Abstract
The male sex steroid, testosterone (T), is synthesized from cholesterol in the testicular Leydig cell under control of the pituitary gonadotropin LH. Unlike most cells that use cholesterol primarily for membrane synthesis, steroidogenic cells have additional requirements for cholesterol, because it is the essential precursor for all steroid hormones. Little is known about how Leydig cells satisfy their specialized cholesterol requirements for steroid synthesis. We show that in mice with a unique hypomorphic androgen mutation, which disrupts the feedback loop governing T synthesis, that genes involved in cholesterol biosynthesis/uptake and steroid biosynthesis are up-regulated. We identify LH as the central regulatory molecule that controls both steroidogenesis and Leydig cell cholesterol homeostasis in vivo. In addition to the primary defect caused by high levels of LH, absence of T signaling exacerbates the lipid homeostasis defect in Leydig cells by eliminating a short feedback loop. We show that T signaling can affect the synthesis of steroids and modulates the expression of genes involved in de novo cholesterol synthesis. Surprisingly, accumulation of active sterol response element-binding protein 2 is not required for up-regulation of genes involved in cholesterol biosynthesis and uptake in Leydig cells.
STEROID HORMONES PLAY critical regulatory roles in diverse aspects of mammalian physiology including the response to stress, development of secondary sexual characteristics, the control of the menstrual cycle, and spermatogenesis. All steroid hormones are derived from cholesterol in a series of step-wise enzymatic modifications. In mammals, there are two major sites of steroid hormone synthesis: the adrenals, which mainly produce aldosterone and corticosteroids, and the gonads, which synthesize the sex steroids. In females, estradiol and progesterone are the major sex steroids, whereas males synthesize primarily testosterone (T) and its more potent metabolite, dihydrotestosterone (DHT). Steroid signaling is mediated primarily through the nuclear hormone receptor class of ligand-dependent transcription factors (1). T exerts its actions by signaling through the androgen receptor (AR) (2,3).
The synthesis of steroid hormones is a highly regulated process. The primary site of T production in males is the Leydig cell, the major cell type that populates the interstitial regions of the testis. LH, secreted by the pituitary, binds to the G protein-coupled LH/chorionic gonadotropin receptor (LHCGR) on the surface of Leydig cells (4). Activation of LHCGR stimulates Leydig cell T output by increasing intracellular concentrations of cAMP, thereby increasing the production of proteins necessary for steroidogenesis. T then reduces the synthesis of LH in the pituitary by directly repressing the expression of the LH β-subunit (5). A number of studies also suggest that T can regulate release of the hypothalamic GnRH, the signal that initially stimulates the pituitary to synthesize and release LH (6). This regulatory loop is referred to as the hypothalamic-pituitary-gonadal (HPG) axis.
Transcription of genes encoding proteins involved in steroidogenesis is stimulated by cAMP. The genes encoding steroidogenic enzymes, including steroidogenic acute regulatory protein (StAR; Star), cytochrome P450 11a1 (Cyp11a1, also known as cytochrome P450 side chain cleavage) and cytochrome P450 17a1 (Cyp17a1), are known targets of cAMP-stimulated transcription (7,8,9). T synthesis in Leydig cells is also regulated by a short feedback loop whereby T represses the transcription of genes involved in steroidogenesis. The effect of T on steroidogenic gene expression is well illustrated by studies of Cyp17a1, where T represses cAMP-stimulated Cyp17a1 mRNA in both primary Leydig cell cultures and in MA-10 Leydig tumor cells (10,11).
The regulation of genes involved in the biosynthesis of cholesterol, the precursor of steroid hormones, is well studied (12). Central to the regulation of intracellular cholesterol levels is the membrane-bound transcription factor, sterol response element-binding protein (SREBP). When concentrations of sterols are low within the cell, SREBP cleavage-activating protein (SCAP) facilitates the cleavage of the membrane-bound SREBP, freeing SREBP to translocate to the nucleus and activate genes encoding de novo cholesterol biosynthetic enzymes and receptors mediating cholesterol uptake (13). There are three related SREBP proteins: SREBP1a, which regulates fatty acid and cholesterol biosynthesis; SREBP1c, which regulates fatty acid synthesis; and SREBP2, which primarily regulates genes involved in cholesterol metabolism (14). Several lines of evidence indirectly suggest that the SREBP pathway is active in Leydig cells. First, Leydig cells robustly express Srebp2 and to a lesser extent Srebp1c but not Srebp1a (15). Second, activation of steroidogenesis with human chorionic gonadotropin (hCG), an agonist of LHCGR, results in the depletion of intracellular cholesterol reserves in Leydig or Leydig tumor cells (16,17). Coincident with the depletion of intracellular lipid pools, there is a transient up-regulation of 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase (HMGCR) activity, the rate-limiting activity in de novo cholesterol biosynthesis, suggesting activation of the SREBP2 pathway.
These circumstantial pieces of evidence implicate the SREBP pathway in the regulation of cholesterol synthesis and uptake in Leydig cells. However, several pieces of evidence argue against a role for SREBP in the regulation of genes involved in cholesterol homeostasis in steroidogenic cells. When the Leydig tumor line MA-10 is treated with 25-hydroxycholesterol (25-OHC), a potent inhibitor of SREBP activation, there is no effect on HMG-CoA synthase (Hmgcs1) expression or de novo cholesterol biosynthesis (18). This study suggests that steroidogenic tissues may not regulate cholesterol acquisition via the SREBP-mediated sterol-sensing pathway.
To understand better the regulation of steroidogenesis and cholesterol homeostasis in the testis, we used a unique genetic model that disrupts feedback regulation in the HPG axis. Our group has previously generated a novel allele of the Ar in the mouse that circumvents the complication of cryptorchidism and sex reversal associated with null mutation of Ar. We generated a hypomorphic allele of Ar, Arinvflox(ex1-neo)/Y, by inserting a neomycin resistance (neoR) cassette into intron 1 (supplemental Fig. 1, published as supplemental data on The Endocrine Society’s Journals Online web site at http://mend.endojournals.org) (19). A cryptic splice acceptor within neoR results in stochastic splicing into the neo open reading frame, leading to reduced Ar mRNA. Despite the severe deficiency in AR compared with wild-type, these animals undergo normal testicular descent, and, like the spontaneous null mutant Artfm, they have elevated levels of serum LH. Unlike the Artfm/Y mouse, Arinvflox(ex1-neo)/Y males produce vast amounts of T in response to the high levels of serum LH. In this study, we use this model to dissect the role of hormonal signaling in the regulation of steroidogenesis and maintenance of cholesterol homeostasis in Leydig cells.
RESULTS
Serum Hormone Profiles Reflect Disruption of the HPG Axis
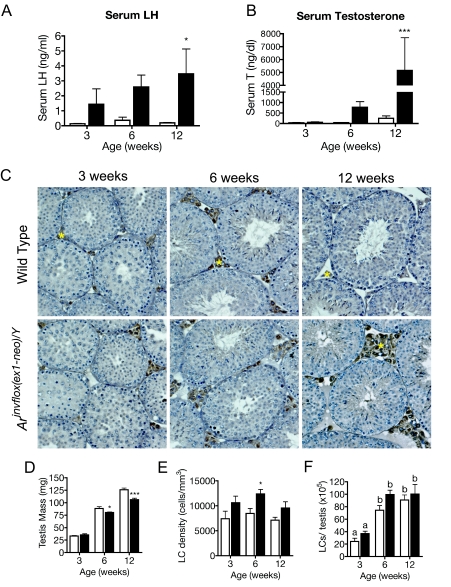
In a previous study, we demonstrated that serum T and LH were dramatically increased in 8-wk-old Arinvflox(ex1-neo)/Y male mice (19). The high levels of LH, produced in response to the disruption of feedback regulation, resulted in high levels of T produced by Arinvflox(ex1-neo)/Y Leydig cells. To better understand the cause and consequence of the changes in the context of pubertal maturation, we measured serum LH and T levels from wild-type and Arinvflox(ex1-neo)/Y mice at 3, 6, and 12 wk of age. In wild-type mice, serum LH levels rise from early to late puberty as expected. However, in Arinvflox(ex1-neo)/Y mice, LH levels appear elevated as early as 3 wk of age and continue to rise into adulthood, when they show a 21-fold elevation above wild-type LH levels (Fig. 1A). Accompanying the rise in serum LH levels, T levels increase into adulthood for both wild-type and Arinvflox(ex1-neo)/Y mice. In Arinvflox(ex1-neo)/Y mice, serum levels of T are approximately 20-fold higher than in adult wild-type mice (Fig. 1B). Thus, the pattern of pubertal increase in serum LH and T is unaffected in Arinvflox(ex1-neo)/Y, but the magnitude of the changes of these hormones is significantly affected.
Figure 1.
Hormonal Profiles and Leydig Cell Numbers during Pubertal Maturation
Hormonal profiles and Leydig cell numbers are shown for wild-type (white bars) and Arinvflox(ex1-neo)/Y (black bars) mice during pubertal maturation. A and B, Serum LH (A) and serum T (A) levels. Each bar represents the mean hormone level for five to 10 animals. *, P < 0.05; **, P < 0.01; ***, P < 0.001 from a two-way ANOVA followed by Bonferroni post test. C, Transverse sections of wild-type and Arinvflox(ex1-neo)/Y testes from animals 3, 6, and 12 wk of age were stained with an antibody against CYP11A1 to mark Leydig cells (brown) and then counterstained with hematoxylin. Yellow asterisks mark CYP11A1+ Leydig cells. D, Testis mass is decreased in Arinvflox(ex1-neo)/Y (black bars) compared with wild-type (white bars). E, Numerical density of Leydig cell nuclei. F, Absolute number of Leydig cells per testis as estimated from numerical density of Leydig cells (LCs) (see Materials and Methods). Each bar represents the mean values for six to eight animals. *, P < 0.05; **, P < 0.01; ***, P < 0.001; letters indicate P < 0.05 for a two-way ANOVA followed by Bonferroni post test.
Leydig Cell Numbers Unaltered in Arinvflox(ex1-neo)/Y
Previous studies suggest that the hormonal milieu of the testis can have a profound effect on Leydig cell numbers (20,21,22). Because increased Leydig cell number might increase T output in Arinvflox(ex1-neo)/Y mice, we examined the histology of the testis for gross signs of Leydig cell hyperplasia (Fig. 1C). Using an antibody against the steroidogenic enzyme CYP11A1 to mark Leydig cells, we observed an apparent increase in the number of Leydig cells populating the interstitial regions of the testis in Arinvflox(ex1-neo)/Y mice. To more definitively assess the Leydig cell numbers during and after pubertal maturation, we used a systematic histological method to estimate Leydig cell number (23). Testis mass was significantly reduced in Arinvflox(ex1-neo)/Y mice by 6 wk of age, probably due to the previously described defect in terminal germ cell differentiation (19). Mass was used to estimate testis volume (see Materials and Methods). As suggested by our initial observations of Arinvflox(ex1-neo)/Y testis histology, the numerical density of Leydig cells is increased in the mutant testis (Fig. 1E). However, the total number of Leydig cells on a per-testis basis is not significantly affected in Arinvflox(ex1-neo)/Y (Fig. 1F). The increase in Leydig cell numerical density was taken into account when analyzing the expression of Leydig cell-specific transcripts (see Materials and Methods).
Genes Involved in Steroidogenesis Are Misregulated in Ar Hypomorphs
Because Leydig cells become increasingly responsive to LH during pubertal maturation, we measured Lhcgr expression in wild-type and Arinvflox(ex1-neo)/Y males at an early-puberty time point (3 wk), a late-puberty time point (6 wk), and adulthood (12 wk) by quantitative RT-PCR (qRT-PCR). We observed that Arinvflox(ex1-neo)/Y Leydig cells express increasing amounts of Lhcgr during pubertal development, reaching 11-fold up-regulation over wild-type by adulthood (Fig. 2A). Elevated levels of Lhcgr mRNA are compatible with an increase in LH-stimulated increase in T production.
Figure 2.
Misregulation of Transcripts Encoding Proteins Involved in T Biosynthesis
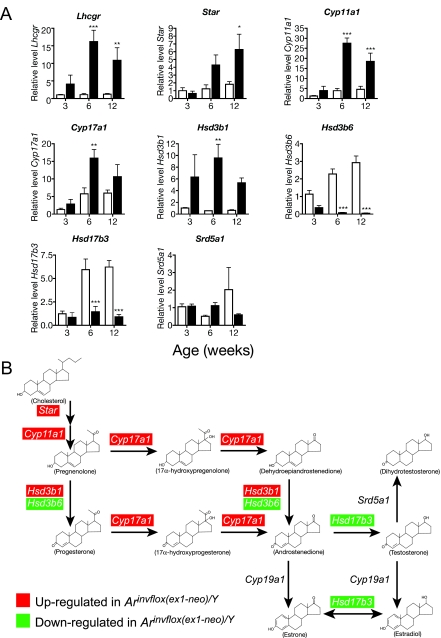
A, Expression of genes involved in steroidogenesis in wild-type (white bars) and Arinvflox(ex1-neo)/Y (black bars) testis. Levels of mRNA were normalized between samples based on the levels of Actb in the case of Northern blots and Rps2 for transcripts analyzed by real-time qRT-PCR. *, P < 0.05; **, P < 0.01; ***, P < 0.001 from a two-way ANOVA followed by Bonferroni post test. B, The pathway of T biosynthesis from cholesterol. Red highlights indicate that the transcript was present at significantly higher levels in Arinvflox(ex1-neo)/Y testes at one or more time points during development, whereas green indicates the transcript was present at lower than wild-type levels at one or more time points.
As suggested by the high levels of T, many transcripts encoding steroidogenic enzymes were highly up-regulated (Fig. 2A). Notably, the transcript encoding the protein that catalyzes the rate-limiting step of steroidogenesis, Star, was up-regulated nearly 5-fold over wild-type levels. However, not all transcripts encoding steroidogenic proteins were up-regulated in Arinvflox(ex1-neo)/Y. Interestingly, Δ5-3β-hydroxysteroid dehydrogenase-6 (Hsd3b6) and hydroxysteroid (17β) dehydrogenase-3 (Hsd17b3) were both significantly down-regulated despite the high levels of LH and Lhcgr. Collectively, the expression of these transcripts shows that the regulation of steroidogenic gene expression is similar in wild-type and Arinvflox(ex1-neo)/Y at 3 wk of age but diverges during puberty into adulthood. By this time, every key transcript in the T biosynthetic pathway is affected (summarized in Fig. 2B).
Up-Regulation of Genes Involved in Cholesterol Uptake and de Novo Biosynthesis
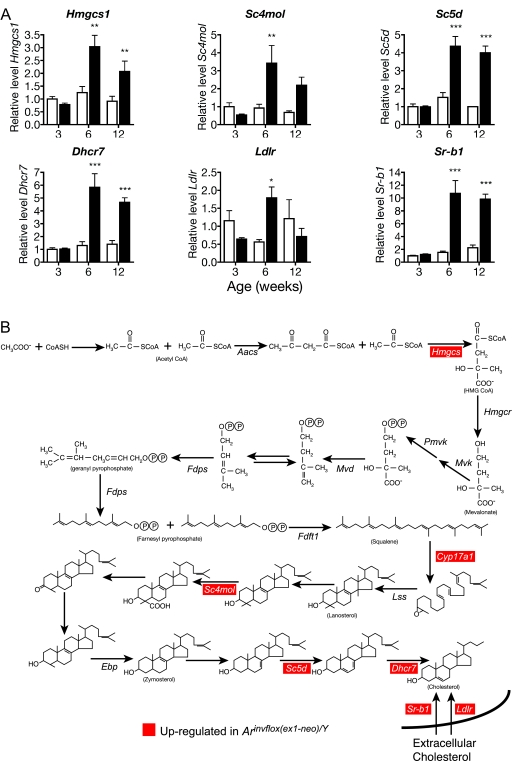
Cholesterol is the essential precursor for all steroid hormones, suggesting that steroidogenic cells have specialized requirements for maintaining proper cholesterol homeostasis. To determine how Leydig cells meet their cholesterol requirements for steroid synthesis, we measured the expression levels of genes involved in cholesterol synthesis and receptor-mediated uptake in wild-type testis and compared them with the levels in the Arinvflox(ex1-neo)/Y testis, which synthesizes approximately 20-fold higher levels of T (Fig. 1B). All genes involved in de novo cholesterol synthesis [Hmgcs1, sterol C4 methyl-oxidase-like (Sc4 mol), sterol C5 desaturase (Sc5d), and 7-dehydrocholesterol reductase (Dhcr7)] that we analyzed showed a statistically significant increase in expression in Arinvflox(ex1-neo)/Y testis compared with wild-type testis by 6 wk of age (Fig. 3A). The two receptors involved in uptake of extracellular cholesterol, the low-density lipoprotein (LDL) receptor (LDLR; Ldlr) and scavenger receptor type B1 (SR-B1; Sr-b1), both showed a significant elevation by 6 wk of age in Arinvflox(ex1-neo)/Y testis. However, Ldlr expression decreased into adulthood, whereas Sr-b1 mRNA remained elevated compared with wild-type. These data suggest that Leydig cells meet their cholesterol needs through both de novo synthesis and uptake of extracellular cholesterol (summarized in Fig. 3B).
Figure 3.
Expression of Genes Involved in Cholesterol Synthesis and Uptake
Expression of genes involved in cholesterol synthesis and uptake in wild-type (white bars) and Arinvflox(ex1-neo)/Y (black bars) testis. A, Each bar represents the relative mean expression level for the given transcript from six individual animals. Levels of mRNA were normalized between samples based on the levels of Actb in the case of Northern blots and Rps2 for transcripts analyzed by real-time qRT-PCR. All mRNA levels are normalized to the density of Leydig cells. *, P < 0.05; **, P < 0.01; ***, P < 0.001 from a two-way ANOVA followed by Bonferroni post test. B, The pathway of synthesis of cholesterol from acetate.
Disruption of Leydig-Cell Lipid Homeostasis in Ar Hypomorphs
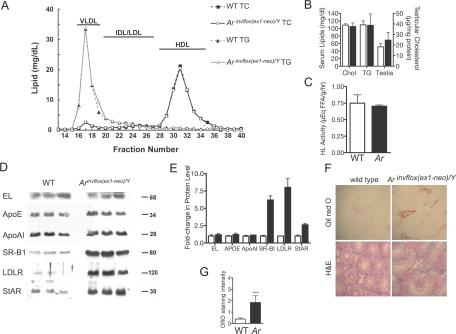
Up-regulation of pathways involved in cholesterol synthesis and uptake in the testes of Arinvflox(ex1-neo)/Y mice is consistent with an enhanced requirement for cholesterol for steroidogenesis. To determine whether the effects of Ar hypomorphism on cholesterol requirements were systemic or restricted to the gonad, we profiled the composition of plasma lipids. Plasma cholesterol and triglyceride levels were similar between genotypes, as were plasma lipid profiles (Fig. 4, A and B). In addition, testicular cholesterol concentrations were similar between the genotypes. These data suggest that alterations in cholesterol requirements in the Arinvflox(ex1-neo)/Y testis is not due to global alternations in lipid homeostasis.
Figure 4.
Global and Testicular Lipid Phenotype of Adult Arinvflox(ex1-neo)/Y Mice
A, Fast-performance liquid chromatography profiles of plasma lipoprotein cholesterol and triglycerides from wild-type and Arinvflox(ex1-neo)/Y mice. Plasma (100 μl) from three mice from each genotype was fractionated by Superose 6 chromatography. Fractions were assayed for cholesterol and triglyceride concentrations using standard colorimetric assays. Horizontal bars indicate lipoprotein distributions. B, Total serum cholesterol and triglycerides were not different between genotypes (n = 3). Testicular cholesterol was slightly elevated in Ar hypomorphs compared with wild-type, the difference was not significantly different. C, Comparison of hepatic lipase activity between genotypes. D, Western blot analysis for EL, ApoE, ApoAI, SR-B1, LDLR, and StAR comparing levels between wild-type and Arinvflox(ex1-neo)/Y total testis extracts. E, Laser-densitometry quantification of protein levels in Western blots shown in D. F, ORO staining in the Leydig cells of Arinvflox(ex1-neo)/Y mice. G, Blindly scored randomly selected sections from Arinvflox(ex1-neo)/Y and wild-type testes. Sections were scored as follows: 0, no staining in Leydig cells; 1, weak/diffuse staining; 2, intermediate staining in some Leydig cells; 3, strong staining in some Leydig cells; 4, strong staining in all Leydig cells. Error bars in all panels reflect sem. ***, Significant difference at P < 0.001, Student’s t test.
Enhanced testicular requirements for cholesterol may also be reflected in an increase in enzymes required for the uptake of high-density lipoprotein (HDL) cholesterol via the selective uptake pathway. Hepatic lipase (HL) and endothelial lipase (EL) both facilitate the flux of cholesterols from the HDL particle. Despite the enhanced cholesterol requirements of the testes of Ar hypomorphs, there was no significant difference in either testicular HL activity (Fig. 4C) or EL protein levels (Fig. 4D, quantified in 4E). Testicular levels of apolipoproteins that mediate uptake of extracellular sources of cholesterol were also unaltered between genotypes (Fig. 4D, quantified in 4E). However, as suggested by its mRNA levels, we observed dramatic up-regulation of SR-B1. LDLR protein was also highly up-regulated despite the absence of an increase in mRNA levels. As suggested by our profiling of genes involved in steroidogenesis, there was an increase in the levels of StAR, the key protein that facilitates the use of cholesterol for steroid synthesis. These observations show that Leydig cell intrinsic components involved in uptake and use of cholesterol are clearly up-regulated.
Because of the low abundance of the steroidogenic Leydig cells within the testis (roughly 3% of cells/testis), alterations in Leydig cell lipid content would not necessarily be observed by measuring total testicular lipid content. Therefore, we used Oil red O (ORO) staining on frozen sections of testis from wild-type and Arinvflox(ex1-neo)/Y mice to assess the relative distribution and abundance of cholesterol. Wild-type animals demonstrated weak, but detectable, ORO staining in Leydig cells, identified definitively in serial sections stained with hematoxylin and eosin (Fig. 4F). Strikingly, there was a dramatic increase in the lipid content of Leydig cells in Arinvflox(ex1-neo)/Y mice. Analysis of blindly scored ORO-stained sections showed a statistically significant increase in the lipid content of Leydig cells of Ar hypomorphs (Fig. 4G). The high lipid content in Arinvflox(ex1-neo)/Y Leydig cells suggests that despite the additional requirements for cholesterol for steroidogenesis, the increase in de novo synthesis and uptake results in abnormal accumulation of lipids.
Regulation of Steroid and Cholesterol Biosynthesis and Uptake by cAMP and DHT
Disrupting the regulation of the HPG axis by reducing Ar message has an impact on both AR and LH signaling. Therefore, changes in gene expression observed in Arinvflox(ex1-neo)/Y mice could be the result of reduced AR signaling, increased LH signaling, or both. To address these possibilities, we used the easily manipulated MA-10 mouse Leydig tumor line (24). MA-10 cells express most genes involved in steroidogenesis but lack Hsd17b3 expression, preventing the synthesis of T. However, in response to LH or its second messenger cAMP, MA-10 cells synthesize large amounts of P and its metabolites.
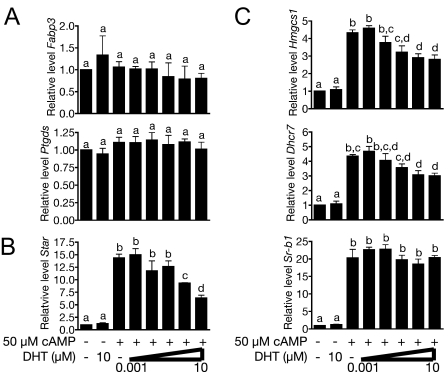
To define which changes in gene expression are due to perturbation of the testicular hormonal milieu, we treated the cells with DHT, 8-Br-cAMP, or both. Expression of markers of Leydig cell maturation is unaffected by treatment with 8-Br-cAMP or DHT (Ptgds and Fabp3, Fig. 5A). This is consistent with their designation as markers of Leydig cell development and not merely targets of AR or LH signaling. In contrast, genes involved in steroid and cholesterol synthesis and uptake were up-regulated in response to treatment with 8-Br-cAMP (Fig. 5, B and C). Interestingly, none of these genes were up-regulated in response to DHT, but in the case of Star, Dhcr7, and Hmgcs1, DHT treatment attenuated cAMP-stimulated gene expression. These data suggest that T signaling modulates cAMP-stimulated cholesterol and steroid synthesis by regulating the expression of target genes.
Figure 5.
Regulation of Steroid Synthesis from de Novo Synthesized Cholesterol by cAMP and DHT in the MA-10 Leydig Tumor Line
A, Genes previously identified as markers of Leydig cell maturation; B, genes required for steroid biosynthesis; C, genes required for cholesterol synthesis and uptake. Letters indicate P < 0.05 for a one-way ANOVA followed by Newman-Keuls post test.
cAMP-Stimulated Expression of Genes Involved in Cholesterol Synthesis and Uptake Is Independent of the Nuclear Recruitment of SREBP2
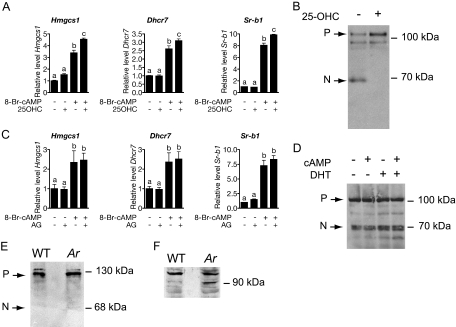
Cholesterol synthesis and uptake is regulated in most mammalian cells by the SREBP2 pathway. To address this possible mechanism of gene regulation, we treated MA-10 cells with cAMP in the presence or absence of 25-OHC, a potent inhibitor of SREBP pathway activation. The addition of 10 μm 25-OHC was unable to suppress 8-Br-cAMP-stimulated expression of Hmgcs1, Dhcr7, or Sr-b1, all known targets of SREBP2 (Fig. 6A). Despite the addition of 25-OHC at a concentration sufficient to block processing of SREBP2 (Fig. 6B), we actually observed a modest increase in the levels of cAMP-stimulated Hmgcs1and Sr-b1 mRNA. These results suggest that SREBP2 processing is not required for the increased expression of genes involved in cholesterol synthesis and uptake.
Figure 6.
cAMP-Stimulated Expression of Genes Involved in Cholesterol Synthesis and Uptake Is Not Mediated by Accumulation of Processed SREBP2
A, Relative mRNA levels in MA-10 cells treated with combinations of 50 μm 8-Br-cAMP and 10 μm 25-OHC. B, These concentrations of 25-OHC are capable of blocking cholesterol-starved HEK-293 cells from activating SREBP2. The 125-kDa precursor form (P) of SREBP2 and 70-kDa nuclear form (N) are visible in whole-cell extracts of lipid-starved HEK-293 cells. These data are representative of duplicate experiments. C, Relative mRNA levels in MA-10 cells treated with combinations of 50 μm 8-Br-cAMP and 5 μg/ml AG. Each bar represents the mean expression level for three independent experiments. Letters indicate P < 0.05 for a one-way ANOVA followed by Newman-Keuls post test. Levels of mRNA were normalized between samples based on the levels of Actb transcript. D, Treatment of MA-10 cells with cAMP or DHT, alone or in combination, has no effect on levels of the 68-kDA nuclear form of SREBP2. These data are representative of experiments performed in triplicate. E, SREBP2 is not processed in Leydig cell-enriched protein extracts from wild-type or Arinvflox(ex1-neo)/Y testes. F, HMGCR levels are elevated in Leydig cell-enriched protein extracts from wild-type or Arinvflox(ex1-neo)/Y testes. Cells used to generate protein extracts in E and F contained more than 60% Leydig cells as determined by 3βHSD cytochemistry.
Because depletion of intracellular cholesterol pools for steroid synthesis could trigger activation of SREBP2, we used a second, independent method to address the possible role of SREBP2 activation of gene expression in response to cAMP stimulation. We tested whether inhibiting steroid synthesis with aminoglutethimide (AG) in cAMP-stimulated MA-10 cells would prevent the up-regulation of cholesterol biosynthetic genes. As observed under treatment of MA-10 cells with 25-OHC, AG did not repress the expression of Hmgcs1, Dhcr7, or Sr-b1 (Fig. 6C). Again, this suggests that depletion of intracellular sterol stores is not required for cAMP-stimulated expression of genes involved in cholesterol uptake and synthesis. Finally, we assessed the levels of processed SREBP2 in untreated MA-10 cells as well as cells treated with DHT or 8-Br-cAMP alone or in combination. MA-10 cells untreated or treated with cAMP, DHT, or both show a constant amount of processed SREBP2 (Fig. 6D). These data indicate that cAMP-stimulated expression of cholesterol biosynthesis and uptake gene expression is unlikely due to the accumulation of active SREBP2.
The absence of a requirement for active SREBP2 accumulation for cAMP-stimulated cholesterol biosynthetic gene expression was surprising. To determine whether these observations made using the MA-10 Leydig tumor model are consistent with in vivo regulation of cholesterol biosynthetic genes, we collected protein extracts from a Leydig cell-enriched population isolated from wild-type and Arinvflox(ex1-neo)/Y mice. The Arinvflox(ex1-neo)/Y mice have elevated levels of LH (Fig. 1A) and increased levels of cAMP in Leydig-enriched cell populations when compared with wild-type-derived Leydig cell populations (data not shown). In the Leydig-enriched protein extracts, we observed no appreciable amount of processed SREBP2 in either wild-type or Arinvflox(ex1-neo)/Y (Fig. 6E), yet Arinvflox(ex1-neo)/Y Leydig cells still retain elevated expression of HMGCR, a well-defined target of SREBP2 (Fig. 6F). These data reaffirm the conclusion that SREBP2 activation and nuclear accumulation is not required for cAMP-mediated induction of genes involved in cholesterol biosynthesis and uptake.
DISCUSSION
Regulation of testicular steroid and cholesterol homeostasis is critical to the fertility of mammals. In this investigation, we showed that disruption of T-mediated feedback regulation within the HPG axis dramatically disrupts the normal steroid and cholesterol homeostatic mechanisms within the testis. We showed that 1) global reduction of AR signaling results in increased serum LH and T, 2) elevated LH levels increases the expression of genes involved in steroid and cholesterol synthesis and receptor-mediated cholesterol uptake, 3) T modulates the expression of these genes, 4) T’s effect on gene expression is associated with suppression of steroids synthesized from de novo sources of cholesterol, and 5) the regulation of genes involved in cholesterol biosynthesis and uptake is not mediated by the canonical SREBP2-mediated sterol-sensing mechanism in Leydig cells.
AR and Regulation of Pubertal Acquisition of Steroidogenic Potential
It has been long recognized that mutations in Ar have effects on the development and physiology of the Leydig cell, including steroidogenesis (reviewed in Ref. 25). Interestingly, we found decreased levels of two steroidogenic enzymes, Hsd17b3 and Hsd3b6. Previous studies have shown that the expression of Hsd3b6 is associated with the pubertal maturation of adult-type Leydig cells (26). Characterization of Leydig cell gene expression in Artfm/Y has shown that AR is required for the onset of Hsd3b6 expression during puberty (27). The absence of Hsd3b6 expression and Insl3 and Ptdgs, transcripts associated with mature adult Leydig cells, led O’Shaughnessy et al. (27) to conclude that AR is required for normal adult Leydig cell differentiation. We have observed a similar phenotype in Arinvflox(ex1-neo)/Y mice, despite the incomplete inactivation of Ar (Fig. 2 and unpublished data). Strikingly, unlike Arinvflox(ex1-neo)/Y mice, Artfm/Y mutants produce low levels of serum T in response to elevated levels of LH (28). Our results show that LH-stimulated T production is separable from the acquisition of markers of Leydig cell maturation.
Levels of Hsd17b3 message are similarly down-regulated in Artfm/Y and Arinvflox(ex1-neo)/Y testes (27), suggesting that high levels of this mRNA are not critical for the synthesis of T. Despite the absence of Hsd3b6 message, there is no significant difference in the levels of 3β-hydroxysteroid dehydrogenase (3βHSD) activity between wild-type and Artfm/Y testis homogenates (29). Although there is a significant reduction of Hsd3b6 mRNA in Arinvflox(ex1-neo)/Y mice, there is a reciprocal up-regulation of another Hsd3b isoform, Hsd3b1. It is unclear whether up-regulation of Hsd3b1 is critical to the high levels of T synthesized in Arinvflox(ex1-neo)/Y as compared with Artfm/Y. A null mutation of Ar has no effect on the level of Hsd3b1 mRNA but results in a severe reduction in Hsd3b6 levels (27). Enzymatically, the key difference between Arinvflox(ex1-neo)/Y and Artfm/Y is probably the levels of CYP17A1. Levels of CYP17A1 activity in testis slices from Artfm/Y mice are reduced greater than 40-fold compared with wild-type, which is accompanied by a significant reduction in Cyp17a1 mRNA (28,29). In contrast, Cyp17a1 mRNA levels are elevated in the testes of Arinvflox(ex1-neo)/Y mice. Another important factor that promotes steroidogenic activity and T production in Arinvflox(ex1-neo)/Y mice is the normal positioning of the testis. Cryptorchidism alone has a strong effect on steroidogenic activities and may contribute to the differences observed between Artfm/Y and Arinvflox(ex1-neo)/Y mutants (28).
Regulation of Cholesterol Synthesis and Uptake in Leydig Cells
Negative feedback regulation of steroidogenic gene expression is well documented for a number of genes (11,30,31,32). However, AR-mediated feedback on cholesterol biosynthetic gene expression has not previously been demonstrated. Our results suggest that T modulates the expression of a subset of genes that are required for de novo cholesterol synthesis, adding an additional layer of regulation on T synthesis that had not previously been appreciated.
Earlier in vitro studies have demonstrated stimulation of genes involved in cholesterol synthesis and uptake in Leydig cells by treatment with hCG or cAMP. In both cultured rat Leydig cells and MA-10 Leydig tumor cells, a large dose of hCG causes a sharp increase, followed by a rapid decline in HMG-CoA reductase activity (16,17). This decline in activity is associated with the depletion of LHCGR at the cell surface, the so-called desensitization effect. Soccio and colleagues (33) suggest that the up-regulation of Hmgcr is due to the depletion of intracellular sterols by steroidogenesis, thus triggering SREBP-mediated up-regulation of de novo cholesterol biosynthesis. Our data offer a different mechanistic explanation for increase in cholesterol biosynthetic gene expression. Treatment of MA-10 cells with 25-OHC to inhibit SREBP activation, or AG to inhibit steroidogenesis, had no effect on cAMP-stimulated expression of Hmgcs1 or Dhcr7 expression. Perhaps more convincingly, cAMP had no effect on the accumulation of processed SREBP2, indicating that increased nuclear SREBP2 is not the mechanism by which cAMP stimulates increased expression of genes involved in de novo cholesterol biosynthesis. The accumulation of neutral lipids in the Leydig cells of Arinvflox(ex1-neo)/Y mice also argues against the SREBP-mediated sterol-sensing pathway activation of the de novo cholesterol synthesis and uptake pathways. Instead, our data strongly support direct activation of these pathways via LH signaling.
Although increased nuclear SREBP2 is not sufficient to explain the up-regulation of cholesterol biosynthetic gene expression, it remains a possibility that SREBP2 is necessary for the expression of these genes. An additional cAMP-dependent cofactor(s) could be required for the maximal cAMP-stimulated expression of SREBP target genes. One candidate for this role is the cAMP response element-binding protein (CREB), which has been shown to be an effective cofactor for SREBP-dependent activation of the Hmgcs1 promoter (34). Alternatively, expression of cholesterol biosynthetic genes could be independent of the SREBP pathway. Cells overexpressing the steroidogenic cell transcription factor SF1 express Hmgcs1, even in the face of treatment with 25-OHC (18). This suggests that in steroidogenic tissues, expression of cholesterol biosynthetic genes may be driven in an SF1-dependent manner, independent of SREBP activation.
Uptake of extracellular sources of cholesterol is mediated by LDLR and SR-B1, both of which are transcriptional targets of SREBP2. Rats treated for 4 d with hCG show a massive up-regulation of SR-B1 expression accompanied by a more modest up-regulation of LDLR in Leydig cells (35). These results are consistent with the very high levels of Sr-b1 and modest change in Ldlr mRNA in Arinvflox(ex1-neo)/Y testes. Despite the modest increase in Ldlr message in the current study, there was a large increase in testicular LDLR protein. This may reflect translational control of the Ldlr mRNA. As we observed for de novo synthesis genes in MA-10 cells, Sr-b1 expression was responsive to cAMP stimulation, and its expression was unaffected by 25-OHC or AG, again suggesting that the up-regulation of these genes does not require additional accumulation of active SREBP2.
Recently, a number of studies have demonstrated the importance of another group of nuclear hormone receptors, the liver X receptors (LXRs), in the maintenance of cholesterol homeostasis in the adrenals and the testis. The null mutation of both Lxrα and Lxrβ leads to profound accumulation of cholesterol in the adrenals and extremely high levels of serum corticosteroids (36). The overt phenotypic similarity between the Lxrα/β−/− adrenals and Leydig cell phenotype of Arinvflox(ex1-neo)/Y share some similar molecular details as well: increased expression of Star and Cyp11a1, which probably lead to increased steroidogenesis. However, there are no changes in the expression of genes involved in either de novo cholesterol synthesis or uptake, but rather the cholesterol accumulation in these mutants is due to dysfunctional cholesterol efflux in the Lxrα/β−/− adrenals. Like the adrenals, the testes of Lxrα/β−/− mice accumulate large amounts of cholesterol (37), as evidenced by increased cholesterol in total testis extracts. We did not observe an increase in cholesterol in total testis extracts but instead observed an increase in neutral lipids in Leydig cells by ORO staining. Surprisingly, the high levels of cholesterol observed in the Lxrα/β−/− testis is not localized in Leydig cells but rather in the Sertoli cells, suggesting that LXRs regulate different aspects of cholesterol homeostasis than the LH-T-AR loop described in this study.
Taken together, our data suggest that LH directly stimulates the transcription of genes involved in steroidogenesis and genes required for cholesterol synthesis and uptake. Also, our data indicate that a subset of cholesterol biosynthetic genes can be down-regulated by T, providing an additional point of feedback regulation in Leydig cells. We have shown that targets of LH signaling are affected in an HPG axis disruption model in vivo. We also have shown that elevated expression levels of cholesterol biosynthesis and uptake genes are associated with high LH levels in vivo and cAMP stimulus in vitro. By disrupting these points of regulation in the Leydig cell, we have significantly perturbed steroid and cholesterol homeostasis in the testis as observed by high levels of serum T and accumulation of lipids in Leydig cells. Lastly, the up-regulation of genes involved in cholesterol homeostasis and steroidogenesis appears to happen in the absence of activation of SREBP2, a major regulator of cholesterol biosynthesis in nongonadal tissues.
MATERIALS AND METHODS
Animals
All animals were housed in a specific pathogen-free environment and cared for under Institutional Animal Care and Use Committee guidelines. Generation of the hypomorphic Ar allele Arinvflox(ex1-neo) has been previously described (19). Male mice carrying the hypomorphic allele and their wild-type littermates were killed by CO2 asphyxiation at the ages of 3, 6, and 12 wk. Animals were genotyped using PCR as previously described (19).
Phenotypic Analysis
Aortic blood was collected from all animals using a heparin-coated 23-gauge needle. Serum T and LH levels were measured by RIA at the University of Virginia Ligand Core facility. RNA and protein from one testis were prepared as previously described (19). The second whole testis was immersion fixed overnight in Bouin’s fixative for histological analysis.
Leydig cell number was determined using the method described by Mori and Christiansen (23). In brief, Bouin’s fixed testes were cut into four sections and embedded in paraffin blocks. Five-micrometer sections were collected in a systematic method from each block to represent different regions of the testis. Leydig cells were unambiguously identified by staining with a rabbit antirat cytochrome P450 side chain cleavage antibody (1:250 of Ab1244; Chemicon, Temecula, CA) and then counterstained with Mayer’s hematoxylin (Sigma Chemical Co., St. Louis, MO). Eight fields were randomly selected from four sections from each testis, and Leydig cell nuclei were counted to estimate nuclear density. Because the density of the testis is approximately 1 mg/ml, testis mass was used as an estimate of testis volume.
Analysis of Gene Expression
For Northern analysis, 15 μg total RNA was run on a 1.5% agarose formaldehyde gel. The RNA was then transferred overnight in 20× SSC to Genescreen Plus hybridization membrane (PerkinElmer, Norwalk, CT) and UV cross-linked. Blots were exposed on a Phosphor Storage Screen (GE Healthcare, Piscataway, NJ) and imaged using Storm Scanner 580 (GE Healthcare). Band intensities were measured using ImageQuant (GE Healthcare).
qRT-PCR was performed using the Brilliant SYBR-Green kit (Stratagene, La Jolla, CA). All cDNAs were synthesized from 1 μg of total RNA using random hexamers and Superscript III (Invitrogen, Carlsbad, CA) in a 20-μl reaction volume. Resulting cDNA was diluted 1:10 in water and subjected to qRT-PCR using a Stratagene MX-3000P thermocycler. Primers for most transcripts are listed in Ref. 38 with the exception of ribosomal protein S2 (Rps2) primer, which is listed in Ref. 39. Novel primers were developed for Ldlr (5′-GAG GAG CAG CCA CAT GGT AT-3′ and 5′-GCT CGT CCT CTG TGG TCT TC-3′). Relative expression levels were calculated as described elsewhere using levels of Rps2 to normalize RNA input (39). Normalized gene expression values were then multiplied by the relative density of Leydig cells in the testis to account for differences in Leydig cell number due to age and genotype.
Lipid and Lipoprotein Characterization
For lipid and lipoprotein characterization, blood was collected from three 12-wk-old mice of each genotype. Individual plasma cholesterol and triglyceride concentrations were determined. In addition, pooled plasma lipoproteins were fractionated using fast protein liquid chromatography as described previously (40). Cholesterol and triglyceride levels in whole plasma and in eluted fractions were determined with standard enzymatic assays for cholesterol (Abbot Spectrum, Abbott Park, IL) and triglyceride (GPO-PAP kit; Boehringer Mannheim, Indianapolis, IN). Lipoprotein cholesterol and triglyceride concentrations were obtained by adding the concentrations of fractions 16–19 (very-low-density lipoprotein), 20–22 (intermediate-density lipoprotein), 23–27 (LDL), and 28–34 (HDL).
Testes were harvested and tissue cholesterol was extracted according to Folch et al. (41). Cholesterol concentrations were determined in quadruplicate using standard colorimetric assays, and protein was measured in duplicate. Cholesterol recovery was determined as previously described with minor modifications (42). One half-testis from three individual mice of each genotype was homogenized in 1 ml chloroform/methanol (2:1, vol/vol) containing trace amounts of [14C]cholesteryl-oleate (as an internal standard for recovery) and was processed as previously described for adrenal cholesterol measurements (43).
ORO Staining
To assess Leydig cell cholesterol content, consecutive frozen sections (10 μm) from testis were stained with either ORO to detect neutral lipids or with hematoxylin and eosin to visualize tissue morphology. Testes from seven wild-type and six Arinvflox(ex1-neo)/Y mice were examined. For each testis, three to five sections were stained with ORO and three to five sections were stained with hematoxylin and eosin.
Cell Culture
MA-10 cells were obtained as a gift from Jose Teixeira (Massachusetts General Hospital, Boston, MA) with the permission of Mario Ascoli (24) and cultured as previously described (32). Cells were pretreated with varying concentrations of DHT (Sigma) for 1 h before exposure to 50 μm 8-Br-cAMP (Sigma). Cells were then cultured for 16 h before harvesting total RNA using 1 ml Trizol (Invitrogen) in accordance with the manufacturer’s protocol.
Human embryonic kidney (HEK-293) cells were cultured in DMEM supplemented with l-glutamine, penicillin, streptomycin, and 10% horse serum. For visualizing SREBP2 by Western blot, cells were cultured for 9 h in medium containing 10% lipoprotein-deficient horse serum and 50 μm mevastatin (Sigma) in the presence or absence of 10 μm 25-OHC (Sigma). Lipoprotein-deficient serum was prepared as previously described (17).
Immunoblotting
Aliquots of testicular protein extract were analyzed by Western blotting as described (43). The concentration of testis protein extracts was measured using the Lowry method. Equal loading between lanes was verified by Ponceau S staining of the Western blot membrane. After transfer, each blot was incubated with rabbit polyclonal antimouse EL antibody (1:500, C18PEP4 no. 3, a gift from Thomas Quertermous, Stanford University), polyclonal rabbit anti-SR-B1 antiserum (1:1000; Novus Biologicals, Littleton, CO), rabbit polyclonal antihuman LDLR antibody (1:10,000, a gift from Sandra Erickson and Janet Boyles, University of California, San Francisco, San Francisco, CA), or rabbit polyclonal antiserum against StAR (1:1000, a gift from Dr. Douglas Stocco, Texas Tech University, Lubbock, TX). Plasma was collected from four to five mice of each genotype. The membranes were incubated with a goat antimouse apolipoprotein E (ApoE) antibody that also reacts with mouse ApoA1 (a gift from Dr. Karl H. Weisgraber, Gladstone Institute of Cardiovascular Disease). Western blots were visualized with enhance chemiluminescence substrate (GE Healthcare).
Protein from MA-10 and HEK-293 cells was collected by scraping cells from 10-cm plates in 500 μl protein lysis buffer [10 mm Tris-HCl (pH 7.6), 100 mm NaCl, 1% SDS]. The lysate was then passed through a 23-gauge needle 11 times and vortexed for 20 min at room temperature. Protein from the cell lysates was quantitated using the BCA assay (Pierce, Rockford, IL). Primary antibodies used were mouse antihuman SREBP2 1C6 (1:25; Santa Cruz Biotechnology, Santa Cruz, CA) and rabbit antimouse SREBP2 (7.5 μg/ml; a gift of Dr. Jay Horton).
Leydig Cell Purification
Purification of mouse Leydig cells was carried out according to the method in Ref. 44 with some modification. Eight testes per genotype were removed, and testicular cells were dispersed by treating the decapsulated testis with collagenase (0.25 mg/ml; Sigma) in M199 medium (Invitrogen) at room temperate for 20 min with gentle shaking. After incubation, the dispersed tissues were diluted with M199, and the solution was filtered. Interstitial cells were precipitated by centrifugation of the filtrate and washed once with M199 and twice with PBS. Enrichment for Leydig cells was estimated by 3βHSD cytochemistry.
Statistical Analysis
All statistical analyses were performed using Prism (version 4.0c; GraphPad Software, San Diego, CA). For comparison between wild-type and mutant animals at each time point, two-way ANOVA was conducted and significance of differences assessed by Bonferroni post test. For comparisons of gene expression between MA-10 cells treated under various regimes, one-way ANOVA followed by Newman-Keuls post test was used.
Supplementary Material
Acknowledgments
We thank Kelly Tysseling for her excellent technical assistance, Glenda Froehlick for assistance in preparing histological sections, Drs. Mario Ascoli and Jose Teixiera for MA-10 cells, and Dr. Jay Horton for SREBP2 antibody. We thank the Ligand Core facility at University of Virginia for their help in measuring serum hormone levels as a part of the Specialized Cooperative Centers Program in Reproductive Research (SCCPRR).
Footnotes
This work was supported by a SCCPRR Grant U54 HD12629 (to R.E.B.) and by R01HL-69775 (to H.L.D).
Disclosure Statement: The authors have nothing to disclose.
First Published Online November 21, 2007
Abbreviations: AG, Aminoglutethimide; ApoE, apolipoprotein E; AR, androgen receptor; DHT, dihydrotestosterone; EL, endothelial lipase; hCG, human chorionic gonadotropin; HDL, high-density lipoprotein; HL, hepatic lipase; HMG-CoA, 3-hydroxy-3-methylglutaryl coenzyme A; HMGCR, HMG-CoA reductase; HPG, hypothalamic-pituitary-gonadal; 3βHSD, 3β-hydroxysteroid dehydrogenase; LDL, low-density lipoprotein; LDLR, LDL receptor; LHCGR, LH/chorionic gonadotropin receptor; LXR, liver X receptor; 25-OHC, 25-hydroxycholesterol; ORO, Oil red O; qRT-PCR, quantitative RT-PCR; SR-B1, scavenger receptor type B1; SREBP, sterol response element-binding protein; StAR, steroidogenic acute regulatory protein; T, testosterone.
References
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schütz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM 1995 The nuclear receptor superfamily: the second decade. Cell 83:835–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CS, Kokontis J, Liao ST 1988 Molecular cloning of human and rat complementary DNA encoding androgen receptors. Science 240:324–326 [DOI] [PubMed] [Google Scholar]
- Lubahn DB, Joseph DR, Sullivan PM, Willard HF, French FS, Wilson EM 1988 Cloning of human androgen receptor complementary DNA and localization to the X chromosome. Science 240:327–330 [DOI] [PubMed] [Google Scholar]
- Dufau ML 1997 The luteinizing hormone receptor. In: Payne AH, Hardy MP, Russell LD, eds. The Leydig cell. Vienna, IL: Cache River Press; 333–350 [Google Scholar]
- Jorgensen JS, Nilson JH 2001 AR suppresses transcription of the LHβ subunit by interacting with steroidogenic factor-1. Mol Endocrinol 15:1505–1516 [DOI] [PubMed] [Google Scholar]
- Tilbrook AJ, Clarke IJ 2001 Negative feedback regulation of the secretion and actions of gonadotropin-releasing hormone in males. Biol Reprod 64:735–742 [DOI] [PubMed] [Google Scholar]
- Clark BJ, Wells J, King SR, Stocco DM 1994 The purification, cloning, and expression of a novel luteinizing hormone-induced mitochondrial protein in MA-10 mouse Leydig tumor cells. Characterization of the steroidogenic acute regulatory protein (StAR). J Biol Chem 269:28314–28322 [PubMed] [Google Scholar]
- Clark AM, Chuzel F, Sanchez P, Saez JM 1996 Regulation by gonadotropins of the messenger ribonucleic acid for P450 side-chain cleavage, P450(17) α-hydroxylase/C17,20-lyase, and 3β-hydroxysteroid dehydrogenase in cultured pig Leydig cells. Biol Reprod 55:347–354 [DOI] [PubMed] [Google Scholar]
- Payne AH, Hales DB 2004 Overview of steroidogenic enzymes in the pathway from cholesterol to active steroid hormones. Endocr Rev 25:947–970 [DOI] [PubMed] [Google Scholar]
- Hales DB, Sha LL, Payne AH 1987 Testosterone inhibits cAMP-induced de novo synthesis of Leydig cell cytochrome P-450(17α) by an androgen receptor-mediated mechanism. J Biol Chem 262:11200–11206 [PubMed] [Google Scholar]
- Burgos-Trinidad M, Youngblood GL, Maroto MR, Scheller A, Robins DM, Payne AH 1997 Repression of cAMP-induced expression of the mouse P450 17α-hydroxylase/C17–20 lyase gene (Cyp17) by androgens. Mol Endocrinol 11:87–96 [DOI] [PubMed] [Google Scholar]
- Horton JD, Goldstein JL, Brown MS 2002 SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest 109:1125–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL 1999 A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc Natl Acad Sci USA 96:11041–11048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Shimomura I, Brown MS, Hammer RE, Goldstein JL, Shimano H 1998 Activation of cholesterol synthesis in preference to fatty acid synthesis in liver and adipose tissue of transgenic mice overproducing sterol regulatory element-binding protein-2. J Clin Invest 101:2331–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fon Tacer K, Kalanj-Bognar S, Waterman MR, Rozman D 2003 Lanosterol metabolism and sterol regulatory element binding protein (SREBP) expression in male germ cell maturation. J Steroid Biochem Mol Biol 85:429–438 [DOI] [PubMed] [Google Scholar]
- Charreau EH, Calvo JC, Nozu K, Pignataro O, Catt KJ, Dufau ML 1981 Hormonal modulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase activity in gonadotropin-stimulated and -desensitized testicular Leydig cells. J Biol Chem 256:12719–12724 [PubMed] [Google Scholar]
- Freeman DA, Ascoli M 1982 Studies on the source of cholesterol used for steroid biosynthesis in cultured Leydig tumor cells. J Biol Chem 257:14231–14238 [PubMed] [Google Scholar]
- Mascaro C, Nadal A, Hegardt FG, Marrero PF, Haro D 2000 Contribution of steroidogenic factor 1 to the regulation of cholesterol synthesis. Biochem J 350(Pt 3):785–790 [PMC free article] [PubMed] [Google Scholar]
- Holdcraft RW, Braun RE 2004 Androgen receptor function is required in Sertoli cells for the terminal differentiation of haploid spermatids. Development 131:459–467 [DOI] [PubMed] [Google Scholar]
- Teerds KJ, Closset J, Rommerts FF, de Rooij DG, Stocco DM, Colenbrander B, Wensing CJ, Hennen G 1989 Effects of pure FSH and LH preparations on the number and function of Leydig cells in immature hypophysectomized rats. J Endocrinol 120:97–106 [DOI] [PubMed] [Google Scholar]
- Hardy MP, Kelce WR, Klinefelter GR, Ewing LL 1990 Differentiation of Leydig cell precursors in vitro: a role for androgen. Endocrinology 127:488–490 [DOI] [PubMed] [Google Scholar]
- Baker PJ, O’Shaughnessy PJ 2001 Role of gonadotrophins in regulating numbers of Leydig and Sertoli cells during fetal and postnatal development in mice. Reproduction 122:227–234 [DOI] [PubMed] [Google Scholar]
- Mori H, Christensen AK 1980 Morphometric analysis of Leydig cells in the normal rat testis. J Cell Biol 84:340–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascoli M 1981 Characterization of several clonal lines of cultured Leydig tumor cells: gonadotropin receptors and steroidogenic responses. Endocrinology 108:88–95 [DOI] [PubMed] [Google Scholar]
- Eacker SM, Braun RE 2007 Androgen receptor in Leydig cell function and development. In: Payne AH, Hardy MP, eds. The Leydig cell in health and disease. Totowa, NJ: Humana Press; 345–361 [Google Scholar]
- Baker PJ, Sha JA, McBride MW, Peng L, Payne AH, O’Shaughnessy PJ 1999 Expression of 3β-hydroxysteroid dehydrogenase type I and type VI isoforms in the mouse testis during development. Eur J Biochem 260:911–917 [DOI] [PubMed] [Google Scholar]
- O’Shaughnessy PJ, Johnston H, Willerton L, Baker PJ 2002 Failure of normal adult Leydig cell development in androgen-receptor-deficient mice. J Cell Sci 115:3491–3496 [DOI] [PubMed] [Google Scholar]
- Murphy L, O’Shaughnessy PJ 1991 Testicular steroidogenesis in the testicular feminized (Tfm) mouse: loss of 17α-hydroxylase activity. J Endocrinol 131:443–449 [DOI] [PubMed] [Google Scholar]
- Murphy L, Jeffcoate IA, O’Shaughnessy PJ 1994 Abnormal Leydig cell development at puberty in the androgen-resistant Tfm mouse. Endocrinology 135:1372–1377 [DOI] [PubMed] [Google Scholar]
- Payne AH, Sha LL 1991 Multiple mechanisms for regulation of 3β-hydroxysteroid dehydrogenase/Δ5–Δ4-isomerase, 17α-hydroxylase/C17–20 lyase cytochrome P450, and cholesterol side-chain cleavage cytochrome P450 messenger ribonucleic acid levels in primary cultures of mouse Leydig cells. Endocrinology 129:1429–1435 [DOI] [PubMed] [Google Scholar]
- Heggland SJ, Signs SA, Stalvey JR 1997 Testosterone decreases 3β-hydroxysteroid dehydrogenase-isomerase messenger ribonucleic acid in cultured mouse Leydig cells by a strain-specific mechanism. J Androl 18:646–655 [PubMed] [Google Scholar]
- Houk CP, Pearson EJ, Martinelle N, Donahoe PK, Teixeira J 2004 Feedback inhibition of steroidogenic acute regulatory protein expression in vitro and in vivo by androgens. Endocrinology 145:1269–1275 [DOI] [PubMed] [Google Scholar]
- Soccio RE, Adams RM, Maxwell KN, Breslow JL 2005 Differential gene regulation of StarD4 and StarD5 cholesterol transfer proteins. Activation of StarD4 by sterol regulatory element-binding protein-2 and StarD5 by endoplasmic reticulum stress. J Biol Chem 280:19410–19418 [DOI] [PubMed] [Google Scholar]
- Dooley KA, Bennett MK, Osborne TF 1999 A critical role for cAMP response element-binding protein (CREB) as a co-activator in sterol-regulated transcription of 3-hydroxy-3-methylglutaryl coenzyme A synthase promoter. J Biol Chem 274:5285–5291 [DOI] [PubMed] [Google Scholar]
- Reaven E, Zhan L, Nomoto A, Leers-Sucheta S, Azhar S 2000 Expression and microvillar localization of scavenger receptor class B, type I (SR-BI) and selective cholesteryl ester uptake in Leydig cells from rat testis. J Lipid Res 41:343–356 [PubMed] [Google Scholar]
- Cummins CL, Volle DH, Zhang Y, McDonald JG, Sion B, Lefrançois-Martinez AM, Caira F, Veyssière G, Mangelsdorf DJ, Lobaccaro JM 2006 Liver X receptors regulate adrenal cholesterol balance. J Clin Invest 116:1902–1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volle DH, Mouzat K, Duggavathi R, Siddeek B, Déchelotte P, Sion B, Veyssière G, Benahmed M, Lobaccaro JM 2007 Multiple roles of the nuclear receptors for oxysterols liver X receptor to maintain male fertility. Mol Endocrinol 21:1014–1027 [DOI] [PubMed] [Google Scholar]
- O’Shaughnessy PJ, Willerton L, Baker PJ 2002 Changes in Leydig cell gene expression during development in the mouse. Biol Reprod 66:966–975 [DOI] [PubMed] [Google Scholar]
- Sadate-Ngatchou PI, Pouchnik DJ, Griswold MD 2004 Follicle-stimulating hormone induced changes in gene expression of murine testis. Mol Endocrinol 18:2805–2816 [DOI] [PubMed] [Google Scholar]
- Dichek HL, Brecht W, Fan J, Ji ZS, McCormick SP, Akeefe H, Conzo L, Sanan DA, Weisgraber KH, Young SG, Taylor JM, Mahley RW 1998 Overexpression of hepatic lipase in transgenic mice decreases apolipoprotein B-containing and high density lipoproteins. Evidence that hepatic lipase acts as a ligand for lipoprotein uptake. J Biol Chem 273:1896–1903 [DOI] [PubMed] [Google Scholar]
- Folch J, Lees M, Sloane Stanley GH 1957 A simple method for the isolation and purification of total lipids from animal tissues. J Biol Chem 226:497–509 [PubMed] [Google Scholar]
- Brown MS, Faust JR, Goldstein JL 1975 Role of the low density lipoprotein receptor in regulating the content of free and esterified cholesterol in human fibroblasts. J Clin Invest 55:783–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichek HL, Agrawal N, El Andaloussi N, Qian K 2006 Attenuated corticosterone response to chronic ACTH stimulation in hepatic lipase-deficient mice: evidence for a role for hepatic lipase in adrenal physiology. Am J Physiol Endocrinol Metab 290:E908–E915 [DOI] [PubMed] [Google Scholar]
- Moraga PF, Llanos MN, Ronco AM 1997 Arachidonic acid release from rat Leydig cells depends on the presence of luteinizing hormone/human chorionic gonadotrophin receptors. J Endocrinol 154:201–209 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.