Abstract
The three-dimensional structure of Corynebacterium 2,5-diketo-d-gluconic acid reductase A (2,5-DKGR A; EC 1.1.1.-), in complex with cofactor NADPH, has been solved by using x-ray crystallographic data to 2.1-Å resolution. This enzyme catalyzes stereospecific reduction of 2,5-diketo-d-gluconate (2,5-DKG) to 2-keto-l-gulonate. Thus the three-dimensional structure has now been solved for a prokaryotic example of the aldo–keto reductase superfamily. The details of the binding of the NADPH cofactor help to explain why 2,5-DKGR exhibits lower binding affinity for cofactor than the related human aldose reductase does. Furthermore, changes in the local loop structure near the cofactor suggest that 2,5-DKGR will not exhibit the biphasic cofactor binding characteristics observed in aldose reductase. Although the crystal structure does not include substrate, the two ordered water molecules present within the substrate-binding pocket are postulated to provide positional landmarks for the substrate 5-keto and 4-hydroxyl groups. The structural basis for several previously described active-site mutants of 2,5-DKGR A is also proposed. Recent research efforts have described a novel approach to the synthesis of l-ascorbate (vitamin C) by using a genetically engineered microorganism that is capable of synthesizing 2,5-DKG from glucose and subsequently is transformed with the gene for 2,5-DKGR. These modifications create a microorganism capable of direct production of 2-keto-l-gulonate from d-glucose, and the gulonate can subsequently be converted into vitamin C. In economic terms, vitamin C is the single most important specialty chemical manufactured in the world. Understanding the structural determinants of specificity, catalysis, and stability for 2,5-DKGR A is of substantial commercial interest.
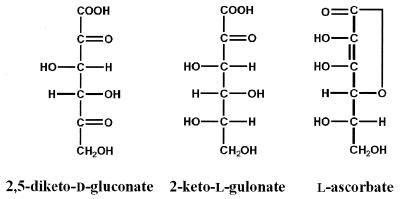
Vitamin C (l-ascorbic acid) is a reducing sugar consisting of a cyclic lactone of a sugar acid (Fig. 1), and it forms an essential part of the diet of human beings. A deficiency of l-ascorbic acid causes scurvy, which is characterized by fragile skin, tendons, and blood vessels. The underlying cause of scurvy is inhibition of post-translational hydroxylation of proline and lysine residues in collagen, an ascorbate-dependent reaction. In economic terms, vitamin C is the single most important specialty chemical manufactured in the world, with annual worldwide consumption exceeding 50,000 tons (45 × 106 kg). Because of its importance, much effort has been focused on developing and improving the industrial synthesis of l-ascorbate. At present, the preferred method for commercial production is the modified Reichstein and Grussner organic synthesis (1). This lengthy procedure, which involves a microbial fermentation and a series of chemical steps, converts d-glucose to 2-keto-l-gulonate (2-KLG). This compound is very stable and can readily be converted into l-ascorbic acid by a simple acid- or base-catalyzed cyclization (see Fig. 1). Therefore, most of the research in this area is dedicated to improved synthesis of 2-KLG by microbial methods (2, 3). An alternative procedure for producing 2-KLG involves the microbial conversion from d-glucose via 2,5-diketo-d-gluconic acid (2,5-DKG; Fig. 1). A tandem fermentation method described by Sonoyama and coworkers (4) first converts d-glucose to 2,5-DKG by using a species of Erwinia and then stereospecifically converts 2,5-DKG to 2-KLG by using Corynebacterium. The enzyme responsible for the latter step has been identified as 2,5-DKG reductase (2,5-DKGR; EC 1.1.1.-). Two variants of this enzyme have been identified: DKGR A (5) and DKGR B (4), also called AKR5C and AKR5D, respectively, in the new proposed nomenclature for the aldo–keto reductase superfamily (6). With the use of recombinant DNA technology, the Sonoyama tandem fermentation method was further improved by cloning and expressing the 2,5-DKGR A gene into a 2,5-DKG-producing strain of Erwinia (5). This manipulation resulted in a genetically modified organism that can directly produce 2-KLG from d-glucose.
Figure 1.
Structures of 2,5-diketo-d-gluconate (2,5-DKG), 2-keto-l-gulonate (2-KLG), and l-ascorbate (vitamin C). The uppermost carbon is C1.
In the production of 2-KLG from a genetically engineered microorganism, the rate-limiting step may be the reduction of 2,5-DKG by 2,5-DKGR (7). Determination of the biophysical and biochemical properties of this enzyme is, therefore, essential in optimizing its catalytic efficiency and hence its utility in the commercial production of l-ascorbic acid. The DKGR B variant has higher catalytic efficiency than the A variant has, and although both exhibit relatively poor thermal stability, DKGR A has significantly higher thermal stability (7). Recently, 2,5-DKGR A from Corynebacterium has been the subject of protein engineering efforts to improve its catalytic properties (7). For these reasons, the A variant was selected for crystallization studies. This enzyme belongs to the aldo–keto reductase superfamily (8) and shares the common (β/α)8-barrel structural motif with other members of this family. 2,5-DKGR A, like other aldo–keto reductases, binds nicotinamide cofactor without a Rossman fold motif (9–11), a structural feature that distinguishes aldo–keto reductases from other oxidoreductases (8). The x-ray crystal structure of 2,5-DKGR A will aid in the elucidation of the structural determinants of specificity, catalysis, and stability for this enzyme.
METHODS
Crystallization of 2,5-DKGR A.
The 2,5-DKGR A gene from Corynebacterium was cloned and expressed in Acetobacter cerinus IFO 3263 (5, 7). The 2,5-DKGR A protein was purified from large-scale cultures of A. cerinus containing expression plasmid ptrp1–35 as described (7). The purified enzyme was dialyzed against distilled water and concentrated to 18.3 mg/ml for crystallization experiments. In an attempt to explore the crystal structure of the enzyme⋅cofactor complex, protein solution was mixed with 5 times molar excess of NADPH (Sigma). All crystallization trials were set up by using the hanging-drop vapor diffusion method (12) in 24-well plastic tissue culture plates (Linbro). The drops were formed by adding 5 μl of precipitant and 5 μl of enzyme⋅cofactor complex solution and were allowed to equilibrate on silane-treated glass coverslips. Experiments were designed on the basis of various combinations of buffers, precipitants, and additives as described in Hampton Crystal Screen I and II (Hampton Research, Riverside, CA). Parallel experiments were performed at room temperature and at 4°C. The drops were checked for crystal growth after a period of 2–3 weeks. Rod-shaped crystals, which diffracted up to 1.9 Å, were observed growing from 1.0 M sodium phosphate/1.0 M potassium phosphate/100 mM Hepes buffer, pH 7.0.
Data Collection and Processing.
A single crystal, approximately 0.15 × 0.15 × 0.5 mm, was mounted in a thin walled quartz capillary tube (Hampton Research) for collecting x-ray data from a Rigaku RU200 rotating anode x-ray source (40 kV, 90 mA, graphite monochromated CuKα radiation) equipped with an R-AXIS II imaging plate system. The data collection parameters are summarized in Table 1. The data were processed by using the denzo (13) software program. One round of auto-indexing yielded a monoclinic P21 unit cell with dimensions a = 35.7 Å, b = 55.8 Å, c = 74.7 Å, and β = 92.2°. The final data set is 85% complete up to 2.1-Å resolution with 14,803 unique reflections and an Rmerge of 11.6% (Table 1).
Table 1.
Crystal, data collection, molecular replacement, and refinement statistics
| Data | Value |
|---|---|
| Crystal data | |
| Space group | P21 |
| Cell constants | a = 35.7 Å, b = 55.8 Å, c = 74.7 Å, β = 92.2° |
| Molecules/asymmetric unit | 1 |
| Matthews’ constant (Vm), Å3/Da | 2.16 |
| Maximum resolution, Å | 1.9 |
| Data collection and processing | |
| Independent reflections | 14,803 |
| Completion, % (to 2.1 Å) | 85 |
| I/σ (overall) | 10.0 |
| I/σ (2.18–2.10 Å) | 2.5 |
| Wilson temperature factor, Å2 | 14.6 |
| Rmerge, % (to 2.1 Å) | 11.6 |
| Molecular replacement | |
| Rotation | α = 11.1°, β = 147.7°, γ = 355.8° |
| Translation | TA = 42.4 Å, TB = —, TC = 89.1 Å |
| Rfactor, % | 49.2 |
| Correlation coefficient | 34.0 |
| Refinement | |
| Rcryst (60–2.1 Å), % | 19.1 |
| Rfree (60–2.1 Å), % | 28.8 |
| rms bond length deviation, Å | 0.01 |
| rms bond angle deviation, ° | 1.9 |
| rms B factor deviation,* Å2 | 1.7 |
*Temperature factor restraint library of Tronrud (19).
Molecular Replacement.
On the basis of sequence comparisons, 2,5-DKGR A is known to have an α8/β8 motif found in many other members of the aldo–keto reductase family. The 1.65-Å crystal structure of human aldose reductase (Protein Data Bank code 1ADS) (9), which shares 38% sequence identity with 2,5-DKGR A, was chosen as a model for performing the molecular replacement search. The NADPH cofactor was not included in the search model. All calculations were done by using the package AMoRe (14) on a Silicon Graphics INDY R5000 workstation. A single solution corresponding to rotation and translation values of α = 111.10°, β = 148.40°, and γ = 355.80° and TA = 42.4 Å and TC = 89.1 Å was obtained, which also exhibited an acceptable packing arrangement of molecules within the unit cell. This solution was subjected to 20 cycles of rigid body refinement in AMoRe, after which it had a crystallographic R factor (Rcryst) of 49.2% and a correlation coefficient of 34.0%. At this stage the amino acid side chains of the search model were replaced with those of 2,5-DKGR A by using the program look (Molecular Applications Group).
Crystallographic Refinement.
The model with correct sequence was subjected to rigid body refinement as implemented in x-plor (15). The entire model was taken as a single rigid domain and after 30 cycles of refinement the Rcryst dropped to 46.1% for 2,183 reflections in the resolution range 10 to 4.0 Å. This refinement was followed by several cycles of least-squares and simulated annealing refinement (10.0 to 2.1 Å, with 13,587 reflections, B factor set to the Wilson plot value of 14.6 Å2), which further reduced the Rcryst to 33.1%. At this point, the data were randomly separated into working (90%) and test (10%) data sets (16). When the test data set was used, Rfree was calculated to be 44.2%. The 2Fo − Fc Fourier maps calculated at this stage revealed clear electron density for about 75% of the molecule, primarily comprising the central β-barrel region. Side chains for which the electron density was ambiguous were replaced by Ala residues. Similarly, surface loop regions for which no clear electron density was observed were deleted. Alternate cycles of refinement using simulated annealed omit and difference Fourier maps and manual model building with the graphics program o (17) resulted in improved electron density maps with the gradual appearance of missing side chains and loops in the model. The Rcryst and Rfree values dropped to 29.0% and 36.8%, respectively. One cycle of grouped B-factor refinement in x-plor decreased the Rcryst to 27.3% and Rfree to 34.5%. At this stage, there was electron density in both the 2Fo − Fc and Fo − Fc maps in the region of the active site where NADPH was observed in the related aldose reductase structure. The NADPH cofactor could readily be modeled into this density, contoured at the 2σ level in the difference density map. Additional rounds of refinement utilized the tnt least-squares refinement package (18) with correlated thermal factor restraints (19). Solvent molecules were added to the structure if there was unambiguous density in the 2Fo − Fc maps, appropriate stereochemistry with hydrogen bonding partners of the protein or cofactor, and refined thermal factors of 60 Å2 or less. A total of 2,122 non-hydrogen atoms, 48 NADPH atoms, and 106 solvent molecules were included in the refined model. Final model building and refinement yielded an Rcryst of 19.1% and Rfree of 28.8% when all data from 60 to 2.1 Å were used. Structure factors and model coordinates have been deposited with the Brookhaven Data Bank (1A80).
RESULTS
Quality of the Model.
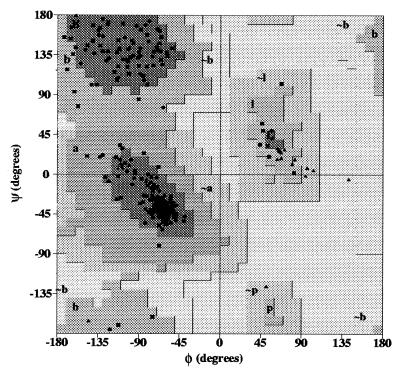
The refinement statistics and other crystallographic properties of 2,5-DKGR A crystals are summarized in Table 1. The stereochemical quality of the 2,5-DKGR A model is demonstrated in the Ramachandran plot (Fig. 2), with 89.5% of residues residing in the most favored regions. All remaining residues are located in additional allowed regions. The rms deviation from ideal bond length is 0.01 Å, and for bond angles, the rms deviation is 1.9°. The average B factors for main-chain and side-chain atoms are 15.9 and 21.7 Å2, respectively.
Figure 2.
Ramachandran plot (29) of the refined structure of 2,5-DKGR A (glycine and proline residues indicated by triangles). Of the 238 non-glycine and non-proline residues, 213 (89.5%) are in most-favored regions of the φ, ψ plot, and none are in disallowed regions.
Overall Structure.
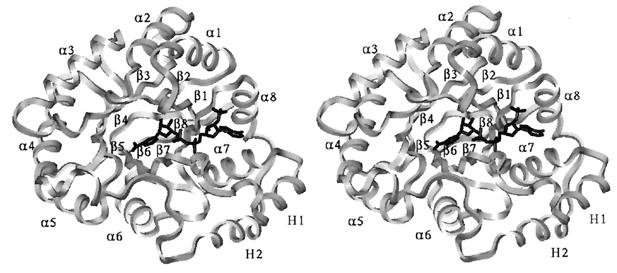
The 2,5-DKGR A folds into a parallel α/β-barrel consisting of eight α-helices and eight β-strands (Fig. 3). This type of structural fold, first observed in triose-phosphate isomerase (20), has since been demonstrated in many other enzymes (21) belonging to the aldo–keto reductase family (9–11). The regular (β/α)8 structure of 2,5-DKGR A is interrupted by two extra helices toward the C-terminal side of the molecule. These helices (named H1 and H2) are packed side by side and have also been observed in other members of the aldo–keto reductase superfamily (9, 21, 22). The helix H1 lies in between β7 and α8 and H2 is found following α8. The N terminus of the molecule folds into a β-hairpin-like structure which sits over the N-terminal face of the central β-barrel. The C-terminal 18 residues form a lip around the edge of the C-terminal face of the central β-barrel, delineating part of the substrate-binding pocket (Fig. 3). The human and porcine aldose reductases are characterized by the presence of long loops (one between β4 and α4 and another between β7 and H1) which are significantly shorter in 2,5-DKGR A. The DKGR model superimposes on the human aldose reductase structure with an rms deviation of 2.6 Å for corresponding Cα atoms.
Figure 3.
Stereo view of the main chain Cα trace of the refined structure of 2,5-DKGR A. View is down the axis of the central β-barrel and shows the arrangement of the outer α-helices and the location of the bound NADPH cofactor (in black).
Mode of Binding of NADPH.
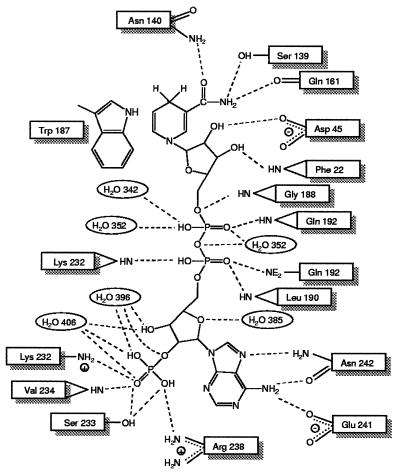
The active site of 2,5-DKGR A is located at the C-terminal face of the barrel, a location that is consistent with other members of the (β/α)8 family (21). The NADPH cofactor binds in an extended conformation in a crevice that extends from the outer edge of the barrel to the inner core. For the adenine and pyrophosphate group of NADPH, this crevice is described by the interface between the strand β7/loop 7 region and β8. The nicotinamide group is positioned deep in the central region of the barrel formed by eight β strands. The complex between NADPH and enzyme is stabilized by a total of 16 hydrogen bonds, 2 ionic bonds, and 1 aromatic stacking interaction with the nicotinamide ring (Fig. 4). The number of interactions is significantly lower compared with the homologous enzyme human aldose reductase, which forms 19 hydrogen bonds, 3 salt bridges, and 1 aromatic stacking with the cofactor (9). As a result of these differences 2,5-DKGR A is expected to bind less tightly to its cofactor compared with human aldose reductase; this looser binding may be responsible for the reported 3-fold higher Km for NADPH seen in 2,5-DKGR A (7) compared with aldose reductase (23).
Figure 4.
Noncovalent interactions between the NADPH cofactor and 2,5-DKGR A. In addition to hydrogen bonding and electrostatic interactions, the side chain of Trp-187 is involved in an aromatic stacking interaction with the nicotinamide ring of the NADPH cofactor.
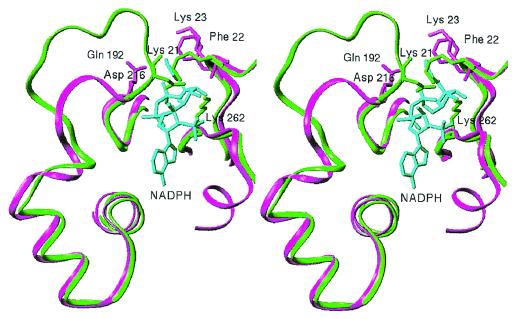
The pyrophosphate group of the NADPH is bound in a crevice on the surface of the barrel. One side of the crevice is lined with residues 188–192 (spanning the β7/loop 7 region), and the other side is lined with residues Phe-22 and Lys-23 (β1/loop 1) and Pro-231 and Lys-232 (β8). The pyrophosphate group is held in place by specific hydrogen bonding interactions with Gly-188, Leu-190, Gln-192, Lys-232, and solvent molecules 342 and 352 (Fig. 4). In the human aldose reductase, there is an extension in the polypeptide region of β7/loop 7 that folds over the pyrophosphate group of the cofactor and essentially forms a short tunnel or a “safety belt” through which the pyrophosphate moiety passes (9). This “safety belt” is stabilized by salt bridges between Asp-216, Lys-21, and Lys-262 (Fig. 5). However, in the crystal structure of 2,5-DKGR A, residue Asp-216 is missing due to a deletion in loop 7, and Lys-23 (similar to Lys-21 in human aldose reductase) is located approximately 7 Å distal to the pyrophosphate group. Therefore, a short tunnel or a “safety belt” type of structural arrangement is missing in 2,5-DKGR A (Fig. 5). As a result of these structural differences, the solvent-accessible area of the bound NADPH cofactor is significantly greater in 2,5-DKGR A than in aldose reductase. The accessible area of the NADPH in complex with 2,5-DKGR A is 103 Å2 (when a 1.4-Å-radius probe is used) (24). This represents approximately 11% of the total accessible area of the unbound cofactor. However, in aldose reductase the accessible surface area of bound cofactor is reduced to 78 Å2. Because the “safety belt” structure in aldose reductase covers the NADPH cofactor, a significant conformational change in the loop 7 region appears necessary for cofactor binding and release (9). The large movement of loop 7 required for the formation and dissociation of the NADPH⋅enzyme complex has also been thought to be responsible for the biphasic kinetics of NADPH binding and release in aldose reductase (25, 26). On the basis of this structure (Fig. 5), we would predict that cofactor binding to 2,5-DKGR A would not involve such structural rearrangements, and the kinetics of cofactor binding would not exhibit a biphasic characteristic.
Figure 5.
Stereo view of the details of the structure of 2,5-DKGR A and aldose reductase in the vicinity of the NADPH cofactor binding site. In aldose reductase (green), the loop 7 region completely covers the NADPH cofactor (cyan) by means of electrostatic interactions between residues Lys-21, Asp-216, and Lys-262. In 2,5-DKGR A (magenta), however, this loop is shorter and does not clamp over the cofactor. In aldose reductase a major rearrangement of the loop 7 region appears necessary to allow the NADPH cofactor to enter and exit the binding cleft.
Active-Site Pocket.
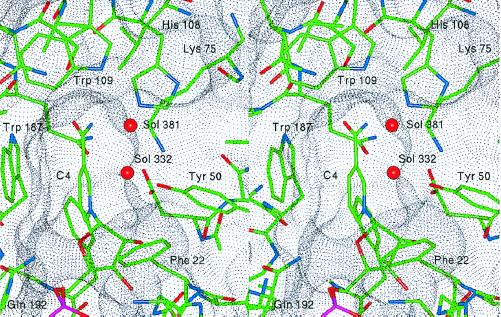
The active-site pocket has dimensions of approximately 5 Å × 6 Å and a depth of about 9 Å (Fig. 6). These dimensions are significantly smaller in comparison to the active-site pocket of human aldose reductase [7 Å × 13 Å and a depth of 10 Å (9)]. The bottom portion of the active-site pocket is composed of residues Phe-22, Asp-45, Ala-47, Tyr-50, Lys-75, Leu-106, Ser-139, Asn-140, and Trp-187. One side of the upper rim of the pocket is mainly formed by aromatic and apolar residues Ile-49, Trp-77, His-108, and Trp-109. The opposite side is lined with the C-terminal residues Ser-271 to Asp-278. Of the residues forming the active-site pocket, Ala-47 and Trp-77 are conserved in all aldo–keto reductases (8). However, amino acid residues at positions 49 and 109 vary and are known to determine the substrate specificity for aldo–keto reductases (8). Because of its location, the C terminus (residues 271–277) may be involved in forming hydrogen bonds with the carbohydrate substrate and may also control the entry and alignment of the substrate in the active site. It has been reported that deleting the C-terminal eight amino acids of the enzyme results in a 40-fold decrease in Vmax (7). Similar mutagenesis studies have demonstrated a critical role for the C-terminal residues in the catalytic activity of aldose reductase (23).
Figure 6.
Stereo view of the active site of 2,5-DKGR A in the region of the binding site for the 2,5-DKG substrate. The van der Waals dot surface indicates the general shape of the substrate-binding pocket. The residues that define the pocket are indicated, as well as two solvent molecules (Sol 332 and 381) that reside within the pocket. These water molecules are postulated to be displaced by specific substrate functional groups (see text).
The active-site pocket of 2,5-DKGR A is also characterized by the presence of two well ordered water molecules, Sol-332 and Sol-381. These water molecules form a chain of hydrogen bonds involving Tyr-50 OH and Trp-109 Nɛ1 groups (Fig. 6). The relative positioning of these water molecules with respect to Tyr-50 OH, Trp-109 Nɛ1, His-108 Nɛ2, and NC4 of NADPH gives some clues regarding the orientation of the substrate carbonyl and hydroxyl groups in the active-site pocket (27). Sol-332, which hydrogen bonds with the hydroxyl group of Tyr-50, is postulated to be displaced by the 5-keto carbonyl group of the substrate. This carbonyl group is the one that undergoes reduction to a hydroxyl group in the enzymatic reaction. The solvent molecule 381 most likely is displaced by the 4-hydroxyl group of the substrate, forming a hydrogen bond with Trp-109 Nɛ1. This interpretation is consistent with the molecular dynamics and enzyme kinetics studies of pentose sugars with the pocket of human aldose reductase (27).
Catalytic Mechanism.
The catalytic mechanism of 2,5-DKGR A appears to be similar to that of aldose reductase and other members of this superfamily (8, 9, 28), constituting a two-step process. The first step involves the transfer of a hydride ion (H− ) from NADPH to the substrate, leaving the oxidized cofactor. This transfer is followed by the transfer of a proton (H+) to the substrate (28). It has been shown that the catalytic activity of 2,5-DKGR A is carried out by stereospecific transfer of the 4-pro-R hydrogen from the nicotinamide ring to the substrate (7). In the crystal structure of 2,5-DKGR A, the nicotinamide ring lies deep in the active-site pocket with the 4-pro-R hydrogen facing into the opening of the pocket where substrate is postulated to bind (Fig. 6). Therefore, a stereospecific hydride ion transfer from C4 of nicotinamide to the substrate carbonyl carbon atom most likely constitutes the mechanism of catalysis of this enzyme. However, concomitant with this, a complete reduction reaction would also require the protonation of the substrate carbonyl oxygen atom from a proton-donating functional group of the enzyme. There are two possible candidates for this activity as seen in the crystal structure of 2,5-DKGR A: Tyr-50 and His-108. Between these two candidates, Tyr-50 is considered to be the most likely proton donor group mainly because of its proximity with the C4 of nicotinamide and its involvement in the hydrogen bonding network with Lys-75, which in turn is hydrogen bonded to Asp-45 (Fig. 6). These interactions could facilitate the removal of a proton from Tyr-50. His-108 is buried in a hydrophobic environment, with Trp-109 on one side and Trp-77 on the opposite side. This does not favor the formation of a charged imidazolium group, and thus the abstraction of a proton. The critical role of Tyr-50 as the proton donor in aldo–keto reductases is supported by site-directed mutagenesis experiments (7, 28).
To improve the catalytic efficiency of 2,5-DKGR A, a series of mutations were made by substituting amino acids at analogous positions from 2,5-DKGR B, which is known to have better catalytic efficiency, albeit lower thermal stability (7). The structural determinants of these active-site mutants can now be identified on the basis of the crystal structure. A single Phe → Tyr substitution in 2,5-DKGR A at position 22 causes an approximately 2.5-fold decrease in Km for 2,5-DKG, from 31.2 to 12.3 μM, whereas the value of kcat is essentially unchanged (7). In the crystal structure, Phe-22 forms part of the substrate-binding pocket (Fig. 6) and is involved in backbone hydrogen bond formation with a cofactor hydroxyl group (Fig. 4). A Tyr residue at this site would be in a position to form a hydrogen bond with the sugar substrate. Thus, the observed decrease in Km may result from an additional enzyme–substrate hydrogen bond interaction. Preliminary model building studies of 2,5-DKG substrate bound in the active site suggest that the C3 substrate hydroxyl group is a potential hydrogen bonding partner.
In another experiment, Gln at position 192 was replaced with Arg (as in the B variant). This mutant exhibits a 6-fold increase in kcat value, from 8.4 sec−1 to 51 sec−1, with an associated 3.5-fold increase in Km (31 mM to 110 mM). In the crystal structure of human aldose reductase a Ser residue at the corresponding position adopts a χ1 angle of −164° and is orientated away from the bound NADPH cofactor. However, in 2,5-DKGR the Gln residue at position 192 adopts an alternative χ1 angle of −65°. This orients the side chain toward the NADPH cofactor, with the side-chain Nɛ1 atom hydrogen bonding with the NO1 oxygen of the cofactor pyrophosphate group (Fig. 6). Replacing Gln with an Arg at this position, and overlaying the Arg Nɛ group with the Gln Nɛ2, would thus replace this enzyme–cofactor hydrogen bond with a salt bridge. Furthermore, preliminary model building studies with bound substrate indicate that the Gln side chain Nɛ2 group at position 192 is also within hydrogen bonding distance of the bound substrate C6 hydroxyl group. Therefore, this side chain appears to interact with both cofactor and substrate. This interaction perturbs the substrate binding affinity but improves overall catalysis.
Another point mutant was constructed in which Ala-271 in the C-terminal tail was replaced with Gly. As mentioned earlier, the C-terminal eight amino acids are important for defining the substrate binding pocket geometry. Replacement of an Ala with a Gly at position 271 may allow a hinge-like rigid body movement of the residues in the C-terminal region. Such movement would alter the geometry of the substrate-binding pocket. Because this mutation increases the Vmax by approximately 2.6-fold (7) the reactive substrate carbonyl and hydroxyl groups are postulated to be more optimally oriented with the catalytic groups of the enzyme.
CONCLUSIONS
The crystal structure of 2,5-DKGR A, a prokaryotic member of the aldo–keto reductase superfamily, reveals both conserved and unique features in comparison with other aldo–keto reductases. Although its amino acid sequence is significantly different from the sequences of eukaryotic aldo–keto reductases, the structural fold remains the same (8). Common to other members of the superfamily, 2,5-DKGR A displays stereospecific binding to NADPH in which the cofactor binds in an extended conformation (9, 10, 22). However, there are distinct differences in the manner in which 2,5-DKGR recognizes NADPH. As observed in the crystal structures, the binding and release of NADPH cofactor necessitates a conformational change in the loop 7 region of human aldose reductase and other aldo–keto reductases (9, 10, 22). However, for 2,5-DKGR A, the association and dissociation of the cofactor are not expected to be accompanied by large backbone movements of loop 7. Consequently, cofactor binding is postulated not to display biphasic kinetics, as is the case with most other aldo–keto reductases (25, 26). The aldo–keto reductases recognize four broad categories of substrates: aldehydes, ketones, monosaccharides, and steroids. The presence of two crystallographically ordered solvent molecules in the active site provides structural clues to the location of a substrate hydroxyl and reactive keto moiety. The 2,5-DKGR model has provided a structural interpretation for three previously described active-site mutants. The basis for the altered catalytic properties of these mutants involves structural determinants of substrate binding, cofactor binding, and binding-pocket geometry. The 2,5-DKGR model will prove useful in the design of other active-site mutants, as well as mutants with enhanced thermal stability. In addition to addressing fundamental questions of catalysis, specificity, and protein stability, mutagenesis studies of 2,5-DKGR may also prove useful in the development of an efficient alternative method for the commercial production of vitamin C.
Acknowledgments
We thank Dr. T. Somasundaram for expert assistance during data collection and Drs. D. L. D. Caspar and B. Bhyravbhatla for helpful discussions. This project was supported in part by the Lucille P. Markey Charitable Trust. D.B.P. and S.A. were supported by National Institutes of Health Grants GM08339, R01AG11525, and R01GM50733.
ABBREVIATIONS
- 2
5-DKG, 2,5-diketo-d-gluconate
- 2-KLG
2-keto-l-gulonate
- 2
5-DKGR, 2,5-DKG reductase
Footnotes
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY 11973 (reference 1A80).
References
- 1.Reichstein T, Grussner A. Helv Chim Acta. 1934;17:311–328. [Google Scholar]
- 2.Kulhanek M. Adv Appl Microbiol. 1970;12:11–33. doi: 10.1016/s0065-2164(08)70581-1. [DOI] [PubMed] [Google Scholar]
- 3.Crawford T C, Crawford S A. Adv Carbohydr Chem Biochem. 1980;37:79–155. doi: 10.1016/s0065-2318(08)60020-7. [DOI] [PubMed] [Google Scholar]
- 4.Sonoyama T, Tani H, Matsuda K, Kageyama B, Tanimoto M, Kobayashi K, Yagi S, Kyotani H, Mitsushima K. Appl Environ Microbiol. 1982;43:1064–1069. doi: 10.1128/aem.43.5.1064-1069.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson S, Marks C B, Lazarus R, Miller J, Stafford K, Seymour J, Light D, Rastetter W, Estell D. Science. 1985;230:144–149. doi: 10.1126/science.230.4722.144. [DOI] [PubMed] [Google Scholar]
- 6.Jez J M, Flynn T G, Penning T M. Biochem Pharmacol. 1997;54:639–647. doi: 10.1016/s0006-2952(97)84253-0. [DOI] [PubMed] [Google Scholar]
- 7.Powers D P. Ph.D. thesis. Piscataway: Univ. of Medicine and Dentistry of New Jersey; 1996. [Google Scholar]
- 8.Jez J M, Bennett M J, Schlegel B P, Lewis M, Penning T M. Biochem J. 1997;326:625–636. doi: 10.1042/bj3260625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson D K, Bohren K M, Gabbay K H, Quiocho F A. Science. 1992;257:81–84. doi: 10.1126/science.1621098. [DOI] [PubMed] [Google Scholar]
- 10.Wilson D K, Nakano T, Petrash J M, Quiocho F A. Biochemistry. 1995;34:14323–14330. doi: 10.1021/bi00044a009. [DOI] [PubMed] [Google Scholar]
- 11.Hoog S S, Pawlowski J E, Alzari P M, Penning T M, Lewis M. Proc Natl Acad Sci USA. 1994;91:2517–2521. doi: 10.1073/pnas.91.7.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jancarik J, Kim S-H. J Appl Crystallogr. 1991;24:409–411. [Google Scholar]
- 13.Otwinowski Z. In: Proceedings of the CCP4 Study Weekend: “Data Collection and Processing,”. Sawyer L, Isaacs N, Bailey S, editors. Daresbury, England: Science and Engineering Research Council Daresbury Laboratory; 1993. pp. 56–62. [Google Scholar]
- 14.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 15.Brunger A, Kuriyan K, Karplus M. Science. 1987;235:458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- 16.Brunger A T. Nature (London) 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 17.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 18.Tronrud D E, Ten Eyck L F, Matthews B W. Acta Crystallogr A. 1987;43:489–501. [Google Scholar]
- 19.Tronrud D E. J Appl Crystallogr. 1996;29:100–104. [Google Scholar]
- 20.Banner D W, Bloomer A C, Petsko G A, Phillips D C, Pogson C I, Wilson I A, Corran P H, Furth A J, Milman J D, Offord R E, et al. Nature (London) 1975;255:609–614. doi: 10.1038/255609a0. [DOI] [PubMed] [Google Scholar]
- 21.Farber G K, Petsko G A. Trends Biochem Sci. 1990;15:228–234. doi: 10.1016/0968-0004(90)90035-a. [DOI] [PubMed] [Google Scholar]
- 22.Borhani D W, Harter T M, Petrash J M. J Biol Chem. 1992;267:24841–24847. doi: 10.2210/pdb1abn/pdb. [DOI] [PubMed] [Google Scholar]
- 23.Bohren K M, Grimshaw C E, Gabbay K H. J Biol Chem. 1992;267:20965–20970. [PubMed] [Google Scholar]
- 24.Zhang X-J, Matthews B W. J Appl Crystallogr. 1995;28:624–630. [Google Scholar]
- 25.Kubiseki T J, Hyndman D J, Morjana N A, Flynn T G. J Biol Chem. 1992;267:6510–6517. [PubMed] [Google Scholar]
- 26.Grimshaw C E, Shahbaz M, Putney C G. Biochemistry. 1990;29:9947–9955. doi: 10.1021/bi00494a027. [DOI] [PubMed] [Google Scholar]
- 27.De Winter H L, von Itzstein M. Biochemistry. 1995;34:8299–8308. doi: 10.1021/bi00026a011. [DOI] [PubMed] [Google Scholar]
- 28.Tarle I, Borhani D W, Wilson D K, Quiocho F A, Petrash J M. J Biol Chem. 1993;268:25687–25693. [PubMed] [Google Scholar]
- 29.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]