Summary
A novel cadmium-inducible gene, cdr-1, was previously identified and characterized in the nematode C. elegans and found to mediate resistance to cadmium toxicity. Subsequently, six homologs of cdr-1 were identified in C. elegans. Here we describe two homologs: cdr-4, which is metal-inducible, and cdr-6, which is non-inducible. Both cdr-4 and cdr-6 mRNAs contain open reading frames of 831 nt and encode predicted 32-kDa integral membrane proteins, which are similar to CDR-1. cdr-4 expression is induced by arsenic, cadmium, mercury, and zinc exposure as well as by hypotonic stress. In contrast, cdr-6 is constitutively expressed at a high level in C. elegans, and expression is not affected by these stressors. Both cdr-4 and cdr-6 are transcribed in post-embryonic pharyngeal and intestinal cells in C. elegans. In addition, cdr-4 is transcribed in developing embryos. Like CDR-1, CDR-4 is targeted to intestinal cell lysosomes in vivo. Inhibition of CDR-4 and/or CDR-6 expression does not render C. elegans more susceptible to cadmium toxicity; however, there is a significant decrease in their lifespan in the absence of metal. Although nematodes in which CDR-4 and/or CDR-6 expression is knocked-down accumulate fluid in the pseudocoelomic space, exposure to hypertonic conditions did not significantly affect growth or reproduction in these nematodes. These results suggest that CDR expression is required for optimal viability but does not function in osmoregulation.
Introduction
The transition metal cadmium is a pervasive and persistent environmental contaminant that is ranked in the top ten on the CERCLA Hazardous Substance Priority List 1. Exposure to cadmium is correlated with a variety of cytotoxic effects and human pathologies 2; 3; 4. To attenuate the cytotoxic effects, this metal can be detoxified by chelation, or exported from the cell or into lysosomes 5; 6; 7; 8. In addition, toxic by-products associated with cadmium exposure can be removed. Furthermore, metal-induced cellular damage can be repaired 9; 10; 11; 12; 13; 14.
Several genomic screens have been performed to identify cadmium responsive genes and cognate metal-responsive regulatory pathways 15; 16; 17; 18; 19. A cadmium-responsive gene, designated cdr-1, was identified and characterized from the nematode Caenorhabditis elegans (C. elegans) 20. CDR-1 is a hydrophobic, 32-kDa, lysosomal, integral membrane protein. The expression of CDR-1 is limited to the intestinal cells of the nematode throughout post-embryonic development. Northern blot, DNA microarray and RPA1 confirmed that cdr-1 transcription is induced >50-fold in response to cadmium exposure, but the steady state level of expression is not affected following exposure to other metals or stressors 19; 20. Inhibition of cdr-1 expression, using RNAi or a strain in which cdr-1 has been deleted (strain RB966; cdr-1 (ok863)), results in increased sensitivity to cadmium exposure, clearly demonstrating a role for this gene in the defense against cadmium-induced toxicity.
BLAST 21; 22 analysis of cdr-1 identified six homologs in C. elegans, designated cdr-2, cdr-3, cdr-4, cdr-5, cdr-6 and cdr-7 23. The seven homologs share a high level of both nucleotide and amino acid sequence identity. Gene expression analysis shows that cdr-1 and cdr-4 transcription is induced by cadmium, cdr-6 is constitutively expressed at a high level compared to the other homologs and it is not cadmium-inducible; and cdr-2, cdr-3, cdr-5 and cdr-7 are expressed at low levels in both control and cadmium-treated nematodes 23. To further characterize the cdr genes, cdr-4 (cadmium-inducible) and cdr-6 (constitutive) were examined. These genes were selected for further investigation because they had the highest levels of amino acid and DNA sequence identity with cdr-1 (∼66%), and they where the two most similar members of the CDR family. Phylogenetic analysis shows that CDR-4 and CDR-6 are most closely related, compared to the other members in the CDR family 23. Similar to cdr-1, the expression of cdr-4 and cdr-6 predominately occurs in the intestinal cells at all post-embryonic developmental stages. However, cdr-4 is also expressed in developing embryos and its transcription can be induced by arsenic, cadmium, mercury, zinc and hypotonic stress. In contrast, cdr-6 transcription is not induced by any of these stressors. Based on the phenotype of fluid accumulation observed when CDR-1 expression was attenuated, it was hypothesized that members of the CDR family may function in osmoregulation 20. Inhibition of cdr-4 and cdr-6 expression under hypertonic conditions did not significantly affect growth, reproduction or lifespan of C. elegans. These results indicate that CDR's are required for optimal viability but that they do not function in osmoregulation.
Experimental Procedures
Growth of C. elegans
The N2 Bristol strain of C. elegans was maintained at 20 °C on NGM agar plates seeded with E. coli strain OP50 as a food source 24. The cdr-4 knockout strain RB966 (cdr-4(ok863)) was obtained from the Caenorhabditis Genetics Center (University of Minnesota). To obtain large quantities of C. elegans, nematodes were grown in liquid S medium, collected, and stored as previously described 25. In the experiments in which nematodes were exposed to metals, they were incubated for 24 h in media supplemented with 100 μM NaAsO2, 100 μM CdCl2, 18 μM HgCl2, or 124 μM ZnCl2 26; 27.
RNA Isolation
Total RNA was isolated from mixed-stage populations of C. elegans exposed to metal, or control, non-exposed nematodes as previously described 20; 25. In the experiment in which nematodes were exposed to osmotic stress, nematodes were grown in S medium, and then transferred to S medium in which the sodium chloride concentration was reduced from 100 mM to 10 mM 28. Where indicated poly(A+) RNA was isolated using Oligotex mRNA Midi Kits following the manufacturer's instructions (Qiagen).
RNase Protection Assay
cDNAs for CDR-1, CDR-4, CDR-6, and myosin light chain loading control were prepared by reverse transcriptase-PCR and then inserted into pGEM-T as previously described 23. Sequences of the primers used in the PCR's, and characteristics of the products are described in Table 1.
TABLE 1.
Probes used in RNase Protection Assay
| Probe Name | PCR Primer Sequence (5′→3′) | Probe Length (bp) | |
|---|---|---|---|
| rpa-cdr-1 | 5′ | TAAAATTCCGTCGCTTCC | 271 |
| 3′ | TCTGTGAAGAATCTCGTCGAGC | ||
| rpa-cdr-4 | 5′ | GTGATCTCTTCAACTTGC | 275 |
| 3′ | AACATCGGTCAGATGAGAACG | ||
| rpa-cdr-6 | 5′ | TCAACGCCGTATTGGTTCCTC | 278 |
| 3′ | CTCAAGAATTTTTGGGAAATCC | ||
| rpa-mlc | 5′ | TTGACAGGAACTGACCCAGAGG | 149 |
| 3′ | ATAGCCTTGACCTCATCCTCG | ||
To synthesize antisense RNA probes, plasmid DNA templates were linearized following digestion with either NcoI or NotI restriction enzymes. Biotin-labeled RNA probes were generated from the linearized plasmids using a MAXIscript™ in vitro Transcription Kit following the manufacturer's protocol (Ambion). In vitro transcription reactions contained SP6 RNA Polymerase when NcoI was used to linearize the template DNA or T7 RNA Polymerase when NotI was used to linearize the template DNA. Gel analysis and purification of probes were performed following the manufacturer's instructions, and the final concentration of each probe was determined from its absorbance at 260 nm.
RNase Protection Assays were performed using RPAIII™ Ribonuclease Protection Assay Kits according to the manufacturer's protocol (Ambion). In each reaction, 10 μg of total RNA from treated or control C. elegans was combined with 800 pg of gene specific probe and 400 pg of loading control probe. Hybridization reactions were incubated at 56 °C for 16 h, after which mixtures were incubated with RNases to degrade unhybridized RNAs. The protected fragments were resolved by 5% acrylamide/8 M urea denaturing polyacrylamide gel electrophoresis. Nucleic acids were transferred to BrightStar-Plus™ (Ambion) by electroblotting. Probes were visualized using BrightStar™ BioDetect™ Nonisotopic Detection Kit (Ambion). Steady-state levels of mRNA expression were normalized to those of the constitutively expressed myosin light chain mRNAs 29.
Preparation and Analysis of cdr-4/lacZ and cdr-6/lacZ Transgenic C. elegans
A ∼3.5-kb fragment of genomic DNA that is immediately 5'of the initiator ATG in cdr-4 was generated by PCR using cosmid K01D12 as template; the 5'-primer 5'CACTGGGCAACAACAAACGAT3' and the 3'-primer 5'GGACACCTCCGTCTACATTC3'. The PCR product was purified and then cloned into pGEM-T. The genomic fragment was excised from pGEM-T following digestion with SphI/SalI. This fragment was then inserted into the C. elegans β-galactosidase expression vector pPD95.03 (AddGene) that had been digested with identical enzymes. The resulting expression vector was designated pcdr-4/lacZ.
A ∼3.5-kb fragment of genomic DNA that is immediately upstream from the initiator ATG in cdr-6 was also generated by PCR using cosmid K01D12 as template, the 5'-primer 5'ACAGCAACACACGATTCTGG3', and the 3'-primer 5'GCCCGAATCTATCACCATTTTGC3'. The PCR product was purified and then inserted into pGEM-T. A cdr-6 expression vector, designated pcdr-6/lacZ was generated by inserting the genomic fragment into pPD95.03 as described above. The pcdr-4/lacZ and pcdr-6/lacZ expression vectors express a form of β-galactosidase that contains the SV40 large T antigen nuclear targeting sequence, which causes the protein to accumulate in nucleus 30.
Transgenic C. elegans were generated following microinjection of a mixture of pcdr-4/lacZ or pcdr-6/lacZ (100 μg/ml) and a plasmid containing the dominant selectable marker gene rol-6 (su1006) into the gonadal syncytia of young adult C. elegans. Transgenic C. elegans were selected and maintained, as described previously 25; 30.
Cell-specific, developmentally regulated patterns of cdr-4 and cdr-6 expression were determined by staining for β-galactosidase as previously described 25. Cells that actively transcribe cdr-4 or cdr-6 were identified from the level of β-galactosidase present in the nuclei.
In Vivo Intracellular Localization of CDR-4
To determine the intracellular location of CDR-4 in vivo, a C. elegans expression vector was constructed in which the expression of a CDR-4-eGFP fusion protein is regulated by a ∼3.5-kb cdr-4 promoter. To generate this vector, full-length cdr-4 cDNA was prepared by PCR using the 5'-primer 5'TCCCCCGGGTTTTCAAAATGGTTTG3', which introduces a SmaI site (underlined) into the reaction product, and the 3'-primer 5'CCGGTACCATAATTGAAATTGTAAAATCGTTAGG3', which introduces a KpnI site (underlined) and converts the stop codon (boldface) to a serine (TGA → TCA) in the reaction product. Following amplification, the DNA was inserted into pGEM-T. The cdr-4 cDNA was subsequently excised following incubation with SmaI and KpnI and then inserted into the C. elegans GFP expression vector pPD117.01 (AddGene), digested with the same enzymes. This product was then digested with SphI and SmaI to remove the mec-7 promoter fragment that is present in pPD117.01, and then a ∼3.5-kb cdr-4 promoter fragment was inserted. The resulting expression vector was designated pcdr-4/CDR-4-eGFP. Transgenic C. elegans that expressed pcdr-4/CDR-4-eGFP were generated as described above. The intracellular location of CDR-4-eGFP was determined by fluorescence microscopy. In the co-localization experiment in which the intestinal cell lysosomes were labeled with rhodamine, C. elegans were fed rhodamine-labeled RITC-dextran as previously described 20; 31. It has been demonstrated that C. elegans accumulate RITC-dextran in intestinal cell lysosomes 31.
RNA Interference
The full-length cDNA fragment of cdr-4 was generated by RT-PCR using the 5'-primer 5'ATGGTTTGTTGTTGCCCAGTG3' and the 3'-primer 5'CGTTAGGATAGATTTCTTTGCG3' and then inserted into pGEM-T. This plasmid was then digested with NcoI and PstI to excise the cdr-4 cDNA, which was then inserted into the double T7 RNA Polymerase vector pPD129.36 (AddGene), that had been digested with the identical restriction enzymes. The resulting plasmid was designated pCDR-4i.
The full-length cDNA fragment of CDR-6 was generated by RT-PCR using the 5' primer 5'ATGGTGTGTTGCTGTCCAG3' and the 3' primer 5'GATGGTGAAATCATTGGGG3' and then inserted into pGEM-T. CDR-6 cDNA was excised following digestion with NcoI and PstI. The fragment was then inserted into pPD129.36 digested with the identical restriction enzymes. The final RNAi vector was designated pCDR-6i. E. coli strain BL21 (DE3) was transformed with the dsRNA expression vectors pCDR-4i and pCDR-6i 32; 33.
RNAi-mediated inhibition of CDR-4 or CDR-6 expression was accomplished by incubating C. elegans on either NGM plates supplemented with 1 mM isopropyl-1-thio-β-D-galactopyranoside seeded with non-transformed E. coli BL21(DE3) (control) or NGM plates supplemented with 1 mM isopropyl-1-thio-β-D-galactopyranoside and 50 μg/ml ampicillin seeded with pCDR-4i- or pCDR-6i-transformed BL21(DE3) 34. L4 nematodes were placed onto the three types of plates and incubated at 22 °C for 40 h. Subsequently, three of the offspring were isolated, and individually cultured and allowed to lay eggs at 22 °C for 24 h before being removed 34. Progeny (F2) were cultured at 22 °C for another 48 h on RNAi plates before lifespan and other RNAi phenotypes were characterized.
Reproduction and Growth Assays
Nematodes were transferred to the sample cup of a COPAS BIOSORT 35 (Union Biometrica Inc., Somerville, MA) and diluted to a concentration of ∼1 nematode/μL. For the reproduction assay, five L4 nematodes were added to each well of a 96-well plate, containing K-medium, chemical (if tested), and OP50 E. coli in a final volume of 50 μL. Nematodes were incubated at 20°C for 48 h, and then the total number of offspring was measured. For the growth assay, twenty-five L1 nematodes were added to each well of a 96-well plate, containing K-medium, chemical (if tested), and OP50 E. coli in a final volume of 50 μL. Nematodes were incubated for 72 h and then length and optical density of individual C. elegans were measured using the COPAS BIOSORT ReFLx. To assess the effects of hypotonic stress, nematodes were placed in wells containing deionized water and E. coli.
Lifespan assay
Lifespan studies were conducted after RNAi exposure to non-functional control, or CDR-4, CDR-6, or CDR-4 and CDR-6 combined. Four replicates of 15 L4 animals each were transferred for each of the RNAi treatments and incubated at 20°C as previously described 19. After 48 h, adults were transferred to fresh NGM plates supplemented with sodium chloride at 0, 100, or 200 mM. The final sodium chloride concentrations in the medium are 50, 150 and 250 mM. Nematodes were gently probed with a platinum wire to determine survival and then transferred every 24 h to fresh NGM plates with corresponding RNAi and salt treatments. Lifespan curves were plotted and median lifespans were calculated using GraphPad Prism software (GraphPad Software, San Diego, CA). Because the lifespans of nematodes not grown under hypertonic conditions were affected by RNAi treatment, proportional hazards regression was used to assess the differences in survival between different RNAi treatment groups.
Results
Sequence Analysis of cdr-4 and cdr-6
The full-length cDNA sequence of CDR-4 is 944 nt, and contains an 831 nt open reading frame. An initiator codon ATG (nt 23−25) lies within the context of the consensus C. elegans translation start site. The 5' end of the CDR-4 mRNA is trans-spliced with the 22-nt SL1 spliced leader. A 3'-untranslated region of 73 nt follows the translation stop codon (nt 856−858) and terminates with a poly(A) tail with a typical polyadenylation signal (AATAAA) (Fig. 1).
Figure 1. Sequences of cdr-4 cDNA and protein.
The nucleotide sequence of the cdr-4 cDNA is shown with the deduced amino acid sequence presented below the corresponding codons. The SL1 sequence is underlined, and the translation start and stop codons are presented in bold. The polyadenylation signal is identified with a double-underline.
The full-length cDNA sequence of CDR-6 is 916 nt and contains an 831 nt open reading frame. An initiator codon ATG (nt 2−4) lies within the context of the consensus C. elegans translation start site. The 5' end of the CDR-6 mRNA is also trans-spliced with the 22-nt SL1 spliced leader. A 3'-untranslated region of 62 nt follows the translation stop codon (nt 833−835), and terminates with a poly(A) tail with the typical polyadenylation signal (Fig. 2).
Figure 2. Sequences of cdr-6 cDNA and protein.
The nucleotide sequence of the cdr-6 cDNA is shown with the deduced amino acid sequence presented below the corresponding codons. The SL1 sequence is underlined, and the translation start and stop codons are presented in bold. The polyadenylation signal is identified with a double-underline.
The open reading frames of CDR-4 and CDR-6 are identical in length (277 amino acids) and are predicted to encode proteins with molecular masses of 32,033 and 31,650, respectively. Pairwise comparison of the deduced amino acid sequences of CDR-4 and CDR-6 shows a high level of identity, 83.39%. CDR-4 and CDR-6 amino acid sequences also have a high level of similarity with the other five members in the CDR gene family 23.
Both CDR-4 and CDR-6 are predicted to be highly hydrophobic, integral membrane proteins with two transmembrane spanning domains 23. PROSITE analysis 36 reveals that both CDR-4 and CDR-6 contain putative cAMP- and cGMP-dependent protein kinase phosphorylation sites, protein kinase C phosphorylation sites, casein kinase II phosphorylation sites, and tyrosine kinase phosphorylation sites. In addition, both are predicted to contain N-glycosylation and N-myristoylation sites (Table 2).
TABLE 2.
Predicted protein motifs
| Motif | Residues | |
|---|---|---|
| CDR-4 | CDR-6 | |
| cAMP- and cGMP-dependent protein kinase phosphorylation | KKDT (42−45) |
KKDT (42−45) |
| Protein kinase C phosphorylation | TIK (29−31) |
TIK (29−31) |
| SIK (215−217) |
SIK (215−217) |
|
| SVR (265−267) | ||
| Casein kinase II phosphorylation | SLPD (121−124) |
TEQE (124−127) |
| SASD (150−153) |
SSVD (150−153) |
|
| TPAD (227−230) |
TPTD (227−230) |
|
| Tyrosine kinase phosphorylation | KSASDSLY (149−156) |
KSSVDMFY (149−156) |
| N-glycosylation | NGTL (90−93) |
NGTL (90−93) |
| N-myristoylation | GQLATV (235−240) |
GLPSAF (164−169) |
| GQLASV (235−240) | ||
Sequence analysis of the proximal 1.0 kb of the 5'-flanking DNA of cdr-4 and cdr-6 identified several putative URE's that have been shown to affect the transcription of mammalian metal/stress-responsive genes: heat shock element (HSE), activation protein-1 (AP-1) binding site, and cyclic AMP regulatory element (CRE) 37; 38; 39. These URE's have also been shown to have similar functions in C. elegans 40; 41; 42. In addition, multiple GATA elements (see below) are present in the promoters of cdr-4 and cdr-6 (Table 3) 43; 44.
TABLE 3.
Putative upstream regulatory elements
| Element | Consensus sequence | Sequence (location)a | |
|---|---|---|---|
| cdr-4 | cdr-6 | ||
| Heat shock | NGAANNTTCNNGAAN | TGGAAATTCTCGAAG (−726 to −712) |
|
| Antioxidant Response | TGA(C/G)TCA | TGAGTAA (−153 to −147) |
TGGCTCA (−340 to −334) |
| TAACTCA (−378 to −372) |
TTACTCA (−865 to −859) |
||
| GATA | (A/T)GATA(A/G) | TGATAT (−288 to −283) |
AGATAG (−268 to −263) |
| CGATAG (−903 to −898) |
TGATAA (−438 to −433) |
||
| TGATAA (−899 to −894) |
|||
| TGATAT (−929 to −924) |
|||
| cAMP Response | TGACGTCA | TCACTTCA (−52 to −45) |
TAACGTCT (−83 to −76) |
| TGTAGTCA (−192 to −185) |
TTTCGTCA (−115 to −108) |
||
| TTCCGTCA (−539 to −532) |
TGAAGTGA (−472 to −465) |
||
| TGTCATCA (−886 to −879) |
|||
Positions are relative to the translation start site.
Effect of Metals on cdr-4 and cdr-6 Transcription
To determine the effects of various metals on the transcription of cdr-4 and cdr-6, C. elegans were exposed to arsenic, cadmium, mercury or zinc. Exposure to cadmium resulted in a 3.0-fold increase in the steady state level of cdr-4 mRNA, and caused a 1.7-fold reduction in the level of cdr-6 mRNA. In nematodes exposed to arsenic, mercury or zinc, the expression of cdr-4 increased 2.1-fold, 3.2-fold and 1.7-fold compared to that observed in control nematodes, respectively (Fig. 3). In contrast, the expression of cdr-6 decreased 1.3-fold, and 1.7-fold in response to arsenic and zinc exposure, and was unaffected by exposure to mercury (Fig. 3).
Figure 3. Effects of metals on cdr-4 and cdr-6 transcription.
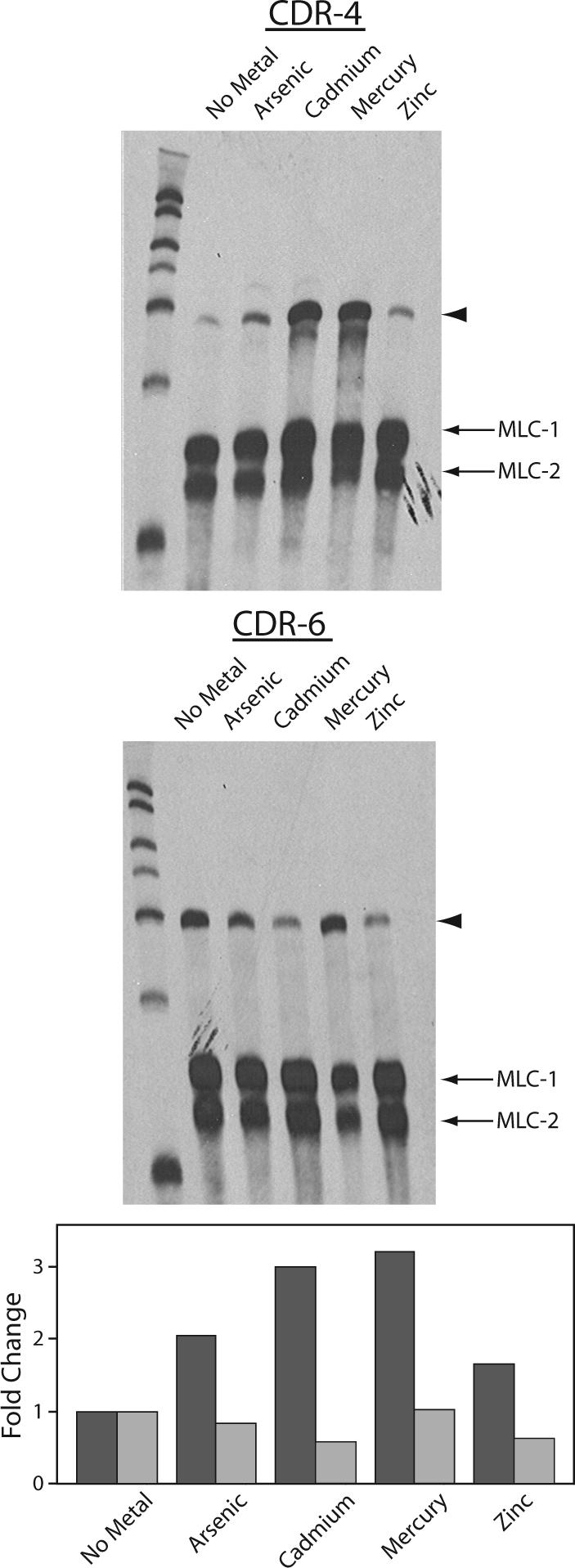
Total RNA was prepared from control, non-treated C. elegans and C. elegans exposed to 100 μM NaAsO2, 100 μM CdCl2, 18 μM HgCl2, or 124 μM ZnCl2 for 24 h. 10 μg of total RNA, 800 pg of a cdr-4 (upper) or cdr-6 (middle) mRNA specific probe, and 400 pg of myosin light chain 1 and 2 (MLC-1 and MLC-2), were hybridized in each reaction. The arrowhead indicates the position of the CDR target. The lower panel presents the fold change, relative to untreated controls, of cdr-4  and cdr-6
and cdr-6  mRNAs in response to metal treatment. The fold change was normalized to the level of myosin light chain mRNA.
mRNAs in response to metal treatment. The fold change was normalized to the level of myosin light chain mRNA.
Cell Specific and Developmental Expression of cdr-4 and cdr-6
The expression patterns of cdr-4 and cdr-6 were investigated in transgenic C. elegans containing cdr-4/lacZ or cdr-6/lacZ transgenes. Several independent lines of transgenic C. elegans containing the cdr-4/lacZ transgene were investigated. cdr-4 expression was observed in all of the intestinal cells of C. elegans in the absence of the exposure to any stressor (Fig. 4). This result is consistent with that observed by RPA in which cdr-4 gene was constitutively expressed in control C. elegans (Fig. 3). Expression was also observed in the terminal bulb and procorpus of the pharynx; likely in the pharyngeal muscle cells; pm3, pm5, pm6, and pm7. The highest level of cdr-4 transcription was observed in pm5, pm6 and pm7. cdr-4 transcription was observed in all post embryonic developmental stages and in developing embryos (Fig. 4). Although cdr-4 transcription is induced by cadmium (Fig. 3), the cell-specific, developmental pattern of expression of cdr-4 was not affected by metal exposure (data not shown).
Figure 4. Cell-specific and developmental expression of cdr-4.
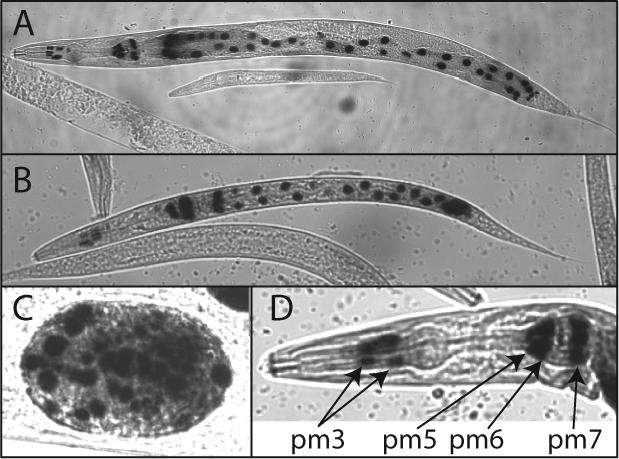
Transgenic C. elegans containing pcdr-4/lacZ were stained for β-galactosidase activity 25. The typical expression pattern of cdr-4 in an adult (panel A), L1 larva (panel B), and embryo (panel C) are shown. An enlarged view of an adult C. elegans pharynx with pharyngeal muscle nuclei identified (panel D).
In transgenic C. elegans containing pcdr-6/lacZ, cdr-6 promoter activity was observed in the intestinal cells, in the absence of the exposure to any stressor, similar to cdr-4 (Fig. 5). Cdr-6 expression was also observed throughout the pharynx: terminal bulb, isthmus, metacarpus and procorpus. Location of the stained nuclei suggests that pharyngeal muscle cells pm3, pm4, pm5 and pm6 express cdr-6. The highest levels of transcription were observed in pm3, pm4, and pm5. In contrast to cdr-4, cdr-6 expression was not observed in the pm7 cells. A low level of cdr-6 transcription was observed in several body wall muscle cells, located in the head and tail regions. Unlike cdr-4, cdr-6 transcription was not observed in developing embryos. Exposure to cadmium, zinc, mercury or arsenic did not affect the cellular patterns of expression of cdr-4 or cdr-6. Constant with the RPA results the amount of β-galactosidase in cdr-4 transcribing cells was greater in the metal-treated nematodes, compared to the non-exposed animals.
Figure 5. Cell-specific and developmental expression of cdr-6.

Transgenic C. elegans containing pcdr-6/lacZ were stained for β-galactosidase activity as previously described 25. The typical expression pattern of cdr-6 in an adult C. elegans (panel A), two, L1 larva (panel B), and an adult pharynx with the pharyngeal muscle cell nuclei identified (panel C).
CDR-4 Protein Localization In Vivo
To determine the intracellular location of CDR-4, transgenic C. elegans were generated that express a CDR-4-eGFP fusion protein whose expression is regulated by the cdr-4 promoter. The CDR-4-eGFP fusion protein was expressed predominately in the intestinal cells of the nematodes. It was concentrated in small punctate structures within these cells (Fig. 6). The size and distribution of these structures were similar to those of lysosomes and of CDR-1-eGFP 20; 31. It has been shown that when a non-fusion form of eGFP is expressed in intestinal cells, it accumulates in the cytoplasm, not in vesicles (unpublished observation). To confirm that CDR-4 was targeted to the lysosomes, the transgenic nematodes were fed RITC-labeled dextran, which labels intestinal cell lysosomes 31. In the double-labeling experiment, both eGFP and rhodamine co-localized to the same structures confirming that CDR-4 is targeted to intestinal cell lysosomes (Fig. 6).
Figure 6. Intracellular location of CDR-4 in vivo.
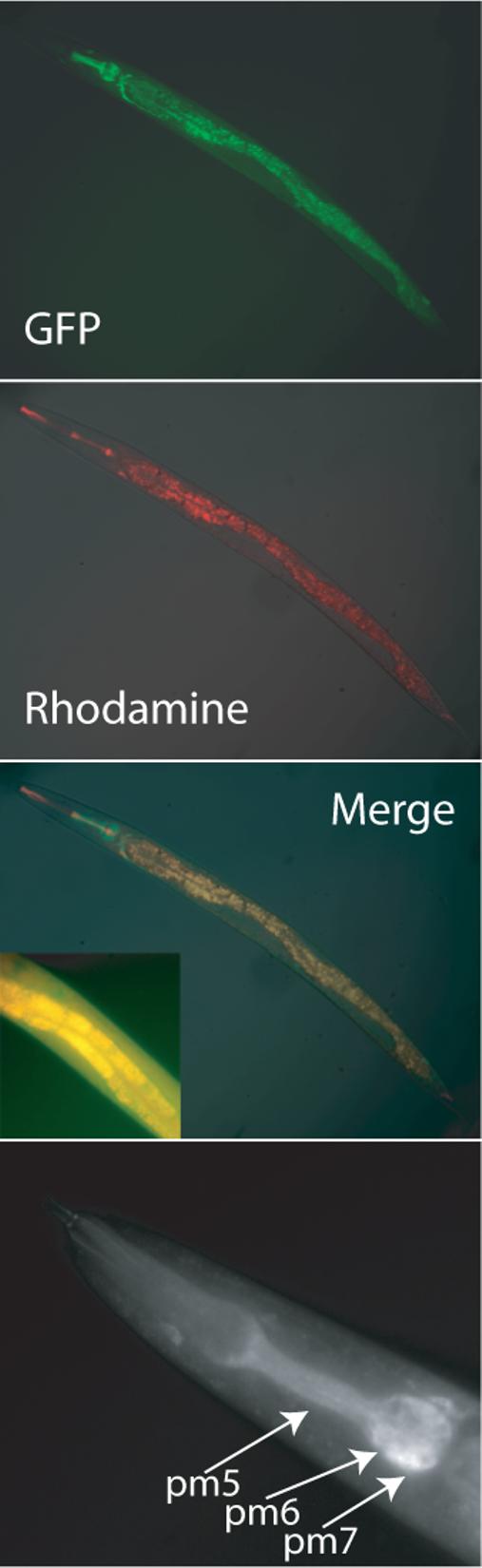
Transgenic C. elegans that contain a cdr-4/CDR-4-eGFP expression vector, were fed RITC-dextran. The location of CDR-4-eGFP (GFP) and RITC-labeled lysosomes (rhodamine) were visualized using the appropriate filter sets. A false-color, composite image (merge) is presented that demonstrates the localization of CDR-4 in lysosomes. A higher magnified (400×) image of the identical transgenic C. elegans is presented in the insert. Locations where the eGFP and rhodamine signals are coincident can be identified by the yellow color. The lower panel indicates the location of CDR-4-eGFP within the pharynx of an adult C. elegans. The location of pharyngeal muscles cells; pm5, pm6, and pm 7; are indicated. Pm5 cells are columnar cells, which extend from the anterior region of the terminal bulb to the posterior of the metacorpus 62.
Effect of Hypotonic Stress on cd-4 and cdr-6 expression
Since CDR-4 and CDR-6 are predicted to function in osmoregulation, the effects of hypotonic stress on the transcription of cdr-4 and cdr-6 were assessed. Exposure to hypotonic stress resulted in a 1.54-fold increase in the steady state level of cdr-4 mRNA. In contrast hypotonic stress did not have any effect on the steady-state level of cdr-6 relative to control. In addition cdr-1 mRNAs were undetectable in both control and hypotonic stressed C. elegans (Fig. 8).
Figure 8. Effects of hypotonic stress on cdr transcription.
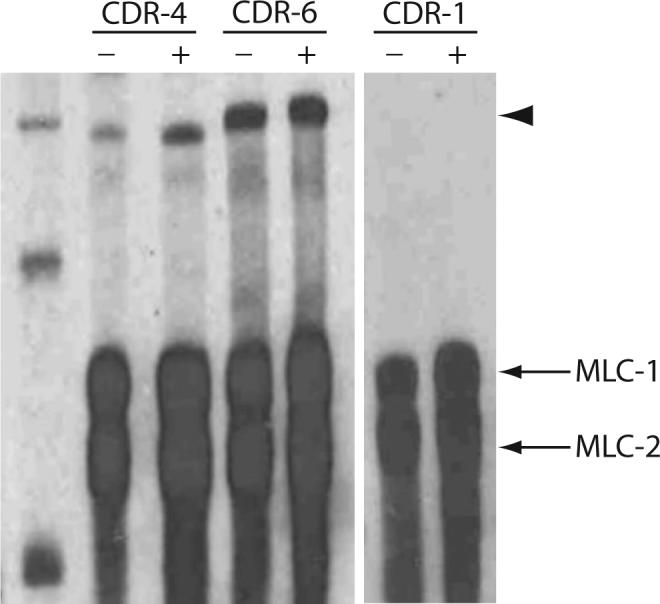
Total RNA was prepared from non-treated C. elegans (−) or C. elegans exposed to a hypotonic stress for 24 h (+). 10 μg of total RNA, 800 pg of cdr-1, cdr-4 or cdr-6 mRNA specific probe, and 400 pg of myosin light chains 1 and 2 (MLC-1 and MLC-2) probe were hybridized in each reaction. The arrowhead indicates the position of the protected product.
Functional Analysis of CDR-4 and CDR-6
The biological function of CDR-4 and CDR-6 was determined using RNAi. Inhibition of CDR-4 or CDR-6 expression resulted in the accumulation of fluid-filled droplets in the pseudocoelom and the tissues (Fig. 7). This phenotype was observed in the absence of metal exposure. Concomitant exposure to cadmium did not significantly affect the magnitude of the RNAi phenotype, nor did the simultaneous inhibition of CDR-4 and CDR-6 expression (results not shown).
Figure 7. Phenotype associated with RNAi of CDR-4 and CDR-6.
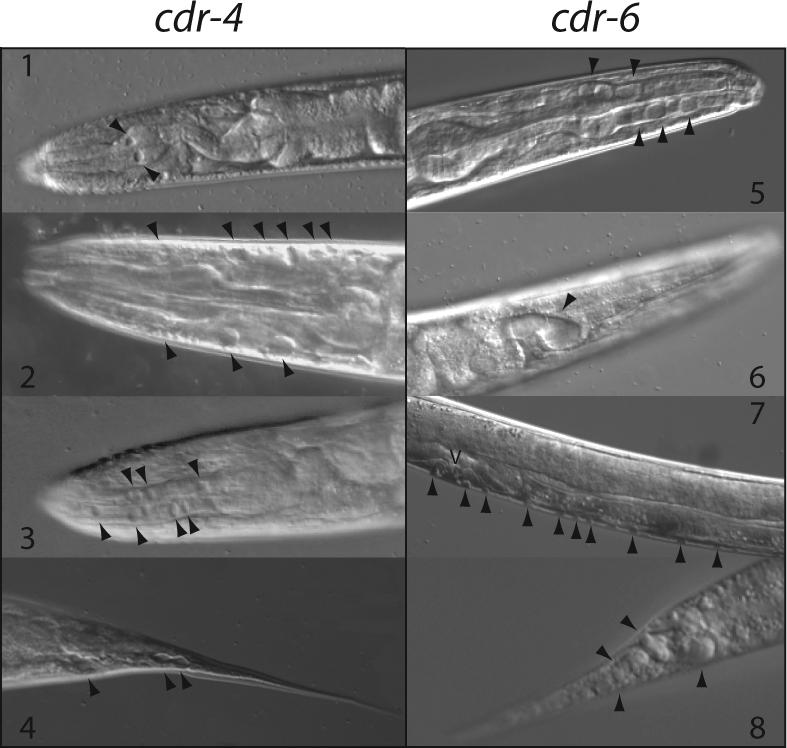
Wild type C. elegans were grown on NGM plates containing 1 mM isopropyl-1-thio-β-D-galactopyranoside and 50 μg/ml ampicillin, seeded with BL21(DE3) containing pCDR-4i (left) or pCDR-6i (right). Panels 1, 3 and 5, adult pharynx with vesicles in the pharyngeal muscle cells; Panels 2 and 6, adult pharynx with vesicle in the pseudocoelomic space between the pharynx and hypodermis; Panel 7, C. elegans larva with vesicles along the body wall in the pseudocoelomic space between the hypodermis and intestine; Panels 4 and 8, adult tail containing fluid filled vesicles. Arrowheads indicate the position of vesicles.
Two assays were used to assess potential differences in reproduction and growth between wild-type and cdr-4(ok863) mutants after exposure to cadmium, hypertonic or hypotonic stress. There were not significant differences between the wild type and the cdr-4 (ok863) nematodes, in the absence of stress. Furthermore, both strains responded similarly to all treatments (result not shown).
To further asses the effects the relation between the cdr genes and osmotic stress, C. elegans lifespan assays were performed. Nematodes grown under isotonic conditions and CDR-4 or CDR-6 RNAi alone had a decreased lifespan compared to the control vector (log rank test; p < 0.0001). Furthermore, the combination of both CDR RNAi's led to an even shorter lifespan than either RNAi treatment alone (Fig. 9). For these reasons, multiple comparisons were performed using proportional hazards regression. Hypertonic conditions led to decreased median lifespans regardless of RNAi treatment. However, when the differences in lifespan without salt treatment were taken into account, nematodes with knocked down cdr-4 and/or cdr-6 expression did not exhibit increased sensitivity in terms of decreased lifespans compared to isosmotic conditions (Fig 9). These results suggest that the CDR's are required for optimal viability but that they may not function in osmoregulation.
Figure 9. Effect of osmotic stress on C. elegans lifespan.
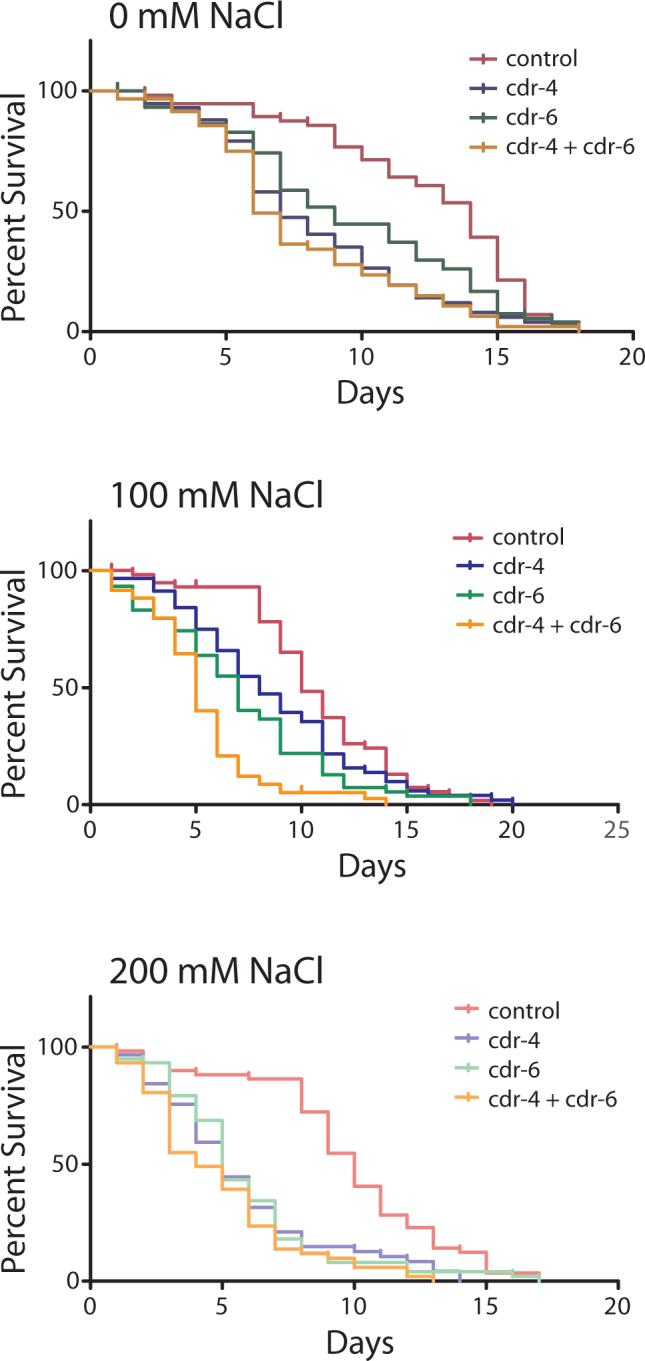
Wild type C. elegans were grown on NGM plates containing 0, 100, or 200 mM added sodium chloride. Plates contained bacteria that expressed, control, CDR-4, CDR-6 or a mixture of CDR-4 and CDR-6 dsRNA. Percent survival was calculated as indicated.
DISCUSSION
A novel cadmium-inducible gene, cdr-1, was identified during a genomic screen for cadmium-responsive genes in C. elegans. cdr-1 encodes a 32-kDa, integral membrane protein that is expressed exclusively in intestinal cells of post-embryonic nematodes following cadmium exposure. The CDR-1 protein is targeted to the intestinal cell lysosomes. The role of cdr-1 in cadmium detoxification was demonstrated when inhibition of its expression using RNAi, or with a cdr-1 null strain, made C. elegans more sensitive to cadmium toxicity. In addition, inhibition of CDR-1 expression causes the accumulation of fluid within the nematode. Therefore, CDR-1 is predicted to function in osmoregulation, possibly as a pump that can transfer ionic material across lysosomal membranes 20.
Six homologs of CDR-1 designated CDR-2, −3, −4, −5, −6 and −7 were identified and characterized 23. Among the seven genes, cdr-1 and cdr-4 transcription significantly increases following exposure to cadmium; however cdr-4 is also constitutively expressed. In contrast, cdr-6 is constitutively expressed in control nematodes, and it is not metal-inducible. Both cdr-4 and cdr-6 mRNAs contain open reading frames of 831 nt, and are trans-spliced with the 22-nt SL1 spliced leader RNA (Figs. 1 and 2). They provide predicted ∼32-kDa, integral membrane proteins with two transmembrane domains. In contrast, CDR-1 is predicted to contain four transmembrane spanning domains and is not trans-spliced. Like CDR-1, both CDR-4 and CDR-6 have multiple, potential post-translational modification sites (Table 3 and 20).
Although the primary and predicted structures of CDR-1, CDR-4, and CDR-6 are similar, there is considerable variation in the ability of different stressors to activate their transcription. Exposure to arsenic, cadmium, mercury and zinc increases the steady state level of cdr-4 mRNA (Fig. 3). This confirms that metals are strong activators of cdr-4 transcription. In contrast, cdr-6 expression is not affected by metal exposure (Fig. 3). Similarly, cdr-1 transcription increases in response to cadmium exposure, however, it is not responsive to other metals 20. The mechanism by which metal exposures induces cdr-1 and cdr-4 transcription remains to be resolved. Analysis of the 1.0-kb promoter regions of cdr-1, cdr-4 and cdr-6 identified consensus sequences for several stress-responsive URE's (Table 2 and 20). Although these genes contain similar URE's, their responsiveness to different environmental stressors is unique. Sequence comparisons among cdr-1, cdr-4, and the metal responsive genes mtl-1 and mtl-2 did not identify conserved DNA sequences, other that GATA elements. This suggests that multiple metal-regulatory processes may control the metal-inducible transcription in C. elegans. This is further supported by the observation that cadmium is the only metal capable of activating cdr-1 transcription, whereas multiple metals are able to affect cdr-4 transcription; similar to that observed for mtl-1 and mtl-2 27. The presence of multiple metal-responsive regulatory pathways has been observed in higher eukaryotes. For example, cadmium can induce transcription via MTF-1, NFκB, Heat shock factor, Nrf2 and by affecting MAPK signaling cascades 45; 46; 47; 48; 49. Detailed analysis of the regulatory regions of the cdr genes and comparisons with other stress responsive genes will reveal the mechanisms for constitutive, metal-inducible and cell-specific transcription.
CDR-1, CDR-4 and CDR-6 have similar primary and predicted structures, cellular expression patterns, and loss-of-function phenotypes; suggesting that they have comparable or redundant biological functions. Variations in their activity may be controlled by the ability of the cognate genes to be expressed in response to environmental stimuli or in specific cell types. For example cdr-1 transcription is induced only by cadmium, whereas cdr-4 transcription is affected by multiple environmental stressors, and cdr-6 is constitutively expressed at a level greater than any other member of the CDR family. Thus, the biological activity of CDR proteins may be controlled at the transcriptional level (i.e., their ability to be transcribed in response to specific biological stimuli or in unique patterns of cellular expression).
Studies using transgenic C. elegans confirm that both cdr-4 and cdr-6 are expressed in intestinal cells (Figs. 4, 5, and 6). This pattern is similar to that of cdr-1; other metal-responsive genes, mtl-1 and mtl-2 25; and several other non-stress responsive genes including the six vitellogenins, vit-1 through vit-6, the cysteine protease cpr-1 and the gut esterase ges-1 42; 50; 51. Intestinal cell-specific transcription in C. elegans is regulated by the binding of the transcription factor ELT-2 to GATA elements in the promoters of these genes 42; 43; 50; 51; 52; 53; 54; 55; 56. GATA-binding transcription factors constitute a family of structurally related proteins that are expressed in distinct developmental and tissue-specific patterns. Their involvement in the regulation of cell-specific transcription has been well established 57. Sequence analysis of the cdr-4 and cdr-6 promoters identified two and four GATA elements, respectively (Table 2). Further analysis of the GATA elements present in cdr-1, cdr-4 and cdr-6 will provide new insights into the mechanisms that govern the constitutive and inducible gene expression through GATA elements and factors in C. elegans.
Unlike cdr-1, cdr-4 and cdr-6 are also expressed in non-intestinal cells; specifically pharyngeal muscles cell (Figs. 4 and 5). cdr-4 and cdr-6 have overlapping patterns of cellular expression within the pharynx: pm3 and pm6. The major difference in pharyngeal expression is that cdr-4 is actively transcribed in the posterior region of the pharynx, in pharyngeal muscle cells pm6 and pm7, while cdr-6 is actively transcribed in the anterior region of the pharynx in cells pm3, pm 4 and pm5. Both cdr-4 and cdr-6 are expressed in the terminal bulb of the pharynx. In addition to muscle, the terminal bulb contains the dorsal and ventral gland cells. These cells contain vesicles and secrete digestive enzymes into the grinder of the pumping pharynx. In addition, the cells have processes that extend into the procorpus and isthmus 58; 59. The expression of cdr-4 and cdr-6 in the pharynx may be related to a biological activity, where these proteins transport material into the gland cells. The functional consequence of the distribution in of cdr-4 and cdr-6 expression remains to be resolved.
CDR-1 is targeted specifically to the intestinal cell lysosomes 20. Similarly, CDR-4 is also targeted to the intestinal cell lysosomes (Fig. 6). A transgene was generated in which the expression of a CDR-6-eGFP fusion protein is regulated by ∼3.5 kb of the cdr-6 promoter. Strains of C. elegans containing this transgene were not viable, which suggests that over-expression of CDR-6 may severely disrupt C. elegans metabolism. This response may not have been observed in CDR-1-eGFP and CDR-4-eGFP expressing transgenic nematodes because the basal levels of the endogenous proteins are significantly lower than that of CDR-6 23 (Fig.3 and 8).
A proposed biological role for CDR-1 was determined using RNAi 20; 23. Using identical protocols, the functions of CDR-4 and CDR-6 were investigated. Nematodes fed CDR-4 dsRNA, CDR-6 dsRNA, or a combination of CDR-4 and CDR-6 dsRNA proliferated and appeared to develop normally. This response was similar to that observed when CDR-1 expression was inhibited in C. elegans grown in the absence of cadmium 20; 23. In the absence of stress, there was a significant decrease in the lifespan of C. elegans fed CDR-4 and/or CDR-6 dsRNA. The addition of cadmium did not significantly enhance the decrease in lifespan. This is in contrast to the effect of inhibiting cdr-1 expression, where blocking cdr-1 expression made the nematodes more susceptible to cadmium toxicity.
Inhibiting cdr-4 or cdr-6 expression causes the nematodes to accumulate fluid-filled droplets in the pseudocoelom and tissues throughout the organism (Fig. 7). This phenotype is similar to that observed when the cells of the C. elegans secretory-excretory system were ablated 28. In addition, similar phenotypes have been observed in hyperactive egl-15 signaling and in clr-1 mutants 60. In all cases, it is proposed that disruption of the biological activity of these genes, or cognate signaling pathways, cause fluid imbalances or disruption in C. elegans osmoregulation. To assess the roles of cdr-4 and cdr-4 in osmoregulation, C. elegans in which the expression of these two genes was attenuated were grown under hypertonic conditions. The lack of response to hypertonic stress suggests that these genes may not be involved in the osmoregulation (Fig. 8),
The CDR family may be part of a new class of transmembrane proteins. Orthologs of the C. elegans cdr genes have been identified in other Caenorhabditi species: C. briggsae and C. remanei. Partial orthologs have also been tentatively identified in other invertebrates 61. To date, orthologs have not been identified in mammals. Further analysis will determine if CDR proteins are unique to lower organisms, or if they have evolved into more complex proteins, which are present in higher organisms.
Footnotes
* This work was supported (in part) by a National Institutes of Health Grant ES09949 (to J.H.F.) and the intramural research program of the NIH, National Institute of Environmental Health Sciences and the National Toxicology Program. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The abbreviations used are: cdr, cadmium-responsive gene; eGFP, enhanced green fluorescent protein; RITC, rhodamine B isothiocyanate; RNAi, RNA interference; RPA, RNase protection assay; and URE, upstream regulatory element.
References
- 1.Agency for Toxic Substances and Disease Registry . CERCLA Priority List of Hazardous Substances. Atlanta, GA: 2006. [Google Scholar]
- 2.Waalkes MP, Coogan TP, Barter RA. Toxicological principles of metal carcinogenesis with special emphasis on cadmium. Crit Rev Toxicol. 1992;22:175–201. doi: 10.3109/10408449209145323. [DOI] [PubMed] [Google Scholar]
- 3.Stohs SJ, Bagchi D. Oxidative mechanisms in the toxicity of metal ions. Free Radic. Biol. Med. 1995;18:321–336. doi: 10.1016/0891-5849(94)00159-h. [DOI] [PubMed] [Google Scholar]
- 4.Koizumi T, Li ZG. Role of oxidative stress in single-dose, cadmium-induced testicular cancer. J. Toxicol. Environ. Health. 1992;37:25–36. doi: 10.1080/15287399209531654. [DOI] [PubMed] [Google Scholar]
- 5.Broeks A, Gerrard B, Allikmets R, Dean M, Plasterk RH. Homologues of the human multidrug resistance genes MRP and MDR contribute to heavy metal resistance in the soil nematode Caenorhabditis elegans. EMBO J. 1996;15:6132–6143. [PMC free article] [PubMed] [Google Scholar]
- 6.Endo T. Transport of cadmium across the apical membrane of epithelial cell lines. Comp Biochem Physiol C Toxicol Pharmacol. 2002;131:223–9. doi: 10.1016/s1532-0456(02)00009-1. [DOI] [PubMed] [Google Scholar]
- 7.Havelaar AC, de Gast IL, Snijders S, Beerens CE, Mancini GM, Verheijen FW. Characterization of a heavy metal ion transporter in the lysosomal membrane. FEBS Lett. 1998;436:223–7. doi: 10.1016/s0014-5793(98)01133-8. [DOI] [PubMed] [Google Scholar]
- 8.Paulsen IT, Saier MH,, Jr. A novel family of ubiquitous heavy metal ion transport proteins. J. Membr. Biol. 1997;156:99–103. doi: 10.1007/s002329900192. [DOI] [PubMed] [Google Scholar]
- 9.Kagi JH, Schaffer A. Biochemistry of metallothionein. Biochemistry. 1988;27:8509–8515. doi: 10.1021/bi00423a001. [DOI] [PubMed] [Google Scholar]
- 10.Hartwig A, Schwerdtle T. Interactions by carcinogenic metal compounds with DNA repair processes: toxicological implications. Toxicol Lett. 2002;127:47–54. doi: 10.1016/s0378-4274(01)00482-9. [DOI] [PubMed] [Google Scholar]
- 11.Jungmann J, Reins HA, Schobert C, Jentsch S. Resistance to cadmium mediated by ubiquitin-dependent proteolysis. Nature. 1993;361:369–371. doi: 10.1038/361369a0. [DOI] [PubMed] [Google Scholar]
- 12.Lee MJ, Nishio H, Ayaki H, Yamamoto M, Sumino K. Upregulation of stress response mRNAs in COS-7 cells exposed to cadmium. Toxicology. 2002;174:109–17. doi: 10.1016/s0300-483x(02)00045-8. [DOI] [PubMed] [Google Scholar]
- 13.Gupta S, Athar M, Behari JR, Srivastava RC. Cadmium-mediated induction of cellular defence mechanism: a novel example for the development of adaptive response against a toxicant. Ind. Health. 1991;29:1–9. doi: 10.2486/indhealth.29.1. [DOI] [PubMed] [Google Scholar]
- 14.Kostic MM, Ognjanovic B, Dimitrijevic S, Zikic RV, Stajn A, Rosic GL, Zivkovic RV. Cadmium-induced changes of antioxidant and metabolic status in red blood cells of rats: in vivo effects. Eur. J. Haematol. 1993;51:86–92. doi: 10.1111/j.1600-0609.1993.tb01598.x. [DOI] [PubMed] [Google Scholar]
- 15.Tan Y, Shi L, Hussain SM, Xu J, Tong W, Frazier JM, Wang C. Integrating time-course microarray gene expression profiles with cytotoxicity for identification of biomarkers in primary rat hepatocytes exposed to cadmium. Bioinformatics. 2006;22:77–87. doi: 10.1093/bioinformatics/bti737. [DOI] [PubMed] [Google Scholar]
- 16.Zhou T, Jia X, Chapin RE, Maronpot RR, Harris MW, Liu J, Waalkes MP, Eddy EM. Cadmium at a non-toxic dose alters gene expression in mouse testes. Toxicol Lett. 2004;154:191–200. doi: 10.1016/j.toxlet.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 17.Regunathan A, Glesne DA, Wilson AK, Song J, Nicolae D, Flores T, Bhattacharyya MH. Microarray analysis of changes in bone cell gene expression early after cadmium gavage in mice. Toxicol Appl Pharmacol. 2003;191:272–93. doi: 10.1016/s0041-008x(03)00163-7. [DOI] [PubMed] [Google Scholar]
- 18.Yamada H, Koizumi S. DNA microarray analysis of human gene expression induced by a non-lethal dose of cadmium. Ind Health. 2002;40:159–66. doi: 10.2486/indhealth.40.159. [DOI] [PubMed] [Google Scholar]
- 19.Cui Y, McBride SJ, Boyd WA, Alper S, Freedman JH. Toxicogenomic analysis of Caenorhabditis elegans reveals novel genes and pathways involved in the resistance to cadmium toxicity. Genome Biol. 2007;8:R122. doi: 10.1186/gb-2007-8-6-r122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liao VH, Dong J, Freedman JH. Molecular characterization of a novel, cadmium-inducible gene from the nematode Caenorhabditis elegans. A new gene that contributes to the resistance to cadmium toxicity. J Biol Chem. 2002;277:42049–59. doi: 10.1074/jbc.M206740200. [DOI] [PubMed] [Google Scholar]
- 21.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 22.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong J, Song MO, Freedman JH. Identification and characterization of a family of Caenorhabditis elegans genes that is homologous to the cadmium-responsive gene cdr-1. Biochim Biophys Acta. 2005;1727:16–26. doi: 10.1016/j.bbaexp.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 24.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freedman JH, Slice LW, Dixon D, Fire A, Rubin CS. The novel metallothionein genes of Caenorhabditis elegans. Structural organization and inducible, cell-specific expression. J. Biol. Chem. 1993;268:2554–2564. [PubMed] [Google Scholar]
- 26.Stringham EG, Candido EPM. Transgenic hsp16-lacZ strains of the soil nematode Caenorhabditis elegans as biological monitors of environmental stress. Environ. Toxicol. Chem. 1994;13:1211–1220. [Google Scholar]
- 27.Cioci LK, Qiu L, Freedman JH. Transgenic strains of the nematode Caenorhabditis elegans as biomonitors of metal contamination. Environ. Toxicol. Chem. 2000;19:2122–2129. [Google Scholar]
- 28.Nelson FK, Riddle DL. Functional study of the Caenorhabditis elegans secretory-excretory system using laser microsurgery. J. Exp. Zool. 1984;231:45–56. doi: 10.1002/jez.1402310107. [DOI] [PubMed] [Google Scholar]
- 29.Cummins C, Anderson P. Regulatory myosin light-chain genes of Caenorhabditis elegans. Mol Cell Biol. 1988;8:5339–49. doi: 10.1128/mcb.8.12.5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mello C, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- 31.Clokey GV, Jacobson LA. The autofluorescent “lipofuscin granules” in the intestinal cells of Caenorhabditis elegans are secondary lysosomes. Mech. Ageing Dev. 1986;35:79–94. doi: 10.1016/0047-6374(86)90068-0. [DOI] [PubMed] [Google Scholar]
- 32.Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- 33.Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- 34.Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2 doi: 10.1186/gb-2000-2-1-research0002. RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pulak R. Techniques for analysis, sorting, and dispensing of C. elegans on the COPAS flow-sorting system. Methods Mol Biol. 2006;351:275–86. doi: 10.1385/1-59745-151-7:275. [DOI] [PubMed] [Google Scholar]
- 36.Sigrist CJ, Cerutti L, Hulo N, Gattiker A, Falquet L, Pagni M, Bairoch A, Bucher P. PROSITE: a documented database using patterns and profiles as motif descriptors. Brief Bioinform. 2002;3:265–74. doi: 10.1093/bib/3.3.265. [DOI] [PubMed] [Google Scholar]
- 37.Tully DB, Collins BJ, Overstreet JD, Smith CS, Dinse GE, Mumtaz MM, Chapin RE. Effects of arsenic, cadmium, chromium, and lead on gene expression regulated by a battery of 13 different promoters in recombinant HepG2 cells. Toxicol Appl Pharmacol. 2000;168:79–90. doi: 10.1006/taap.2000.9014. [DOI] [PubMed] [Google Scholar]
- 38.Alam J. Multiple elements within the 5' distal enhancer of the mouse heme oxygenase-1 gene mediate induction by heavy metals. J. Biol. Chem. 1994;269:25049–25056. [PubMed] [Google Scholar]
- 39.Williams GT, Morimoto RI. Maximal stress-induced transcription from the human HSP70 promoter requires interactions with the basal promoter elements independent of rotational alignment. Mol Cell Biol. 1990;10:3125–36. doi: 10.1128/mcb.10.6.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eto K, Takahashi N, Kimura Y, Masuho Y, Arai K, Muramatsu MA, Tokumitsu H. Ca(2+)/Calmodulin-dependent protein kinase cascade in Caenorhabditis elegans. Implication in transcriptional activation. J. Biol. Chem. 1999;274:22556–22562. doi: 10.1074/jbc.274.32.22556. [DOI] [PubMed] [Google Scholar]
- 41.Hahn JS, Hu Z, Thiele DJ, Iyer VR. Genome-wide analysis of the biology of stress responses through heat shock transcription factor. Mol Cell Biol. 2004;24:5249–56. doi: 10.1128/MCB.24.12.5249-5256.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Britton C, McKerrow JH, Johnstone IL. Regulation of the Caenorhabditis elegans gut cysteine protease gene cpr- 1: requirement for GATA motifs. J. Mol. Biol. 1998;283:15–27. doi: 10.1006/jmbi.1998.2093. [DOI] [PubMed] [Google Scholar]
- 43.Moilanen LH, Fukushige T, Freedman JH. Regulation of metallothionein gene transcription. Identification of upstream regulatory elements and transcription factors responsible for cell-specific expression of the metallothionein genes from Caenorhabditis elegans. J. Biol. Chem. 1999;274:29655–29665. doi: 10.1074/jbc.274.42.29655. [DOI] [PubMed] [Google Scholar]
- 44.Simon MC. Gotta have GATA. Nat Genet. 1995;11:9–11. doi: 10.1038/ng0995-9. [DOI] [PubMed] [Google Scholar]
- 45.Jeong EM, Moon CH, Kim CS, Lee SH, Baik EJ, Moon CK, Jung YS. Cadmium stimulates the expression of ICAM-1 via NF-kappaB activation in cerebrovascular endothelial cells. Biochem Biophys Res Commun. 2004;320:887–92. doi: 10.1016/j.bbrc.2004.05.218. [DOI] [PubMed] [Google Scholar]
- 46.Liu RY, Corry PM, Lee YJ. Potential involvement of a constitutive heat shock element binding factor in the regulation of chemical stress-induced hsp70 gene expression. Mol Cell Biochem. 1995;144:27–34. doi: 10.1007/BF00926737. [DOI] [PubMed] [Google Scholar]
- 47.Alam J, Wicks C, Stewart D, Gong P, Touchard C, Otterbein S, Choi AM, Burow ME, Tou J. Mechanism of heme oxygenase-1 gene activation by cadmium in MCF-7 mammary epithelial cells. Role of p38 kinase and Nrf2 transcription factor. J Biol Chem. 2000;275:27694–702. doi: 10.1074/jbc.M004729200. [DOI] [PubMed] [Google Scholar]
- 48.Hung JJ, Cheng TJ, Lai YK, Chang MD. Differential activation of p38 mitogen-activated protein kinase and extracellular signal-regulated protein kinases confers cadmium-induced HSP70 expression in 9L rat brain tumor cells. J Biol Chem. 1998;273:31924–31. doi: 10.1074/jbc.273.48.31924. [DOI] [PubMed] [Google Scholar]
- 49.Saydam N, Steiner F, Georgiev O, Schaffner W. Heat and heavy metal stress synergize to mediate transcriptional hyperactivation by metal-responsive transcription factor MTF-1. The Journal of Biological Chemistry. 2003;278:31879–31883. doi: 10.1074/jbc.M302138200. [DOI] [PubMed] [Google Scholar]
- 50.Spieth J, Blumenthal T. The Caenorhabditis elegans vitellogenin gene family includes a gene encoding a distantly related protein. Mol Cell Biol. 1985;5:2495–501. doi: 10.1128/mcb.5.10.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kennedy BP, Aamodt EJ, Allen FL, Chung MA, Heschl MF, McGhee JD. The gut esterase gene (ges-1) from the nematodes Caenorhabditis elegans and Caenorhabditis briggsae. J Mol Biol. 1993;229:890–908. doi: 10.1006/jmbi.1993.1094. [DOI] [PubMed] [Google Scholar]
- 52.Hawkins MG, McGhee JD. elt-2, a second GATA factor from the nematode Caenorhabditis elegans. J. Biol. Chem. 1995;270:14666–14671. doi: 10.1074/jbc.270.24.14666. [DOI] [PubMed] [Google Scholar]
- 53.Egan CR, Chung MA, Allen FL, Heschl MF, Van Buskirk CL, McGhee JD. A gut-to-pharynx/tail switch in embryonic expression of the Caenorhabditis elegans ges-1 gene centers on two GATA sequences. Dev Biol. 1995;170:397–419. doi: 10.1006/dbio.1995.1225. [DOI] [PubMed] [Google Scholar]
- 54.MacMorris M, Broverman S, Greenspoon S, Lea K, Madej C, Blumenthal T, Spieth J. Regulation of vitellogenin gene expression in transgenic Caenorhabditis elegans: short sequences required for activation of the vit-2 promoter. Mol. Cell. Biol. 1992;12:1652–1662. doi: 10.1128/mcb.12.4.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fukushige T, Hawkins MG, McGhee JD. The GATA-factor elt-2 is essential for formation of the Caenorhabditis elegans intestine. Dev Biol. 1998;198:286–302. [PubMed] [Google Scholar]
- 56.Aamodt EJ, Chung MA, McGhee JD. Spatial control of gut-specific gene expression during Caenorhabditis elegans development. Science. 1991;252:579–82. doi: 10.1126/science.2020855. [DOI] [PubMed] [Google Scholar]
- 57.Evans T. Regulation of cardiac gene expression by GATA-4/5/6. Trends Card Med. 1997;7:75–83. doi: 10.1016/S1050-1738(97)00010-8. [DOI] [PubMed] [Google Scholar]
- 58.Albertson DG, Thomson JN. The pharynx of Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1976;275:299–325. doi: 10.1098/rstb.1976.0085. [DOI] [PubMed] [Google Scholar]
- 59.Hall DH, Altun ZF. Atlas of C. elegans Anatomy - An illustrated Handbook. 2004 [Google Scholar]
- 60.Huang P, Stern MJ. FGF signaling functions in the hypodermis to regulate fluid balance in C. elegans. Development. 2004;131:2595–604. doi: 10.1242/dev.01135. [DOI] [PubMed] [Google Scholar]
- 61.2006. WormBase Release WS160.
- 62.White J. The Anatomy. In: Wood WB, editor. The Nematode Caenorhabditis elegans. Cold Spring Harbor Press; Cold Spring Harbor, NY: 1985. pp. 81–122. [Google Scholar]




