Abstract
Molecular and biochemical mechanisms that modulate the production of eumelanin or pheomelanin pigments involve the opposing effects of two intercellular signaling molecules, α-melanocyte stimulating hormone (MSH) and agouti signal protein (ASP). ASP is an antagonist of MSH signaling through the melanocyte-specific MSH receptor, although its mechanism(s) of action is controversial. We previously have reported significant down-regulation of all known melanogenic genes during the eumelanin to pheomelanin switch in murine hair follicle melanocytes and in cultured melanocytes treated with recombinant ASP. To identify factors that might be involved in the switch to pheomelanogenesis, we screened ASP-treated melanocytes by using differential display and identified three up-regulated genes: a DNA replication control protein, a basic helix–loop–helix transcription factor, and a novel gene. We have simultaneously identified six down-regulated genes in ASP-treated melanocytes; two of those encode tyrosinase and TRP2, melanogenic genes known to be down-regulated during pheomelanogenesis, which provide good internal controls for this approach. These results suggest that there are complex mechanisms involved in the switch to pheomelanin production, and that these modulated genes might be involved in the pleiotropic changes seen in yellow mice, including the change in coat color.
Keywords: agouti, melanogenesis, pheomelanin, pigmentation, gene expression
In animals, melanocytes can produce two types of melanin, black/brown eumelanin and red/yellow pheomelanin; coat color is determined by the relative proportions of these two melanins in the hair (1–3). The eumelanin/pheomelanin ratio is regulated in mice by two loci, termed agouti and extension, which encode agouti signal protein (ASP) and the melanocyte-stimulating hormone (MSH) receptor, respectively (reviewed in refs. 4 and 5). Wild-type mice have a darker dorsal coat color, consisting of black hairs with subapical yellow stripes, and a lighter ventral coat color. In the skin, ASP is produced by dermal papillae cells, and it acts as a paracrine factor to control whether eumelanin or pheomelanin is produced by follicular melanocytes (6–8). In wild-type agouti mice that carry the A allele, eumelanin is produced by all follicular melanocytes in the first 4 days, presumably because of constitutive expression of MSH. Transient expression of ASP from days 4–6 of the hair growth cycle switches follicular melanocytes to the production of pheomelanin rather than eumelanin; after that, agouti gene expression is turned off, and eumelanin is produced again. Analyses of protein and mRNA expression during such hair growth have shown that tyrosinase-related protein (TRP) 1, TRP2, Pmel17/silver, and the protein encoded at the pinkeyed-dilution locus are completely suppressed during pheomelanogenesis, whereas expression of tyrosinase is reduced but not eliminated (9, 10). This pattern of expression is consistent with the fact that tyrosinase is required for both types of pigment synthesis, but expression of these other melanogenic proteins is required only for eumelanin synthesis.
Mutations at the agouti locus can elicit the production of all yellow or all black hair, depending on whether the mutation leads to overexpression/hyperfunction or nonexpression/nonfunction of ASP, respectively (11–13). For example, the dominant lethal yellow mutation (Ay) results in the production of completely yellow hairs, whereas the nonagouti black (a) leads to the production of completely black hairs. In addition, mice carrying dominant agouti mutations exhibit adult onset obesity, hyperinsulinemia, noninsulin-dependent diabetes, and susceptibility to a variety of spontaneous and/or induced tumors. Several studies have shown that ASP acts as a competitive antagonist of the melanocortin 1 receptor, and the switch between eumelanogenesis and pheomelanogenesis involves the opposing effects of ASP and MSH as ligands for that receptor (14–17). Other groups have shown that some effects of ASP might be mediated by an alteration in calcium channels, suggesting that ASP also may function through other, as yet undefined, receptors to elicit its pleiotropic effects (18–21). Whatever mechanism(s) are involved, agouti, MSH, and melanocortin 1 receptor interact to regulate human pigmentation as well because all three are similarly expressed and functional in human skin and melanocytes (22–26).
We have reported that melanocytes that normally produce eumelanin exclusively in vivo begin to produce significant amounts of pheomelanin when cultured in vitro and that pheomelanogenesis can be further stimulated by in vitro treatment with purified recombinant ASP (27). After treatment with ASP, the expression of genes encoding tyrosinase and other melanogenic proteins is suppressed in cultured melanocytes, which also exhibit other physiologic features characteristic of pheomelanogenesis in vivo, thus providing an in vitro model for characterization of mechanisms and genes involved in this switch. So far, a number of melanosomal proteins involved in eumelanogenesis, as enzymes or as structural components, have been identified by cloning genes responsible for mouse coat colors (5, 28). However, little currently is known about components that actively regulate the pheomelanogenic pathway. To understand mechanisms and genes important to pheomelanogenesis, we have used two-dimensional gel electrophoresis and differential display in our melanocyte culture model to identify proteins and genes modulated during pheomelanogenesis.
MATERIALS AND METHODS
Cell Culture.
The melan-a melanocyte line (29), derived from C57BL/6J nonagouti black mice (a/a B/B C/C), was a kind gift from Dorothy Bennett (St. George’s Hospital Medical School, London). Melanocytes were cultured at 37°C with 10% CO2 in minimal essential medium containing penicillin, streptomycin, sodium pyruvate, nonessential amino acids, 5% fetal bovine serum, 200 nM phorbol-12-myristate-13 acetate, and 100 μM 2-mercaptoethanol at pH 6.9 until reaching semiconfluence, as reported previously (29). Typically, 1.5 × 106 cells were seeded in 15-cm diameter dishes and allowed to attach for 24 hr; treatment with 10 nM recombinant murine ASP (30) then was initiated for 4 days as previously described (27). Controls were cultured in the absence of ASP, but with similar volumes of storage buffer (20 mM Pipes, pH 6.8/50 mM NaCl).
Animals.
C57BL/6J nonagouti black mice (a/a B/B C/C) and C57BL/6J lethal yellow mice (Ay/a B/B C/C) were purchased from The Jackson Laboratory. Lethal yellow mice were maintained by matings between heterozygous lethal yellow (Ay/a) male and black (a/a) female mice.
Metabolic Labeling and Two-Dimensional Gel Electrophoresis.
Melan-a melanocytes were cultured with or without 10 nM ASP for 4 days as detailed above, then cultured for 18 hr with [35S]methionine (1,175 Ci/mmol, ICN). After metabolic labeling, cells were harvested, solubilized in 1% Nonidet P-40, 0.01% SDS in 0.1 M Tris⋅HCl buffer, pH 7.2 for 1 hr at 4°C, then separated by two-dimensional gel electrophoresis (31). Radioactive bands were visualized by using a PhosphorImager 425 (Molecular Dynamics).
RNA Extraction and Differential Display.
Total cellular RNA was isolated from melan-a cells and from 14-day newborn mouse skin by using an RNeasy total RNA isolation kit (Qiagen) according to the manufacturer’s instructions. Differential display was performed as described in ref. 32 with modifications. After treatment with DNase I (Sigma), 0.2 μg of total RNA was reverse-transcribed with three different one-base-anchored oligo(dT) primers (H-T11A, H-T11C, and H-T11G) containing HindIII restriction sites. Four separate reverse-transcription reactions were performed for each RNA sample by using 0.2 μg of total RNA in 1× reverse-transcription buffer, 10 mM DTT, 20 μM of each dNTP, and 1 μM of primers H-T11A, H-T11C, or H-T11G. The solutions were heated to 65°C for 5 min and cooled to 42°C for 10 min, after which 200 units of Superscript II reverse transcriptase (Life Technologies, Gaithersburg, MD) were added. After incubation at 42°C for 50 min, the mixtures were heated to 75°C for 5 min before storage at −20°C.
PCR of reverse-transcription reactions was carried out in quadruplicate. PCR mixtures contained 0.1 volume of reverse-transcription reaction, 1× PCR buffer, 2 μM each of dGTP, dTTP, and dCTP, 0.2 μM HindIII restriction site-containing an arbitrary 13-mer (GenHunter, Brookline, MA), 0.2 μM of the respective H-T11V (V = A, C or G) oligonucleotide, 0.5 unit of AmpliTaq DNA polymerase (Perkin–Elmer), and 10 μCi of α-[35S]dATP (1,200 Ci/mmol, ICN). Each PCR amplification was carried out on a thermal cycler (Perkin–Elmer GeneAmp PCR System 2400) for a total of 40 cycles at 94°C for 15 sec, at 40°C for 2 min, and at 72°C for 30 sec, followed by 5-min postextension at 72°C. Radiolabeled PCR amplification products were analyzed by electrophoresis in denaturing 6% polyacrylamide gels, and the dried gels were exposed to x-ray film. Four independent samples from ASP-treated cells and from controls were compared side by side on gels to confirm the reproducibility of banding patterns. PCR product bands of interest ranging from 200 to 800 bp were recovered from sequencing gels and reamplified in a 40-cycle PCR in the absence of isotope.
Northern Analysis and Subcloning.
Total RNA (5–10 μg) was denatured, electrophoresed through 1% agarose gels, and then blotted to Nytran membranes (Schleicher & Schuell) using a TurboBlotter System (Schleicher & Schuell) and UV light crosslinking (UV Stratalinker 1800, Stratagene). Radioactive probes were generated by using a Prime-It RmT random prime labeling kit (Stratagene) with α-[32P]dCTP (ICN).
For differential display, Northern blot analysis first was performed using probes generated directly from PCR reamplification. Those samples demonstrating differential expression in Northern blot analysis were subcloned by using the TA Cloning kit (InVitrogen) according to the manufacturer’s instructions, and the Northern blot results were reconfirmed (Northern blots shown in Fig. 3 were derived from these subclones). A cDNA probe specific for glyceraldehyde-3-phospho-dehydrogenase (CLONTECH) was used to standardize RNA loading on the blots.
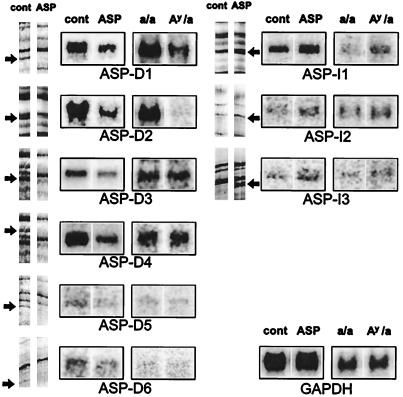
Figure 3.
Three ASP up-regulated bands and six ASP down-regulated bands were identified by differential display. Northern blot analysis of the differentially displayed bands using total RNAs from melan-a cells (left of each group) and from mouse skin samples (right of each group). Five micrograms of total RNA from cultured melan-a cells, with or without ASP treatment, were blotted to the lanes as marked on the left; 10 μg of total RNA from skins of sibling lethal yellow and black mice are blotted on the right as marked. The same blots reprobed with glyceraldehyde-3-phospho-dehydrogenase (GAPDH) are shown as a loading control.
For evaluating expression patterns of genes in vivo, 10 μg of total RNAs from skins of sibling 14-day newborn lethal yellow and nonagouti black mice were blotted and hybridized with 32P-labeled probes. Each blot was scanned with a PhosphorImager 425 (Molecular Dynamics) and quantified by using ImageQuant software (Molecular Dynamics). The results of Northern blot analysis are expressed as mean percentage of the control ± SD. Except where noted, experiments were performed by using a minimum of four different RNA samples.
cDNA Library Screening.
cDNA clones of larger sizes were isolated by cDNA library screening. A cDNA library was synthesized from total RNA obtained from melan-a cells in the pSPORT 1 plasmid (Life Technologies) using a CapFinder cDNA Library Construction Kit and a CapFinder PCR cDNA Synthesis Kit (CLONTECH) with modification of PCR primers. Oligonucleotide probes complementary to cDNA segments obtained by differential display were used to isolate plasmids containing longer cDNAs with the GeneTrapper cDNA Positive selection system (Life Technologies). Isolated positive clones were transformed into Escherichia coli and confirmed with radiolabeled cDNA probes obtained from differential display, as described above. Several positive clones were identified and purified with a QIAfilter plasmid kit (Qiagen), and the longest ones subsequently were sequenced.
DNA Sequencing and DNA Sequence Homology Search.
DNA sequencing from the TA cloning vector used the AmpliCycle Sequencing Kit (Perkin–Elmer). Gene database searches were performed at the National Center for Biotechnology Information by using the blast network service.
RESULTS AND DISCUSSION
Modulation of Melanocyte Proteins During Pheomelanogenesis.
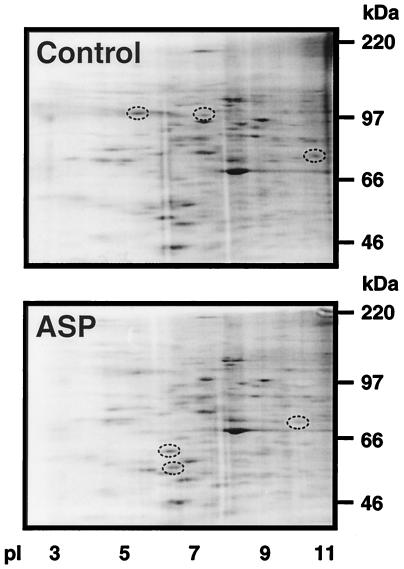
We initially examined protein expression in eumelanogenic and pheomelanogenic melanocytes by using metabolic labeling and two-dimensional gel electrophoresis (Fig. 1). Banding patterns over a wide range of acrylamide concentrations reproducibly demonstrated that not only was the expression of several proteins down-regulated after treatment with ASP, as expected based on our previous studies (9, 27), but that additional spots could be readily identified that represented proteins up-regulated during this process. There also were quantitative differences in the intensities of many of the bands visible in the autoradiographs. These results suggested that the regulation of the pheomelanogenic switch was much more complex than was thought originally, and a molecular approach (differential display) subsequently was used to identify and clone gene(s) that were involved.
Figure 1.
Comparison of two-dimensional gel patterns of control and ASP-treated melanocytes; proteins were metabolically labeled as detailed in Materials and Methods, then separated on 10.5% acrylamide gels and visualized by autoradiography. Circles indicate bands that were reproducibly up- or down-regulated in several independent experiments.
Differential Display.
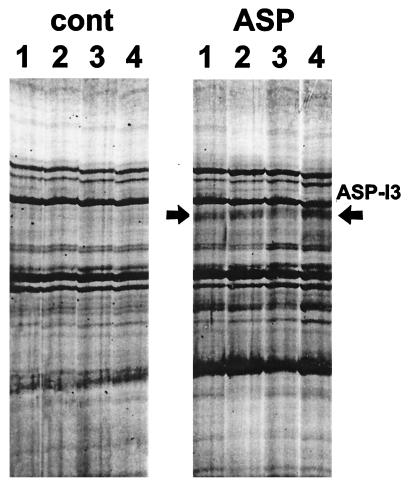
Total RNA was isolated from untreated or from ASP-treated melanocytes, reverse-transcribed, and analyzed by PCR in the presence of α-[35S]dATP. The PCR fragments were displayed on 6% DNA sequencing gels and autoradiographed. A total of 24 5′-arbitrary primers (H-AP1 through H-AP24) were screened against three one-base-anchored oligo 3′(dT) primers (H-T11A, H-T11C, and H-T11G). Each differential display lane yielded 100–150 discrete bands, allowing evaluation of more than 10,000 RNA species thought to represent about 50% of the repertoire of an estimated 15,000–20,000 cellular mRNAs.
The patterns of amplified PCR products from four independent samples of mRNAs from control or ASP-treated melanocytes showed virtually identical banding patterns with each primer set (Fig. 2). Among the 24 primer sets, obvious differences in the reverse transcription–PCR displays were reproducibly found for only 40 bands in all four experiments (in this evaluation we considered only bands that were comparably modulated in all four independent samples). Those 40 bands were further evaluated by using Northern blot analysis with random-labeled cDNA probes; no signal could be detected for nine of those bands, and no significant differences in expression were observed for 22 of the other bands (data not shown). However, significant and reproducible differences in expression were confirmed for the remaining nine bands (Fig. 3). All nine of those PCR fragments also detected other nondifferentially expressed mRNA species caused by heterogeneity of the PCR fragments (not shown), and therefore, each of those fragments was subcloned individually into the TA cloning vector. Several pure clones from each subcloning reaction were used again as probes in Northern blot analysis to identify which clones were differentially expressed. Nine distinctive cDNAs subcloned from those bands showed changes in expression compatible with the results of the initial differential display, although the magnitude of altered expression had been exaggerated by differential display in several of those cDNA clones.
Figure 2.
Example of the reproducibility of patterns generated by reverse transcription–PCR from mRNAs of control and ASP-treated melanocytes in four different experiments. Results from a single primer set are shown as an example, but comparably reproducible patterns were observed for the other 23 primer sets. The arrows point to bands that were modified in expression after ASP treatment.
Three bands (ASP-I1, ASP-I2, and ASP-I3) were up-regulated after ASP treatment, whereas six bands (ASP-D1 through ASP-D6) were down-regulated (Fig. 3). The size of those mRNAs, and other characteristics of their expression, in ASP-treated melanocytes are summarized in Table 1. Some bands (e.g., ASP-D1), which appeared by differential display to be quantitatively turned off or on in response to ASP treatment, were shown in subsequent Northern blot analysis to be expressed at detectable levels even though the pattern of up- or down-regulation was maintained. This discrepancy is no doubt caused by the exponential amplification of PCR that enhances differences in mRNA populations.
Table 1.
Characteristics of genes modulated by ASP
| Band name | ASP treatment, % control ± SD | P vs. control | Probe size, bp | mRNA size, kb | DNA sequence homology by blast search | Sequence identity |
|---|---|---|---|---|---|---|
| Down-regulated genes | ||||||
| ASP-D1 | 62 ± 25 | <0.02 | 298 | 1.8 | Murine tyrosinase | 99% |
| ASP-D2 | 63 ± 18 | <0.01 | 366 | 2.6 | Murine TRP2 | 98% |
| ASP-D3 | 84 ± 12 | <0.05 | 608 | 4.8 | Murine dystroglycan | 98% |
| ASP-D4 | 68 ± 21 | <0.02 | 614 | 1.8 | β-subunit of murine TCP- containing chaperonin complex | 99% |
| ASP-D5 | 77 ± 18 | <0.05 | 696 | 2.6 | Homology to an EST 3′-directed murine cDNA | 100% |
| ASP-D6 | 80 ± 13 | <0.05 | 409 | 6.6 | Human KIA A0261 | 71% |
| Up-regulated genes | ||||||
| ASP-I1 | 178 ± 18 | <0.02 | 340 | 2.3 | Murine MCM6 | 99% |
| ASM-I2 | 196 ± 28 | <0.02 | 704 | 6.5 | Murine ITF2 | 100% |
| ASP-13 | 133 ± 18 | <0.02 | 410 | 6.2 | Homology to 3 ESTS human retina cDNAs | 90% |
The expression patterns of these genes in melanocytes in vivo were evaluated by Northern blot analysis using total RNA samples obtained from skins of sibling pheomelanogenic lethal yellow and nonagouti black mice. All three ASP up-regulated genes (ASP-I1, ASP-I2, and ASP-I3) showed significant increases in expression in yellow mouse skin (Fig. 3). Among the ASP down-regulated genes, ASP-D1 and ASP-D2 hybridized to two distinctive mRNA transcripts of approximate sizes 1.8 and 2.6 kilobases, which by visual inspection showed more than a 2- and 10-fold higher expression, respectively, in nonagouti black mouse skin. Although ASP-D3, ASP-D4, ASP-D5, and ASP-D6 appeared to detect mRNA transcripts similar to those identified by Northern blot analysis using RNAs from melan-a cells, no significant differences were observed in Northern blot analysis between lethal yellow and black mouse skin. However, it should be noted that melanocytes represent a very minor population of cells in the skin (<1%) and thus quantification, and even detection, of melanocyte-specific transcripts is quite difficult.
Sequence Analysis.
These nine differentially expressed genes then were sequenced and identified by a blast search (Table 1). All bands were found to have the expected primers at their 5′ and 3′ ends. Screening of a melan-a melanocyte cDNA library followed by a blast search revealed that seven of the nine bands matched perfectly (more than 98% identical) throughout the lengths of their reamplified PCR products to previously reported genes or to expressed sequence tags (ESTs). Among the ASP down-regulated genes identified were two melanogenic enzymes (tyrosinase and TRP2), and two cytoskeleton-related genes [dystroglycan (33, 34) and the β-subunit of TCP1 containing the chaperonin complex (35)]. ASP-D5 had complete sequence homology (100%) to a 3′ mouse EST (GenBank accession no. D18565), whereas band ASP-D6 had 71% homology to the human KIA A0261 gene.
One of the ASP up-regulated bands (ASP-I1) showed 99% homology to the mouse MCM6 gene (36–38), which is the murine homolog of the yeast mis gene, which encodes a novel member of the CDC46/minichromosome maintenance protein family required for the onset of DNA synthesis. The second ASP up-regulated gene (ASP-I2) corresponded to the murine ITF2 gene, and ASP-I3 had 90% homology to three human retina cDNAs in the EST database (GenBank accession nos. W26416, W28248, and W28288).
Conclusions and Implications.
Our findings show that differential display can rapidly identify genes altered in expression during the switch from eumelanogenesis to pheomelanogenesis. Among the down-regulated genes, we have independently identified two genes encoding melanogenic enzymes (tyrosinase and TRP2), which were previously reported to be down-regulated during pheomelanogenesis (9, 27). In addition to providing an excellent internal control for the reliability and versatility of this technique, the fact that two of the five genes known to be so down-regulated (TRP1, Pmel17/silver, and pinkeyed-dilution are the others) suggests that so far we may have identified fewer than 50% of the overall number of genes involved in this pigmentary switch at the melanosomal level.
The identification of tyrosinase and TRP2 as down-regulated genes during pheomelanogenesis was expected based on earlier studies, but similar regulation of genes encoding dystroglycan and TCP1 were a surprise. It is beyond the scope of this study to further characterize the role(s) of those gene products but one might easily envision their functions. For example, mammalian dystroglycans are important to linkages between the cytoskeleton and the basal lamina, particularly laminin (33). Melanocytes bind laminin and fibronectin, and such molecules are thought to play important roles in localization of melanocytes at the dermal/epidermal border, and in the disruption of that localization after transformation of melanocytes to melanoma cells. Interestingly, MSH has been shown to affect the interaction of melanocytes with laminin (39), a finding consistent with the down-regulation of the dystroglycan gene after treatment with ASP. The role of TCP-1 is similarly provocative in this context; TCP-1 is the β-subunit of a cytosolic chaperone involved in protein folding (35). Calnexin, another molecular chaperone, has been shown to be important to the folding and processing of tyrosinase and TRP1 (40); it is an interesting possibility that the TCP-1 chaperone system might be involved with similar processes on other melanogenic proteins. The further characterization of the identity and function of ASP-D5 and ASP-D6 must await further sequence analysis.
In contrast, the identification of three genes up-regulated during pheomelanogenesis suggests that there are at least several determinants involved during this redirection of differentiation. These three genes encode a potential DNA replication control protein (the MIS5 homolog), a basic helix–loop–helix transcription factor (ITF2), and an additional gene expressed in retinal cells that has an as yet unknown function (it should be noted that melanocytes comprise the retinal pigment epithelium). These genes probably are involved in the various mutational changes seen in lethal yellow mice as well as in the coat color change.
The characterization of the specific functions of these regulatory genes in controlling pheomelanogenesis and related cellular events, and the biochemical mechanisms involved, will be an exciting challenge. A particularly interesting gene should prove to be ITF2; another basic helix–loop–helix (bHLH) transcription factor encoded by the microphthalmia locus (termed MITF) has been shown to regulate the TRP gene family (41–43). Such transcription factors are functional in gene regulation only after dimerization, and heterodimerization of bHLH transcription factors often plays a key role in their gene recognition and function. An interesting possibility would arise if ITF2 forms a heterodimer with MITF that might affect the latter’s activation of melanogenic genes; such a scenario might imply that ITF2 expression during pheomelanogenesis blocks the activating function of MITF on the TRP family. Such a hypothesis is consistent with observations of protein and gene expression during the eumelanogenic to pheomelanogenic switch.
Because differential display is applicable to investigating pheomelanogenic gene expression in wild-type agouti mice during the programmed shift from eumelanogenesis to pheomelanogenesis during hair growth, it should be possible in the future to use this system to characterize the mechanisms involved in abnormalities seen in lethal yellow mice by comparing the gene expression in vivo with our in vitro studies. The interaction of ASP and melanocortin 1 receptor also is thought to play an important role in regulating human pigmentation (22–26). The commitment to produce pheomelanin rather than eumelanin in human skin has important negative implications for photoprotection of that tissue and is closely correlated with dramatically increased risk for photocarcinogenesis. It is thus very important to identify pheomelanogenic determinants in mammals and to identify biochemical mechanisms involved in this switch. Although enzymes and/or factors that influence the pheomelanogenic pathway are unknown at this time, characterization of the function(s) of novel gene products should provide important clues necessary to understand the regulation of this important biochemical pathway.
ABBREVIATIONS
- ASP
agouti signal protein
- EST
expressed sequence tag
- MSH
α-melanocyte stimulating hormone
- TRP
tyrosinase-related protein
References
- 1.Prota G. Melanins and Melanogenesis. New York: Academic; 1992. pp. 1–290. [Google Scholar]
- 2.Prota G, Lamoreux M L, Muller J, Kobayashi T, Napolitano A, Vincenzi R, Sakai C, Hearing V J. Pigment Cell Res. 1995;8:153–163. doi: 10.1111/j.1600-0749.1995.tb00657.x. [DOI] [PubMed] [Google Scholar]
- 3.Ito S. In: Biochemistry and Physiology of Melanin. Levine N, editor. Boca Raton, FL: CRC; 1993. pp. 33–60. [Google Scholar]
- 4.Barsh G S. Am J Hum Genet. 1995;57:743–747. [PMC free article] [PubMed] [Google Scholar]
- 5.Barsh G S. Trends Genet. 1996;12:299–305. doi: 10.1016/0168-9525(96)10031-7. [DOI] [PubMed] [Google Scholar]
- 6.Bultman S J, Michaud E J, Woychik R P. Cell. 1992;71:1195–1204. doi: 10.1016/s0092-8674(05)80067-4. [DOI] [PubMed] [Google Scholar]
- 7.Miller M W, Duhl D M J, Vrieling H, Cordes S P, Ollmann M M, Winkes B M, Barsh G S. Genes Dev. 1993;7:454–467. doi: 10.1101/gad.7.3.454. [DOI] [PubMed] [Google Scholar]
- 8.Millar S E, Miller M W, Stevens M E, Barsh G S. Development (Cambridge, UK) 1995;121:3223–3232. doi: 10.1242/dev.121.10.3223. [DOI] [PubMed] [Google Scholar]
- 9.Kobayashi T, Vieira W D, Potterf B, Sakai C, Imokawa G, Hearing V J. J Cell Sci. 1995;108:2301–2309. doi: 10.1242/jcs.108.6.2301. [DOI] [PubMed] [Google Scholar]
- 10.Lamoreux M L, Zhou B K, Rosemblat S, Orlow S J. Pigment Cell Res. 1995;8:263–270. doi: 10.1111/j.1600-0749.1995.tb00673.x. [DOI] [PubMed] [Google Scholar]
- 11.Perry W L, Copeland N G, Jenkins N A. BioEssays. 1994;16:705–707. doi: 10.1002/bies.950161002. [DOI] [PubMed] [Google Scholar]
- 12.Siracusa L D. Trends Genet. 1994;10:423–428. doi: 10.1016/0168-9525(94)90112-0. [DOI] [PubMed] [Google Scholar]
- 13.Vrieling H, Duhl D M J, Millar S E, Miller K A, Barsh G S. Proc Natl Acad Sci USA. 1994;91:5667–5671. doi: 10.1073/pnas.91.12.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu D, Willard D, Patel I R, Kadwell S, Overton L, Kost T, Luther M, Chen W, Woychik R P, Wilkison W O, Cone R D. Nature (London) 1994;371:799–802. doi: 10.1038/371799a0. [DOI] [PubMed] [Google Scholar]
- 15.Blanchard S G, Harris C O, Ittoop O R R, Nichols J S, Parks D J, Truesdale A T, Wilkison W O. Biochemistry. 1995;34:10406–10411. doi: 10.1021/bi00033a012. [DOI] [PubMed] [Google Scholar]
- 16.Siegrist W, Willard D H, Wilkison W O, Eberle A N. Biochem Biophys Res Commun. 1996;218:171–175. doi: 10.1006/bbrc.1996.0030. [DOI] [PubMed] [Google Scholar]
- 17.Siegrist W, Drozdz R, Cotti R, Willard D H, Wilkison W O, Eberle A N. J Recept Signal Trans Res. 1997;17:75–98. doi: 10.3109/10799899709036595. [DOI] [PubMed] [Google Scholar]
- 18.Willard D H, Bodnar W, Harris C, Kiefer L, Nichols J S, Blanchard S, Hoffman C, Moyer M, Burkhart W, Weiel J, et al. Biochemistry. 1995;34:12341–12346. doi: 10.1021/bi00038a030. [DOI] [PubMed] [Google Scholar]
- 19.Zemel M B, Kim J H, Woychik R P, Michaud E J, Kadwell S H, Patel I R, Wilkison W O. Proc Natl Acad Sci USA. 1995;92:4733–4737. doi: 10.1073/pnas.92.11.4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perry W L, Nakamura T, Swing D A, Secrest L, Eagleson B, Hustad C M, Copeland N G, Jenkins N A. Genetics. 1996;144:255–264. doi: 10.1093/genetics/144.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim J H, Kiefer L L, Woychik R P, Wilkison W O, Truesdale A T, Ittoop O R R, Willard D, Nichols J S, Zemel M B. Am J Physiol. 1997;272:E379–E384. doi: 10.1152/ajpendo.1997.272.3.E379. [DOI] [PubMed] [Google Scholar]
- 22.Kwon H Y, Bultman S J, Loffler C, Chen W J, Furdon P J, Powell J G, Usala A L, Wilkison W, Hansmann I, Woychik R P. Proc Natl Acad Sci USA. 1994;91:9760–9764. doi: 10.1073/pnas.91.21.9760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson B D, Ollmann M M, Kang L, Stoffel M, Bell G I, Barsh G S. Hum Mol Gen. 1995;4:223–230. doi: 10.1093/hmg/4.2.223. [DOI] [PubMed] [Google Scholar]
- 24.Valverde P, Healy E, Jackson I J, Rees R L, Thody A J. Nat Genet. 1995;11:328–330. doi: 10.1038/ng1195-328. [DOI] [PubMed] [Google Scholar]
- 25.Hunt G, Todd C, Thody A J. Mol Cell Endocrinol. 1996;116:131–136. doi: 10.1016/0303-7207(95)03708-x. [DOI] [PubMed] [Google Scholar]
- 26.Suzuki I, Tada A, Ollmann M, Barsh G S, Im S, Lamoreux M L, Hearing V J, Nordlund J J, Abdel-Malek Z A. J Invest Dermatol. 1997;108:838–842. doi: 10.1111/1523-1747.ep12292572. [DOI] [PubMed] [Google Scholar]
- 27.Sakai C, Ollmann M, Kobayashi T, Abdel-Malek Z A, Muller J, Vieira W D, Imokawa G, Barsh G S, Hearing V J. EMBO J. 1997;16:3544–3552. doi: 10.1093/emboj/16.12.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Furumura M, Sakai C, Abdel-Malek Z A, Barsh G S, Hearing V J. Pigment Cell Res. 1996;9:191–203. doi: 10.1111/j.1600-0749.1996.tb00109.x. [DOI] [PubMed] [Google Scholar]
- 29.Bennett D C, Cooper P J, Hart I R. Int J Cancer. 1987;39:414–418. doi: 10.1002/ijc.2910390324. [DOI] [PubMed] [Google Scholar]
- 30.Ollmann M M, Lamoreux M L, Wilson B D, Barsh G S. Genes Dev. 1998;12:316–330. doi: 10.1101/gad.12.3.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoffman W L, Dowben R M. Anal Biochem. 1978;89:540–549. doi: 10.1016/0003-2697(78)90383-4. [DOI] [PubMed] [Google Scholar]
- 32.Liang P, Pardee A B. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 33.Deys K A, Bowe M A, Leszyk J D, Fallon J R. J Biol Chem. 1995;270:25956–25959. doi: 10.1074/jbc.270.43.25956. [DOI] [PubMed] [Google Scholar]
- 34.Yang B, Jung D, Motto D, Meyer J, Koretzky G, Campbell K P. J Biol Chem. 1995;270:11711–11714. doi: 10.1074/jbc.270.20.11711. [DOI] [PubMed] [Google Scholar]
- 35.Kubota H, Hynes G, Carne A, Ashworth A, Willison K. Curr Biol. 1994;4:89–99. doi: 10.1016/s0960-9822(94)00024-2. [DOI] [PubMed] [Google Scholar]
- 36.Kimura H, Ohtomo T, Yamaguchi M, Ishii A, Sugimoto K. Genes Cells. 1996;1:977–993. doi: 10.1046/j.1365-2443.1996.840284.x. [DOI] [PubMed] [Google Scholar]
- 37.Sykes D E, Weiser M M. Gene. 1995;163:243–247. doi: 10.1016/0378-1119(95)00297-j. [DOI] [PubMed] [Google Scholar]
- 38.Takahashi K, Yamada H, Yanagida M. Mol Biol Cell. 1994;5:1145–1158. doi: 10.1091/mbc.5.10.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hunt G, Donatien P D, Cresswell J E, Thody A J. Ann NY Acad Sci. 1993;680:549–551. doi: 10.1111/j.1749-6632.1993.tb19735.x. [DOI] [PubMed] [Google Scholar]
- 40.Jimbow K, Gomez P F, Toyofuku K, Chang D, Miura S, Tsujiya H, Park J S. Pigment Cell Res. 1997;10:206–213. doi: 10.1111/j.1600-0749.1997.tb00486.x. [DOI] [PubMed] [Google Scholar]
- 41.Yasumoto K, Yokoyama K, Shibata K, Tomita Y, Shibahara S. Mol Cell Biol. 1994;14:8058–8070. doi: 10.1128/mcb.14.12.8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yasumoto K, Mahalingam H, Suzuki H, Yoshizawa M, Yokoyama K, Shibahara S. J Biochem. 1995;118:874–881. doi: 10.1093/jb/118.5.874. [DOI] [PubMed] [Google Scholar]
- 43.Bertolotto C, Bille K, Ortonne J P, Ballotti R. J Cell Biol. 1996;134:747–755. doi: 10.1083/jcb.134.3.747. [DOI] [PMC free article] [PubMed] [Google Scholar]