Abstract
Estrogen has direct and indirect effects on mitochondrial activity, but the mechanisms mediating these effects remain unclear. Others reported that long-term estradiol (E2) treatment increased nuclear respiratory factor-1 (NRF-1) protein in cerebral blood vessels of ovariectomized rats. NRF-1 is a transcription factor that regulates the expression of nuclear-encoded mitochondrial genes, e.g. mitochondrial transcription factor A (TFAM), that control transcription of the mitochondrial genome. Here we tested the hypothesis that E2 increases NRF-1 transcription resulting in a coordinate increase in the expression of nuclear- and mitochondrial- encoded genes and mitochondrial respiratory activity. We show that E2 increased NRF-1 mRNA and protein in MCF-7 breast and H1793 lung adenocarcinoma cells in a time-dependent manner. E2-induced NRF-1 expression was inhibited by the estrogen receptor (ER) antagonist ICI 182,780 and actinomycin D but not by phosphoinositide-3 kinase and MAPK inhibitors, indicating a genomic mechanism of E2 regulation of NRF-1 transcription. An estrogen response element (ERE) in the NRF-1 promoter bound ERα and ERβ in vitro, and E2 induced ERα and ERβ recruitment to this ERE in chromatin immunoprecipitation assays in MCF-7 cells. The NRF-1 ERE activated reporter gene expression in transfected cells. Small interfering RNA to ERα and ERβ revealed that ERα mediates E2-induced NRF-1 transcription. The E2-induced increase in NRF-1 was followed by increased TFAM and the transcription of Tfam-regulated mitochondrial DNA-encoded COI and NDI genes and increased mitochondrial biogenesis. Knockdown of NRF-1 blocked E2 stimulation of mitochondrial biogenesis and activity, indicating a mechanism by which estrogens regulate mitochondrial function by increasing NRF-1 expression.
EXPERIMENTAL EVIDENCE demonstrates that liver and brain mitochondria of normal, but not ovariectomized (Ovx), female rats generate less reactive oxygen species (ROS) and have higher respiratory potential resulting from decreased oxidative damage (1). The decrease in ROS and higher respiratory potential may help explain the observed increased longevity of females in most mammalian species (2). Although the sex differences in mitochondrial function are likely mediated by estrogens, the mechanism(s) underlying these effects remain ill defined. Therefore, a goal in the present study was to elucidate one of the pathways that may contribute to the observed estrogen-regulated increase in mitochondrial function.
Classical intracellular estrogen action is mediated by estrogen receptors (ERs) via regulation of gene transcription. There are two subtypes of ER: ERα and ERβ. In an estrogen-responsive cell, the vast majority of ER resides within the nucleus where ERα, but not ERβ, is complexed with the heat-shock protein 90 chaperonin complex when a ligand is not present (3,4). Once activated by estradiol (E2) or other estrogen-like compounds, ERs dimerize and bind to estrogen response elements (EREs) located in the promoters or distal enhancer regions of target genes (5). The majority of estrogen-sensitive genes do not contain palindromic EREs; instead, single or multiple imperfect or half-site EREs regulate the E2 response (6). In addition, ER binds directly to other DNA-bound transcription factors, e.g., Sp1 or AP1, via a tethering mechanism, resulting in increased transcription (7). ER mediates its genomic effects through interactions with coactivators that recruit chromatin remodeling complexes and increase gene transcription (8).
In addition to its genomic effects that occur within 3–6 h after estrogen administration, ER mediates rapid, membrane-initiated, nongenomic effects of E2 that occur within seconds to minutes after treatment in a cell type-dependent manner (9). Nongenomic effects of ER have been best characterized in endothelial cells in which E2 rapidly inhibits calcium influx (10), increases intracellular Ca2+ (11) and cAMP (12), and stimulates nitric oxide (NO) release resulting in vasodilation (13) and thus accounting in part for the observed cardioprotective effects of estrogens. The membrane-associated ERs that mediate these nongenomic effects are transcribed from the same messages used to generate nuclear ERα and ERβ and are inhibited by the ER antagonist ICI 182,780 (14). Membrane-associated ER conveys these rapid intracellular effects by activating G-coupled proteins, adenylate cyclase production, and inositol phosphate production (14). By activating networks including the phosphoinositide-3 kinase (PI3K) and MAPK, nongenomic ER rapidly induces a number of changes within the cell including changes in gene expression through phosphorylation of transcription factors and coregulators (15,16).
The role of estrogens in mitochondrial function is well established from studies with ERα knockout (ERKO) and Ovx rodents. In a model of cardiac ischemia-reperfusion, the mitochondria of ERKO mice contained noticeable ultrastructural damage and a decrease in mitochondrial respiratory chain (MRC) function (17). A similar study comparing Ovx or E2-supplemented Ovx rats showed that myocardial mitochondrial damage and decreased MRC function in the Ovx rats was ameliorated by E2 (18). These studies suggest an important role for ER in maintaining mitochondrial structure and function in the ischemia-reperfusion model.
Estrogens may exert direct or indirect effects on mitochondrial function in a variety of tissues, most notably in mediating the neuroprotective effects of estrogen (19). ERβ was localized to mitochondria in several brain cell types as well as primary cardiomyocytes (20). Mitochondrial localization of ERβ was demonstrated by a variety of complementary techniques including immunocytochemistry, subcellular fractionation, and mass spectrometry (20). ERα and ERβ have also been detected in mitochondria of MCF-7 human breast cancer cells (21), and treatment with E2 enhanced ER mitochondrial localization in a concentration- and time-dependent manner (22). An E2-dependent increase in mRNA levels of mitochondrial DNA (mtDNA)-encoded genes cytochrome c oxidase subunits I and II (MTCOI and MTCOII) was detected, suggesting a direct effect of E2 on mtDNA gene transcription (22). However, the intra-mitochondrial localization of ER is controversial, and one group reported that ERβ was not detectable in mouse liver mitochondria as analyzed by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (23). The mitochondrial localization of ER may be cell specific because ERβ was recently identified in human heart mitochondrial proteins by MALDI-TOF (24).
The transcription factor nuclear respiratory factor-1 (NRF-1) plays a key role in integrating the transcription of nuclear- and mitochondrial-encoded genes (25). NRF-1 homodimerizes and binds to palindromic NRF-1 sites in the promoters of nuclear-encoded mitochondrial genes (26). NRF-1 interacts with the coactivators Peroxisome proliferator activated receptor γ coactivator-1 (PGC-1)α and PGC-1 related coactivator (PRC) to regulate target genes involved in mitochondrial respiratory function (27,28). NRF-1 target genes include subunits of the five MRC complexes, assembly factors for the respiratory apparatus, parts of the mtDNA transcription and replication machinery, mitochondrial and cytosolic enzymes of heme biosynthesis, components of mitochondrial protein import, and three key mtDNA transcription factors, mitochondrial transcription factor A (Tfam) and mitochondrial transcription factor B types 1 and 2 (TFB1M and TFB2M, mtTFB1 and mtTFB2, hereafter referred to as TFBs) (29). Tfam and TFBs transcribe the mitochondrial genome, resulting in an increase in mitochondrial-encoded subunits of the MRC (29). NRF-1 plays an essential role in integrating nucleo-mitochondrial interactions (30). The NRF-1 knockout mouse showed a decrease in mtDNA that resulted in embryonic lethality, further demonstrating that NRF-1 is essential for viability (31).
Expression of NRF-1 is increased by a variety of environmental stimuli and hormones (32,33,34). Thyroid hormone (T3) increases NRF-1 gene expression through a hormone-responsive element in the NRF-1 promoter (35). Although glucocorticoids and estrogens have both been suggested to play a critical role in mitochondrial up-regulation and mitochondrial gene expression, no one has evaluated whether these hormones directly regulate NRF-1 transcription. Interestingly, a recent study revealed that NRF-1 is up-regulated in cerebral blood vessels of Ovx rats chronically treated with E2, suggesting that estrogen may regulate NRF-1 transcription (36).
Coordinate expression of mitochondrial- and nuclear-encoded genes has been proposed to be mediated by hormone receptors within mitochondria inducing expression of mtDNA-encoded genes (37). Here we tested the hypothesis that estrogen increases mitochondrial activity and mtDNA-encoded mRNA expression through ER-mediated up-regulation of NRF-1 gene transcription. We report for the first time that NRF-1 mRNA is up-regulated by E2 and that this is a direct transcriptional effect mediated by ER in a breast cancer and lung cancer cell line.
RESULTS
E2-Induced NRF-1 Transcription Is Mediated by Genomic ER
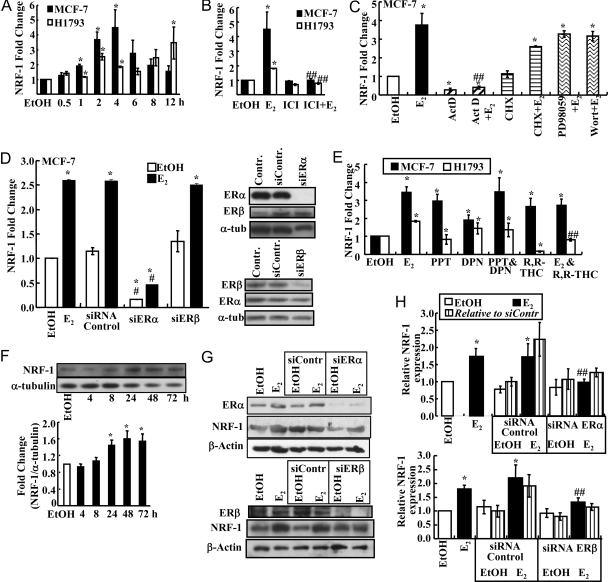
Although treatment of Ovx rats with E2 increased NRF-1 in cerebral blood vessels (36), no one has examined whether this increase in NRF-1 was a primary estrogen response or a result of a long-term alterations in gene expression patterns. To determine whether NRF-1 is a direct E2 target gene, estrogen-responsive H1793 human lung (38) and MCF-7 human breast adenocarcinoma (39) cells were treated with E2 and NRF-1 expression measured by quantitative real-time-PCR (QRT-PCR) (Fig. 1A). NRF-1 mRNA expression was significantly up-regulated by E2 in both cell lines, with earlier and higher induction in MCF-7 than H1793 cells, which also showed an apparent second phase of induction at 12 h (Fig. 1A). Dose-response experiments revealed maximal NRF-1 mRNA induction with 10 nm E2 after 4 h of treatment in MCF-7 cells (data not shown).
Figure 1.
ERα and ERβ Mediate the E2-Induced Increase in NRF-1 mRNA Expression in MCF-7 and H1793 Cells in a Genomic Manner
A, B, C, E, and H, NRF-1 expression as determined by QRT-PCR as described in MATERIALS AND METHODS; A, MCF-7 and H1793 cells were treated with EtOH or 10 nm E2 for the time indicated and as described in MATERIALS AND METHODS; B, MCF-7 and H1793 cells were pretreated with 100 nm ICI 182,780 for 6 h before addition of 10 nm E2 for 4 h; C, MCF-7 cells were pretreated with 10 μg/ml ActD or CHX or with 50 μm PD98059 or 50 nm wortmannin for 1 h before incubation with 10 nm E2 for 4 h; D, MCF-7 cells were transfected with control siRNA or siRNA targeting ERα (siERα) or ERβ (siERβ) for a total of 48 h, as described in MATERIALS AND METHODS, and then treated with EtOH or E2 for 4 h. NRF-1mRNA expression was determined by Q-RT-PCR. Representative Western blots (40 μg WCE per lane) for ERα, ERβ, and α-tubulin in the EtOH-treated MCF-7 cells are shown. Similar results were observed in E2-treated cells. E, MCF-7 or H1793 cells were treated with EtOH, 10 nm E2, 10 nm DPN, 10 nm PPT, or 100 nm R,R-THC, alone or in combination as indicated, for 4 h. F, A representative Western blot (40 μg WCE per lane) is shown for NRF-1 expression in MCF-7 cells treated with 10 nm E2 for the indicated times. The bar graph summarizes NRF-1 protein normalized to α-tubulin from the same blot from three separate experiments. G, MCF-7 cells were either transfected with control siRNA, siERα, or siERβ for 48 h and then treated with EtOH or E2 for 48 h or not transfected and treated with EtOH or E2 for 48 h. H, Quantitation of the NRF-1 protein relative to β-actin in the same blot relative to 48-h EtOH values. As indicated, the striped bars are NRF-1 normalized to siRNA control EtOH NRF-1/β-actin values. Values with error bars are the average of three to six separate experiments ± sem. *, P < 0.05 compared with EtOH; ##, significantly different from the E2 alone value.
ICI 182,780 is a well-established antagonist of genomic ER that both prevents coactivator recruitment and enhances ER proteasomal degradation (40). To determine whether the E2-induced increase in NRF-1 is mediated directly by ER, MCF-7 and H1793 cells were pretreated with ICI 182,780 for 6 h before E2 treatment. ICI 182,780 blocked the E2-induced increase in NRF-1 mRNA, indicating that ER mediated this response (Fig. 1B).
NRF-1 Is a Primary Estrogen-Responsive Gene Mediated by Genomic ER
The transcriptional inhibitor actinomycin D (ActD) and protein synthesis inhibitor cycloheximide (CHX) were used to determine whether the E2-ER-mediated increase in NRF-1 was a direct effect of ER at the genomic level or required synthesis of a secondary estrogen-responsive protein. Notably, ActD, but not CHX, inhibited the E2-induced increase in NRF-1 mRNA (Fig. 1C), indicating that the de novo expression of an E2-induced protein was not required for increased NRF-1 transcription. We conclude that NRF-1 is a primary E2-responsive gene.
To determine whether the E2-induced increase in NRF-1 is mediated by nongenomic ER activity, MCF-7 cells were pretreated for 1 h with the MAPK (MEK) and PI3K inhibitors PD98059 and wortmannin, respectively. Neither inhibitor altered the E2-induced increase in NRF-1 (Fig. 1C), indicating that the E2 response is mediated by genomic ER activity and not nongenomic/membrane-initiated activation of the PI3K/Akt and MAPK signaling pathways.
Small Interfering (siRNA) to ERα But Not ERβ Inhibits E2-Induced NRF-1 Expression in MCF-7
Because ERα and ERβ proteins are expressed in MCF-7 (38,41) (see also supplemental Fig. 2, published as supplemental data on The Endocrine Society’s Journals Online web site at http://mend.endojournals.org) and H1793 cells (38), the observed ER-dependent up-regulation of NRF-1 by E2 could be mediated by both or either subtype. To examine the contribution of each ER subtype to the E2-induced NRF-1 transcription, MCF-7 cells were transfected with control/nonspecific siRNA or siRNA targeting ERα or ERβ for 48 h followed by treatment with ethanol (EtOH) or 10 nm E2 for 4 h. Control siRNAs did not affect basal or E2-induced NRF-1 transcription (Fig. 1D). Knockdown of ERα reduced basal and E2-stimulated NRF-1 mRNA by 84 and 89%, respectively. In contrast, knockdown of ERβ did not alter basal NRF-1 or E2-induced NRF-1 mRNA expression (Fig. 1D). Subtype-specific siRNAs reduced ERα and ERβ protein levels by about 85 and 75%, respectively. Together these data indicate that ERα mediates the E2-induced transcription of NRF-1 in MCF-7 cells.
ERα- and ERβ-Selective Agonists Increase NRF-1 Transcription
To further address the roles of ERα and ERβ in regulating NRF-1 transcription, cells were treated with concentrations of the ERα- and ERβ-selective agonists, propyl pyrazole triol (PPT) (42) and diarylpropionitrile (DPN) (43), that selectively activate each respective ER subtype. PPT induced the same increase in NRF-1 as E2, and DPN yielded about 50% of the E2 increase in NRF-1 in MCF-7. These data indicate that NRF-1 is transcriptionally regulated by agonist-occupied ERα in MCF-7 cells. When MCF-7 cells were treated with PPT and DPN, NRF-1 induction was identical to PPT alone, indicating a saturated response (Fig. 1E). DPN increased NRF-1 to the same extent as E2 in H1793 cells, whereas PPT had no effect, indicating an ERβ-mediated transactivation. These data agree with the higher expression of ERβ than ERα in H1793. R,R-tetrahydrochrysene (R,R-THC), an ERα agonist/ERβ antagonist (44), stimulated NRF-1 transcription in MCF-7 and had no effect when combined with E2. However, R,R-THC inhibited basal and E2-induced NRF-1 expression in H1793 cells. Overall, we conclude that the induction of NRF-1 in response to E2 appears to be ERα subtype selective in MCF-7 and that ERβ is responsible for E2-induced NRF-1 transcription in H1793 lung adenocarcinoma cells.
NRF-1 Protein Expression Is Increased by E2
Western blot analysis of NRF-1 protein expression after E2 treatment revealed a time-dependent increase in NRF-1 in MCF-7 cells with statistical significance reached after 24 h of E2 treatment (Fig. 1F). These results agree with the increase seen in NRF-1 mRNA expression with E2 treatment but are delayed in time, as seen for other E2-target genes (45).
The effect of knockdown of ERα and ERβ on NRF-1 protein expression was also examined. ERα and ERβ proteins were reduced about 85 and 95% by their respective siRNAs (Fig. 1G). siERα reduced E2-induced NRF-1 protein by about 45%, whereas siERβ reduced E2-induced NRF-1 protein by about 24% (Fig. 1, G and H). The siERβ-mediated decrease in NRF-1 correlates with an approximately 20% reduction in ERα protein in the MCF-7 cells 72 h after transfection (data not shown).
The Expression of Tfam; Complex IV, Cytochrome c Oxidase Subunit I (COI, MTCO1); and NADH Dehydrogenase Subunit I (NDI) mRNAs Are Increased by E2
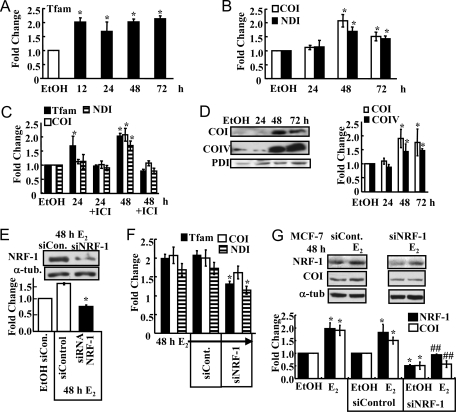
NRF-1 stimulates transcription of the mitochondrial transcription factor Tfam by binding to an NRF-1 response element in the Tfam promoter (46). Subsequently, Tfam increases the transcription of mtDNA-encoded gene targets (30). The expression of Tfam and two mitochondrial-encoded mRNAs, COI and NDI, were analyzed by QRT-PCR to determine whether the E2-induced increase in NRF-1 resulted in a downstream increase in the transcription of nuclear-encoded Tfam and mitochondrial-encoded Tfam target genes. Tfam mRNA was increased 12–72 h after E2 treatment (Fig. 2A). The expression of Tfam-regulated, mtDNA-encoded COI and NDI mRNAs was increased 48–72 h after E2 treatment (Fig. 2B). Furthermore, this increase and that of Tfam was inhibited by ICI 182,780, indicating an ER dependence (Fig. 2C). To further examine the downstream effects of E2-induced NRF-1 up-regulation, the expression of secondary gene targets COI (mtDNA-encoded, MTCOI) and cytochrome c oxidase (COIV, nuclear-encoded) proteins was evaluated compared with protein disulfide isomerase (PDI) control (Fig. 2D). COI and COIV are both subunits of complex IV in the MRC. COI is transcriptionally regulated by Tfam and TFBs, whereas COIV is a direct target of NRF-1 (30). E2 induced a significant increase in both proteins after 48 and 72 h (Fig. 2D).
Figure 2.
NRF-1 Target Gene Expression Is Increased in E2-Treated MCF-7 Cells
A–C, MCF-7 cells were treated with EtOH or 10 nm E2 for the indicated times, and QRT-PCR was performed for Tfam (A) or COI and NDI (B and C). Where indicated, cells were pretreated with ICI 182,780 for 6 h (C). D, A representative Western blot of COI and COIV protein expression in 10 nm E2-treated MCF-7 cells is shown above a summary graph of the data normalized to PDI in the same blots. E, A representative Western blot of NRF-1 protein expression in cells transfected with control siRNA (siCon.) or siRNA targeting NRF-1 (siNRF-1) is shown above a summary graph of data normalized to α-tubulin (α-tub.) in blots from three separate experiments ± sem. F, mRNA expression levels of Tfam, COI, and NDI were measured after 48 h treatment with 10 nm E2 in MCF-7 cells transfected with control siRNA or siNRF-1. G, A representative Western blot of NRF-1 and COI protein expression in cells transfected with control siRNA or siNRF-1 and treated with EtOH or 10 nm E2 for 48 h. The membrane was stripped and reprobed for α-tubulin. Below is a summary graph of the Western data normalized to each protein in EtOH-treated cells. Values are the average of three to six separate experiments ± sem. *, P < 0.05 compared with EtOH; ##, significantly different from the E2 alone value without siNRF-1.
To address the importance of the E2-induced increase in NRF-1 expression in the observed NRF-1-stimulated downstream nuclear and mitochondrial effects, NRF-1 expression was reduced by siRNA knockdown and followed by EtOH or E2 treatment. NRF-1 protein expression was decreased by 50% compared with control siRNA (Fig. 2E). QRT-PCR was performed to examine secondary gene expression in parallel with experiments reported in Fig. 2, A–C. siRNA knockdown of NRF-1 resulted in a significant decrease in Tfam, a direct NRF-1 target, and in secondary gene target NDI (Fig. 2F). The decrease in COI was not statistically significant, indicating that E2-regulated factors in addition to NRF-1 contribute to COI expression. However, knockdown of NRF-1 reduced basal and E2-induced COI protein expression (Fig. 2G). These experiments indicate that the E2-induced increase in NRF-1 results in a downstream increase in its nuclear-encoded target gene TFAM and COIV and mtDNA-encoded Tfam target genes MTCOI and MTNDI in an ER-dependent manner in MCF-7 cells.
E2 Activates NRF-1 Promoter Function
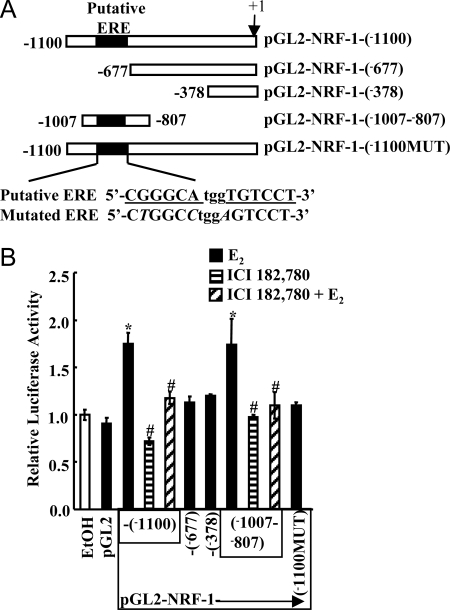
The Gene2Promoter program (47) predicted the NRF-1 promoter to be within the first −600-bp region of the NRF-1 start site. It was previously reported that a −1000-bp region contained the NRF-1 gene promoter based on transient transfections in COS, HeLa, and L6 cells (48). Using MatInspector (49), an extended −1100-bp region 5′ of the NRF-1 start site was examined for putative transcription factor binding sites. The MatInspector program selected an area −963 to −944 as a canonical palindromic ERE with a matrix similarity of 0.831. To investigate the putative ERE in E2 activation of NRF-1 transcription, we used luciferase assays. A series of constructs containing a wild-type NRF-1 promoter, deletions of the ERE, and mutations in the putative ERE were cloned into the pGL2-luciferase reporter plasmid (Fig. 3A). E2 stimulated an increase in luciferase activity from the pGL2-NRF-1 (−1100) and (−1007 to −807) constructs, which contain the putative ERE but not the pGL2-NRF-1 (−677), (−378), or (−1007 to −807) constructs lacking the ERE, indicating that the region from −1007 to −807 containing the putative ERE is estrogen responsive (Fig. 3B). Mutagenesis was performed to eliminate the putative ERE from the pGL2-NRF-1 (−1100) construct to further evaluate the role of the putative ERE in the estrogen response. As demonstrated in Fig. 3B, when nucleotides within the core ERE were mutated within the pGL2-NRF-1 (−1100) construct, E2 did not stimulate luciferase activation. Treatment with ICI 182,780 had no effect on luciferase activity from the full-length promoter construct or pGL2-NRF-1 (−1007 to −807); however, cotreatment of cells with E2 and ICI 182,780 ablated the E2-induced increase in luciferase activity. These results indicate that the E2-induced reporter activity is mediated by ER interaction with the NRF-1 promoter region containing the putative ERE.
Figure 3.
E2 Stimulates Luciferase Reporter Activity from the NRF-1 Promoter in MCF-7 Cells
A, The diagram represents the pGL2-basic luciferase expression vectors containing various lengths of 5′-flanking regions of the human NRF-1 gene promoter and a construct in which the ERE sequence was mutated. The ERE is underlined in the wild-type putative NRF-1 ERE. The nucleotides in bold are different from the consensus ERE (6), and the nucleotides in italics indicate mutations compared with the wild-type NRF-1 ERE in the mutated pGL2-NRF-1-(−1100MUT) construct. B, MCF-7 cells were transfected with the indicated pGL2-NRF-1 promoter constructs and treated with EtOH, 10 nm E2, ICI 182,780, or ICI 182,780 plus E2 for 24 h as described in MATERIALS AND METHODS. Values are the mean ± sem of four. *, Significantly different from the ETOH control; #, significantly different from the E2 treated sample with the same construct.
ER Binds the Putative ERE in the NRF-1 Promoter in Vivo and in Vitro
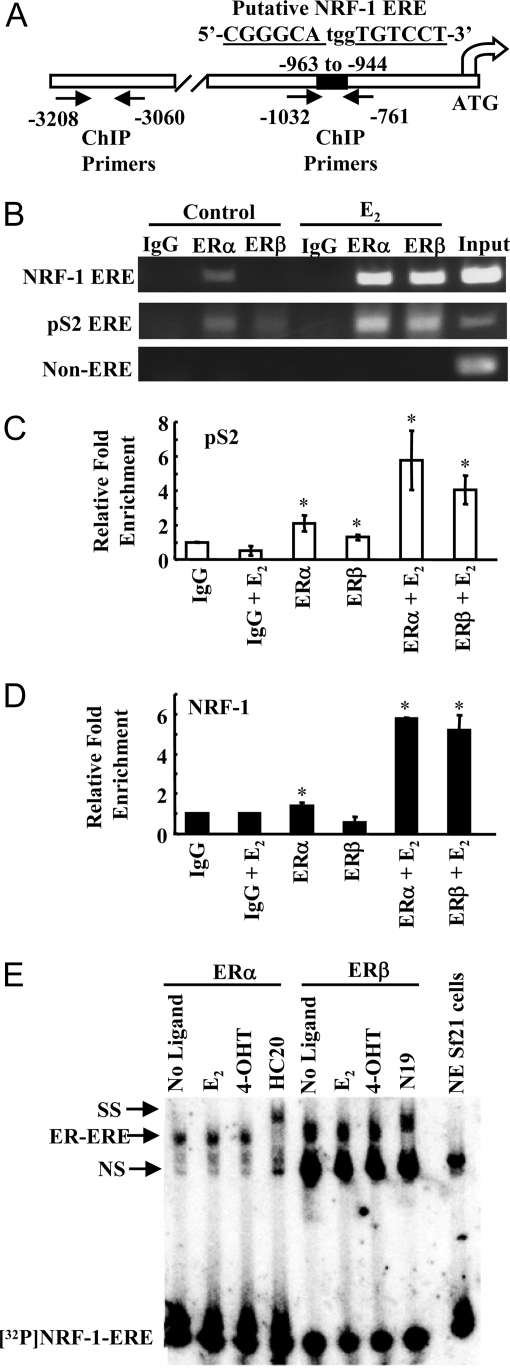
Chromatin immunoprecipitation (ChIP) assays were used to evaluate whether ERα and ERβ bound directly to a putative ERE identified in silico and in the promoter deletion transient transfection assays in the NRF-1 promoter (−963 to −944) (Fig. 4A). As a positive control for ChIP, the ERE-containing, E2-regulated human pS2 gene (50) was examined in parallel. A distal region of the NRF-1 promoter (−3060 to −3208) was used as a negative control. Before E2 treatment, ERα was present on both the NRF-1 and pS2 ERE-containing regions, and ERβ was observed on the pS2 ERE but not on the NRF-1 ERE. E2 increased ERα and ERβ binding to the pS2 and NRF-1 EREs (Fig. 4B). No binding of ERα or ERβ was observed in the distal NRF-1 promoter, demonstrating the specificity of the interaction of each ER with the NRF-1 ERE region.
Figure 4.
ERα and ERβ Bind to the Putative NRF-1 ERE
A, Diagram of the NRF-1 promoter and putative ERE (−963 to −944) with the location of the ChIP primers. ERE half-sites are underlined and letters in bold indicate deviations from the consensus ERE. B, MCF-7 cells were treated with EtOH or E2 for 1 h, and ChIP was performed as described in MATERIALS AND METHODS. C, QRT-PCR was performed for ERα and ERβ occupancy on the pS2 ERE (C) and NRF-1 ERE (D) in ChIP samples as described in MATERIALS AND METHODS. Relative promoter enrichment compared with IgG is plotted. Values are the average and sd of two separate experiments. *, P < 0.05 for control. E, Baculovirus-expressed ERα and ERβ were incubated with 32P-labeled NRF-1 ERE in the presence of E2, 4-hydroxytamoxifen (4-OHT), or no ligand, as indicated. EMSA was performed as described in MATERIALS AND METHODS. NS, Nonspecific binding; SS, supershift with the indicated ER antibodies.
QRT-PCR was performed on the ChIP samples to confirm the enrichment of ERα and ERβ recruitment observed in response to E2 (Fig. 4B). In agreement with previous reports, QRT-PCR demonstrated that ERα occupied the pS2 promoter by in the absence of E2, and E2 increased ERα recruitment (Fig. 4C). Similarly, ERβ also bound the pS2 promoter in the absence of ligand, and recruitment was significantly increased by E2. In agreement with the results in Fig. 4B, ERα but not ERβ occupied the NRF-1 ERE in the absence of ligand. E2 significantly increased the recruitment of both ERα and ERβ to the NRF-1 ERE (Fig. 4D).
The putative NRF-1 ERE (5′-CGGGCAtggTGTCCT-3′) differs from the palindromic consensus ERE (5′-AGGTCAgagTGACCT-3′) by two base pair changes in the 5′ half-site and one base pair change in the 3′ half-site indicated in bold. These changes, while reducing the affinity of ER for the ERE, do not eliminate the site. In fact, all three of the changes in the ERE site in the NRF-1 promoter occur at nucleotide positions previously identified as lower in importance to the binding of ERα (5,6). EMSAs indicated that recombinant ERα and ERβ bind directly to the putative ERE in the NRF-1 promoter without ligand and in the presence of E2 and 4-hydroxytamoxifen (Fig. 4C).
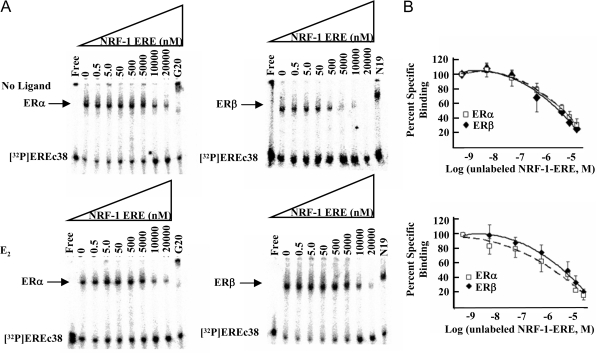
Deviation from the perfect consensus ERE correlates with decreasing affinity of ER-ERE binding (6). To determine the Ki of the ER-NRF-1-ERE interaction, competition EMSAs were performed (Fig. 5A). As shown, the NRF-1-ERE had lower binding affinity for ER than EREc38. The binding affinity was similar for unliganded ERα and ERβ with a Ki = 668 ± 76.2 and 699 ± 71 nm, respectively, and 1056 ± 98 and 895 ± 51 nm for E2-ERα and -ERβ, respectively, indicating lower affinity of E2-ERα for the NRF-1-ERE than E2-ERβ in vitro. ERα binds the nonconsensus ERE at position −1211 in the human c-Fos promoter that has two nucleotide changes in the 5′ half-site and a 13-bp palindrome with a Kd of 328 ± 38 nm, and ERβ binds this ERE with a Kd = 240 ± 24 nm (51).
Figure 5.
ERα and ERβ Bind the NRF-1 ERE with Reduced Affinity in Comparison with a Palindromic ERE
A, Competitive EMSAs were performed using unliganded (top) or E2-liganded ERα (left) and ERβ (right) incubated with 1.1 nm [32P]EREc38 plus increasing concentrations of unlabeled NRF-1-ERE as indicated and described in MATERIALS AND METHODS. ERα and ERβ antibodies G20 and N19, respectively, were included in the indicated binding reactions to demonstrate the specificity of the bound complex (SS). B, Competitive binding curves are shown for ERα (□) and ERβ (♦), and the IC50 was calculated as described in MATERIALS AND METHODS.
Mitochondrial Biogenesis Is Increased by E2
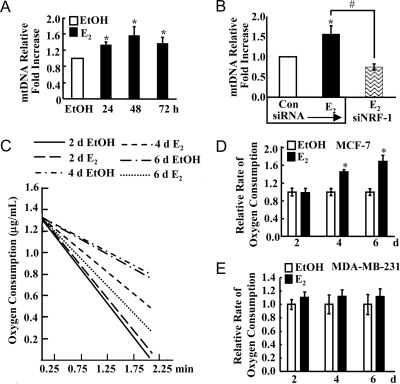
We examined whether E2 increased mitochondrial biogenesis by measuring two endpoints: mtDNA copy number and oxygen consumption. mtDNA copy number was assayed by semiquantitative PCR using primers that recognized a 250-bp region of the mtDNA normalized to primers for the 18S gene. E2 significantly increased mtDNA copy number after 24, 48, and 72 h (Fig. 6A). Because Tfam is essential for mtDNA replication (52), these data are consistent with the increase in E2-induced Tfam mRNA beginning at 12 h. Notably, siRNA knockdown of NRF-1 inhibited the E2-induced increase in mtDNA (Fig. 6B), indicating that the E2-induced increase in mitochondrial biogenesis is mediated by NRF-1 and not directly by E2.
Figure 6.
E2 Increases Mitochondrial Biogenesis
A, MCF-7 cells were treated with EtOH or 10 nm E2 for the indicated time. A summary graph of four experiments evaluating mtDNA normalized to 18S expression by semiquantitative PCR is shown. B, MCF-7 cells were transfected with control/nonspecific siRNA or siRNA targeting NRF-1 and treated with EtOH or 10 nm E2 for 48 h. A summary graph of three experiments evaluating mtDNA normalized to 18S expression by semiquantitative PCR is shown. C, Oxygen consumption was measured in MCF-7 cells treated with EtOH or 10 nm E2 for the indicated number of days, and representative slopes are shown. D, The average rate of oxygen consumption from three to six separate experiments is shown ± sem relative to EtOH values. E, Oxygen consumption was measured in MDA-MB-231 cells treated with EtOH or 10 nm E2. The average rate of O2 consumption from three to four separate experiments is shown ± sem relative to EtOH values. *, P < 0.05 compared with EtOH; #, significantly different from the E2-treated sample without siNRF-1.
Oxygen consumption was used as a measure of MRC function in E2-treated MCF-7 cells. A significant increase in oxygen consumption was seen after 4 and 6 d of E2 treatment (Fig. 6, C and D). This is consistent with the up-regulation of COI and COIV proteins beginning at 48 h, which, in concert with increased expression of MRC proteins resulting from NRF-1, Tfam, and TFB activity, would lead to an increase in oxygen consumption in the cell.
To further examine the contribution of ERα to the observed E2-induced increase in rates of oxygen consumption, identical experiments were performed in MDA-MB-231 cells, i.e. an E2-insensitive, ERα-negative breast cancer cell line (Fig. 6, C and D). E2 did not increase oxygen consumption in this estrogen-independent cell line (Fig. 6E).
DISCUSSION
Although it is clear that estrogens have protective effects on mitochondrial function in multiple cell types and rodent models, the mechanisms mediating these effects remain undefined and are likely to be multifaceted. Several theories have been postulated regarding the action of ER on mitochondrial structure and function (21). Here we report that E2 stimulates mitochondrial function through a genomic mechanism of ER action involving direct ERα and ERβ interaction with a nonconsensus ERE in the NRF-1 promoter. Knockdown experiments indicated that E2 stimulates NRF-1 transcription through ERα and not ERβ in MCF-7 cells. E2 increases transcription of NRF-1, which is an established integrator of nuclear and mitochondrial interactions (30). The increase in NRF-1 mRNA and protein with E2 treatment agrees with the report that NRF-1 protein was increased in the cerebral blood vessels of E2-treated Ovx rats (36). Although the present study was under review, a newly published microarray profiling study of genes regulated by E2 in aortas from Ovx vs. E2-supplemented Ovx αERKO, βERKO, and wild-type mice identified more than 18 nuclear-encoded MRC subunits that are E2-ERα regulated (53). Importantly, NRF-1 mRNA was decreased in the αERKO but not βERKO mice, indicating that E2-ERα regulates NRF-1 in mouse aorta. These in vivo data support the results of our siRNA experiments indicating that ERα and not ERβ was responsible for the E2-induced NRF-1 in MCF-7 cells. Notably, NRF-1 was down-regulated by E2-ERβ and genes including an NRF-1 response element in the −1-kb promoter were also down-regulated in αERKO mice, indicating that ERβ actively represses a subset of MRC genes (53). The authors speculated that ERα and ERβ directly regulate NRF-1 transcription through an ERE in the promoter and call for studies along these lines. A caveat of this study is the 1-wk E2 treatment; thus, many identified genes are unlikely to be direct E2 targets, but rather secondary or tertiary targets. Nonetheless, together with the previous report regarding E2 up-regulation of NRF-1 in rat cerebral blood vessels, these data provide physiological relevance to the molecular characterization of E2-induced NRF-1 transcription reported here. These results may explain previously observed increases in mitochondrial activity in response to E2, but they do not rule out a role for ER action within mitochondria. A role for nongenomic E2 action in mediating NRF-1 transcription was eliminated because inhibitors of the two dominant nongenomic estrogen signaling pathways in MCF-7 cells, MEK and PI3K (54,55), had no effect on E2-induced NRF-1 expression. Likewise, E2 increased the expression of genes regulated by NRF-1, Tfam mRNA, and subsequently, in a time-delayed manner, mtDNA-encoded Tfam-regulated COI and NDI mRNA. These data are in agreement with a report demonstrating a 2-fold increase in COI mRNA with E2 treatment in MCF-7 cells (22); however, there was a time difference noted between that study and our results. We detected an increase in COI at 48 h, whereas the previous report (22) indicated this increase at 12 and 24 h after E2 treatment of MCF-7 cells. The use of a 10-fold higher E2 concentration (100 nm, nonphysiological) in the previous study may account for the difference in induction times.
Subsequent to the increase in NRF-1 and secondary gene expression, including COI and COIV proteins, an increase in mtDNA and oxygen consumption was observed. The increase in mtDNA in concert with COI and NDI suggests a pathway by which E2-induced NRF-1 and subsequent increased Tfam activates both transcription and mtDNA replication. In addition to the observed increase in the expression of these genes as reported here, microarray analyses of E2-regulated genes in MCF-7 cells have identified increases in mRNAs encoding genes involved in a variety of functions within mitochondria (supplemental Table 1, published as supplemental data on The Endocrine Society’s Journals Online web site at http://mend.endojournals.org). In parallel to the increase in mitochondrial gene expression, E2 increased oxygen consumption, which is an index of MRC activity. Whether this increase is due to increased MRC activity or an increase in mitochondrial biogenesis leading to more mitochondria per cell remains to be determined.
Importantly, we add NRF-1 to the list of genes regulated by nonpalindromic, low-affinity ER-ERE binding (6). Despite the low binding affinity of ER for the NRF-1 ERE as measured by EMSA, ChIP revealed that the putative NRF-1 ERE is occupied by both ERα and ERβ in response to E2, similar to the pS2 promoter, an established ER target gene (56). Furthermore, the luciferase assays using the NRF-1 promoter deletion and ERE-mutation constructs demonstrate that the NRF-1 ERE is responsible for E2-induced luciferase reporter activity from the human NRF-1 promoter using endogenous ER in MCF-7 cells.
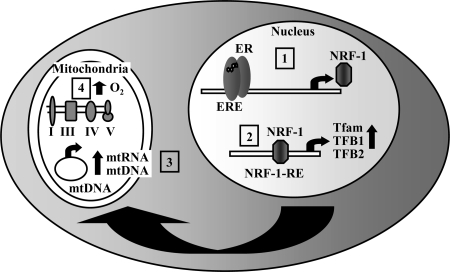
In summary, we have identified NRF-1 as a primary estrogen target gene regulated by direct ER interaction with an ERE in the promoter. The E2-stimulated increase in NRF-1 was followed in a time-dependent manner by increases in NRF-1 regulated Tfam mRNA transcription and then, in turn, by Tfam-regulated COI and NDI mRNA expression. In concert with these changes in mRNA and protein expression, oxygen consumption and mtDNA copy number were increased by E2, indicating an increase in mitochondrial biogenesis. Together these results suggest a mechanism to explain, in part, an important role for E2 in up-regulating mitochondrial activity in estrogen-responsive cells (Fig. 7). These data with the work of other investigators may contribute an explanation for the observed E2-mediated decrease in mitochondrial ROS production and the well-established enhanced longevity in females in most mammalian species (1,2).
Figure 7.
Model of E2-Induced Mitochondrial Activity via Up-Regulation of NRF-1 Expression
1) E2-activated ER binds to the ERE in the NRF-1 gene promoter leading to an increase in NRF-1 gene transcription and protein expression. 2) NRF-1 subsequently binds to its response elements and increases the transcription of its nuclear-encoded target genes including the mitochondrial transcription factors Tfam and TFBs (TF1M and TFB2M). 3) Tfam and TFBs are imported into mitochondria where Tfam increases mtDNA replication. Tfam and TFBs increase transcription from the mtDNA. 4) The resulting increase in MRC proteins leads to an increase in O2 consumption through complex IV.
MATERIALS AND METHODS
Chemicals
E2, 4-hydroxytamoxifen, PD98059, wortmannin, ActD, and CHX were purchased from Sigma-Aldrich (St. Louis, MO). PPT, DPN, and ICI 182,780 were purchased from Tocris (Ellisville, MO).
Antibodies
Antibodies were purchased as follows: ERα (HC-20) and ERβ (H-150) from Santa Cruz Biotechnology (Santa Cruz, CA), ERβ (PA1-311 and MA1-23217) from Affinity Bioreagents (Golden, CO), NRF-1 from Rockland Scientific (Gilbertsville, PA), COI and COIV from Mitoscience (Eugene, OR), ERα (AER320) and α-tubulin from NeoMarkers (Freemont, CA), and PDI and β-actin from Sigma-Aldrich.
Cell Culture and Treatment
MCF-7 and H1793 cells were purchased from American Type Culture Collection (Manassas, VA). MCF-7 cells and H1793 cells were maintained as described (38). Before treatment, the cells were placed in phenol red-free media supplemented with 5% dextran-coated charcoal stripped fetal bovine serum (DCC-FBS) for 72–96 h. EtOH was used as the vehicle control for all experiments. Cells were treated with various concentrations of E2 as indicated in the figures. For the indicated experiments, MCF-7 cells were pretreated with 100 nm ICI 182,780 for 6 h before E2 treatment. For other experiments, cells were pretreated for 1 h with 10 μg/ml ActD, 10 μg/ml CHX, 50 μm PD98059, or 50 nm wortmannin before E2 treatment for 4 h.
RNA Isolation, RT-PCR, and QRT-PCR
RNA was extracted using Trizol (Invitrogen, Carlsbad, CA). The high-capacity cDNA archive kit (PE Applied Biosystems, Foster City, CA) was used to reverse transcribe total RNA using random hexamers. TaqMan primers and probes for NRF-1, Tfam, and 18S rRNA were purchased as Assays-on-Demand Gene Expression Products (PE Applied Biosystems). Measurement of COI, NDI, and 18S using SYBR Green (PE Applied Biosystems) was performed as previously described (57). The expression of each target gene was determined in triplicate in three to six separate experiments and normalized using 18S. QRT-PCR for NRF-1 and 18S Assays-on-Demand was performed in the ABI PRISM 7900 Sequence Detection System 2.1 (PE Applied Biosystems) using relative quantification. QRT-PCR for COI and 18S was performed using absolute quantification. Analysis and fold differences were determined using the comparative cycle threshold (CT) method. Fold change was calculated from the ΔΔCT values with the formula 2−ΔΔCT, and data are presented as relative to expression in EtOH-treated cells.
Protein Isolation
Whole-cell extracts (WCE) were collected in 300 μl modified radioimmunoprecipitation buffer (as described in Ref. 38), and membrane preparations were prepared as described previously (58,59). Protein concentrations were determined using DC Protein Assay (Bio-Rad Laboratories, Hercules, CA).
siRNA Knockdown
For silencing of ERα and ERβ, siRNA knockdown was performed in MCF-7 cells using siRNA duplexes obtained from either New England Biolabs (ERα ShortCut siRNA Mix, i.e. a heterogeneous mixture of 21–23 bp siRNA for ERα) or Invitrogen (ESR2 Stealth Select 3 RNAi). Control siRNA from each company was used in parallel with the ER-subtype-specific siRNA. The cells were transfected with the recommended reagent according to the manufacturer’s protocol. Twenty-four hours after transfection, the medium was replaced with phenol red-free Iscove’s MEM (IMEM) with 5% DCC-FBS, and 24 h later (48 h after transfection with siRNA), the cells were treated with 10 nm E2 or EtOH for 4 h, and total RNA or WCE was harvested.
For knockdown of NRF-1, MCF-7 cells were cultured in six-well plates until reaching 60% confluency. The cells were transfected with siRNA targeting NRF-1 (Santa Cruz Biotechnology) according to the manufacturer’s protocol. Forty-eight hours after transfection, the cells were treated with 10 nm E2 for 48 h, and then cells were harvested for RNA, WCE, and genomic DNA as described above.
Western Blot Analysis
Protein (20–40 μg) was separated on 8–15% polyacrylamide SDS gels and electroblotted onto polyvinylidene difluoride membranes with data quantitated as previously described (38). Un-Scan-It (Silk Scientific, Orem, UT) was used to quantitate the integrated OD (IOD) for each band. The IOD for NRF-1 was divided by the concordant α-tubulin or β-actin IOD in the same blot. The EtOH value was set to 1 for comparisons.
Cloning of Human NRF-1 Promoter and Deletion Constructs for Luciferase Reporter Assays
Primers were designed to the −1100-bp region of the NRF-1 promoter using Primer3 (forward GAGTCTCCTGACGACACTAA, reverse GAAGTTCTACTCAGAGCGGC). The −1100-bp fragment of hNRF-1 (NM_005011) was cloned from human genomic DNA (Roche, Indianpolis, IN) into the pCR-Blunt II-Topo vector (Invitrogen). After sequencing, restriction digestion (XhoI/SacI) was used to subclone the −1100 promoter fragment into the pGL2-basic-luciferase vector (Promega, Madison, WI). Primers were then designed to clone two smaller fragments of the NRF-1 promoter at approximately 300-bp intervals. The primers designed resulted in a −677-bp fragment (forward TATCTGGCACAGCACGAGAC, reverse GAAGTTCTACTCAGAGCGGC) and a −378 fragment (forward CGCCGCTTCTCCGGGGCGTC, reverse GAAGTTCTACTCAGAGCGGC). Primers were also designed to clone the 200-bp region surrounding the putative NRF-1 ERE (forward GAGTCTCCTGACGACACTAA, reverse TAACGGTGGAGACACACGAG). The −677-, −378-, and 200-bp fragments were inserted into the pCR-Blunt II-Topo vector, sequenced, and subsequently digested with XhoI and SacI and placed into the pGL2-basic-luciferase plasmid. The plasmids are named after their corresponding fragments as noted in Fig. 3A.
The pGL2-NRF-1-(−1100MUT) mutant ERE was generated using the QuikChange Mutagenesis Kit (Stratagene, La Jolla, CA) according to the manufacturer’s protocol using the following primers: forward CCCCTCTGCCTGGCCTGGAGTCCTGTAAT and reverse GCCTCCCAAAGTGCTGGTATTACAGGA. The primers introduced two base pair changes into the 5′ half-site and one base pair change in the 3′ half-site, abolishing the putative palindromic ERE present in the NRF-1 promoter. Presence of the correct mutation was confirmed by sequencing.
Luciferase Assays
For luciferase assays, cells were plated in 24-well plates at a density of 1.5 × 104 cells per well in phenol red-free OPTI-MEM I reduced serum medium (Invitrogen) supplemented with 10% DCC-FBS, 1% penicillin/streptomycin. Transient transfection was performed using FuGene6 (Roche). Each well received 250 ng of a pGL2-pro-luciferase reporter (Promega) and 5 ng of a Renilla luciferase reporter (pRL-tk) from Promega. Twenty-four hours after transfection, triplicate wells were treated with EtOH (vehicle control) or 10 nm E2. The cells were harvested 30 h after treatment using Promega’s Passive Lysis buffer. Luciferase and Renilla luciferase activities were determined using Promega’s Dual Luciferase assay in a Plate Chameleon luminometer (BioScan, Washington, DC) (38). Firefly luciferase was normalized by Renilla luciferase to correct for transfection efficiency. Fold induction was determined by dividing the averaged normalized values from each treatment by the EtOH value for each transfection condition within that experiment. Values were averaged from multiple experiments as indicated in the figure legend.
ChIP Assay
MCF-7 cells were transferred to phenol red-free IMEM with 10% DCC-FBS 72 h before treatment. ChIP assays were performed as described previously (56). In brief, 4 × 106 cells per immunoprecipitation were treated with 2.5 μm α-amanitin for 2 h. The medium was then removed and replaced with medium containing 10 nm E2 or EtOH for 60 min. Chromatin was cross-linked using 1.5% formaldehyde for 5 min at 37 C, and the cells were collected after two washings with PBS. Subsequent incubation and washes were completed as described previously (56). The extracts were incubated with anti-ERα antibody (HC-20), anti-ERβ (H-150) antibody, or normal rabbit IgG (all from Santa Cruz). After elution of the antibody-protein complexes for 30 min in 1% SDS with 0.1 m NaHCO3, the DNA was purified using PCR Clean-Up Kit (QIAGEN, Valencia, CA) and probed for target sequences. Primers for the putative NRF-1 ERE probed the −761- to −1032-bp region, i.e. forward primer GGTCCCAGGACTCAAAACAA and reverse primer CAGGTGCCTGAGAAGTAGGG. As a positive control, primers flanking the established ERE in the human pS2 (Trefoil Factor 1; TFF1) gene promoter (56) were used. As a negative control, primers were designed to a distal region (−3060 to −3208) of the NRF-1 promoter, forward primer ACCCTTGTGGAAACAGCATC and reverse primer AAACGGACTGGGCTTACCTT.
QRT-PCR with ChIP Samples
ChIP was performed as described above. QRT-PCR of 3 μl purified immunoprecipitated DNA was done with the primers for NRF-1 and pS2 listed above using SYBR Green Master Mix. Relative promoter enrichment compared with IgG is plotted (60).
ERα and ERβ Proteins
Recombinant human ERα (61) and Flag-tagged recombinant human ERβ1 (62) were expressed by baculovirus infection of Sf21 cells. The concentrations of ERα and ERβ were determined by quantitating specific [3H]E2 binding in a hydroxyapatite assay (63) and were 150 and 95 fmol/μg protein for ERα and ERβ, respectively.
EMSA and ER-NRF-1-ERE Ki Determination
The DNA oligonucleotide sequences used in EMSA were as follows: putative NRF1-ERE from the human NRF-1 promoter 5′-GGAAGCCGGGCATGGTGTCCTGT-3′ and a consensus EREc38 (64). The ERE is underlined with nonconsensus ERE nucleotides in bold. EMSA was performed and quantitated in a Packard Instruments Instant Imager and with Packard Imager for Windows version 2.04 as previously described (65).
To measure the Ki in competition assays, reactions included 1.2 nm ERα or 1.5 nm ERβ, 1.1 nm [32P]EREc38 (5′-CCAGGTCAGAGTGACCTGAGCTAAAATAACACATTCAG-3′) (64), 100 nm E2, and 0.5–20,000 nm unlabeled NRF-1-ERE (66). The concentrations of free and ER-bound [32P]EREc38 were fit to the one-site binding model (determination coefficient R2 > 0.93 and 0.98 for ERα and ERβ without ligand and 0.97 and 0.94 with E2). The IC50 was determined from the Pseudo-Hill plot: log %/(100 − %) = nlog{[I] + nlogIC50, where % = percent competition of specific binding and I = competitor. The Ki was calculated from the IC50, using the equation of Cheng and Prusoff (67).
Mitochondrial Biogenesis
Semiquantitative PCR analysis of mtDNA relative to the 18S rRNA gene (nuclear encoded) was one measure of mitochondrial biogenesis in 10 nm E2-treated MCF-7 cells. DNA was purified using the Wizard Genomic DNA Isolation Kit (Promega), and 1 ng total DNA was subjected to PCR for a 282-bp segment of the mtDNA (6115–6396) and a 150-bp segment of the 18S gene for 25 cycles. The products were resolved on a 2% agarose gel, and quantitation was performed as described previously (68).
Oxygen consumption was measured using 5 × 106 cells suspended in a total volume of 0.5 ml IMEM containing 5% DCC-FBS using a Strathkelvin Instruments Electrode. Rates of O2 consumption were calculated using 782 System version 3.0 (Strathkelvin Instruments, Glasgow, UK) from four to six replicates.
Statistics
Statistical analyses were performed using GraphPad Prism (San Diego, CA). Two-tailed Student’s t tests were used for the pair-wise comparison of experimental groups. Statistical significance was defined at ≥95% confidence interval or P ≤ 0.05. In each figure, asterisks indicate significantly different from EtOH (P < 0.05). Bar graphs represent the mean ± sem for the number of independent experiments indicated in each figure legend.
Supplementary Material
Acknowledgments
We thank Drs. Barbara J. Clark and John W. Eaton for their suggestions for this manuscript and Joshua Thornburg and Kristen Nelson for their assistance with the oxygen consumption experiments. We thank Jacque H. Caruthers for assistance with the karyotyping and cytogenetic analysis of the MCF-7 cells used in these experiments.
Footnotes
This work was supported by NIH Grant R01 DK53220 (to C.M.K.) and American Heart Association predoctoral fellowship 315087B (to K.A.M.).
Disclosure Statement: The authors have nothing to disclose.
First Published Online November 29, 2007
Abbreviations: ActD, Actinomycin D; ChIP, chromatin immunoprecipitation assay; COI, cytochrome c oxidase subunit I (complex IV, subunit I); COIV, cytochrome c oxidase subunit IV (complex IV, subunit IV); CHX, cycloheximide; CT, cycle threshold; DCC-FBS, dextran-coated charcoal stripped fetal bovine serum; DPN, diarylpropionitrile; E2, estradiol; ER, estrogen receptor; ERE, estrogen receptor element; ERKO, ERα knockout; EtOH, ethanol; IOD, integrated OD; IMEM, Iscove’s MEM; MRC, mitochondrial respiratory chain; mtDNA, mitochondrial DNA; NDI, NADH dehydrogenase subunit I; NRF-1, nuclear respiratory factor 1; Ovx, ovariectomized; PDI, protein disulfide isomerase; PI3K, phosphoinositide-3 kinase; PPT, propyl pyrazole triol; QRT-PCR, quantitative real-time PCR; ROS, reactive oxygen species; R,R-THC, R,R-tetrahydrochrysene; siRNA, small interfering RNA; Tfam, mitochondrial transcription factor A; TFBs, mitochondrial transcription factor B types 1 and 2; WCE, whole-cell extracts.
References
- Borras C, Sastre J, Garcia-Sala D, Lloret A, Pallardo FV, Vina J 2003 Mitochondria from females exhibit higher antioxidant gene expression and lower oxidative damage than males. Free Radic Biol Med 34:546–552 [DOI] [PubMed] [Google Scholar]
- Vina J, Borras C, Gambini J, Sastre J, Pallardo FV 2005 Why females live longer than males: control of longevity by sex hormones. Sci Aging Knowledge Environ 2005:pe17 [DOI] [PubMed] [Google Scholar]
- Kuiper GG, Carlsson B, Grandien J, Enmark E, Haggblad J, Nilsson S, Gustafsson JA 1997 Comparison of the ligand binding specificity and transcript tissue distribution of estrogen receptors α and β. Endocrinology 138:863–870 [DOI] [PubMed] [Google Scholar]
- Pratt WB, Toft DO 1997 Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr Rev 18:306–360 [DOI] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, Fox EA, Silver PA, Brown M 2005 Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122:33–43 [DOI] [PubMed] [Google Scholar]
- Klinge CM 2001 Estrogen receptor interaction with estrogen response elements. Nucleic Acids Res 29:2905–2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paech K, Webb P, Kuiper GG, Nilsson S, Gustafsson J, Kushner PJ, Scanlan TS 1997 Differential ligand activation of estrogen receptors ERα and ERβ at AP1 sites. Science 277:1508–1510 [DOI] [PubMed] [Google Scholar]
- McKenna NJ, O’Malley BW 2002 Combinatorial control of gene expression by nuclear receptors and coregulators. Cell 108:465–474 [DOI] [PubMed] [Google Scholar]
- Nilsson S, Gustafsson JA 2002 Estrogen receptor action. Crit Rev Eukaryot Gene Expr 12:237–257 [DOI] [PubMed] [Google Scholar]
- Mueck AO, Seeger H, Lippert TH 1996 Calciumantagonistic effect of natural and synthetic estrogens: investigations on a nongenomic mechanism of direct vascular action. Int J Clin Pharmacol Ther 34:424–426 [PubMed] [Google Scholar]
- Goetz RM, Thatte HS, Prabhakar P, Cho MR, Michel T, Golan DE 1999 Estradiol induces the calcium-dependent translocation of endothelial nitric oxide synthase. Proc Natl Acad Sci USA 96:2788–2793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhat MY, Lavigne MC, Ramwell PW 1996 The vascular protective effects of estrogen. FASEB J 10:615–624 [PubMed] [Google Scholar]
- Kauser K, Rubanyi GM 1997 Potential cellular signaling mechanisms mediating upregulation of endothelial nitric oxide production by estrogen. J Vasc Res 34:229–236 [DOI] [PubMed] [Google Scholar]
- Razandi M, Pedram A, Greene GL, Levin ER 1999 Cell membrane and nuclear estrogen receptors (ERs) originate from a single transcript: studies of ERα and ERβ expressed in Chinese hamster ovary cells. Mol Endocrinol 13:307–319 [DOI] [PubMed] [Google Scholar]
- Bian Z, Nilsson S, Gustafsson JA 2001 Selective estrogen receptor modulators and coronary heart disease. Trends Cardiovasc Med 11:196–202 [DOI] [PubMed] [Google Scholar]
- Kelly MJ, Levin ER 2001 Rapid actions of plasma membrane estrogen receptors. Trends Endocrinol Metab 12:152–156 [DOI] [PubMed] [Google Scholar]
- Zhai P, Eurell TE, Cooke PS, Lubahn DB, Gross DR 2000 Myocardial ischemia-reperfusion injury in estrogen receptor-α knockout and wild-type mice. Am J Physiol Heart Circ Physiol 278:H1640–H1647 [DOI] [PubMed] [Google Scholar]
- Zhai P, Eurell TE, Cotthaus R, Jeffery EH, Bahr JM, Gross DR 2000 Effect of estrogen on global myocardial ischemia-reperfusion injury in female rats. Am J Physiol Heart Circ Physiol 279:H2766–H2775 [DOI] [PubMed] [Google Scholar]
- Wise PM 2002 Estrogens and neuroprotection. Trends Endocrinol Metab 13:229–230 [DOI] [PubMed] [Google Scholar]
- Yang SH, Liu R, Perez EJ, Wen Y, Stevens Jr SM, Valencia T, Brun-Zinkernagel AM, Prokai L, Will Y, Dykens J, Koulen P, Simpkins JW 2004 Mitochondrial localization of estrogen receptor β. Proc Natl Acad Sci USA 101:4130–4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedram A, Razandi M, Wallace DC, Levin ER 2006 Functional estrogen receptors in the mitochondria of breast cancer cells. Mol Biol Cell 17:2125–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JQ, Delannoy M, Cooke C, Yager JD 2004 Mitochondrial localization of ERα and ERβ in human MCF7 cells. Am J Physiol Endocrinol Metab 286:1011–1022 [DOI] [PubMed] [Google Scholar]
- Schwend T, Gustafsson JA 2006 False positives in MALDI-TOF detection of ERβ in mitochondria. Biochem Biophys Res Commun 343:707–711 [DOI] [PubMed] [Google Scholar]
- Yang SH, Prokai L, Simpkins JW 2006 Correspondence regarding Schwend and Gustafsson, “False positives in MALDI-TOF detection of ERβ in mitochondria”. Biochem Biophys Res Commun 345:917–918 [DOI] [PubMed] [Google Scholar]
- Evans MJ, Scarpulla RC 1989 Interaction of nuclear factors with multiple sites in the somatic cytochrome c promoter. Characterization of upstream NRF-1, ATF, and intron Sp1 recognition sequences. J Biol Chem 264:14361–14368 [PubMed] [Google Scholar]
- Virbasius CA, Virbasius JV, Scarpulla RC 1993 NRF-1, an activator involved in nuclear-mitochondrial interactions, utilizes a new DNA-binding domain conserved in a family of developmental regulators. Genes Dev 7:2431–2445 [DOI] [PubMed] [Google Scholar]
- Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, Spiegelman BM 1999 Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 98:115–124 [DOI] [PubMed] [Google Scholar]
- Vercauteren K, Pasko RA, Gleyzer N, Marino VM, Scarpulla RC 2006 PGC-1-related coactivator: immediate early expression and characterization of a CREB/NRF-1 binding domain associated with cytochrome c promoter occupancy and respiratory growth. Mol Cell Biol 26:7409–7419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly DP, Scarpulla RC 2004 Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev 18:357–368 [DOI] [PubMed] [Google Scholar]
- Scarpulla RC 2002 Nuclear activators and coactivators in mammalian mitochondrial biogenesis. Biochim Biophys Acta 1576:1–14 [DOI] [PubMed] [Google Scholar]
- Huo L, Scarpulla RC 2001 Mitochondrial DNA instability and peri-implantation lethality associated with targeted disruption of nuclear respiratory factor 1 in mice. Mol Cell Biol 21:644–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron R, Ren JM, Cadman KS, Moore IK, Perret P, Pypaert M, Young LH, Semenkovich CF, Shulman GI 2001 Chronic activation of AMP kinase results in NRF-1 activation and mitochondrial biogenesis. Am J Physiol Endocrinol Metab 281:E1340–E1346 [DOI] [PubMed] [Google Scholar]
- Murakami T, Shimomura Y, Yoshimura A, Sokabe M, Fujitsuka N 1998 Induction of nuclear respiratory factor-1 expression by an acute bout of exercise in rat muscle. Biochim Biophys Acta 1381:113–122 [DOI] [PubMed] [Google Scholar]
- Xia Y, Buja LM, Scarpulla RC, McMillin JB 1997 Electrical stimulation of neonatal cardiomyocytes results in the sequential activation of nuclear genes governing mitochondrial proliferation and differentiation. Proc Natl Acad Sci USA 94:11399–11404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Pena A, Escriva H, Handler AC, Vallejo CG 2002 Thyroid hormone increases transcription of GA-binding protein/nuclear respiratory factor-2 α-subunit in rat liver. FEBS Lett 514:309–314 [DOI] [PubMed] [Google Scholar]
- Stirone C, Duckles SP, Krause DN, Procaccio V 2005 Estrogen increases mitochondrial efficiency and reduces oxidative stress in cerebral blood vessels. Mol Pharmacol 68:959–965 [DOI] [PubMed] [Google Scholar]
- Chen JQ, Yager JD 2004 Estrogen’s effects on mitochondrial gene expression: mechanisms and potential contributions to estrogen carcinogenesis. Ann NY Acad Sci 1028:258–272 [DOI] [PubMed] [Google Scholar]
- Dougherty SM, Mazhawidza W, Bohn AR, Robinson KA, Mattingly KA, Blankenship KA, Huff MO, McGregor WG, Klinge CM 2006 Gender difference in the activity but not expression of estrogen receptors α and β in human lung adenocarcinoma cells. Endocr Relat Cancer 13:113–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs KA, Wickramasinghe NS, Cochrum RK, Watts MB, Klinge CM 2006 Decreased chicken ovalbumin upstream promoter transcription factor II expression in tamoxifen-resistant breast cancer cells. Cancer Res 66:10188–10198 [DOI] [PubMed] [Google Scholar]
- Wijayaratne AL, McDonnell DP 2001 The human estrogen receptor-α is a ubiquitinated protein whose stability is affected differentially by agonists, antagonists, and selective estrogen receptor modulators. J Biol Chem 276:35684–35692 [DOI] [PubMed] [Google Scholar]
- Fuqua SA, Schiff R, Parra I, Friedrichs WE, Su JL, McKee DD, Slentz-Kesler K, Moore LB, Willson TM, Moore JT 1999 Expression of wild-type estrogen receptor β and variant isoforms in human breast cancer. Cancer Res 59:5425–5428 [PubMed] [Google Scholar]
- Stauffer SR, Coletta CJ, Tedesco R, Nishiguchi G, Carlson K, Sun J, Katzenellenbogen BS, Katzenellenbogen JA 2000 Pyrazole ligands: structure-affinity/activity relationships and estrogen receptor-α-selective agonists. J Med Chem 43:4934–4947 [DOI] [PubMed] [Google Scholar]
- Meyers MJ, Sun J, Carlson KE, Marriner GA, Katzenellenbogen BS, Katzenellenbogen JA 2001 Estrogen receptor-β potency-selective ligands: structure-activity relationship studies of diarylpropionitriles and their acetylene and polar analogues. J Med Chem 44:4230–4251 [DOI] [PubMed] [Google Scholar]
- Sun J, Meyers MJ, Fink BE, Rajendran R, Katzenellenbogen JA, Katzenellenbogen BS 1999 Novel ligands that function as selective estrogens or antiestrogens for estrogen receptor-α or estrogen receptor-β. Endocrinology 140:800–804 [DOI] [PubMed] [Google Scholar]
- Silberstein GB, Van Horn K, Hrabeta-Robinson E, Compton J 2006 Estrogen-triggered delays in mammary gland gene expression during the estrous cycle: evidence for a novel timing system. J Endocrinol 190:225–239 [DOI] [PubMed] [Google Scholar]
- Virbasius JV, Scarpulla RC 1994 Activation of the human mitochondrial transcription factor A gene by nuclear respiratory factors: a potential regulatory link between nuclear and mitochondrial gene expression in organelle biogenesis. Proc Natl Acad Sci USA 91:1309–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherf M, Klingenhoff A, Werner T 2000 Highly specific localization of promoter regions in large genomic sequences by PromoterInspector: a novel context analysis approach. J Mol Biol 297:599–606 [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan L, Scarpulla RC 1995 Structure, expression, and chromosomal assignment of the human gene encoding nuclear respiratory factor 1. J Biol Chem 270:18019–18025 [DOI] [PubMed] [Google Scholar]
- Quandt K, Frech K, Karas H, Wingender E, Werner T 1995 MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res 23:4878–4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry M, Nunez AM, Chambon P 1989 Estrogen responsive element of the human pS2 gene is an imperfectly palindromic sequence. Proc Natl Acad Sci USA 86:1218–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinge CM 2003 Estrogen response element sequence as an allosteric regulator of estrogen receptor action. ChemTracts Biochem Mol Biol 16:1–18 [Google Scholar]
- Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA 1998 Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet 18:231–236 [DOI] [PubMed] [Google Scholar]
- O’Lone R, Knorr K, Jaffe IZ, Schaffer ME, Martini PGV, Karas RH, Bienkowska J, Mendelsohn ME, Hansen U 2007 Estrogen receptors α and β mediate distinct pathways of vascular gene expression, including genes involved in mitochondrial electron transport and generation of reactive oxygen species. Mol Endocrinol 21:1281–1296 [DOI] [PubMed] [Google Scholar]
- Lim KT, Cosgrave N, Hill AD, Young LS 2006 Nongenomic oestrogen signalling in oestrogen receptor negative breast cancer cells: a role for the angiotensin II receptor AT1. Breast Cancer Res 8:R33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhang S, Safe S 2006 Activation of kinase pathways in MCF-7 cells by 17β-estradiol and structurally diverse estrogenic compounds. J Steroid Biochem Mol Biol 98:122–132 [DOI] [PubMed] [Google Scholar]
- Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F 2003 Estrogen receptor-α directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115:751–763 [DOI] [PubMed] [Google Scholar]
- Piechota J, Tomecki R, Gewartowski K, Szczesny R, Dmochowska A, Kudla M, Dybczynska L, Stepien PP, Bartnik E 2006 Differential stability of mitochondrial mRNA in HeLa cells. Acta Biochim Pol 53:157–168 [PubMed] [Google Scholar]
- Kulakosky PC, Klinge CM 2003 Maximizing production of estrogen receptor β with the baculovirus expression system. Biotechniques 34:334–338, 340–343 [DOI] [PubMed] [Google Scholar]
- Pitson SM, Moretti PA, Zebol JR, Lynn HE, Xia P, Vadas MA, Wattenberg BW 2003 Activation of sphingosine kinase 1 by ERK1/2-mediated phosphorylation. EMBO J 22:5491–5500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Matthews J, Tujague M, Wan J, Strom A, Toresson G, Lam EWF, Cheng G, Gustafsson JA, Dahlman-Wright K 2007 Estrogen receptor β2 negatively regulates the transactivation of estrogen receptor α in human breast cancer cells. Cancer Res 67:3955–3962 [DOI] [PubMed] [Google Scholar]
- Klinge CM, Studinski-Jones AL, Kulakosky PC, Bambara RA, Hilf R 1998 Comparison of tamoxifen ligands on estrogen receptor interaction with estrogen response elements. Mol Cell Endocrinol 143:79–90 [DOI] [PubMed] [Google Scholar]
- Klinge CM, Jernigan SC, Mattingly KA, Risinger KE, Zhang J 2004 Estrogen response element-dependent regulation of transcriptional activation of estrogen receptors α and β by coactivators and corepressors. J Mol Endocrinol 33:387–410 [DOI] [PubMed] [Google Scholar]
- Pavlik EJ, Coulson PB 1976 Hydroxylapatite “batch” assay for estrogen receptor: increased sensitivity over present receptor assays. J Steroid Biochem 7:357–368 [DOI] [PubMed] [Google Scholar]
- Klinge CM, Peale Jr FV, Hilf R, Bambara RA, Zain S 1992 Cooperative estrogen receptor interaction with consensus or variant estrogen responsive elements in vitro. Cancer Res 52:1073–1081 [PubMed] [Google Scholar]
- Ramsey TL, Risinger KE, Jernigan SC, Mattingly KA, Klinge CM 2004 Estrogen receptor β isoforms exhibit differences in ligand-activated transcriptional activity in an estrogen response element sequence-dependent manner. Endocrinology 145:149–160 [DOI] [PubMed] [Google Scholar]
- Tyulmenkov VV, Klinge CM 2001 A mathematical approach to predict the affinity of estrogen receptors α and β binding to DNA. Mol Cell Endocrinol 182:109–119 [DOI] [PubMed] [Google Scholar]
- Cheng Y, Prusoff WH 1973 Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 percent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol 22:3099–3108 [DOI] [PubMed] [Google Scholar]
- Sumanasekera WK, Zhao L, Ivanova M, Morgan DD, Noisin EL, Keynton RS, Klinge CM 2006 Effect of estradiol and dihydrotestosterone on hypergravity-induced MAPK signaling and occludin expression in human umbilical vein endothelial cells. Cell Tissue Res 324:243–253 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.