Abstract
In situ estrogen synthesis is implicated in tumor cell proliferation through autocrine or paracrine mechanisms especially in postmenopausal women. Several recent studies demonstrated activity of aromatase, an enzyme that plays a critical role in estrogen synthesis in breast tumors. Proline-, glutamic acid-, and leucine-rich protein-1 (PELP1/MNAR) is an estrogen receptor (ER) coregulator, and its expression is deregulated in breast tumors. In this study, we examined whether PELP1 promotes tumor growth by promoting local estrogen synthesis using breast cancer cells (MCF7) that stably overexpress PELP1. Immunohistochemistry revealed increased aromatase expression in MCF7-PELP1-induced xenograft tumors. Real-time PCR analysis showed enhanced activation of the aromatase promoter in MCF7-PELP1 clones compared with MCF7 cells. Using a tritiated-water release assay, we demonstrated that MCF7-PELP1 clones exhibit increased aromatase activity compared with control MCF-7 cells. PELP1 deregulation uniquely up-regulated aromatase expression via activation of aromatase promoter I.3/II, and growth factor signaling enhanced PELP1 activation of aromatase. PELP1-mediated induction of aromatase requires functional Src and phosphatidylinositol-3-kinase pathways. Mechanistic studies revealed that PELP1 interactions with ER-related receptor-α and proline-rich nuclear receptor coregulatory protein 2 lead to activation of aromatase. Immunohistochemistry analysis of breast tumor array showed increased expression of aromatase in ductal carcinoma in situ and node-positive tumors compared with no or weak expression in normal breast tissue. Fifty-four percent (n = 79) of PELP1-overexpressing tumors also overexpressed aromatase compared with 36% (n = 47) in PELP1 low-expressing tumors. Our results suggest that PELP1 regulation of aromatase represents a novel mechanism for in situ estrogen synthesis leading to tumor proliferation by autocrine loop and open a new avenue for ablating local aromatase activity in breast tumors.
MAMMARY TUMORIGENESIS IS accelerated by the action of ovarian hormones, and approximately 70% of breast tumors are estrogen (E2) receptor (ER) positive at the time of presentation. Endocrine therapy is the most important component of adjuvant therapy for patients with early-stage ER-positive breast cancer (1). The biological functions are mediated by the ER, a ligand-dependent transcription factor that modulates gene transcription via direct recruitment to the target gene chromatin (classical pathway) (2,3). The ER can also modulate gene expression via its interactions with other transcription factors (nonclassical pathway). In addition, the ER also participates in cytoplasmic and membrane-mediated signaling events (nongenomic signaling) and generally involves the stimulation of the Src kinase, MAPK, and phosphatidylinositol-3-kinase (PI3K) (4,5). ER signaling requires coregulatory proteins, and their composition in a given cell determines the magnitude and specificity of the ER signaling (6,7).
Aromatase (Cyp19), a key enzyme involved in E2 synthesis (8), appears to be expressed in breast tumors, and locally produced E2 might act in a paracrine or autocrine fashion (9). Furthermore, breast tumors from postmenopausal women are shown to contain higher amounts of E2 than would be predicted from levels circulating in plasma (10). Expression of the aromatase gene is under the control of several distinct and tissue-specific promoters; however, the coding region of aromatase transcripts and the resulting protein is identical (11). In disease-free breast, aromatase expression is directed via distal 1.4 promoter, whereas aromatase expression is shown to be activated via PII and 1.3 in adipose tissue in breast bearing a tumor (11,12). The molecular mechanism by which breast tumors enhance aromatase expression is not completely understood.
Recently, aromatase inhibitors that inhibit peripheral E2 synthesis are shown to be more effective in enhancing the survival of postmenopausal women with ER-positive breast cancer (13). Emerging evidence suggests that tumors use adaptive mechanisms for growth after the initiation of first-line endocrine therapy. Hypersensitivity to E2, local E2 synthesis, excessive nongenomic signaling, and excessive ER-growth factor signaling cross talk are suggested as some of the possible means by which tumor cells acquire resistance (14). Accumulating evidence also suggests that a variety of different factors may regulate expression and activity of aromatase under pathological conditions, and local production of E2 may enhance tumor growth and may also interfere with hormone therapy (15).
Proline-, glutamic acid-, and leucine-rich protein-1 (PELP1) is a novel ER coactivator (16). PELP1 is also referred to as a modulator of nongenomic actions of ER (MNAR) (17). PELP1 interacts with the ER and modulates its genomic and nongenomic functions (18). PELP1 promotes E2-mediated cell proliferation by sensitizing cells to G1→S progression via its interactions with the pRb pathway (19). In the nuclear compartment, PELP1 interacts with histones and histone-modifying enzymes and thus plays a role in chromatin remodeling for ligand-bound ERs (20). Recent evidence also suggests that PELP1 couples ER to several signaling axes, such as Src-MAPK, PI3K-Akt, and epidermal growth factor receptor-signal transducer and activator of transcription 3 (EGFR-STAT3) (18). PELP1 expression is deregulated in breast cancer, and emerging evidence suggests that PELP1 has oncogenic potential and that its deregulation promotes hormone independence (21). However, mechanisms by which PELP1 promote tumorigenesis is elusive.
Here we provide new evidence that deregulation of the ER coactivator PELP1 promotes in situ synthesis of E2 in breast tumor epithelial cells. PELP1-overexpressing tumors showed increased expression of aromatase. Furthermore, we show that PELP1 also cooperates with growth factor signaling components in the activation of the aromatase promoter in breast cancer cells. These results suggest that PELP1 signaling and its interactions with growth factor signaling components and orphan nuclear receptors may locally influence the in situ production of E2 and thus promote tumor growth by promoting ER autocrine signaling.
RESULTS
PELP1 Overexpression Correlates with Increased Expression of Aromatase
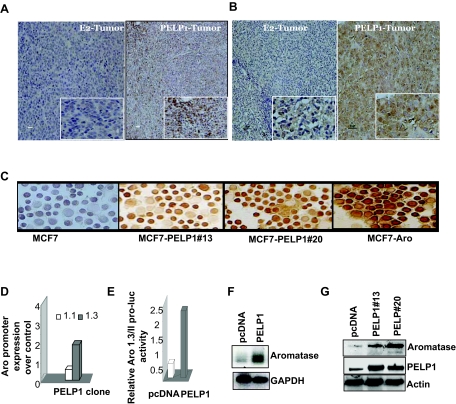
Emerging evidence suggest that breast tumors in postmenopausal women have potential to acquire ability to produce local E2 by activating expression of aromatase. Recent studies from our laboratory showed that the ER coregulator PELP1 functions as a potential protooncogene, and its deregulation in MCF7 breast cancer epithelial cells promotes hormone-independent growth of these cells in a xenograft model (21). To examine whether PELP1 deregulation promotes aromatase expression, we determined whether the expression of aromatase in tumors is induced by PELP1 overexpression. MCF7 tumors induced by E2 supplementation were used as a control. We have used two different monoclonal antibodies (mAb) in immunohistochemistry (IHC) that recognize distinct epitopes on aromatase (Fig. 1, A and B) (22,23,24). Results of IHC showed that sections from PELP1-induced xenograft tumors had increased aromatase expression compared with the control E2-induced MCF7 tumors (Fig. 1, A and B). To further confirm these results, we have examined the expression of aromatase in PELP1-overexpressing MCF7 model cells. MCF7 cells stably expressing aromatase were used as a positive control. Cytospin sections from two different PELP1 stable clones (PELP clone 20 or PELP clone 13) showed higher expression of aromatase as compared with parental MCF7 cells, suggesting deregulation of PELP1 promotes increased expression of aromatase (Fig. 1C).
Figure 1.
PELP1 Deregulation Promotes Aromatase Expression
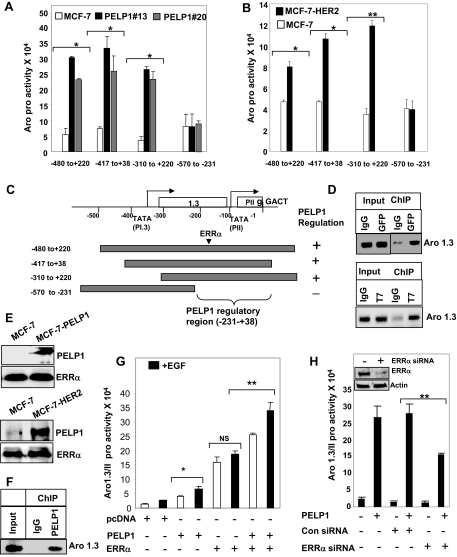
A and B, Immunohistological examination of aromatase expression in mouse xenograft tumor sections from E2-induced or PELP1-induced tumors: A, sections stained with Serotec mAb; B, sections stained with a Novartis mAb. C, Cytospin slides were prepared from control MCF7 and MCF7-PELP1 clones (13 or 20), and expression of aromatase was analyzed by immunohistochemistry. MCF7 Aro cells were used as a positive control. D, Total RNA was isolated from MCF7 and MCF7 PELP1 clone 13 cells and real-time PCR analysis of aromatase gene expression was performed using exon 1.3- and 1.1-specific primers. E, MCF7 and MCF7-PELP1 clone 13 cells were transiently transfected with Aro-1.3/II luciferase reporter, and 48 h later, reporter activity was analyzed. F, Total RNA was extracted from MCF7 and MCF7-PELP1 clone 13 cells, and expression of aromatase RNA was analyzed by RT-PCR. G, Total lysates from MCF7, MCF7-PELP clone 13, and MCF7-PELP1 clone 20 cells were analyzed for aromatase expression using Western blot analysis.
PELP1 Regulates Aromatase Expression through the Aromatase Promoter I.3/II
Expression of the aromatase gene is under the control of several distinct and tissue-specific promoters. To identify the promoter by which PELP1 deregulation enhances aromatase gene expression, we performed real-time RT-PCR analysis using exon-specific primers (25). Results from studies using exon I.1-, I.3-, or I.4-specific primers showed that MCF7-PELP1 clones have 2.5-fold increased levels of exon I.3 transcript compared with MCF7-pcDNA clones. (Fig. 1D). No significant changes in the activity of exon I.1 or I.4 promoters were observed (data not shown). In reporter gene assays using Aro-1.3 luciferase, MCF7-PELP1 cells showed a 5-fold increase in the reporter gene activity (Fig. 1E). In RT-PCR analysis, MCF7-PELP1 cells showed significant increase in total aromatase transcript levels relative to MCF7-pcDNA cells, which had negligible or low expression of the total aromatase transcript (Fig. 1F). Western blot analysis showed that MCF7-PELP1 clones have increased levels of aromatase compared with the aromatase levels in the control MCF7-pcDNA (Fig. 1G). Collectively, these results suggest that PELP1 deregulation has potential to regulate the aromatase gene expression via the I.3 promoter.
PELP1 Deregulation Promotes Aromatase Activity
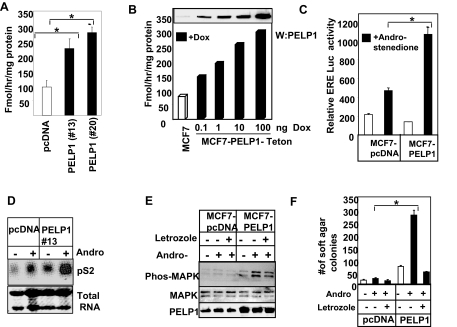
To establish the physiological significance of PELP1 regulation of aromatase gene expression, we examined whether MCF7-PELP1 model cells had increased basal aromatase activity. Aromatase activity was determined using the tritiated-water release assay (26). The basal level of aromatase activity was found to be three times higher in PELP1 model cells (Fig. 2A). To confirm that the increase in aromatase activity seen in MCF7-PELP1 is indeed due to alteration in PELP1 levels, we used Teton-PELP1 model cells in which PELP1 expression is under the control of a Tet-inducible promoter (20). PELP1 expression in these cells can be induced by addition of doxycycline in a dose-dependent manner. Results from these studies showed that induction of PELP1 expression in Teton-PELP1 model cells increases aromatase activity in a PELP1 dose-dependent manner (Fig. 2B).
Figure 2.
PELP1 Deregulation Promotes Aromatase Activity and in Situ Estrogen Synthesis
A, Aromatase activity in control MCF7 cells and MCF7-PELP1 clones was measured by tritiated-water release assay. Each experimental condition was measured in triplicate, and aromatase activity was expressed in picomoles per milligram of protein per hour (mean ± sem). B, Tet-inducible PELP1 model cells were treated with increasing doses of doxycycline to induce variable levels of PELP1, and 24 h later, aromatase activity was measured; inset shows induction of PELP1 expression as measured by Western blot analysis. C, MCF7 and MCF7-PELP1 clone cells cultured in DCC medium were transiently transfected with ER reporter gene. After 48 h, the cells were treated with or without the aromatase substrate androstenedione for 12 h, and then reporter gene activity was measured. D, MCF7 and MCF7-PELP1 clones cells were cultured in DCC medium for 2 d and treated with androstenedione for 24 h, and expression of the ER target gene pS2 was measured by Northern blot analysis. E, MCF7 and MCF7-PELP1 clone cells were treated with androstenedione for 24 h and with or without letrozole, and activation of MAPK pathway was measured using Western blot analysis with phosphorylated antibodies. F, MCF7 and MCF7-PELP1 clone cells were plated in soft agar in the presence or absence of androstenedione and with or without letrozole. Colony formations were counted after 21 d. *, P < 0.05; **, P < 0.001. These experiments were repeated three times; the average of the results is shown in the graph.
PELP1 Deregulation Promotes in Situ Estrogen Synthesis
To examine whether increased aromatase expression in PELP1 clones translates to local E2 synthesis, we have examined whether addition of the aromatase substrate androstenedione activates the E2-ER signaling pathway in these model cells. The addition of androstenedione substantially increased ER reporter gene activity in MCF7-PELP1 cells compared with MCF7-pcDNA cells (Fig. 2C). Similarly, addition of androstenedione significantly increased the expression of the ER target gene PS2 (Fig. 2D). Furthermore, androstenedione addition increased MAPK activation in MCF7-PELP1 clones (Fig. 2E). Parental MCF7 cells expressing pcDNA demonstrated low or weak potential to form soft-agar colonies even in the presence of androstenedione (Fig. 2F). However, addition of androstenedione substantially increased the ability of PELP1 model cells to grow in an anchorage-independent manner. Treatment of PELP1 model cells with letrozole, an aromatase inhibitor, abolished the PELP1-mediated increase in soft-agar colony formation (Fig. 2F), implicating in situ E2 synthesis through increased aromatase expression in MCF7-PELP1 clones. Collectively, these results suggest that aromatase induced in PELP1 clones is functional and has potential to induce local E2 synthesis, leading to activation of the E2-ER signaling pathway in an autocrine manner.
Growth Factor Signaling Promotes PELP1 Regulation of Aromatase Expression
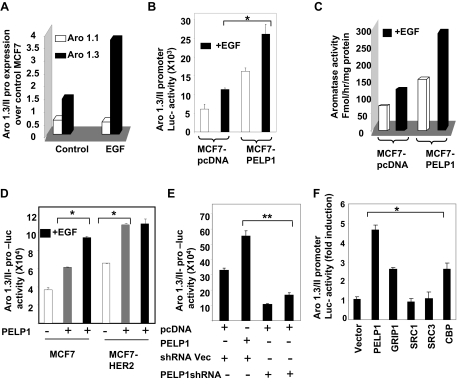
Because growth factor signaling has been shown to modulate PELP1 functions via tyrosine phosphorylation (18,27), we examined whether growth factor signaling enhances PELP1 regulation of the aromatase promoter. MCF7-pcDNA and MCF7-PELP1 cultured in dextran-coated charcoal (DCC)-treated serum were treated with or without epidermal growth factor (EGF), and the expression of aromatase was analyzed by real-time PCR (Fig. 3A) and Aro-1.3/II reporter gene assays (Fig. 3B). In both of these assays, MCF7-PELP1 clones exhibited higher basal Aro-1.3 expression compared with MCF7-pcDNA, and growth factor stimulation resulted in a further 2.5-fold increase in aromatase expression. Similarly, growth factor stimulation of MCF7-PELP1 also resulted in a 2- to 3-fold induction of aromatase activity as compared with MCF7-pcDNA cells (Fig. 3C).
Figure 3.
EGF Enhances PELP1-Mediated Activation of Aromatase
A, MCF7 and MCF7-PELP1 cells were cultured in serum-free medium for 24 h and then treated with or without 100 ng/ml EGF. Total RNA was isolated, and real-time PCR analysis of aromatase gene expression was performed using aromatase exon 1.3- and 1.1-specific primers. B, MCF7 and MCF7-PELP1 cells were transiently transfected with Aro-1.3/II luciferase reporter. After 48 h, cells were cultured in serum-free medium for 24 h, after which they were treated with or without 100 ng EGF/ml for 12 h, and luciferase activity was measured. C, MCF7 and MCF7-PELP1 clone cells were treated with or without EGF, and aromatase activity in control MCF7 and MCF7-PELP1 clones was measured by tritiated-water release assay. D, MCF7 and MCF7-HER2 cells were transiently transfected with Aro-1.3/II reporter gene along with or without PELP1 and treated with or without EGF for 12 h. Luciferase activity was measured. E, MCF7-HER2 cells were cotransfected with Aro-1.3/II reporter along with or without PELP1 shRNA vectors. After 72 h, luciferase reporter activity was measured. F, MCF7 cells were cotransfected with Aro-1.3/II reporter along with PELP1, GRIP1, SRC1, SRC3, and CBP expression vectors. After 72 h, cells were treated with 100 ng/ml EGF for 12 h, and then luciferase reporter activity was measured. *, P < 0.05; **, P < 0.001. Data shown are the means ± se from three independent experiments performed in triplicate wells.
HER2 Signaling Enhances PELP1 Regulation of the Aromatase Promoter
Deregulation of HER2 signaling or expression occurs in a subset of breast tumors and recent studies showed that HER2 deregulation promotes aromatase expression (28). We examined whether PELP1 participates in HER2-mediated activation of aromatase expression using Aro-1.3 luciferase reporter gene assays in MCF7-HER2 model cells that overexpress HER2. As shown in previous studies, MCF7-HER2 exhibited higher Aro-1.3 reporter gene activity compared with MCF7 cells. Cotransfection of PELP1 into MCF7-HER2 cells further increased the reporter activity to a level similar to that observed when MCF7 cells were treated with growth factor. However, no further increase in Aro-1.3 reporter activity was observed when MCF7-HER2 cells expressing PELP1 were treated with EGF (Fig. 3D). However, EGF treatment of MCF7 cells overexpressing PELP1 showed increased aromatase reporter activity to a level similar to PELP1 overexpression in MCF7-HER2 cells. To further examine the significance of PELP1 in HER2-mediated activation of aromatase, we used two different PELP1 short hairpin RNAs (shRNAs) that target PELP1 at different regions and down-regulate endogenous PELP1 expression (Fig. 3F). Cotransfection of both PELP1 shRNAs substantially reduced Aro-1.3/II promoter activity in MCF7-HER2 cells compared with shRNA vector transfected controls, indicating that PELP1 plays a critical role in HER2-mediated activation of aromatase. We then examined whether other ER coregulators also contribute to aromatase up-regulation in growth factor signaling-stimulated MCF-7 cells. We have performed aromatase 1.3/II reporter gene assays by cotransfecting coactivators PELP1, GRIP1, SRC1, SRC3, and CBP (Fig. 3F). The results showed that coregulators have different abilities to affect the aromatase reporter. Of all the coregulators tested, PELP1 showed the highest fold of induction, whereas SRC1 and SRC3 show low or no effect on the reporter activity in these cells. On the contrary, CBP and GRIP show moderate ability to modulate aromatase reporter activity. Together, these results suggest that either growth factor signaling or HER2 overexpression alone is sufficient to modulate PELP1 functions in the activation of aromatase in breast cancer cells.
PELP1-Mediated Induction of Aromatase Expression Is Independent of the Estrogen Receptor
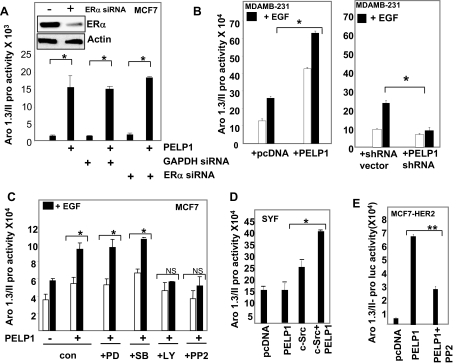
Because PELP1 functions as a coregulator of the ER, we next examined whether PELP1 regulation of aromatase involves the ER. To examine this possibility, we transfected MCF7 cells with ER-specific small interfering RNA (siRNA) or GAPDH-specific siRNA. After 3 d, cells were transfected with Aro-1.3/II luciferase reporter along with or without PELP1 expression vector. Western analysis showed that ER siRNA down-regulated 70–80% of the ER. Down-regulation of ER did not affect PELP1-mediated activation of aromatase reporter (Fig. 4A), suggesting that the PELP1-mediated increase in aromatase promoter activity is independent of the ER. To further confirm these findings, we repeated these assays in ERα-negative MDA-MB-231 cells. Results showed that PELP1 was able to activate Aro-1.3/II promoter in these cells, confirming that PELP1 activation of aromatase does not require ERα (Fig. 4B, left panel). Because EGF also activated aromatase reporter gene in MDA-MB-231 cells and because these cells express endogenous PELP1, we examined whether endogenous PELP1 plays a role in EGF-mediated activation of aromatase promoter. To test this possibility, we have cotransfected MDA-MB-231 cells with Aro-1.3/II promoter luciferase reporter along with control or PELP1-specific shRNA. EGF did not significantly enhance aromatase reporter gene activity in PELP1 shRNA-transfected cells, whereas EGF promoted reporter gene activation in control shRNA-transfected cells (Fig. 4B, right panel). Collectively, these results suggest that PELP1 can activate aromatase promoter independent of ER and also participates in EGF-mediated activation of aromatase gene.
Figure 4.
PELP1 Modulation of Aromatase Requires Nongenomic Signaling
A, MCF7 cells were transiently transfected either with ERα siRNA or control siRNA. After 72 h, cells were transfected with Aro-1.3/II-Luc, β-Gal reporter gene along with or without PELP1, and 48 h later, reporter gene activation was measured. Inset shows Western analysis of ERα in control and siRNA-transfected cells. B, MDA-MB-231 cells were cotransfected with the Aro-1.3/II-Luc reporter with or without PELP1 (left panel) and with or without PELP1 shRNA (right panel). After 48 h, cells were stimulated with or without EGF (100 ng/ml) for 12 h, and luciferase activity was measured. C, MCF7 cells were cotransfected with Aro-1.3/II luciferase reporter with or without PELP1. After 48 h, cells were pretreated for 1 h with p42/44MAPK inhibitor (PD98059, 20 μm), p38MAPK inhibitor (SB203580, 40 μm), Src inhibitor PP2 (10 μm), or PI3K inhibitor (LY294002, 20 μm) and then stimulated with EGF for 12 h. Luciferase activity was measured. D, Src-negative SYF cells were transiently cotransfected with Aro-1.3/II along with or without PELP1 and c-Src expression vectors. After 48 h, luciferase activity was measured. E, MCF7-HER2 cells were cotransfected with an Aro-1.3/II luciferase reporter and β-Gal reporter gene along with PELP1. After serum starvation for 48 h, cells were treated with or without 10 μm Src inhibitor PP2 for 12 h. Luciferase activity was measured. In all reporter gene assays, parental vectors were used as controls, and the total amount of the DNA in the transfection was kept constant by adding appropriate empty vectors. β-Gal values were used to normalize luciferase activity for transfection efficiency. *, P < 0.05; **, P < 0.001. Data shown are the means ± se from three independent experiments performed in triplicate wells.
PELP1-Mediated Induction of Aromatase Expression Requires Functional Src Signaling
To identify signaling pathways that are involved in PELP1-mediated induction of aromatase expression, we pretreated MCF7 cells with various signaling inhibitors that block specific pathways: PD98059, MAPK/ERK inhibitor; PP2, the Src family tyrosine kinase inhibitor; LY-294002, the PI3K inhibitor; SB203580; and the p38MAPK inhibitor. MCF-7 cells were transiently transfected with PELP1 along with Aro-1.3 luciferase reporter gene, and EGF was added after pretreatment with various inhibitors. Luciferase assay results showed that PELP1-induced aromatase promoter activity was abolished by pretreatment with c-Src or PI3K pathway inhibitors, whereas pretreatment of MAPK pathway inhibitors had no effect on PELP1-induced aromatase expression (Fig. 4C). These results suggest that functional c-Src and PI3K pathways are required for PELP1-mediated induction of aromatase. To further establish a causal relationship of the c-Src pathway in PELP1 regulation of aromatase, we used c-Src, fyn, yes null fibroblast cells (SYF cells). These model cells are widely used by many investigators to establish the role of Src kinases (29). Cotransfection of PELP1 along with Aro-1.3 luciferase reporter failed to activate Aro-1.3 promoter in SYF cells; however, c-Src cotransfection along with PELP1 into SYF cells restored PELP1 ability to activate Aro-1.3/II promoter (Fig. 4D). Similarly, HER2 regulation of PELP1-mediated activation of aromatase was also abolished by pretreatment of MCF7-HER2 cells with the Src inhibitor PP2 (Fig. 4E). Collectively, these findings suggest that c-Src signaling plays a vital role in PELP1-mediated induction of aromatase.
Proximal Region of Aromatase 1.3 Promoter Confers PELP1 Inducibility
To identify the PELP1-responsive region in the aromatase promoter, we used several deletions of the aromatase promoter reporter gene (Fig. 5C). These constructs were transfected into MCF7, MCF-PELP clone 20, or MCF-PELP clone 13 cells that were treated with EGF. Luciferase assay results showed significant up-regulation of −417/+38, −480/+220, and −310/+220 construct activity in both MCF-PELP1 model cells, whereas the construct −570/−231 showed no induction compared with the parental MCF7 cell line (Fig. 5A). These results suggest that 269 bases located in the −231/+38 Aro-P1.3/II promoter are required for PELP1 regulation of aromatase. Because our earlier studies showed that HER2 signaling regulated PELP1-mediated activation of aromatase, we repeated these deletion studies using MCF7-HER2 cells. The results showed that HER2 signaling also required Aro-1.3/II −231/+38 region for PELP1-mediated activation of aromatase (Fig. 5B). These results further demonstrate that PELP1 regulates transcriptional activity of aromatase through the promoter I.3/II, and the sequences between −231/+38 play an important role in PELP1-mediated induction of aromatase.
Figure 5.
PELP1/MNAR Is Recruited to the Aromatase Chromatin and Interacts with ERRα
A, MCF7 and MCF7-PELP1 clone cells were transiently transfected with various deletions of Aro-1.3/II luciferase and β-Gal reporter gene and 48 h later treated with 100 ng/ml EGF for 12 h. The reporter gene activation was measured. B, MCF7 and MCF7-HER2 clones were transiently transfected with various deletions of Aro-1.3/II luciferase and β-Gal reporter gene along with PELP1, and 48 h later, reporter gene activation was measured. Data shown are the means ± se from three independent experiments performed in triplicate wells. *, P < 0.05; **, P < 0.001. C, Schematic representation of aromatase deletion constructs used. D, MCF7 cells expressing GFP-PELP1 (upper panel) or T7-PELP1 (bottom panel) were treated with 100 ng/ml EGF for 1 h, and ChIP was performed with IgG, GFP, or T7-tagged antibody. PELP1 recruitment to the aromatase chromatin was analyzed by primers spanning the aromatase 1.3/II region. E, MCF7 and MCF7-PELP1 cells were treated with 100 ng/ml EGF, and nuclear extracts were subjected to immunoprecipitation using an ERRα antibody, followed by Western blot analysis using a PELP1 antibody (upper panel). Nuclear extracts from MCF7 and MCF7-HER2 cells were subjected to immunoprecipitation using an ERRα antibody, and PELP1 presence in the immunoprecipitates was analyzed by Western blot analysis (bottom panel). F, MCF7-HER2 cells were cross-linked with formaldehyde, and ChIP was performed with either IgG or PELP1 antibody. PELP1 recruitment to the aromatase chromatin was analyzed by primers spanning the aromatase 1.3/II region. G, MDA-MB-231 cells were transiently transfected with Aro-1.3/II luciferase and β-Gal reporter gene along with ERRα and PELP1, and 48 h later, reporter gene activation was measured. H, MDA-MB-231 cells were transiently transfected either with ERRα siRNA or control siRNA. After 72 h, cells were transfected with Aro-1.3/II luciferase and β-Gal reporter gene along with PELP1, and 48 h later, reporter gene activation was measured. Inset shows Western blot analysis of ERRα in control and siRNA-transfected cells *, P < 0.05; **, P < 0.001; NS, nonsignificant. Data shown are the means ± se from three independent experiments performed in triplicate wells.
PELP1 Is Recruited to Aromatase Promoter and Interacts with E2-Related Receptor-α (ERRα)
To establish whether PELP1 gets specifically recruited to the aromatase promoter I.3/II region, chromatin immunoprecipitation (ChIP) analysis was performed using primers that span −231/+38. MCF7-PELP1 clones were treated with EGF, ChIP was performed using the green fluorescent protein (GFP) antibody. The results showed that PELP1 is specifically recruited to the −231/+38 region (Fig. 5D, upper panel). We have also confirmed PELP1 recruitment using different epitope-tagged PELP1 (T7 tag) (Fig. 5D, bottom panel). The region spanning −231/+38 has been reported to possess binding regions for ERRα, BRCA1, and a transcriptional silencer element (S1) (30). Because PELP1 lacks a DNA-binding domain, we examined whether PELP1 interacts with BRCA1 or ERRα using immunoprecipitation. We failed to see any PELP1 in BRCA1 immunoprecipitates (data not shown); however, ERRα immunoprecipitation showed detectable levels PELP1 (Fig. 5E, upper panel). Similarly, immunoprecipitation of ERRα in HER2 cells, but not in MCF7 cells, showed detectable levels of PELP1 (Fig. 5E, bottom panel). We examined whether endogenous PELP1 is recruited to the aromatase promoter in MCF7-HER2 cells by performing ChIP using the PELP1 antibody. The results showed that PELP1 is specifically recruited to the −231/+38 region (Fig. 5F). We then examined whether PELP1 cooperates with ERRα in the activation of aromatase gene using reporter gene assays. Cotransfection of ERRα and PELP1 significantly enhanced the Aro-1.3/II luciferase activity compared with ERRα or PELP1 alone (Fig. 5G). To further confirm the role of ERRα in PELP1-mediated activation of aromatase promoter, we have used ERRα-specific siRNA to down-regulate ERRα expression. The ERRα-specific siRNA but not control siRNA reduced 60–70% of the endogenous ERRα. Under these conditions, PELP1 mediated up-regulation of aromatase promoter was also reduced to 50% in ERR siRNA-transfected cells compared with the levels seen in control siRNA-transfected cells (Fig. 5H). These results suggest that PELP1 is recruited to the aromatase promoter and promotes activation of Aro-1.3/II promoter via interactions with the ERRα.
PELP1 Enhances Aromatase Expression via ERRα-Proline-Rich Nuclear Receptor Coregulatory Protein 2 (PNRC2) Interactions
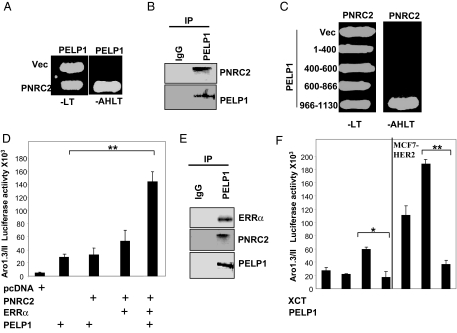
During the course of this investigation, our ongoing screen to identify PELP1-interacting proteins using a yeast two-hybrid screen identified PNRC2 (31) as a PELP1-interacting protein. The specificity of the PELP1 and PNRC2 interactions was verified by cotransfection of PELP1 and PNRC2 plasmids in yeast cells. Cotransfection of PELP1 and PNRC2 constructs conferred the ability to grow in medium lacking adenosine, histidine, tryptophan, and leucine on transformed colonies, confirming the interaction of these proteins as observed in the library screen (Fig. 6A). Immunoprecipitation revealed that these proteins formed a complex in vivo (Fig. 6B). Further analysis of interaction in this yeast-based screen using various deletions of PELP1 and PNRC2 revealed that PELP1 interacts with PNRC2 via its C-terminal 966-1130 amino acids, whereas PNRC2 interacts with PELP1 via its N-terminal 84 amino acids (Fig. 6C). PNRC2 functions as a coactivator of several nuclear receptors, including ERRα, and participates in the activation of aromatase in conjunction with ERRα (32). Because PELP1 interacted with ERRα, we further examined whether PELP1 and PNRC2 act synergistically with ERRα to modulate aromatase promoter activation. In reporter gene assays using MCF-7 cells, cotransfection of PELP1, ERRα, and PNRC2 significantly increased the activation of Aro-1.3 luciferase reporter compared with PNRC2:PELP1 or ERRα:PNRC2 cotransfections (Fig. 6D). To confirm that PELP1 indeed interacted and formed functional complexes in vivo with ERRα and PNRC2, we performed immunoprecipitation. The results showed that PELP1 did form functional complexes with ERRα and PNRC2 in vivo (Fig. 6E). To further establish that PELP1 modulates aromatase promoter activity via the ERRα pathway, we have used XCT790, a potent and specific inverse agonist of ERRα that inhibits the activity of ERRα (33). Pretreatment of both MCF-7 and MCF7-HER2 model cells with XCT790 significantly reduced PELP1 ability to modulate aromatase promoter in luciferase assays (Fig. 6F). Collectively, these results suggest that PELP1 modulates aromatase expression by functioning as a coactivator of ERRα/PNRC2 complex.
Figure 6.
PELP1 Interacts with PNRC2
A, Yeast cells were transfected with a control Gal4-activation domain vector (Vec) or GAD-PNRC2, along with a Gal4-DNA-binding domain vector or GBD-PELP1. Growth was recorded after 72 h on selection plates lacking leucine and tryptophan (−LT) or adenosine, histidine, leucine, and tryptophan (−AHLT). B, MCF7 cells were treated with 100 ng/ml EGF, and nuclear extracts were subjected to immunoprecipitation using a control IgG or a PELP1/MNAR antibody, followed by Western blot analysis using a PNRC2 antibody. C, Gal4 activation domain fusions of PELP1 deletion mutants were used to determine the PNRC2-binding region in PELP1. Positive interactions were selected on agar plates lacking adenine, histidine, leucine, and tryptophan (−AHLT). D, MCF-7 cells were cotransfected with an Aro-1.3/II luciferase reporter, with or without various combinations of PELP1/MNAR, PNRC2, and ERRα. After 48 h, luciferase activity was measured. E, MCF7 cells expressing GFP-PELP1 were treated with 100 ng/ml EGF, and nuclear extracts were immunoprecipitated with the GFP antibody. The presence of ERRα and PNRC2 in the immunoprecipitates was analyzed by Western blot analysis. F, MCF-7 and MCF-7-HER2 cells were cotransfected with Aro-1.3/II luciferase reporter, with or without PELP1/MNAR. After 48 h, cells were treated with XCT790 (5 μm) for 24 h, and luciferase activity was measured. *, P < 0.05; **, P < 0.001. Data shown are the means ± se from three independent experiments performed in triplicate wells.
PELP1 and Aromatase Are Co-Overexpressed in a Subset of Breast Tumors
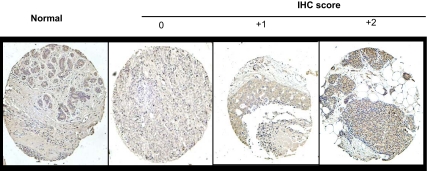
We recently found that PELP1 expression is deregulated in node-positive and advanced-grade breast tumors (21). Because results from this study indicated that PELP1 deregulation promoted aromatase expression in breast epithelial cells, we investigated whether aromatase expression is deregulated in breast tumors and whether its expression correlates with PELP1 expression using a breast cancer tissue microarrays (TMAs) as a proof of principle study. IHC analysis of the breast tumor array showed increased expression of aromatase in ductal carcinoma in situ and node-positive tumors compared with no or weak expression in normal breast tissue (Fig. 7). Statistical analysis revealed that aromatase expression was positively correlated with cancer stage and node status (Table 1). No significant correlation of aromatase expression with the ER, progesterone receptor, or patient age was observed. Interestingly, 54% (n = 79) of PELP1-overexpressing tumors also overexpressed aromatase compared with 36% (n = 47) in PELP1 low-expressing tumors (Table 1). Collectively, these results suggested that deregulation of aromatase expression occurs in advanced-stage and node-positive tumors and that co-overexpression of PELP1 and aromatase may occur in a subset of tumors.
Figure 7.
The Expression of Aromatase in Node-Negative and Node-Positive Breast Tumors
A, Sections were stained with aromatase antiserum. Sections were scored according to IHC intensity in a range from 0–2, in which 0 indicated no expression; 1, low expression; and 2, high expression. A representative sample of each score is shown with the respective aromatase staining.
Table 1.
Distribution of Aromatase Staining and Clinicopathological Factors in Invasive Breast Cancer
| Patient No. | Aromatase Staining
|
P Value | |||
|---|---|---|---|---|---|
| 0 | 1 | 2 | |||
| Age | |||||
| <50 yr | 53 | 12 (22) | 18 (34) | 23 (44) | 0.309 |
| >50 yr | 105 | 25 (24) | 24 (22) | 56 (54) | |
| Stage | |||||
| T1 (<20 mm) | 86 | 26 (30) | 27 (32) | 33 (39) | 0.005 |
| T2 (20–50 mm) | 72 | 11 (15) | 15 (21) | 46 (64) | |
| Grade | |||||
| 1 | 33 | 11 (33) | 8 (24) | 14 (42) | 0.667 |
| 2 | 97 | 20 (21) | 26 (27) | 51 (52) | |
| 3 | 28 | 6 (23) | 8 (27) | 14 (50) | |
| Case type | |||||
| Node negative | 52 | 24 (46) | 12 (23) | 16 (31) | 0.001 |
| Node positive | 53 | 6 (11) | 12 (23) | 35 (66) | |
| Metastatic disease | 53 | 7 (13) | 18 (34) | 28 (53) | |
| ER | |||||
| Positive | 102 | 24 (24) | 26 (26) | 52 (52) | 0.586 |
| Negative (unknown = 4) | 52 | 11 (21) | 16 (31) | 25 (48) | |
| PR | |||||
| Positive | 57 | 17 (31) | 12 (22) | 27 (48) | 0.496 |
| Negative (unknown = 2) | 99 | 20 (20) | 29 (29) | 50 (51) | |
| PELP1 | 0.008 | ||||
| 1 | 11 | 5 (45) | 1 (9) | 5 (46) | |
| 2 | 51 | 17 (33) | 17 (33) | 17 (33) | |
| 3 | 96 | 15 (16) | 24 (25) | 57 (60) | |
DISCUSSION
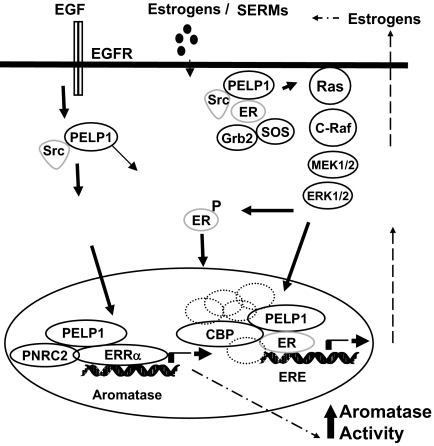
Mammary tumorigenesis is accelerated by the action of ovarian hormones. Emerging findings suggest that abnormal expression of the aromatase enzyme in breast cancer cells and surrounding stroma may influence the breast tumor growth and maintenance (9,34). Inhibition of the aromatase enzyme is considered to be an effective approach in reducing the growth-stimulatory effects of E2. Currently, several aromatase inhibitors are in clinical use, and they appear to be very beneficial to postmenopausal women (12). Recent studies using laboratory models suggest that long-term use of aromatase inhibitors also leads to development of hormone resistance (15,35). However, very little is known about the mechanisms by which tumors acquire local E2 production. Here, we identify one signaling pathway that is used by breast cancer cells to induce local E2 synthesis. We found that growth factor signaling promoted ER coactivator PELP1 to form a complex with ERRα and PNRC2, leading to activation of aromatase expression in breast epithelial cells. This results in the formation of a positive feedback loop locally synthesizing E2, resulting in the activation of E2-ER signaling in breast cancer cells (Fig. 8).
Figure 8.
Schematic Representation of Potential PELP1/MNAR Autocrine Signaling Loop in Breast Cancer Cells
Growth factor signaling and/or activation of nongenomic signaling pathways promote ER coactivator PELP1 to form a complex with ERRα and PNRC2, leading to activation of aromatase expression promoting formation of a positive feedback loop locally synthesizing E2, resulting in the activation of E2-ER-PELP1 signaling in breast cancer cells.
Our findings demonstrate that PELP1 regulates transcriptional activity of the aromatase gene by recruiting to the silencer region located between promoter I.3 and II (−231/+38) of the aromatase gene. The region spanning −231/+38 has been reported to possess binding regions for ERRα, BRCA1, and a transcriptional silencer element (S1) (36,37,38). In this study, we found that growth factor signaling promotes PELP1 recruitment to the aromatase chromatin and its formation of a complex with ERRα and PNRC2. Earlier studies have shown that PELP1/MNAR colocalizes with acetylated histones and interacts with acetyltransferases, and deacetylases; thus, PELP1 recruitment to aromatase chromatin may play a role in chromatin remodeling (18,39). This facilitates up-regulation of aromatase transcription. Because Aro-1.3/II promoter region (−231/+38) contains several sites for corepressors, it is also possible that PELP1 recruitment to this region may facilitate de-repression of aromatase promoter I.3/II, leading to augmentation of aromatase expression in mammary gland tumor tissues.
Evolving evidence suggests that intratumor production of E2 in postmenopausal women occurs via aromatization of steroids, such as androstenedione and testosterone into E2, and this enzymatic reaction is catalyzed by aromatase enzyme (34,40) The regulation of aromatase expression is complex and controlled by activation of distinct promoters in a tissue-specific manner. Substantial evidence suggests that aromatase promoter I.3 and II are the main promoters that regulate aromatase expression in breast tumors (41). In normal breast tissue, proximal aromatase 1.3/II promoters remain quiescent via inhibitor recruitment to the silencer region, and aromatase promoter I.4 plays an important role in its expression (42). In malignant breast epithelial tissue, promoter I.3 and II regions are occupied by multiple transcriptional enhancers as a result of activation of multiple signaling pathways to increase aromatase expression (41,42). In this study, we found that PELP1 overexpression or growth factor signaling enhances PELP1 recruitment to the silencer regions of the promoter I.3/II, suggesting PELP1 could be one of those factors that promote aromatase expression via activation of the 1.3/II promoters. Further increased association of aromatase expression in PELP1-overexpressing cells also suggests that this pathway may contribute to local E2 synthesis in pathological conditions. In this study, we have performed IHC analysis of tumor arrays as a proof of principle to examine whether a correlation seen in cell culture also occurs in breast tumors. Coexpression of PELP1 and aromatase was observed in a subset of tumors. Future studies using a larger number of samples are warranted to examine whether PELP1 could serve as a prognostic or diagnostic marker.
Nuclear receptor (NR) coregulators are proteins that interact with NRs to modulate NR transactivation functions (6,43). Recent evidence suggests that coregulators function as major regulators and coordinators of hormone receptor physiology, which includes the determination of tissue specificity of hormonal action, the integration of membrane and nuclear signaling, the integration of growth factor and physiological signaling to NRs, and the coordination of cell motility (6). Several ER-coregulator proteins are differentially expressed in tumors (2,44,45). Dysregulation of these coregulators could influence target gene expression and participate in the development of hormone-responsive cancers. The ER and ER-coactivators are targets of growth factor signaling, and their phosphorylation is suggested to have a role in hormone resistance (46). Our results suggest that the ER coregulator PELP1 has potential to activate aromatase expression leading to local E2 synthesis in breast epithelial cells. Earlier studies have shown that PELP1 deregulation in MCF7 cells promote tumor growth in the absence of exogenous E2. PELP1 regulation of aromatase expression could be one possible mechanism by which PELP1 promotes tumor growth by promoting an autocrine ER signaling loop.
The orphan ERRα is a member of the NR family structurally most related to the canonical ER and has been shown to modulate E2 signaling in some contexts (47). Earlier studies found that ERRα up-regulates aromatase expression via the I.3/II promoters in MCF7 cells (48). A recent study using laser capture microdissection of breast tumor samples demonstrated positive correlation of ERRα expression with aromatase levels (49). In addition, the NR coregulator PNRC2 was shown to enhance the transcriptional activation mediated by ERRα (50). Our results suggests that deregulation of PELP1 can promote local E2 synthesis through its interaction with the orphan NR ERRα and PNRC2 complex. In addition, these findings also suggest a possibility that PELP1 functions as an adaptor protein coupling orphan receptors with the growth factor signaling axis.
The mechanisms of resistance to the hormone therapy remain elusive, and many resistant tumors retain the ER. In recent years, hormone-resistant tumors have been targeted with aromatase inhibitors, such as letrozole, in an adjuvant setting. Even though these new treatments appear successful, emerging data suggest that tumors evade this treatment by developing adaptive hypersensitivity manifested as hormone-independent tumorigenesis through increased nongenomic signaling and growth factor signaling cross-talk (35,46,51). Deregulation of HER2 expression/signaling has emerged as the most significant factor in the development of hormone resistance. HER2 is an oncogene that has been shown to be overexpressed, amplified, or both in several human malignancies, including breast tumors. ER expression occurs in about 50% of HER2-positive breast cancers, and cross-talk between the ER and HER2 pathways promotes endocrine therapy resistance (52). ER-coregulators are targeted by excessive ER-HER2 cross-talk leading to hormone resistance in a subset of breast tumors (53). Recent studies also demonstrated that HER2 status plays an important role in tumor-induced aromatase activity via the cyclooxygenase-2 pathway (28). Furthermore, HER2 overexpression can also promote ligand-independent recruitment of coactivator complexes to E2-responsive promoters and thus may play a role in the development of letrozole resistance (35). Earlier studies showed that PELP1 interacts with HER2 signaling components, and HER signaling promotes tyrosine phosphorylation of PELP1 (27,54). Our results suggest that HER2 signaling enhances PELP1 recruitment to the aromatase promoter, promotes its interactions with ERRα and PELP1, and plays a critical role in HER2-mediated induction of aromatase expression.
Protooncogene c-Src is a multifunctional intracellular tyrosine kinase implicated in the regulation of a variety of processes, including proliferation, differentiation, survival, and motility (55). Src interacts with multiple cellular factors including HER2 and EGFR. The ER and breast tumors overexpress Src kinase (56). Emerging evidence suggests that PELP1 acts as a scaffolding protein coupling ER with Src kinase leading to activation of ER-Src-MAPK pathway (17,57,58). Our results suggest that nongenomic signaling mediated by c-Src kinase have the potential to modulate aromatase gene expression in breast cancer cells by recruiting coregulator proteins such as PELP1. Our results using kinase inhibitors suggested that Src kinase plays a critical role in the PELP1 activation of the aromatase gene. These findings suggest a possibility that combinational therapy of Src inhibitors along with letrozole may delay resistance and increase the therapeutic potential in hormone-resistant tumors.
MATERIALS AND METHODS
Cell Cultures and Reagents
Generation and characterization of MCF7 clones overexpressing PELP1 (MCF7-PELP1 clone 20 and clone 13) were described earlier (19). Generation and characterization of PELP1-Teton cells were described earlier (20). MCF7 human breast cancer cells and MCF7-PELP1 clones (PELP clone 20, PELP clone 13) were maintained in RPMI plus 10% fetal calf serum (FCS) for routine culture and 10% DCC FCS for treatment conditions. MCF-Aro cells were maintained in RPMI plus 10% FCS (26). SYF (Src-, Yes-, Fyn-negative) cells were purchased from American Type Culture Collection (Manassas, VA). MCF7 cells that stably overexpress full-length HER2 cDNA (MCF-7/HER2) were kindly provided by Dr. Kaladar B. Reddy (Wayne State University, Detroit, MI). Antibodies against vinculin, the steroid hormone 17β-estradiol, β-nicotinamide adenine dinucleotide 2′-phosphate reduced tetrasodium salt, progesterone, XCT790, and dextran-treated charcoal were purchased from Sigma Chemical Co. (St. Louis, MO). ICI 182,780 was purchased from Tocris (Ballwin, MO). The anti-T7-epitope antibody was purchased from Novagen (Milwaukee, WI). Antibodies against phospho-MAPK were purchased from Cell Signaling (Beverly, MA). The antibody for the GFP tag was purchased from Clontech (Palo Alto, CA). The antibody for the Src kinase inhibitor PP2 was purchased from Calbiochem (La Jolla, CA). [1β-3H(N)]Androst-4-ene-3,17-dione was purchased from PerkinElmer (Waltham, MA). The antibody for ERRα (1:1000) was purchased from Upstate (Chicago, IL), and the antibody for PNRC2 (1:1000) was purchased from ProteinTech (Chicago, IL). ERα, ERRα, GDPDH siRNAs were purchased from Dharmacon (Chicago, IL). PELP1/MNAR-specific shRNA (SureSilencing shRNA plasmids, catalog item KH19454N), and control shRNA vector were purchased from SuperArray (Frederick, MD).
Reporter Gene Assays
Plasmids for pGL-3-promoter I.1, pGL-3-promoter I.3/II, I.3/II deletions (−175/+525, −5/+525, −251/+74) were described earlier (59). For ease of convenience, we have used new numbering to refer to these constructs using the transcriptional start site as +1. The new numbers are 1.3/II (−417 to +38), −175/+525 (−480/+220), 5/+525 (−310/+220), and −251/+74 (−570/−230). The pGL-3 basic reporter vector served as a negative control. For transient transfections, MCF7 cells were cotransfected with 100 ng PELP1 along with Aro-1.3 luciferase reporter using the FuGENE6 method (Roche Diagnostics Corp., Indianapolis, IN). The siRNAs were transfected using Oligofectamine according to the manufacturer’s protocol (Invitrogen, Carlsbad, CA). After 24 h of transfection, cells were serum starved for 48 h and then treated with or without EGF for 12 h. For some assays, MCF7 cells stably expressing PELP1 (MCF7-PELP1) or HER2 (MCF7-HER2) were used. Empty parental vectors were used in the reporter gene assays as controls, and the total amount of the DNA in the transfections was kept constant by adding appropriate empty vectors. A small aliquot (10 ng) of β-galactosidase reporter gene was also cotransfected along with all the reporter genes used in the study and β-galactosidase values were used to normalize luciferase activity for transfection efficiency. Each transfection was performed in triplicate and repeated at least three times. At the end of the experiment, cells were lysed in passive-lysis buffer, and the luciferase assay was performed using a luciferase reporter assay kit (Promega, Madison, WI).
Aromatase Assay
Aromatase activity was determined using 3H2O release assay as described (59). Cells were grown in six-well cell culture plates, washed twice with PBS, and then incubated with 1 ml serum-free medium containing 500 nm progesterone and 100 nm [3H]androst-4-ene-3,17-dione as substrate. After a 12-h incubation at 37 C, the reaction mixture was removed and extracted with an equal volume of chloroform to extract unused substrate and further treated with dextran-treated charcoal. After centrifugation, the 3H2O-containing supernatants were counted in a liquid scintillation counter. The protein concentrations were determined after dissolving cells in 0.5 m NaOH by the method of Bradford (Bio-Rad, Hercules, CA). Aromatase activity was calculated as femtomoles per milligram protein per hour.
Real-Time PCR, Northern Blotting, and Soft-Agar Colony Formation
Cells were harvested with TRIzol reagent (Life Technologies, Carlsbad, CA), and total RNA was isolated according to the manufacturer’s manual. Real-time PCR was performed using aromatase exon-specific primers as described (25). Northern blots were hybridized in QuickHyb hybridization solution (Amersham Pharmacia Biotech, Piscataway, NJ) and, using 32P-labeled pS2 cDNA as a probe, exposed to a PhosphorImager screen. Soft-agar colony-growth assays were performed as previously described (21).
ChIP
ChIP analysis was performed as described previously (20). MCF7-PELP1 or MCF7 cells expressing GFP-PELP1 were cross-linked by adding formaldehyde, and the chromatin was subjected to immunoprecipitation using either the T7 or the GFP antibody. Isotype-specific IgG was used as a control. DNA was resuspended in 50 μl Tris-EDTA buffer and used for PCR amplification using the following primers: forward gctttcaattgggaatgcac and reverse agctcctgttgcttcagagg.
Cytospin Preparation
For cytospin slides, MCF7 and MCF7-PELP1 (2 × 105) cells were washed in cold 2% FCS-PBS twice and diluted in 100 μl ice-cold 1% BSA-PBS. Cells were aliquoted (100 μl each) into appropriate wells of a cytospin slide and spun at 3000 rpm for 3 min to form a smear. The cytospin smears were examined for normal morphology using phase-contrast microscopy and were dried in desiccators overnight before fixing in 10% neutral-buffered formalin for 12 h. The slides were then processed for immunohistochemistry.
TMAs
The TMAs used in this study were obtained from the Cooperative Breast Cancer Tissue Resource of the National Cancer Institute. Those TMA arrays are designed by National Cancer Institute statisticians to provide high statistical power and are suitable for use in the investigation of the differences in prevalence of potential markers in three stages of invasive breast cancer: node-negative, node-positive, and metastatic disease. Each TMA consists of 288 0.6-mm cores taken from paraffin-embedded specimens that represent a total of 252 breast cancer and normal breast tissue specimens plus 36 controls.
Immunohistochemistry
IHC of TMA was performed as described. The PELP1 antibody was developed in our laboratory. It was well characterized and has been used in IHC research in published studies (16,27,57). Commercially available aromatase mAb was obtained from Serotec (Oxford, UK). As a control in some assays, aromatase mAb 677 generated by Novartis was used. The TMA sections were then incubated with the polyclonal rabbit antihuman-PELP1 antiserum (1:500), the aromatase mAb (1:50, Serotec, MCA2077S) (22), or the aromatase mAb (1:1000; Novartis no. 677) (23,24) and was incubated overnight at room temperature. The preparation of negative controls was accomplished by replacing the primary antibody with control rabbit or mouse IgG. The finding that no cells or less than 10% of the cells were immunoreactive was considered to be a negative result, and the finding that more than 10% of the cells were immunoreactive was considered a positive result. The sections were scored by three evaluators blinded to the patient’s clinical status. PELP1 and aromatase staining was performed according to the established method (27,57), and the results were classified as follows: 0, no expression; 1, weak expression; more than 2, high expression. Clinical correlates obtained from the Cooperative Breast Cancer Tissue Resource were used to correlate aromatase with various clinicopathological variables, including patient age, cancer stage, grade, and case type, ER, and PR. Associations among variables were analyzed with the χ2 and Spearman’s rank correlation coefficients. All tests were performed with SPSS software (SPSS Inc., Chicago, IL).
Acknowledgments
We thank Dr. Dimple Chakravarty, and Binoj C. Nair for critical reading of the manuscript.
Footnotes
This work was supported by National Institutes of Health National Cancer Institute Grants CA095681 (R.K.V.) and P30CA54174 (RRT).
Disclosure Statement: The authors have nothing to disclose.
First Published Online December 13, 2007
Abbreviations: ChIP, Chromatin immunoprecipitation; DCC, dextran-coated charcoal; E2, estrogen; EGF, epidermal growth factor; ER, estrogen receptor; ERR, estrogen-related receptor; FCS, fetal calf serum; GFP, green fluorescent protein; IHC, immunohistochemistry; mAb, monoclonal antibody; MNAR, modulator of nongenomic actions of estrogen receptor; NR, nuclear receptor; PELP1, proline-, glutamic acid-, and leucine-rich protein-1; PI3K, phosphatidylinositol-3-kinase; PNRC2, proline-rich nuclear receptor coregulatory protein 2 shRNA, short hairpin RNA; siRNA, small interfering RNA; TMA, tissue microarray.
References
- Moy B, Goss PE 2006 Estrogen receptor pathway: resistance to endocrine therapy and new therapeutic approaches. Clin Cancer Res 12:4790–4793 [DOI] [PubMed] [Google Scholar]
- McDonnell DP, Norris JD 2002 Connections and regulation of the human estrogen receptor. Science 296:1642–1644 [DOI] [PubMed] [Google Scholar]
- Jensen EV, Jordan VC 2003 The estrogen receptor: a model for molecular medicine. Clin Cancer Res 9:1980–1989 [PubMed] [Google Scholar]
- Losel R, Wehling M 2003 Nongenomic actions of steroid hormones. Nat Rev Mol Cell Biol 4:46–56 [DOI] [PubMed] [Google Scholar]
- Bjornstrom L, Sjoberg M 2005 Mechanisms of estrogen receptor signaling: convergence of genomic and nongenomic actions on target genes. Mol Endocrinol 19:833–842 [DOI] [PubMed] [Google Scholar]
- Lonard DM, O’Malley BW 2006 The expanding cosmos of nuclear receptor coactivators. Cell 125:411–414 [DOI] [PubMed] [Google Scholar]
- Collingwood TN, Urnov FD, Wolffe AP 1999 Nuclear receptors: coactivators, corepressors and chromatin remodeling in the control of transcription. J Mol Endocrinol 23:255–275 [DOI] [PubMed] [Google Scholar]
- Simpson ER, Mahendroo MS, Means GD, Kilgore MW, Hinshelwood MM, Graham-Lorence S, Amarneh B, Ito Y, Fisher CR, Michael MD 1994 Aromatase cytochrome P450, the enzyme responsible for estrogen biosynthesis. Endocr Rev 15:342–355 [DOI] [PubMed] [Google Scholar]
- Lu Q, Nakmura J, Savinov A, Yue W, Weisz J, Dabbs DJ, Wolz G, Brodie A 1996 Expression of aromatase protein and messenger ribonucleic acid in tumor epithelial cells and evidence of functional significance of locally produced estrogen in human breast cancers. Endocrinology 137:3061–3068 [DOI] [PubMed] [Google Scholar]
- James VH, McNeill JM, Lai LC, Newton CJ, Ghilchik MW, Reed MJ 1987 Aromatase activity in normal breast and breast tumor tissues: in vivo and in vitro studies. Steroids 50:269–279 [DOI] [PubMed] [Google Scholar]
- Bulun SE, Sebastian S, Takayama K, Suzuki T, Sasano H, Shozu M 2003 The human CYP19 (aromatase P450) gene: update on physiologic roles and genomic organization of promoters. J Steroid Biochem Mol Biol 86:219–224 [DOI] [PubMed] [Google Scholar]
- Jordan VC, Brodie AM 2007 Development and evolution of therapies targeted to the estrogen receptor for the treatment and prevention of breast cancer. Steroids 72:7–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaklamani VG, Gradishar WJ 2005 Adjuvant therapy of breast cancer. Cancer Invest 23:548–560 [DOI] [PubMed] [Google Scholar]
- Pietras RJ 2006 Biologic basis of sequential and combination therapies for hormone-responsive breast cancer. Oncologist 11:704–717 [DOI] [PubMed] [Google Scholar]
- Sabnis G, Goloubeva O, Jelovac D, Schayowitz A, Brodie A 2007 Inhibition of the phosphatidylinositol 3-kinase/Akt pathway improves response of long-term estrogen-deprived breast cancer xenografts to antiestrogens. Clin Cancer Res 13:2751–2757 [DOI] [PubMed] [Google Scholar]
- Vadlamudi RK, Wang RA, Mazumdar A, Kim Y, Shin J, Sahin A, Kumar R 2001 Molecular cloning and characterization of PELP1, a novel human coregulator of estrogen receptor α. J Biol Chem 276:38272–38279 [DOI] [PubMed] [Google Scholar]
- Wong CW, McNally C, Nickbarg E, Komm BS, Cheskis BJ 2002 Estrogen receptor-interacting protein that modulates its nongenomic activity-crosstalk with Src/Erk phosphorylation cascade. Proc Natl Acad Sci USA 99:14783–14788 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Vadlamudi RK, Kumar R 2007 Functional and biological properties of the nuclear receptor coregulator PELP1/MNAR. Nucl Recept Signal 5:e004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasenthil S, Vadlamudi RK 2003 Functional interactions between the estrogen receptor coactivator PELP1/MNAR and retinoblastoma protein. J Biol Chem 278:22119–22127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair SS, Mishra SK, Yang Z, Balasenthil S, Kumar R, Vadlamudi RK 2004 Potential role of a novel transcriptional coactivator PELP1 in histone H1 displacement in cancer cells. Cancer Res 64:6416–6423 [DOI] [PubMed] [Google Scholar]
- Rajhans R, Nair S, Holden AH, Kumar R, Tekmal RR, Vadlamudi RK 2007 Oncogenic potential of the nuclear receptor coregulator proline-, glutamic acid-, leucine-rich protein 1/modulator of the nongenomic actions of the estrogen receptor. Cancer Res 67:5505–5512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirianni R, Chimento A, Malivindi R, Mazzitelli I, Andò S, Pezzi V 2007 Insulin-like growth factor-I, regulating aromatase expression through steroidogenic factor 1, supports estrogen-dependent tumor Leydig cell proliferation. Cancer Res 67:8368–8377 [DOI] [PubMed] [Google Scholar]
- Sasano H, Edwards DP, Anderson TJ, Silverberg SG, Evans DB, Santen RJ, Ramage P, Simpson ER, Bhatnagar AS, Miller WR 2003 Validation of new aromatase monoclonal antibodies for immunohistochemistry: progress report. J Steroid Biochem Mol Biol 86:239–244 [DOI] [PubMed] [Google Scholar]
- Sasano H, Anderson TJ, Silverberg SG, Santen RJ, Conway M, Edwards DP, Krause A, Bhatnagar AS, Evans DB, Miller WR 2005 The validation of new aromatase monoclonal antibodies for immunohistochemistry: a correlation with biochemical activities in 46 cases of breast cancer. J Steroid Biochem Mol Biol 95:35–39 [DOI] [PubMed] [Google Scholar]
- Wang X, Chen S 2006 Aromatase destabilizer: novel action of exemestane, a food and drug administration-approved aromatase inhibitor. Cancer Res 66:10281–10286 [DOI] [PubMed] [Google Scholar]
- Zhou DJ, Pompon D, Chen SA 1990 Stable expression of human aromatase complementary DNA in mammalian cells: a useful system for aromatase inhibitor screening. Cancer Res 50:6949–6954 [PubMed] [Google Scholar]
- Vadlamudi RK, Balasenthil S, Broaddus RR, Gustafsson JA, Kumar R 2004 Deregulation of estrogen receptor coactivator proline-, glutamic acid-, and leucine-rich protein-1/modulator of nongenomic activity of estrogen receptor in human endometrial tumors. J Clin Endocrinol Metab 89:6130–6138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbaramaiah K, Howe LR, Port ER, Brogi E, Fishman J, Liu CH, Hla T, Hudis C, Dannenberg AJ 2006 HER-2/neu status is a determinant of mammary aromatase activity in vivo: evidence for a cyclooxygenase-2-dependent mechanism. Cancer Res 66:5504–5511 [DOI] [PubMed] [Google Scholar]
- Klinghoffer RA, Sachsenmaier C, Cooper JA, Soriano P 1999 Src family kinases are required for integrin but not PDGFR signal transduction. EMBO J 18:2459–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Yu B, Zhou D, Chen S 2002 Regulation of aromatase promoter activity in human breast tissue by nuclear receptors. Oncogene 21:2854–2863 [DOI] [PubMed] [Google Scholar]
- Zhou D, Chen S 2001 PNRC2 is a 16 kDa coactivator that interacts with nuclear receptors through an SH3-binding motif. Nucleic Acids Res 29:3939–3948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Ye JJ, Li Y, Lui K, Chen S 2006 The molecular basis of the interaction between the proline-rich SH3-binding motif of PNRC and estrogen receptor α. Nucleic Acids Res 34:5974–5986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Hinshelwood MM, Giguere V, Mendelson CR 2006 Estrogen related receptor-αenhances surfactant protein-A gene expression in fetal lung type II cells. Endocrinology 147:5187–5195 [DOI] [PubMed] [Google Scholar]
- Miller WR, Anderson TJ, Jack WJ 1990 Relationship between tumour aromatase activity, tumour characteristics and response to therapy. J Steroid Biochem Mol Biol 37:1055–1059 [DOI] [PubMed] [Google Scholar]
- Shin I, Miller T, Arteaga CL 2006 ErbB receptor signaling and therapeutic resistance to aromatase inhibitors. Clin Cancer Res 12:1008s–1012s [DOI] [PubMed] [Google Scholar]
- Ghosh S, Lu Y, Katz A, Hu Y, Li R 2007 Tumor suppressor BRCA1 inhibits a breast cancer-associated promoter of the aromatase gene (CYP19) in human adipose stromal cells. Am J Physiol Endocrinol Metab 292:E246–E252 [DOI] [PubMed] [Google Scholar]
- Lu M, Chen D, Lin Z, Reierstad S, Trauernicht AM, Boyer TG, Bulun SE 2006 BRCA1 negatively regulates the cancer-associated aromatase promoters I.3 and II in breast adipose fibroblasts and malignant epithelial cells. J Clin Endocrinol Metab 91:4514–4519 [DOI] [PubMed] [Google Scholar]
- Yang C, Zhou D, Chen S 1998 Modulation of aromatase expression in the breast tissue by ERRα-1 orphan receptor. Cancer Res 58:5695–5700 [PubMed] [Google Scholar]
- Mishra SK, Mazumdar A, Vadlamudi RK, Li F, Wang RA, Yu W, Jordan VC, Santen RJ, Kumar R 2003 MICoA, a novel metastasis-associated protein 1 (MTA1) interacting protein coactivator, regulates estrogen receptor-α transactivation functions. J Biol Chem 278:19209–19219 [DOI] [PubMed] [Google Scholar]
- Bulun SE, Price TM, Aitken J, Mahendroo MS, Simpson ER 1993 A link between breast cancer and local estrogen biosynthesis suggested by quantification of breast adipose tissue aromatase cytochrome P450 transcripts using competitive polymerase chain reaction after reverse transcription. J Clin Endocrinol Metab 77:1622–1628 [DOI] [PubMed] [Google Scholar]
- Bulun SE, Lin Z, Imir G, Amin S, Demura M, Yilmaz B, Martin R, Utsunomiya H, Thung S, Gurates B, Tamura M, Langoi D, Deb S 2005 Regulation of aromatase expression in estrogen-responsive breast and uterine disease: from bench to treatment. Pharmacol Rev 57:359–383 [DOI] [PubMed] [Google Scholar]
- Simpson ER, Clyne C, Rubin G, Boon WC, Robertson K, Britt K, Speed C, Jones M 2002 Aromatase: a brief overview. Annu Rev Physiol 64:93–127 [DOI] [PubMed] [Google Scholar]
- Smith CL, O’Malley BW 2004 Coregulator function: a key to understanding tissue specificity of selective receptor modulators. Endocr Rev 25:45–71 [DOI] [PubMed] [Google Scholar]
- Anzick SL, Kononen J, Walker RL, Azorsa DO, Tanner MM, Guan XY, Sauter G, Kallioniemi OP, Trent JM, Meltzer PS 1997 AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science 277:965–968 [DOI] [PubMed] [Google Scholar]
- Barnes CJ, Vadlamudi RK, Kumar R 2004 Novel estrogen receptor coregulators and signaling molecules in human diseases. Cell Mol Life Sci 61:281–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiff R, Massarweh SA, Shou J, Bharwani L, Arpino G, Rimawi M, Osborne CK 2005 Advanced concepts in estrogen receptor biology and breast cancer endocrine resistance: implicated role of growth factor signaling and estrogen receptor coregulators. Cancer Chemother Pharmacol 56(Suppl 1):10–20 [DOI] [PubMed] [Google Scholar]
- Stein RA, McDonnell DP 2006 Estrogen-related receptor α as a therapeutic target in cancer. Endocr Relat Cancer 13(Suppl 1):S25–S32 [DOI] [PubMed] [Google Scholar]
- Chen S, Itoh T, Wu K, Zhou D, Yang C 2002 Transcriptional regulation of aromatase expression in human breast tissue. J Steroid Biochem Mol Biol 83:93–99 [DOI] [PubMed] [Google Scholar]
- Miki Y, Suzuki T, Tazawa C, Yamaguchi Y, Kitada K, Honma S, Moriya T, Hirakawa H, Evans DB, Hayashi S, Ohuchi N, Sasano H 2007 Aromatase localization in human breast cancer tissues: possible interactions between intratumoral stromal and parenchymal cells. Cancer Res 67:3945–3954 [DOI] [PubMed] [Google Scholar]
- Zhou D, Quach KM, Yang C, Lee SY, Pohajdak B, Chen S 2000 PNRC: a proline-rich nuclear receptor coregulatory protein that modulates transcriptional activation of multiple nuclear receptors including orphan receptors SF1 (steroidogenic factor 1) and ERRα1 (estrogen related receptor α-1). Mol Endocrinol 14:986–998 [DOI] [PubMed] [Google Scholar]
- Gururaj AE, Rayala SK, Vadlamudi RK, Kumar R 2006 Novel mechanisms of resistance to endocrine therapy: genomic and nongenomic considerations. Clin Cancer Res 12:1001s–1007s [DOI] [PubMed] [Google Scholar]
- Marcom PK, Isaacs C, Harris L, Wong ZW, Kommarreddy A, Novielli N, Mann G, Tao Y, Ellis MJ 2007 The combination of letrozole and trastuzumab as first or second-line biological therapy produces durable responses in a subset of HER2 positive and ER positive advanced breast cancers. Breast Cancer Res Treat 102:43–49 [DOI] [PubMed] [Google Scholar]
- Shou J, Massarweh S, Osborne CK, Wakeling AE, Ali S, Weiss H, Schiff R 2004 Mechanisms of tamoxifen resistance: increased estrogen receptor-HER2/neu cross-talk in ER/HER2-positive breast cancer. J Natl Cancer Inst 96:926–935 [DOI] [PubMed] [Google Scholar]
- Manavathi B, Nair SS, Wang RA, Kumar R, Vadlamudi RK 2005 Proline-, glutamic acid-, and leucine-rich protein-1 is essential in growth factor regulation of signal transducers and activators of transcription 3 activation. Cancer Res 65:5571–5577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevino JG, Summy JM, Gallick GE 2006 SRC inhibitors as potential therapeutic agents for human cancers. Mini Rev Med Chem 6:681–687 [DOI] [PubMed] [Google Scholar]
- Russello SV, Shore SK 2004 SRC in human carcinogenesis. Front Biosci 9:139–144 [DOI] [PubMed] [Google Scholar]
- Vadlamudi RK, Manavathi B, Balasenthil S, Nair SS, Yang Z, Sahin AA, Kumar R 2005 Functional implications of altered subcellular localization of PELP1 in breast cancer cells. Cancer Res 65:7724–7732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barletta F, Wong CW, McNally C, Komm BS, Katzenellenbogen B, Cheskis BJ 2004 Characterization of the interactions of estrogen receptor and MNAR in the activation of cSrc. Mol Endocrinol 18:1096–1108 [DOI] [PubMed] [Google Scholar]
- Zhou D, Chen S 1999 Identification and characterization of a cAMP-responsive element in the region upstream from promoter 1.3 of the human aromatase gene. Arch Biochem Biophys 371:179–190 [DOI] [PubMed] [Google Scholar]