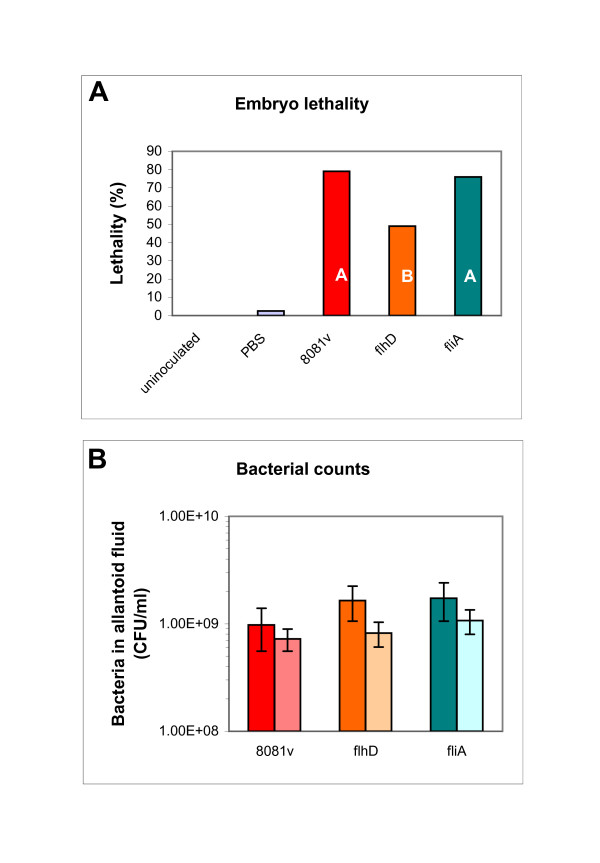
The pathogenic potential of the Y. enterocolitica 8081v strain and its isogenic flhD and fliA mutants was determined with the ELA. Fig. 7A presents the lethality of the inoculated embryos, calculated over three experiments. This includes the two control groups of embryos (uninoculated and PBS) as a comparison. Lethality of the embryos inoculated with the 8081v strain was highest at 79%, lethality caused by the fliA mutant was similar (76%). The flhD mutant exhibited reduced lethality of 49%. When comparing 8081v to the flhD mutant, the odds ratio resulting from the logistic regression analysis for dead embryos was 0.189. This indicates that the likelihood of death was approximately 5 times higher for embryos that were inoculated with the 8081v strain than with the flhD mutant strain. Comparison of the fliA mutant to 8081v yielded an odds ratio of 0.772, indicating that there was no statistically significant difference between the 8081v strain and the fliA mutant. This result was confirmed with the Duncan's multiple group comparison, which found two significantly different groups. The A group contains the 8081v strain and the fliA mutant, the flhD mutant forms its own group B (Fig. 7A).
Figure 7.
Lethality of embryos inoculated with Y. enterocolitica strains (flhD and fliA mutants). The ELA was performed with Y. enterocolitica 8081v, as well as with the flhD and fliA mutants in 8081v. Control groups of embryos were either uninoculated or inoculated with PBS. Embryos were inoculated with 108 CFU of the Y. enterocolitica strains. Lethality of the infected embryos was determined for five consecutive days. The experiment was performed three times. Panel A combines the number of dead embryos across the three experiments for each inoculum. Data are expressed as percentage of the total number of embryos for that inoculum. White capital letters in the Y. enterocolitica data bars represent the different groups from the Duncan's multiple group comparison. Panel B shows the bacterial counts in the allantoic fluid for dead (dark bars) and live embryos (light bars). Data are expressed as mean over the three replicate experiments and error bars indicate 1 s.
Fig. 7B represents the bacterial counts from the allantoic fluid of the embryos that were inoculated with the Y. enterocolitica strains. The average of bacterial counts for each test group of embryos was between 9 × 108 and 1.7 × 109 bacteria/ml of allantoic fluid for dead embryos and 6 × 108 to 1 × 109 bacteria/ml of allantoic fluid for living embryos. While living embryos contained generally somewhat lower numbers of bacteria, differences in growth between the four strains were minor and did not parallel differences in lethality (Fig. 7A). In addition, growth of the strains in liquid LB was very similar for all three strains (8081v, 1.28 gen/h; 8081v fliA, 1.25 gen/h; 8081v flhD, 1.2 gen/h). Obviously, differences in lethality (Fig. 7A) can not be explained by differences in bacterial growth (Fig. 7B). This is consistent with a previous study using the ELA for E. coli strains [20].
The difference in the lethality caused by 8081v and 8081c parallels observations from a mouse model [43]. In this model, the 8081v strain killed mice within 5 days of oral infection and the 8081c strain was unable to kill mice or establish systemic infection [44]. The association of flagella with infection has been demonstrated for several gram-negative bacteria [45-47]. Specifically, mutants of avian E. coli, defective in flagella synthesis, were significantly less able to persist in one day old chicken [48]. Also, flagella were required for persistent infection of chickens by E. coli O157:H7 [49]. This could explain our results with the flhD mutant, but not with the fliA mutant, as neither mutant synthesizes flagella. Results from the fliA mutant parallel a previously published experiment with mice [2].
Our data indicate that the effect of the flhD mutant on virulence is not due to genes that are regulated by FlhD/FlhC in a FliA dependent manner (such as flagellin genes and plasmid-encoded virulence genes), but instead indicates the genes that are regulated by FlhD/FlhC in a FliA independent manner (class II, class N II; Fig. 4)). One of such genes is the flhB gene that encodes one protein of the flagellar specific type III secretion system that is responsible for the transport of certain flagellar proteins [for reviews, please, see [7,50]] and a number of virulence factors [51].
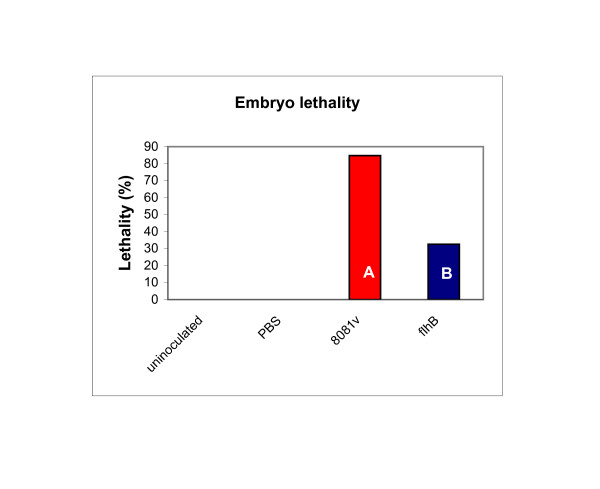
We repeated the ELA, comparing the lethality caused by a flhB mutant to that caused by 8081v (Fig. 8). In three experiments, the odds ratio of lethality for the flhB mutant relative to 8081v was 0.069, indicating that the odds ratio of dying from 8081v was 14 to 15 times higher than for the flhB mutant. The Duncan's multiple group comparison yielded two statistically different groups. These results are similar to the flhD mutant and consistent with a hypothesis that the lethality phenotype of the flhD mutant was due to genes that are regulated by FlhD/FlhC in a FliA independent manner, such as the ones that encode the flagellar type III secretion system.
Figure 8.
Lethality of embryos inoculated with Y. enterocolitica strains (flhB mutant). The ELA was performed with Y. enterocolitica 8081v and its isogenic flhB mutant. Control groups of embryos were either uninoculated or inoculated with PBS. Embryos were inoculated with 108 CFU of the Y. enterocolitica strains. Lethality of the infected embryos was determined for five consecutive days. The experiment was performed three times. The Figure combines the number of dead embryos across the three experiments for each inoculum. Data are expressed as percentage of the total number of embryos for that inoculum. White capital letters in the Y. enterocolitica data bars represent the different groups from the Duncan's multiple group comparison.
Interestingly, possession of the flhD operon might not be the only requirement for virulence of Y. enterocolitica. The genes also may have to be properly regulated. First, cultures of 8081v that had grown only into mid-exponential phase (OD600 of 0.5 to 0.9) had a much reduced lethality (30%) when compared to cultures that had grown into stationary phase (OD600 higher than 1.0). It is possible that these cultures had not reached their maximum motility yet. Second, a plasmid that permitted constitutive expression of flhD did not complement the flhD mutant (odds ratio of 1.266 between 8081v flhD and 8081v flhD pGY17). FlhD/FlhC is a positive regulator of early virulence genes and a repressor of late virulence genes. Therefore, constitutive expression of the flhD operon would not allow for the expression of the late virulence genes. Apparently, full virulence of Y. enterocolitica requires flhD to be regulated in a way that allows for expression of the early virulence genes at the time of infection, and expression of the late virulence genes later during the infection process.
Conclusion
Our phenotypic analysis yielded pleiotropic phenotypes of Y. enterocolitica flhD and fliA mutants. These correlate with previously and newly described gene regulations. Phenotypes include growth differences (between the wild-type cells and the mutants) on several nutrients used as sole carbon or nitrogen sources. Among the phenotypes that can easily be explained by the respective gene regulation is the deficiency of the mutants to grow on L-histidine. This phenotype is likely caused by the simultaneous reduction in the expression level of the hut genes. A dramatic increase in the expression level of the opp1 operon in the mutants probably explains differences between wild-type cells and mutants in the ability to grow on several dipeptides. Pyruvate was postulated as a check-point for the flux of carbon that is affected by FlhD/FlhC.
The second investigated phenotype was virulence, as determined with a chicken embryo model. In this model, flhD mutants, but not fliA mutants, caused a reduced lethality among the embryos when compared to their isogenic wild-type strain. The phenotype of the fliA mutant is consistent with a previous mouse model [2]. The phenotype of the flhD mutant correlates with previous gene regulation [12]. The observation that a mutant in flhB also exhibited a reduced embryo lethality indicates that the lethality phenotype of the flhD mutant might be due to the lack of the flagellar type III secretion system.
Methods
Strains and growth conditions
The two E. coli isolates, V1 and A4, are poultry isolates that were extensively characterized and used to establish and refine the ELA [17-19]. These two isolates were used as positive (virulent; V1) and negative (avirulent; A4) controls. Y. enterocolitica 8081 is one of the American serotypes of Y. enterocolitica, which are human pathogens causing systemic infections more frequently than gastroenteritis [52,53]. The serotype O:8 is virulent for mice, the infection affects the mesenteric lymph nodes, the liver and the spleen [43]. The 8081v strain contains the pYV virulence plasmid, whereas the 8081c strain has been cured of this plasmid [54,55]. The remaining Y. enterocolitica strains are derivatives of 8081v, containing mutations in the flagellar regulator operons, flhD, fliA, and flhB [14]. The flhD operon encodes the master regulator complex FlhD/FlhC [1,3]. The fliA gene encodes the flagellar specific sigma factor FliA [2,10]. The flhB operon encodes components of the flagellar type III secretion system. The mutants were constructed through insertional inactivation of the respective gene in a reverse genetics approach, using the streptomycin Ω cassette. All Y. enterocolitica strains have been kindly provided by Dr. Scott A. Minnich (University of Idaho, Moscow ID). Plasmid pGY17 (flhD and flhC, [1]) was kindly provided by Dr. Virginia L. Miller (Washington University School of Medicine, St. Louis MO) and conjugated into 8081v. Selective antibiotics were 10 μg/ml tetracycline to select for the presence of the plasmid and 40 μg/ml of nalidixic acid to counter-select the E. coli donor strain. The resulting strain SMH999 exhibits wild-type levels of motility and possesses the pYV virulence plasmid.
Cultures of all bacterial isolates and strains were maintained as freezer stocks (-70°C) in 1 ml of Luria Bertani broth (LB: 1 % tryptone broth, 0.5 % NaCl, and 0.5 % yeast extract), containing 15% glycerol or DMSO. Y. enterocolitica 8081v strains were inoculated into liquid LB straight from the freezer stock and incubated over night at 25°C in order to avoid plasmid loss. All other strains were plated onto agar plates prior to the experiment. For Phenotype MicroArrays, bacteria were plated onto R2A plates, as recommended by the manufacturer. For all other experiments, bacteria were inoculated into liquid LB and incubated overnight at the designated temperature.
Phenotype MicroArrays
Phenotype MicroArrays™ (PMs) were obtained from Biolog (Hayward, CA). These are 96 well plates with a different nutrient dried to the bottom of each well [56]. The media are designed to test a multiplicity of cellular phenotypes. An indicator dye turns purple upon transferral of electrons that are produced during respiration. In this study, we used PM1, PM3 and PM10 plates. PM1 plates contain carbon sources, PM3 plates contain nitrogen sources and PM10 plates were designed for pH screening. The plates were inoculated with 100 μl of a cell solution that was standardized with the 85% turbidity standard. Cell suspensions for PM1 and PM3 plates were prepared in IF-0 GN Base inoculation fluid, cell suspensions for PM10 plates in IF-10 GN Base. Sodium pyruvate (20 mM) was added as a carbon source to cell suspensions that were used for PM3 plates. Dye A was used as an indicator dye for all plates as recommended by the manufacturer.
Growth was monitored with a Synergy AT plate reader from Biotek Instruments (Winooski, VT) at 630 nm. Growth at 25°C was analyzed after 24 h, a second reading was taken at 48 h only for PM3 plates. Data for cytosine, uridine, met-ala, and ala-leu are presented from the 48 h reading (Fig. 1B), all other data are from the 24 h reading. Growth at 4°C was analyzed after 240 h. The OD630 of the wild-type culture was divided by the OD630 of the mutant after subtraction of the background in field A1 which contained only the medium without a nutrient. Growth ratios above 1 indicate nutrients that provide better conditions for the wild-type cells. Growth ratios below 1 indicate nutrients that provide better conditions for the mutant. Each experiment was done at least three times and means and standard deviations were determined. A threshold level of a significant difference in growth was defined as growth above 2 or below 0.5, in agreement with previous work [12,16,57]. Only nutrients that allowed one of the strains to grow to an OD630 of at least 0.3 were considered for analysis.
Quantitative PCR
For each experiment, RNA was isolated from three independently grown bacterial cultures. RNA was isolated with the hot phenol/SDS method [58], final cleaning of the RNA was performed with an RNeasy mini column (Qiagen, Valencia, CA). RNA samples were treated with DNaseI (Qiagen) twice. RNA was converted to cDNA using the protocol described by Patrick O. Brown [64].
The qPCR was performed with the iQ SYBR Green Supermix from BioRad (Hercules, CA). The reaction mixture contained 30 – 60 ng cDNA, 50 mM KCl, 20 mM Tris-HCl pH 8.4, 0.2 mM of each dNTP, 0.625 U iTaq DNA polymerase, 3 mM MgCl2, SYBR Green I, 10 nM fluorescein, and 100 nM of each primer (Table 2). Primer pairs were previously described [12,16] or newly designed with Beacon Designer 4.0 (Premier Biosoft International, Palo Alto CA). The enzyme was activated for 3 min at 95°C. Reactions were performed in triplicate at 94°C for 30 sec, 55°C for 30 sec and 72°C for 1 min (50 ×) and monitored in an iCycler iQ qPCR detection system (BioRad). Melting curves were obtained for each reaction and generally contained single peaks. The cDNA dilution (0.01 ng to 10 ng) curves indicated increases in the PCR product concentrations between 1.85 and 2.1 fold per cycle.
Different techniques to normalize expression ratios obtained from qPCR experiments have been discussed [59,60]. In this study, qPCR data were normalized with three housekeeping genes, dnaE, polA, and gapA. These three genes are generally perceived as constitutively expressed with respect to numerous conditions [61,62]. Their expression levels were not affected by FlhD/FlhC or FliA previously [12,16]. Differential threshold cycle crossings (ΔTc) for each gene were corrected by the average difference in Tc for the three housekeeping genes to yield (ΔTcnorm). Expression ratios were calculated as 2-ΔTcnorm. All expression ratios are expressed as mean and standard deviation, obtained from the triplicate measurements of cDNA samples that were obtained from the three independently grown bacterial cultures.
Statistical analysis of the data after the second normalization was performed with the z-test which identifies significant differences between a sample mean (expression ratios) and a set number of 1 (fold). Based upon the low p-values that we obtained for all of our genes, we believe that all these genes are regulated by FlhD/FlhC and FliA.
Embryo lethality assay
Overnight cultures grown at 25°C in LB were washed once with phosphate buffered saline (PBS: NaCl, 8 g/l; KCl, 0.2 g/l; Na2HPO4, 1.44 g/l; K2HPO4, 0.24 g/l; pH 7.2) and diluted in 100 μl of PBS. The number of bacteria used for inoculation was 106 CFU for the E. coli strains and 108 CFU for the Y. enterocolitica strains. The dose response curves for the Y. enterocolitica strains were done between 105 and 2 × 108 CFU. The number of bacteria inoculated into the embryos was verified using the viable count method [18]. The presence of the pYV virulence plasmid in the inocula derived from the 8081v series of strains was confirmed with congo red plates [63]. Bacteria containing the pYV plasmid yielded small pin-point colonies of dark red color on these plates, whereas bacteria that had lost the plasmid yielded larger, less colorful colonies.
Specific pathogen free chicken embryos were purchased from Sunrise Farms, Inc. (Catskill, NY). Upon arrival, the embryos were incubated in a Sportsman incubator, model #1502 (G.Q.F. MFG. CO, Savannah, GA) at 37°C. Humidity was kept constant at 86%. On day twelve after setting, the allantoic fluids of the embryonated eggs were inoculated with the above indicated number of bacteria. Each strain was tested in the ELA in two (dose response curve) or three to four (all other experiments) independent experiments with test groups of embryos of at least 5 (dose response curve) or 10 (all other experiments). Two control groups of embryos (uninoculated and PBS-inoculated embryos) were included in each experiment. After inoculation, incubation was continued for a maximum of seven days. Humidity and temperature of the incubator were checked daily. The embryos were candled each day and the number of deaths was recorded. Data from day 1 were omitted since we observed random death across all test groups of embryos, probably due to trauma from the inoculation. Lethality was expressed as percent of the total number of embryos in each test group. The allantoic fluid of dead embryos was cultured. At the end of the experiment, all remaining eggs were cultured for bacteria. Recovered bacterial counts in the allantoic fluid were typically in the 108 to 109 CFU/ml range for all bacteria used.
Statistical analysis of the ELA data
Because percentage data is not a continuous variable nor is it normally distributed, the percent lethality resulting from each isolate was subjected to a logistic regression analysis. Specifically, odds were calculated for each strain as (p/(1-p)) with p representing the percentage of lethality as determined in the ELA. This was done for each experiment individually. Odds ratios across multiple experiments were determined, using the 8081v strain as baseline reference strain for the logistic regression analysis. Odds ratios presented in the Results section are the result of this regression.
Lethality of several test groups of embryos that were inoculated with the Y. enterocolitica strains was also compared with a Duncan's Multiple Group Comparison. This test determines whether differences in lethality between the test groups are statistically significant. All statistical analyses were performed on SAS v 9.1 (SAS Institute Inc., Cary, NC, USA.).
Preparation of chicken embryo organs
Organs were prepared and tested for bacteria as described [40]. Briefly, we removed livers, hearts, and spleens from 4 to 5 surviving embryos from two of the ELA experiments. All of these embryos had been infected with the 8081v wild-type strain and incubated for five days post infection. Organs were washed twice with 1 ml of PBS and weighed. Livers were minced and stomached in 10 ml of PBS, hearts and spleens in 5 ml. Stomaching was performed in a Stomacher 400 Circulator from Seward (West Sussex, Great Britain) for 2 min. A total of 10 μl of suspension was removed from each bag and serially diluted in steps of 1:10. Bacteria were plated onto LB plates. Bacterial counts were determined after incubation at 25°C overnight and calculated as CFU/g of organ.
Abbreviations
CFU: Colony forming units; DMSO: Dimethylsulfoxide; ELA: Embryo lethality array; LB: Luria Bertani broth; PBS: Phosphate buffered saline; PCR: Polymerase chain reaction; PM: Phenotype MicroArray; qPCR: Quantitative PCR; TCA: Tricarboxylic acid.
Authors' contributions
MKT is a graduate student in the laboratory of BMP. She performed the PM10 arrays, the qPCR, the flhD and fliA ELA, and the dose response curves. NJC and JGY are undergraduate students in the laboratory of BMP. They performed the PM3 and PM1 arrays, respectively. SMH is a research associate in the laboratory of BMP. She did the complementation experiment. PSG and BMP are assistant professors in the Department of Veterinary and Microbiological Sciences. PSG has more than 10 years of experience with the ELA and taught everybody the technique. She also performed some of the allantoic fluid inoculations and helped interpreting the statistical analysis of the ELA data. BMP designed and supervised the project and wrote the manuscript. She also performed the flhB ELA, the organ preparations, and the remaining allantoic fluid inoculations.
All authors have read and approved the manuscript.
Acknowledgments
Acknowledgements
The authors thank Dr. Scott A. Minnich (University of Idaho, Moscow, ID) and Dr. Virginia L. Miller (Washington University, St. Louis MO) for providing strains and plasmids, the researchers from the Sanger Center (Cambridge, UK) for providing their sequence data prior to publication, and Justin Jerdee, Heather Vinson, and Tracey Solseng (all North Dakota State University, Fargo ND) for excellent technical assistance. In addition, we thank Curt Doetkett and Chia-Hao Chang (North Dakota State University, Fargo ND) for the statistical analysis of the ELA data and Dr. Philip Matsumura (University of Illinois at Chicago, Chicago IL) for helpful discussions.
The ELA experiments were supported by the National Research Initiative of the USDA Cooperative State Research, Education and Extension Service, grant number #2006-35604-16675. A previous grant from the United States Poultry and Egg Association Subaward No. RR211-248/1575717 provided the equipment needed for the ELA. Phenotype Microarrays were purchased from a Disease Surveillance grant from USDA/APHIS, grant number #8557-5140-5156. NJC was supported by ND EPSCoR grant EPS-0447679.
Contributor Information
Megan K Townsend, Email: megan.townsend@ndsu.edu.
Nathan J Carr, Email: nathan_carr3@yahoo.com.
Jyoti G Iyer, Email: jiyer@purdue.com.
Shelley M Horne, Email: sm.horne@ndsu.edu.
Penelope S Gibbs, Email: penelope.gibbs@ndsu.edu.
Birgit M Prüß, Email: birgit.pruess@ndsu.edu.
References
- Young GM, Smith MJ, Minnich SA, Miller VL. The Yersinia enterocolitica motility master regulatory operon, flhDC, is required for flagellin production, swimming motility, and swarming motility. J Bacteriol. 1999;181:2823–2833. doi: 10.1128/jb.181.9.2823-2833.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iriarte M, Stainier I, Mikulskis AV, Cornelis GR. The fliA gene encoding sigma 28 in Yersinia enterocolitica. J Bacteriol. 1995;177:2299–2304. doi: 10.1128/jb.177.9.2299-2304.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett DH, Frantz BB, Matsumura P. Flagellar transcriptional activators FlbB and FlaI: gene sequences and 5' consensus sequences of operons under FlbB and FlaI control. J Bacteriol. 1988;170:1575–1581. doi: 10.1128/jb.170.4.1575-1581.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Fleming RT, Westbrook EM, Matsumura P, McKay DB. Structure of the Escherichia coli FlhDC Complex, a Prokaryotic Heteromeric Regulator of Transcription. J Mol Biol. 2006;355:798–808. doi: 10.1016/j.jmb.2005.11.020. [DOI] [PubMed] [Google Scholar]
- Aldridge P, Hughes KT. Regulation of flagellar assembly. Curr Opin Microbiol. 2002;5:160–165. doi: 10.1016/S1369-5274(02)00302-8. [DOI] [PubMed] [Google Scholar]
- Chilcott GS, Hughes KT. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar typhimurium and Escherichia coli. Microbiol Mol Biol Rev. 2000;64:694–708. doi: 10.1128/MMBR.64.4.694-708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnab RM. Flagella and Motility. In: Neidhardt FC, Ingraham JL, Brooks Low K, Magasanik B, Schaechter M, Umbarber HE, editor. Escherichia coli and Salmonella: Cellular and Molecular Biology. Washington, D.C.: American Society for Microbiology; 1996. pp. 123–145. [Google Scholar]
- Prüß BM. FlhD, a transcriptional regulator in bacteria. Recent Res Devel Microbiol. 2000;4:31–42. [Google Scholar]
- Liu X, Matsumura P. The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J Bacteriol. 1994;176:7345–7351. doi: 10.1128/jb.176.23.7345-7351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi K, Kutsukake K, Suzuki H, Iino T. Gene fliA encodes an alternative sigma factor specific for flagellar operons in Salmonella typhimurium. Mol Gen Genet. 1990;221:139–147. doi: 10.1007/BF00261713. [DOI] [PubMed] [Google Scholar]
- Liu X, Matsumura P. An alternative sigma factor controls transcription of flagellar class-III operons in Escherichia coli: gene sequence, overproduction, purification and characterization. Gene. 1995;164:81–84. doi: 10.1016/0378-1119(95)00480-T. [DOI] [PubMed] [Google Scholar]
- Horne SM, Prüß BM. Global gene regulation in Yersinia enterocolitica: effect of FliA on the expression levels of flagellar and plasmid-encoded virulence genes. Arch Microbiol. 2006;185:115–126. doi: 10.1007/s00203-005-0077-1. [DOI] [PubMed] [Google Scholar]
- Kapatral V, Minnich SA. Co-ordinate, temperature-sensitive regulation of the three Yersinia enterocolitica flagellin genes. Mol Microbiol. 1995;17:49–56. doi: 10.1111/j.1365-2958.1995.mmi_17010049.x. [DOI] [PubMed] [Google Scholar]
- Kapatral V, Olson JW, Pepe JC, Miller VL, Minnich SA. Temperature-dependent regulation of Yersinia enterocolitica class III flagellar genes. Mol Microbiol. 1996;19:1061–1071. doi: 10.1046/j.1365-2958.1996.452978.x. [DOI] [PubMed] [Google Scholar]
- Barker CS, Prüß BM. FlhD/FlhC, a global transcriptional regulator in Escherichia coli. In: Prüß BM, editor. Global regulatory networks in enteric bacteria. Trivandrum, Kerala, India: Research Signpost; 2005. pp. 13–30. [Google Scholar]
- Kapatral V, Campbell JW, Minnich SA, Thomson NR, Matsumura P, Prüß BM. Gene array analysis of Yersinia enterocolitica FlhD and FlhC: regulation of enzymes affecting synthesis and degradation of carbamoylphosphate. Microbiol. 2004;150:2289–2300. doi: 10.1099/mic.0.26814-0. [DOI] [PubMed] [Google Scholar]
- Gibbs PS, Maurer JJ, Nolan LK, Wooley RE. Prediction of chicken embryo lethality with the avian Escherichia coli traits complement resistance, colicin V production, and presence of the increased serum survival gene cluster (iss) Avian Dis. 2003;47:370–379. doi: 10.1637/0005-2086(2003)047[0370:POCELW]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Gibbs PS, Wooley RE. Comparison of the intravenous chicken challenge method with the embryo lethality assay for studies in avian colibacillosis. Avian Dis. 2003;47:672–680. doi: 10.1637/7011. [DOI] [PubMed] [Google Scholar]
- Gibbs PS, Petermann SR, Wooley RE. Comparison of several challenge models for studies in avian colibacillosis. Avian Dis. 2004;48:751–758. doi: 10.1637/7176-030404R. [DOI] [PubMed] [Google Scholar]
- Wooley RE, Gibbs PS, Brown TP, Maurer JJ. Chicken embryo lethality assay for determining the virulence of avian Escherichia coli isolates. Avian Dis. 2000;44:318–324. doi: 10.2307/1592546. [DOI] [PubMed] [Google Scholar]
- Nolan LK, Wooley RE, Brown J, Spears KR, Dickerson HW, Dekich M. Comparison of a complement resistance test, a chicken embryo lethality test, and the chicken lethality test for determining virulence of avian Escherichia coli. Avian Dis. 1992;36:395–397. doi: 10.2307/1591518. [DOI] [PubMed] [Google Scholar]
- Sanger http://www.sanger.ac.uk/
- Pfam http://www.sanger.ac.uk/Software/Pfam/
- Amsler CD, Cho M, Matsumura P. Multiple factors underlying the maximum motility of Escherichia coli as cultures enter post-exponential growth. J Bacteriol. 1993;175:6238–6244. doi: 10.1128/jb.175.19.6238-6244.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalir S, McClure J, Pabbaraju K, Southward C, Ronen M, Leibler S, Surette MG, Alon U. Ordering genes in a flagella pathway by analysis of expression kinetics from living bacteria. Science. 2001;292:2080–2083. doi: 10.1126/science.1058758. [DOI] [PubMed] [Google Scholar]
- Prüß BM, Matsumura P. Cell cycle regulation of flagellar genes. J Bacteriol. 1997;179:5602–5604. doi: 10.1128/jb.179.17.5602-5604.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattiau P, Cornelis GR. Identification of DNA sequences recognized by VirF, the transcriptional activator of the Yersinia yop regulon. J Bacteriol. 1994;176:3878–3884. doi: 10.1128/jb.176.13.3878-3884.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis G, Sluiters C, de Rouvroit CL, Michiels T. Homology between virF, the transcriptional activator of the Yersinia virulence regulon, and AraC, the Escherichia coli arabinose operon regulator. J Bacteriol. 1989;171:254–262. doi: 10.1128/jb.171.1.254-262.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert de Rouvroit C, Sluiters C, Cornelis GR. Role of the transcriptional activator, VirF, and temperature in the expression of the pYV plasmid genes of Yersinia enterocolitica. Mol Microbiol. 1992;6:395–409. doi: 10.1111/j.1365-2958.1992.tb01483.x. [DOI] [PubMed] [Google Scholar]
- Skurnik M, Toivanen P. LcrF is the temperature-regulated activator of the yadA gene of Yersinia enterocolitica and Yersinia pseudotuberculosis. J Bacteriol. 1992;174:2047–2051. doi: 10.1128/jb.174.6.2047-2051.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde JR, Fox JM, Minnich SA. Thermoregulation in Yersinia enterocolitica is coincident with changes in DNA supercoiling. Mol Microbiol. 1994;12:187–199. doi: 10.1111/j.1365-2958.1994.tb01008.x. [DOI] [PubMed] [Google Scholar]
- Rohde JR, Luan XS, Rohde H, Fox JM, Minnich SA. The Yersinia enterocolitica pYV virulence plasmid contains multiple intrinsic DNA bends which melt at 37 degrees C. J Bacteriol. 1999;181:4198–4204. doi: 10.1128/jb.181.14.4198-4204.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis GR, Sluiters C, Delor I, Geib D, Kaniga K, Lambert de RC, Sory MP, Vanooteghem JC, Michiels T. ymoA, a Yersinia enterocolitica chromosomal gene modulating the expression of virulence functions. Mol Microbiol. 1991;5:1023–1034. doi: 10.1111/j.1365-2958.1991.tb01875.x. [DOI] [PubMed] [Google Scholar]
- Schmiel DH, Young GM, Miller VL. The Yersinia enterocolitica phospholipase gene yplA is part of the flagellar regulon. J Bacteriol. 2000;182:2314–2320. doi: 10.1128/JB.182.8.2314-2320.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young GM, Badger JL, Miller VL. Motility is required to initiate host cell invasion by Yersinia enterocolitica. Infect Immun. 2000;68:4323–4326. doi: 10.1128/IAI.68.7.4323-4326.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill WJ, Magasanik B. Genetic and metabolic control of histidase and urocanase in Salmonella typhimurium, strain 15–59. J Biol Chem. 1969;244:5392–5402. [PubMed] [Google Scholar]
- Goldberg RB, Magasanik B. Gene order of the histidine utilization (hut) operons in Klebsiella aerogenes. J Bacteriol. 1975;122:1025–1031. doi: 10.1128/jb.122.3.1025-1031.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Sano H, Seki S, Oda M, Fujimura M, Fujita Y. Cloning and sequencing of a 29 kb region of the Bacillus subtilis genome containing the hut and wapA loci. Microbiology. 1995;141:337–343. doi: 10.1099/13500872-141-2-337. [DOI] [PubMed] [Google Scholar]
- Plumbridge J, Vimr E. Convergent pathways for utilization of the amino sugars N-acetylglucosamine, N-acetylmannosamine, and N-acetylneuraminic acid by Escherichia coli. J Bacteriol. 1999;181:47–54. doi: 10.1128/jb.181.1.47-54.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mildiner-Earley S, Miller VL. Characterization of a novel porin involved in systemic Yersinia enterocolitica infection. Infect Immun. 2006;74:4361–4365. doi: 10.1128/IAI.00154-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottone EJ. Yersinia enterocolitica: a panoramic view of a charismatic microorganism. CRC Crit Rev Microbiol. 1977;5:211–241. doi: 10.3109/10408417709102312. [DOI] [PubMed] [Google Scholar]
- Cover TL, Aber RC. Yersinia enterocolitica. N Engl J Med. 1989;321:16–24. doi: 10.1056/NEJM198907063210104. [DOI] [PubMed] [Google Scholar]
- Carter PB. Pathogenicity of Yersinia enterocolitica for mice. Infect Immun. 1975;11:164–170. doi: 10.1128/iai.11.1.164-170.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowe F, O'Gaora P, Maskell D, Cafferkey M, Dougan G. Virulence, persistence, and immunogenicity of Yersinia enterocolitica O:8 aroA mutants. Infect Immun. 1989;57:3234–3236. doi: 10.1128/iai.57.10.3234-3236.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik-Kale P, Raphael BH, Parker CT, Joens LA, Klena JD, Quinones B, Keech AM, Konkel ME. Characterization of Genetically Matched Isolates of Campylobacter jejuni Reveals that Mutations in Genes Involved in Flagellar Biosynthesis Alter the Organism's Virulence Potential. Appl Environ Microbiol. 2007;73:3123–3136. doi: 10.1128/AEM.01399-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa NE, Lauriano CM, McGee R, Klose KE. Phosphorylation of the flagellar regulatory protein FlrC is necessary for Vibrio cholerae motility and enhanced colonization. Mol Microbiol. 2000;35:743–755. doi: 10.1046/j.1365-2958.2000.01745.x. [DOI] [PubMed] [Google Scholar]
- Eaton KA, Suerbaum S, Josenhans C, Krakowka S. Colonization of gnotobiotic piglets by Helicobacter pylori deficient in two flagellin genes. Infect Immun. 1996;64:2445–2448. doi: 10.1128/iai.64.7.2445-2448.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Ragione RM, Sayers AR, Woodward MJ. The role of fimbriae and flagella in the colonization, invasion and persistence of Escherichia coli O78:K80 in the day-old-chick model. Epidemiol Infect. 2000;124:351–363. doi: 10.1017/S0950268899004045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best A, La Ragione RM, Sayers AR, Woodward MJ. Role for flagella but not intimin in the persistent infection of the gastrointestinal tissues of specific-pathogen-free chicks by shiga toxin-negative Escherichia coli O157:H7. Infect Immun. 2005;73:1836–1846. doi: 10.1128/IAI.73.3.1836-1846.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizawa SI. Flagellar assembly in Salmonella typhimurium. Mol Microbiol. 1996;19:1–5. doi: 10.1046/j.1365-2958.1996.344874.x. [DOI] [PubMed] [Google Scholar]
- Young GM, Schmiel DH, Miller VL. A new pathway for the secretion of virulence factors by bacteria: the flagellar export apparatus functions as a protein-secretion system. Proc Natl Acad Sci USA. 1999;96:6456–6461. doi: 10.1073/pnas.96.11.6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford WD, Noce PS, Gutman LT. Pathologic features of enteric infection with Yersinia enterocolitica. Arch Pathol. 1974;98:17–22. [PubMed] [Google Scholar]
- Carniel E, Mazigh D, Mollaret HH. Expression of iron-regulated proteins in Yersinia species and their relation to virulence. Infect Immun. 1987;55:277–280. doi: 10.1128/iai.55.1.277-280.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portnoy DA, Falkow S. Virulence-associated plasmids from Yersinia enterocolitica and Yersinia pestis. J Bacteriol. 1981;148:877–883. doi: 10.1128/jb.148.3.877-883.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portnoy DA, Moseley SL, Falkow S. Characterization of plasmids and plasmid-associated determinants of Yersinia enterocolitica pathogenesis. Infect Immun. 1981;31:775–782. doi: 10.1128/iai.31.2.775-782.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner BR, Gadzinski P, Panomitros E. Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res. 2001;11:1246–1255. doi: 10.1101/gr.186501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüß BM, Campbell JW, Van Dyk TK, Zhu C, Kogan Y, Matsumura P. FlhD/FlhC is a regulator of anaerobic respiration and the Entner-Doudoroff pathway through induction of the methyl-accepting chemotaxis protein Aer. J Bacteriol. 2003;185:534–543. doi: 10.1128/JB.185.2.534-543.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang SE, Daniels DL, Blattner FR. Global regulation of gene expression in Escherichia coli. J Bacteriol. 1993;175:2026–2036. doi: 10.1128/jb.175.7.2026-2036.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong ML, Medrano JF. Real-time PCR for mRNA quantitation. Biotechniques. 2005;39:75–85. doi: 10.2144/05391RV01. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De PK, Pattyn F, Poppe B, Van RN, De PA, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:1–11. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller AC, McEllistrem MC, Stine OC, Morris JG, Jr, Boxrud DJ, Dixon B, Harrison LH. Multilocus sequence typing reveals a lack of diversity among Escherichia coli O157:H7 isolates that are distinct by pulsed-field gel electrophoresis. J Clin Microbiol. 2003;41:675–679. doi: 10.1128/JCM.41.2.675-679.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro JA. Differential action and differential expression of DNA polymerase I during Escherichia coli colony development. J Bacteriol. 1992;174:7262–7272. doi: 10.1128/jb.174.22.7262-7272.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahmanyar M, Cavanaugh DC. Plague manual. World Health Organization; 1976. [Google Scholar]
- RNA to cDNA protocol by Patrick O. Brown http://cmgm.Stanford.edu/pbrown/protocols/aadUTPCoupling Procedure.htm