Abstract
Nanoflow electrospray ionization has been used to introduce intact Escherichia coli ribosomes into the ion source of a mass spectrometer. Mass spectra of remarkable quality result from a partial, but selective, dissociation of the particles within the mass spectrometer. Peaks in the spectra have been assigned to individual ribosomal proteins and to noncovalent complexes of up to five component proteins. The pattern of dissociation correlates strongly with predicted features of ribosomal protein–protein and protein–RNA interactions. The spectra allow the dynamics and state of folding of specific proteins to be investigated in the context of the intact ribosome. This study demonstrates a potentially general strategy to probe interactions within complex biological assemblies.
Mass spectrometry is well established as a powerful and rapidly expanding technique for studying the covalent structures of biological macromolecules. The development of electrospray ionization (ESI) has extended the reach of mass spectrometry into the realm of noncovalent interactions (1). ESI techniques can generate gas-phase ions from solutions in which proteins are in their native state, allowing investigation of their structure and dynamics and events associated with their folding (2). Combined with the sensitivity and generous molecular weight limits of modern mass analyzers, ESI holds the promise of allowing studies of noncovalent structure in large multimolecular complexes (3, 4). Here we report on the successful use of ESI mass spectrometry to initiate investigations of ribosomes from Escherichia coli.
The pivotal role of the ribosome in the translation of genetic information into biological activity has made it the subject of intensive research over the past 30 years, revealing much about its structure and function (5–7). Most of this research has been directed toward E. coli ribosomes. These 2.5-MDa particles contain 54 proteins, many of which are posttranslationally modified, and three large RNA molecules (also posttranscriptionally modified), which comprise the majority of the mass of the ribosome. The largest RNA molecule is thought to be a ribozyme participating in the peptidyltransferase reaction (8, 9). Intact (70S) ribosomes consist of two noncovalently associated subunits: a small (30S) subunit and a large (50S) subunit, each of which is stable in isolation. Interactions within and between the subunits determine and regulate the process of protein biosynthesis within the cell.
MATERIALS AND METHODS
Ribosomes.
Ribosomes were harvested from E. coli strain MRE600 by using standard protocols (10); no salt washing steps were used. Stock solutions then were buffer-exchanged to a final concentration of 50–500 nM in 10 mM ammonium chloride, 1 mM 2-mercaptoethanol, pH 6.5 by using Centricon 100 concentrators. Any monomeric proteins that dissociated from the ribosome during this exchange protocol passed through the membrane. Denatured ribosomes were prepared by suspension of stock ribosomes in 50% acetic acid at 0°C for 30 min (11), and buffer exchange was carried out as for intact ribosomes.
Electrospray Mass Spectrometry.
Solutions were kept at 4°C throughout buffer exchange and immediately before electrospray analysis. Nanoflow capillaries were prepared as described previously(12). The high viscosity of the ribosomal solutions necessitated preparation of capillaries with large tips (by manually breaking the drawn end under a stereomicroscope) and 10–20 psi of backing pressure during electrospraying. The samples were introduced into a Micromass Platform II quadrupole mass spectrometer operating in positive ion mode using a nanoflow probe. Capillary voltages of 1.3–1.5 kV and cone voltages of 60–110 V typically were used. Mass spectra were obtained at 20°C without heating in the interface, and no organic cosolvents were used to enhance electrospray analysis. Conventional ESI yielded spectra of similar appearance although of substantially reduced signal to noise. Spectra represent averages over 5–10 acquisitions. Hydrogen/deuterium (H/D) exchange experiments were performed by diluting stock solutions of ribosomes by a factor of 103 into D2O buffers (measured pH 6.4) and concentrating the resulting solutions with Centricon 100 concentrators.
Data Processing.
MaxEnt processing (13) was used to survey initially the spectra and provide candidate species; reported m/z values for assigned proteins are calculated by conventional deconvolution of the charge-state distributions.
RESULTS AND DISCUSSION
Mass Spectra of Intact 70S Ribosomes and 50S Subunits.
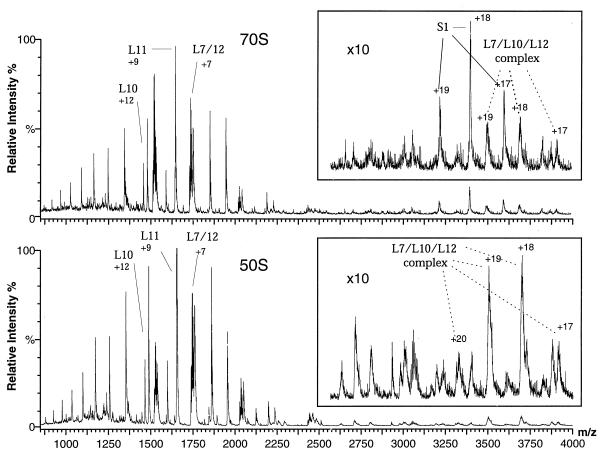
Fig. 1 (Upper) shows positive ion mass spectra obtained from an aqueous solution at pH 6.4 of intact 70S E. coli ribosomes introduced into the nanoflow electrospray ion source of a quadrupole mass spectrometer. The particles give rise to very high-quality spectra consisting of sets of overlapping charge-state distributions. Deconvolution of the m/z values shows that the peaks all correspond to species having masses in the range of 10–70 kDa. Although the peaks arise from many different proteins, the most intense are from just five of the component proteins of the ribosome; four of these are located in the 50S subunit (L7/L12, L10, and L11) and one in the 30S subunit (S1). Observation of the signals indicates that these proteins have dissociated selectively from the ribosome after introduction of the particles into the mass spectrometer. Repeating the experiment with isolated 50S subunits results in similar spectra (Fig. 1, Lower) to those of the intact ribosome, except for the absence of peaks from protein S1. All observed masses (Table 1) are in excellent agreement with the known amino acid sequences and posttranslational modifications.
Figure 1.
Positive ion ESI mass spectra of aqueous solutions of intact 70S ribosomes and 50S subunits at pH 6.4. Only the most intense peak for each assigned protein is labeled (along with the total charge for that peak); each protein gives rise to a charge-state distribution of 4–10 identifiable peaks. (Upper) Intact 70S ribosomes. (Lower) 50S subunits. The spectra are highly reproducible; ca. 100 experiments with 16 separately prepared solutions of ribosomes yielded essentially identical spectra. Control spectra of acid denatured ribosomes show no peaks, confirming that only intact ribosomes are retained during the buffer exchange and that the proteins observed in the mass spectra dissociate from the ribosomes after introduction into the mass spectrometer.
Table 1.
Comparison of predicted and observed masses for ribosomal proteins present in mass spectra of intact 70S ribosomes, 50S subunits, and 30S subunits
| Protein | Predicted, Da | Observed 70S, Da | Observed 50S, Da | Observed 30S, Da |
|---|---|---|---|---|
| L7 | 12,206.0 | 12,207.4 ± 2.7 | 12,203.5 ± 1.0 | |
| L10 | 17,580.4 | 17,582.7 ± 3.8 | 17,578.4 ± 3.2 | |
| L11 | 14,872.4 | 14,871.0 ± 2.3 | 14,868.3 ± 1.9 | |
| L12 | 12,164.0 | 12,165.3 ± 2.7 | 12,162.4 ± 2.8 | |
| S1 | 61,239.1 | 61,207.5 ± 18.6 | 61,205.3 ± 12.9 | |
| S2 | 26,612.5 | 26,615.3 ± 4.2 | ||
| S3 | 25,852.0 | 25,853.4 ± 3.7 | ||
| S5 | 17,514.2 | 17,508.9 ± 2.2 | ||
| S10 | 11,735.6 | 11,733.2 ± 2.3 |
L7 is N-acetyl L12. N-methyl lysine variants of L7 and L12 (30–50% prevalence) also were observed at mass values 12–15 Da higher than the unmodified species. Observed mass is the average of the masses calculated for each recorded charge state of a given protein; uncertainties are 1 SD of the mean.
The spectra of both 70S ribosomes and 50S subunits also contain a series of broad peaks from species whose masses are substantially greater than the largest component proteins; these have been found to result from noncovalent complexes between proteins. One set of peaks, from a component with a mass of 66,533 ± 40 Da, is particularly intense and can be identified as arising from a complex having two copies of L7, two of L12, and one of L10. This group of proteins corresponds to a well-established feature of ribosomes, visible in electron micrographs as a flexible stalk-like protuberance on the 50S subunit (6, 14). The observation of this complex in the spectra shows that these proteins dissociate together from the intact particle within the ion source of the mass spectrometer. The details of the dissociation process are not known; however, increases in the nozzle/skimmer cone voltage result in a decrease of the signals from the complex relative to those of its component proteins; this observation suggests that the complex dissociates within the mass spectrometer and provides additional evidence to support our assignment of the composition of this noncovalent species.
That the interactions between the component proteins are sufficiently strong to maintain this complex after dissociation from the intact ribosome is supported by the fact that the same complex is stable in aqueous solution (15). Interestingly, models of the 50S subunit indicate that L11, the only other 50S protein giving rise to intense signals in the mass spectra, interacts strongly with the L7/L10/L12 group of proteins (16). Moreover, the peaks arising from the L7/L10/L12 complex are not only broad but are of somewhat higher mass (≈180 Da) than predicted from the masses of the component proteins. Both of these features are characteristic of the presence of additional water molecules (17, 18); the mass indicates that an average of 10 such molecules are tightly bound in the complex. Other broad peaks in the spectra at high m/z (Fig. 1) correspond to noncovalent protein complexes whose charge-state distributions apparently extend beyond the m/z range of our spectrometer. Although these are not yet assigned they indicate that groups of proteins other than L7/L10/L12 can remain associated after release from the intact particles.
Mass Spectra of 30S Subunits.
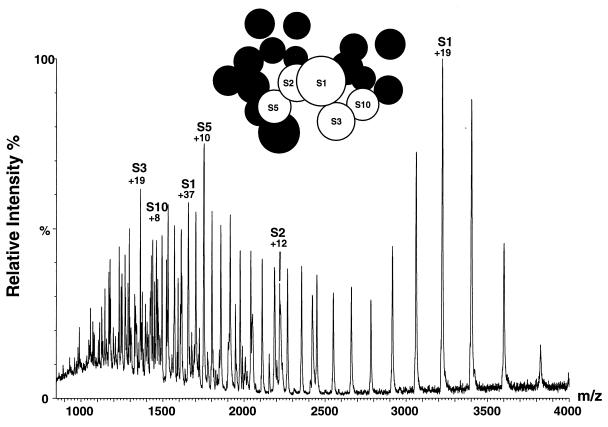
Although the signals from the 50S proteins are closely similar in the spectra of both the isolated subunits and the intact 70S particles, a different situation is found for the 30S proteins. In the intact ribosome, intense peaks from 30S proteins are observed only for S1. In the spectra of isolated 30S subunits, however, intense signals from an additional four proteins are clearly evident (Fig. 2). The factors influencing the intensity of peaks from different proteins are complex and can include suppression of signals by other components. Nevertheless, the greater intensity of peaks from 30S proteins in the absence of the 50S subunits strongly suggests that interactions between the 30S proteins are weaker in the isolated subunits than in intact ribosomes‖. All of the proteins that dissociate readily in the 30S subunits are in close proximity to each other, as indicated in the model of 30S topology obtained from neutron scattering measurements (20) (see Fig. 2). Interestingly, however, they are not located at the subunit interface. Proteins S3, S5, and S10 in particular are known to interact strongly with each other, forming a stable trimer in solution (21). Although the latter has not been detected directly in the mass spectra, the results for both 30S and 50S subunits suggest that groups of strongly interacting proteins may dissociate from the particles together, some of which subsequently split up into monomeric species.
Figure 2.
Positive ion ESI mass spectra of aqueous solutions of 30S subunits at pH 6.4. Ninety-five percent of the peaks observed in this spectrum have been assigned to various charge states of the five 30S proteins indicated; only the most intense peak of the charge-state distribution for each protein is labeled (along with the total charge for that particular peak). (Inset) Representation of the topology of the 30S E. coli ribosomal subunit, derived from neutron scattering measurements of partially deuterated ribosomes (20). Proteins observed in the mass spectrometer are shown in labeled, white circles; unobserved proteins are represented as unlabeled, black circles.
The majority of ribosomal proteins are basic and many contain stretches of 10 or more amino acids where at least half of the residues are lysine or arginine. Such regions, also present in histones and viral coat proteins, are thought to mediate interactions with negatively charged nucleic acids (22). The proteins observed in the mass spectra all lack these regions and are among the most acidic of ribosomal proteins; they thus are expected to have relatively weak interactions with the ribosomal RNA. Moreover, the proteins that dissociate most readily from the ribosome at pH 6.4 in the mass spectrometer all are among the first released into solution by the addition of LiCl (1.0 M for 50S subunits, 3.5 M for 30S) (23). Dissociation in the mass spectrometer, therefore, appears to occur most readily for those groups of proteins whose ionic interactions with RNA are weakest. This finding is consistent with investigations of protein–small molecule complexes, which indicate that ionic interactions are more strongly preserved in the gas phase than are hydrophobic interactions (24, 25). Appearance of proteins in the mass spectra, therefore, may reflect not just their proximity in the structure but the nature of their interactions with each other and with RNA molecules.
Characterization of Individual Ribosomal Proteins.
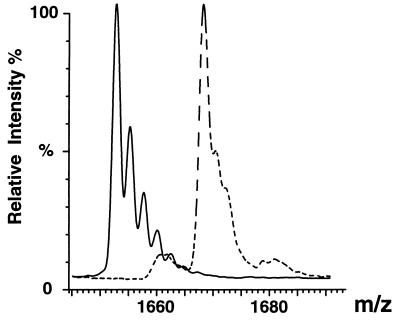
As well as providing a means to probe the interactions between the components of the ribosome, the observation of distinct peaks in the mass spectrometer provides an opportunity to investigate the properties of individual proteins. Many of these are at least partially unfolded in isolation (26), and little is known about their structure and dynamics in the intact particle. A powerful means of probing the latter is through hydrogen/deuterium (H/D) exchange experiments (27), which can be monitored by mass spectrometry as a consequence of the resulting changes in mass (28–31). We have carried out such experiments on 50S subunits, and the numerous mass shifts indicate that exchange protection occurs in many proteins. We have analyzed the results in some detail for two proteins, L10 and L11 (Fig. 3). L10 retains 55 ± 6 protons after 6 hr of exchange in D2O (pH 6.5, 4°C), and L11 90 ± 8 protons. The extent of protection for these proteins is consistent with both being highly folded in the ribosomal particle. Interestingly, NMR spectroscopy of a C-terminal fragment of protein L11 shows the RNA binding domain to be unfolded in the absence of rRNA (32). In the ribosome the extent of protection of L11 is extremely high for a protein of this molecular mass (14,872 Da) and is close to that found for the similarly sized protein lysozyme in crystals rather than in solution (33). This degree of protection suggests that tight packing within the intact ribosome restrains the conformational fluctuations that give rise to hydrogen exchange in an isolated protein in solution.
Figure 3.
Hydrogen/deuterium (H/D) exchange of ribosomal protein L11 in intact ribosomes. ESI mass spectra of 50S subunits in H2O (solid trace) and after 6 hr of exchange in D2O (dotted trace). m/z values for the +9 charge state of L11 are shown in both traces, indicating the increase in mass resulting from chemical exchange of protons for deuterons. Deuterons have been incorporated in an average of 154 of 244 exchangeable sites, indicating that 90 sites are protected from exchange.
The conformational state of a protein also is reflected in its charge-state distribution observed in the mass spectrum (29, 34). The charge-state distributions of all of the proteins except for S1 are typical of those from folded proteins. In the case of S1, the charge states show a striking bimodal distribution (Fig. 2). One of these sets of peaks, centered at 3,200 m/z, is typical of a globular protein. The second, an extensive set of highly charged species centered at 1,650 m/z, is, however, typical of an unfolded state of a protein. There are therefore at least two distinct conformations of S1 within the population of intact ribosomes studied here. This observation may be related to the fact that S1 is known to undergo a transition from a protease-sensitive state to a protected state as a result of a conformational change triggered by mRNA binding (35).
CONCLUSIONS
The electrospray technique has been found previously to be capable of generating gas phase ions from biological assemblies with masses of up to 40 MDa, some of which have been shown to maintain functional integrity during passage through the mass spectrometer (36). Measurement of the spectra of such species, however, is beyond the capabilities of many mass spectrometers**. In the present work we have shown that mass spectrometry still can be used in their study provided that dissociation to smaller units can be achieved. We propose that the dissociation of the large assemblies in the mass spectrometer at least in part reflects the nature of the interactions between the constituent macromolecules. Whatever the mechanism of dissociation, however, the identification of complexes of proteins provides direct evidence for the interactions between their components in the intact particle. Moreover, observation of the component species enables their properties to be probed by using established methods. We believe therefore that the approach described here will have wide applicability in the investigation of other biologically important macromolecular complexes. In the context of the ribosome, we have developed a sensitive assay for studying protein interactions within the ribosome and conformational changes in specific proteins. We plan to use this approach to contrast the properties of isolated proteins with those of ribosome-bound proteins and to monitor specific conformational changes of proteins in actively synthesizing ribosomes.
Acknowledgments
We thank Michael Groß and Andrew Miranker for helpful discussions. The Oxford Centre for Molecular Sciences is supported by the Engineering and Physical Sciences Research Council, the Biotechnology and Biological Sciences Research Council, and the Medical Research Council (U.K.). The research of C.M.D. is supported in part by an International Research Scholars award from the Howard Hughes Medical Institute. D.R.B. is supported by a Hitchings-Elion Fellowship from the Burroughs Wellcome Fund. C.V.R. is a Royal Society University Research Fellow.
ABBREVIATION
- ESI
electrospray ionization
Footnotes
Although ribosomal subunit association generally is thought to require Mg2+, adducts with this cation reduce resolution and sensitivity of ESI mass spectra; we therefore excluded it from the buffers in the present study. Fluorescence measurements of ethidium bromide binding to intact ribosomes and isolated subunits have, however, led to the proposal that subunits interact even in the absence of Mg2+ (19).
Globular proteins generally have higher mass/charge ratios than unfolded proteins (17). Similarly, assemblies of proteins have higher mass/charge ratios than their individual components; for the L7/L10/L12 complex, for example, the charge-state distribution is centered around z = 18, rather than z = 38, which would be predicted from the sum of the species from the component proteins. The lack of detectable ions for intact large complexes therefore is likely to arise in part from mass/charge ratios too high for detection in conventional spectrometers.
References
- 1.Katta V, Chait B. J Am Chem Soc. 1991;113:8534–8535. [Google Scholar]
- 2.Miranker A, Robinson C V, Radford S E, Dobson C M. FASEB J. 1996;10:93–101. doi: 10.1096/fasebj.10.1.8566553. [DOI] [PubMed] [Google Scholar]
- 3.Fitzgerald M C, Chernushevich I, Standing K G, Whitman C P, Kent S B H. Proc Natl Acad Sci USA. 1996;93:6851–6856. doi: 10.1073/pnas.93.14.6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robinson C V, Gross M, Eyles S J, Ewbank J J, Mayhew M, Hartl F U, Dobson C M, Radford S E. Nature (London) 1994;372:646–651. doi: 10.1038/372646a0. [DOI] [PubMed] [Google Scholar]
- 5.Matheson A T. Biochem Cell Biol. 1995;73:739–1227. [PubMed] [Google Scholar]
- 6.Frank J, Zhu J, Penczek P, Li Y H, Srivastava S, Verschoor A, Radermacher M, Grassucci R, Lata R K, Agrawal R K. Nature (London) 1995;376:441–444. doi: 10.1038/376441a0. [DOI] [PubMed] [Google Scholar]
- 7.Rodnina M V, Savelsburgh A, Katunin V I, Wintermeyer W. Nature (London) 1997;385:37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- 8.Noller H F, Hoffarth V, Zimniak L. Science. 1992;256:1416–1419. doi: 10.1126/science.1604315. [DOI] [PubMed] [Google Scholar]
- 9.Spahn C M T, Schafer M A, Krayevsky A A, Nierhaus K H. J Biol Chem. 1996;271:32857–32862. doi: 10.1074/jbc.271.51.32857. [DOI] [PubMed] [Google Scholar]
- 10.Spedding G. In: Ribosomes and Protein Synthesis: A Practical Approach. Spedding G, editor. Oxford: Oxford Univ. Press; 1990. pp. 1–27. [Google Scholar]
- 11.Kerlavage A R, Cooperman B S. Biochemistry. 1986;25:8002–8010. doi: 10.1021/bi00372a032. [DOI] [PubMed] [Google Scholar]
- 12.Wilm M, Mann M. Anal Chem. 1996;68:1–8. doi: 10.1021/ac9509519. [DOI] [PubMed] [Google Scholar]
- 13.Ferrige A G, Seddon M J, Green B N, Jarvis S A, Skilling J. Rapid Commun Mass Spectrom. 1992;6:707–711. [Google Scholar]
- 14.Strycharz W, Nomura M, Lake J. J Mol Biol. 1978;126:123–126. doi: 10.1016/0022-2836(78)90355-8. [DOI] [PubMed] [Google Scholar]
- 15.Petterson I, Hardy S J S, Liljas A. FEBS Lett. 1976;64:135–138. doi: 10.1016/0014-5793(76)80267-0. [DOI] [PubMed] [Google Scholar]
- 16.Walleczek J, Schuler D, Stoffler-Meilicke M, Brimacombe R, Stoffler G. EMBO J. 1988;7:3571–3576. doi: 10.1002/j.1460-2075.1988.tb03234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loo J A. J Mass Spec. 1995;30:180–183. [Google Scholar]
- 18.Light-Wahl K J, Schwartz B L, Smith R D. J Am Chem Soc. 1994;116:5271–5278. [Google Scholar]
- 19.Bonincontro A, Briganti G, Giansanti A, Pedone F, Risuelo G. Biochim Biophys Acta. 1993;1174:27–30. doi: 10.1016/0167-4781(93)90088-u. [DOI] [PubMed] [Google Scholar]
- 20.Capel M S, Engelman D M, Freeborn B R, Kjeldgaard M, Langer J A, Ramakrishnan V, Schindler D G, Schneider D K, Schoenborn B P, Sillers I-Y, et al. Science. 1987;238:1403–1406. doi: 10.1126/science.3317832. [DOI] [PubMed] [Google Scholar]
- 21.Rohde M F, Aune K C. Biochemistry. 1975;14:4344–4350. doi: 10.1021/bi00690a031. [DOI] [PubMed] [Google Scholar]
- 22.Liljas A. Int Rev Cytol. 1991;124:103–135. doi: 10.1016/s0074-7696(08)61525-9. [DOI] [PubMed] [Google Scholar]
- 23.Nierhaus K. In: Ribosomes and Protein Synthesis: A Practical Approach. Spedding G, editor. Oxford: IRL; 1990. pp. 161–188. [Google Scholar]
- 24.Robinson C V, Chung E W, Kragelund B B, Knudsen J, Aplin R T, Poulsen F M, Dobson C M. J Am Chem Soc. 1996;118:8646–8653. [Google Scholar]
- 25.Wu Q, Gao J, Joseph-McCarthy D, Sigal G B, Bruce J E, Whitesides G M, Smith R D. J Am Chem Soc. 1997;119:1157–1158. [Google Scholar]
- 26.Littlechild J, Malcolm A, Paterakis K, Ackermann I, Dijk J. Biochim Biophys Acta. 1987;913:245–255. doi: 10.1016/0167-4838(87)90336-0. [DOI] [PubMed] [Google Scholar]
- 27.Englander S W, Sosnick T R, Englander J J, Mayne L. Curr Opin Struct Biol. 1996;6:18–23. doi: 10.1016/s0959-440x(96)80090-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katta V, Chait B T. Rapid Commun Mass Spectrom. 1991;5:214–217. doi: 10.1002/rcm.1290050415. [DOI] [PubMed] [Google Scholar]
- 29.Przybylski M, Glocker M O. Angew Chem Int Ed Engl. 1996;35:807–826. [Google Scholar]
- 30.Miranker A, Robinson C V, Radford S E, Aplin R T, Dobson C M. Science. 1993;262:896–900. doi: 10.1126/science.8235611. [DOI] [PubMed] [Google Scholar]
- 31.Dharmasiri K, Smith D L. Anal Chem. 1996;68:2340–2344. doi: 10.1021/ac9601526. [DOI] [PubMed] [Google Scholar]
- 32.Markus M A, Hinck A P, Huang S R, Draper D E, Torchia D A. Nat Struct Biol. 1997;4:70–77. doi: 10.1038/nsb0197-70. [DOI] [PubMed] [Google Scholar]
- 33.Pedersen T G, Sigurskjold B W, Andersen K V, Kjaer M, Poulsen F M, Dobson C M, Redfield C. J Mol Biol. 1991;218:413–426. doi: 10.1016/0022-2836(91)90722-i. [DOI] [PubMed] [Google Scholar]
- 34.Chowdhury S K, Katta V, Chait B T. J Am Chem Soc. 1990;112:9012–9013. [Google Scholar]
- 35.Subramanian, A. R. (1984) Trends Biol. Sci., 491–494.
- 36.Siuzdak G, Bothner B, Yeager M, Brugidou C, Fauquet C M, Hoey K, Chang C-M. Chem Biol. 1996;3:45–48. doi: 10.1016/s1074-5521(96)90083-6. [DOI] [PubMed] [Google Scholar]