Summary
Ribosomal protein mutations, termed Minutes, have been instrumental in studying the coordination of cell and tissue growth in Drosophila. Although abundant in flies, equivalent defects in mammals are relatively unknown. Belly spot and tail (Bst) is a semidominant mouse mutation that disrupts pigmentation, somitogenesis and retinal cell fate determination. Here, we identify Bst as a deletion within the Rpl24 riboprotein gene. Bst significantly impairs Rpl24 splicing and ribosome biogenesis. Bst/+ cells have decreased rates of protein synthesis and proliferation, and are outcompeted by wild-type cells in C57BLKS↔ROSA26 chimeras. Bacterial artificial chromosome (BAC) and cDNA transgenes correct the mutant phenotypes. Our findings establish Bst as a mouse Minute and provide the first detailed characterization of a mammalian ribosomal protein mutation.
Keywords: Mouse, Genetics, Minute, Ribosome, Cell cycle, Retina, Bst
Introduction
The Minutes are an intriguing group of eukaryotic mutations that have been valuable in elucidating concepts of cell autonomy, compartmental development and cell competition (Garcia-Bellido et al., 1973; Morata and Ripoll, 1975; Moreno et al., 2002; Stern and Tokunaga, 1971). First described in Drosophila melanogaster more than 80 years ago, this dominantly inherited class of mutants comprises over 50 loci scattered across the Drosophila genome (Lambertsson, 1998; Schultz, 1929). Despite the large degree of locus heterogeneity, Minutes are remarkably similar, sharing three principal phenotypes – a 2–3 day delay in larval development, short thin bristles and recessive lethality. Variable phenotypes include small body size, female sterility, large or rough eyes, and abnormal wings. The haploinsufficiency and nonadditivity of these mutations, and their phenotypic similarity, led Schultz (Schultz, 1929) to propose that each Minute represents a unique member of a common metabolic or biochemical pathway. This prescient prediction was confirmed, as all of the molecularly cloned Minute loci to date encode ribosomal proteins (Lambertsson, 1998).
Although riboprotein mutations have also been identified in Escherichia coli and Saccharomyces cerevisiae (Warner, 1999), little is known about riboprotein mutations in mammals. The mammalian ribosome comprises 79 ribosomal proteins and four rRNAs, which combine in equimolar ratios to form the small (40S) and large (60S) subunits. Each riboprotein is encoded by a single gene. Despite the large number of riboprotein genes (Uechi et al., 2001), only one is known to be mutated in mammals. RPS19, a component of the small ribosomal subunit, is mutated in a quarter of humans with Diamond–Blackfan anemia (DBA, MIM205900), a congenital red blood cell hypoplasia (Draptchinskaia et al., 1999). In addition to anemia, these patients commonly have low birth weight, small stature, and craniofacial and skeletal defects. Aside from reduced somatic growth, it is unclear how these tissue-specific phenotypes result from mutations in a gene that is universally required for protein synthesis.
Belly spot and tail (Bst) is a semidominant, homozygous lethal mutation. Heterozygotes have decreased pigmentation and a kinked tail (Southard and Eicher, 1977). Bst/+ mice also have a significant retinal abnormality, characterized by delayed closure of the choroid fissure, marked deficiency of ganglion cells and subretinal neovascularization (Rice et al., 1995; Rice et al., 1997; Smith et al., 2000; Tang et al., 1999). Accordingly, Bst has been proposed as a model for human optic nerve atrophy and age-related macular degeneration.
Here, we report that the riboprotein gene Rpl24 is mutated in Bst/+ mice. The mutation impairs Rpl24 mRNA splicing and L24 production, which in turn affects ribosome biogenesis, protein synthesis and the cell cycle. Bst/+ cells have a significant growth disadvantage in chimeras that is similar to the classical cell competition effect observed in Drosophila Minute somatic mosaics. These findings establish Bst as a mouse Minute.
Materials and methods
Bst phenotyping
C57BLKS Bst/+ and +/+ mice were weighed daily from P5 to P21 and at 8 weeks of age. Skeletal preparations were stained with Alcian Blue and Alizarin Red (McLeod, 1980) and examined at P0 and P5. Paraffin sections (3 μm) of adult eyes were immunostained with a neurofilament (NF160) monoclonal antibody (NN18, 1:500, Sigma) as described (Brown et al., 2001).
Bst mapping
Mouse strains were obtained from The Jackson Laboratory (Bar Harbor, ME). An intersubspecific backcross was performed between (C57BLKS Bst/+ × CAST/Ei) F1 and C57BLKS +/+ mice. N2 progeny were scored at weaning for white spotting, white hind feet and tail kinks, and were genotyped using MIT markers (Dietrich et al., 1992) and four new microsatellites:
-
D16Ero1
Forward: 5′-CTTTCTCTAATCATGGCAAAAACA-3′
Reverse: 5′-TGAGCAAGACAATCACAGAGTATG-3′
-
D16Ero2
Forward: 5′-GAGCAAAATTTACGATGTGATTTG-3′
Reverse: 5′-ATCTCTATTGCACAAAGATGGACA-3′
-
D16Ero4
Forward: 5′-ACATAGTCAACAGACCGGACCT-3′
Reverse: 5′-ATGTTCTACTCGGTCTCTCCATCT-3′
-
D16Ero7
Forward: 5′-GCATTTAGTCAACACCATCAGAAT-3′
Reverse: 5′-CAAGCAAGCATTTCATACAAAAGT-3′.
PCR products were separated using 6% polyacrylamide sequencing or 4% Metaphor agarose gels. To confirm Bst genotypes, critical recombinants and N2 mice with low expressivity were test-crossed to C57BLKS. Opa1 was genotyped using PCR primers 5′-GAGCAGAAGAGCCCTGGAC-3′ and 5′-CAATGGAGGGTGGATGTTTC-3′ and a BlpI polymorphism in the 3′UTR. To type Hes1, a genomic segment corresponding to the terminus of bacterial artificial chromosome (BAC) clone RP24-110I11 was amplified using primers 5′-TTATTTGTTTCTACGACCCAGCTT-3′ and 5′-ACTGGTCTTGAACTCCTAGACTCC-3′ and scored for a single-strand conformational polymorphism (SSCP) by MDE gel electrophoresis (FMC Bioproducts).
Rpl24 genotyping
The Bst allele was typed by PCR, using upstream primer 5′-TTTGCAGCGCACATACGAG-3′ which overlaps the first intron branchpoint deletion and downstream primer 5′-GCTGACTCACATTTTGCATTAAGA-3′ within the third exon. The wild-type allele was amplified using upstream primer 5′-CTCTTTGCAGCGCACATACTAAC-3′, which overlaps the intact first intron branchpoint and downstream primer 5′-GGAAAACCTGCAGTTAACAAATTC-3′ within the second intron. PCR parameters were 3 minutes at 95°C, followed by 40 cycles (30 seconds at 95°C, 1 minute at 56°C, 1 minute at 72°C) and 7 minutes extension at 72°C.
Reverse transcriptase PCR
Total RNA was extracted using Trizol reagent (Invitrogen) from livers of C57BLKS+/+, C57BLKS Bst/+ and SPRET/Ei adult mice; livers of P21 weanlings and embryonic day (E) 18.5 embryos derived from a (C57BLKS Bst/+ × CAST/Ei) F1 × SPRET/Ei cross; and heads of co-isogenic C57BLKS Bst/+ and +/+ E13.5 embryos. First strand cDNA synthesis was primed using random hexamers. To evaluate Bst candidate genes, 40 cycles of RT-PCR were performed on E13.5 head and body RNA using gene-specific primers. To evaluate Bst effects on Rpl24 splicing, three RT-PCRs were performed (designated A, B and C in Fig. 3D,E). These reactions share a common downstream primer (5′-CTGTCTTCTTTGATGCCTGCTTAG-3′), which was 32P or HEX end-labeled. The upstream primers were 5′-GCAGGCCGACATCTATCAC-3′ (A), 5′-GGATGGCTCCTCTTTGCAG-3′ (B) and 5′-ACATCTATCACCATGAAGGTCGAG-3′ (C). The upstream primer in reaction C spans the exon 1–2 junction and only amplifies from correctly spliced Rpl24 mRNA. PCR parameters were 3 minutes at 95 °C, followed by 35–40 cycles (30 seconds at 95°C, 1 minute at 56°C, 1 minute at 72°C) and 7 minutes at 72°C. To eliminate heteroduplex products in reaction C, a final 10-minute extension step was performed after adding fresh PCR reagents. Samples were then digested overnight with AluI at 37°C. 32P-labeled products were denatured at 95°C, separated on 6% polyacrylamide sequencing gels and analyzed using a Phosphorimager (Amersham Biosciences). HEX-labeled products were separated on an ABI sequencer.
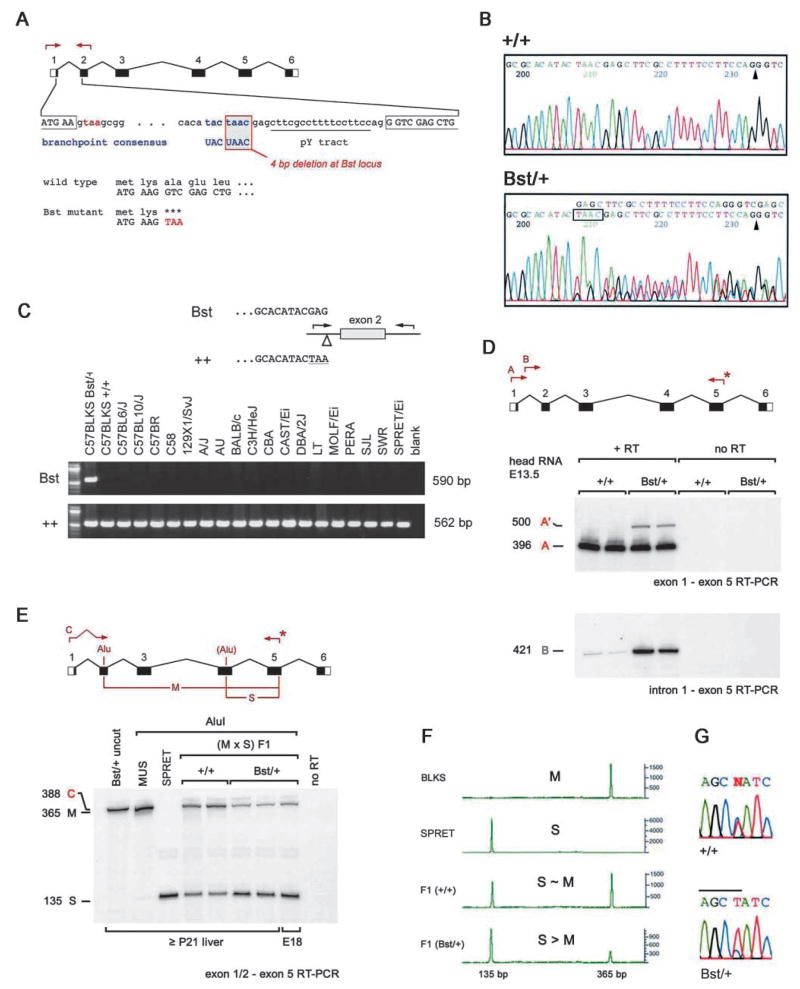
Fig. 3.
The Bst mutation. (A) Map of the Rpl24 gene showing the informative PCR, the 4 bp deletion within the intron 1 branchpoint, the first codons of the open reading frame, and the premature stop codon in Bst (red). (B) Sequence chromatograms comparing +/+ and Bst/+ PCR products. The exon 2 splice acceptor (arrowheads) and 4 bp Bst deletion (boxed) are indicated. (C) Allele-specific PCR assay used to distinguish Bst and wild-type Rpl24 alleles. The deletion is unique to the C57BLKS Bst/+ strain. (D) Bst causes abnormal Rpl24 splicing with retention of intron 1. RT-PCR was performed using the indicated primers. A common downstream primer (*) was end-labeled with 32P using T4 DNA kinase. Product A originates from correctly spliced Rpl24 transcripts, whereas A′ and B indicate inclusion of intron 1. The relatively low abundance of product A′ is due to PCR competition and nonsense-mediated decay. The low level of product B in the wild-type lanes most probably reflects amplification from hnRNA. (E) Some Bst transcripts are correctly spliced. RT-PCR was performed using (Bst/+ × SPRET) F1 RNA and the indicated primers. The upstream primer spans the exon 1–2 junction. Product C (388 bp) was end-labeled and digested with AluI to distinguish between musculus (M, 365 bp) and spretus (S, 135 bp) alleles. The molar ratio of M and S products is 1.27 for +/+ mice and 0.26 for Bst/+ littermates, giving a normalized expression level for Bst of 0.21 (±0.02). (F) Similar fluorometric assay performed using a HEX-labeled reverse primer and ABI sequencer following AluI digestion. The Bst expression level is 0.25 (±0.03) relative to wild-type musculus Rpl24. (G) Sequence of correctly spliced Rpl24 cDNAs amplified from F1 mice, showing the informative AluI site in exon 4 (overlined, sense strand). The normalized ratio of Bst to wild-type peak areas is 0.22 (±0.01).
Rpl24 transgenic analysis
BAC clone RP23-10L10 (accession no. AZ117821) contains the 5.2 kb Rpl24 transcription unit flanked by 93 kb 5′ and 83 kb 3′ genomic DNA. Purified BAC DNA was injected into pronuclei of (C57BL/6 × SJL) F2 eggs, generating 17 transgenic founders. The integrity of the BAC ends was assessed in founders by PCR, using primer pairs that span the vector-insert junctions:
-
10L10 Prox-2
Forward: 5′-TCGAGCTTGACATTGTAGGACTAT-3′
Reverse: 5′-AGGGTGGTTTCAGTGTGTCG-3′
-
10L10 Dist-1
Forward: 5′-GATGTTCATGTTCATGTCTCCTTC-3′
Reverse: 5′-CCCTCAATCTCATTAACCTTCTGT-3′.
To confirm the presence of Rpl24 within each transgene, we crossed founders to SJL mice and compared the ratio of SJL and C57 alleles in offspring using a Sau3A1 RFLP within intron 4 or a microsatellite marker (D16Ero9) located 1 kb upstream of Rpl24. D16Ero9 was typed using primers 5′-CTGGCTAGCATAGCTTTTCTTTTT-3′ and 5′-GGAAAGGTGAGCAGGTTAAGAATA-3′. To evaluate transgene correction, founders were crossed to C57BLKS Bst/+ mice. The resulting progeny were examined for Bst phenotypes and assessed using BAC-specific and Rpl24 allele-specific PCRs.
The human RPL24 cDNA was amplified by PCR from IMAGE clone 5609137 (Open Biosystems) using an upstream primer with an engineered BamHI site (5′-CGTGGATCCGTCGCCATGAAGGTCGAGCTGTG-3′) and a downstream primer with an engineered EcoRI site (5′-TTTAGAATTCTAATCTGCCAGTTTAGCGTTTTCC-3′). Following digestion, the resulting 0.5 kb BamHI-EcoRI fragment was subcloned into BglII- and EcoRI-digested pBROAD3 (Invivogen). The final construct (R26-huL24) contains the entire 471 bp RPL24 coding sequence, is transcribed from the 1.9 kb murine ROSA26 promoter, and terminates in a β-globin polyadenylation site (Fig. 4D). The 3.0 kb transgene was excised with PacI, gel purified and injected into pronuclei of coisogenic C57BLKS Bst/+ and +/+ eggs. Transgenic mice were identified by PCR using primers 5′-ACAGGTGTGAAACAGGAAGAGAAC-3′ and 5′-GGGAACAAAGGAACCTTTAATAGA-3′, which selectively amplify the human cDNA construct.
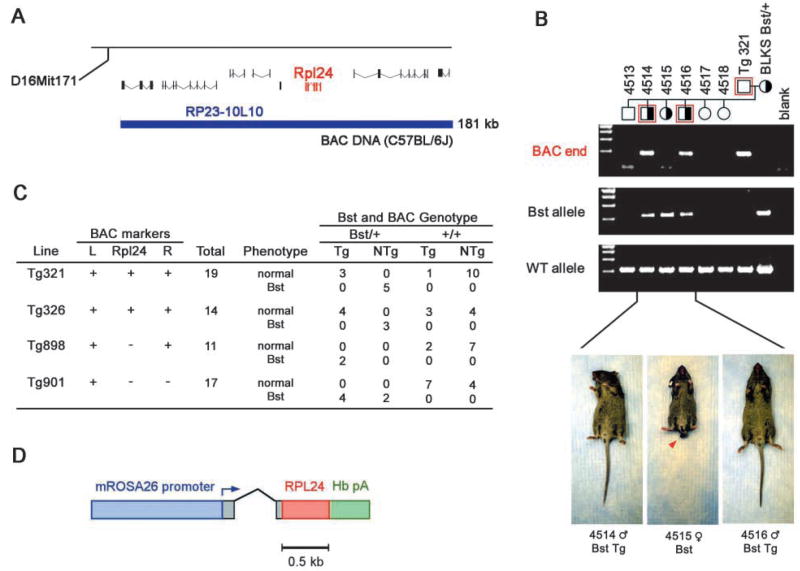
Fig. 4.
Transgene correction of the Bst phenotype. (A) BAC clone RP23-10L10 (blue) contains Rpl24 and five other genes, and spans 181 kb (Ensembl mouse genome database, v. 12.3.1). (B) Founder Tg321 carries an intact RP23-10L10 transgene, which suppresses external Bst phenotypes in all doubly heterozygous offspring (compare mice 4514 and 4516 with 4515). Genotypes were determined using BAC end (red) and allele-specific Rpl24 PCRs. (C) Correction data summary. Four BAC transgenic founders were mated to C57BLKS Bst/+ mice. Tg321 and Tg326, which are intact, correct Bst phenotypes but Tg898 or Tg901, which lack Rpl24, do not. (D) Human RPL24 transgene (R26-huL24). The cDNA is ubiquitously expressed using the murine ROSA26 promoter.
Pulse-labeled rRNA analysis
Sex-matched adult littermates were fasted for 48 hours and re-fed for 2 hours. This fasting protocol decreases liver mass twofold (Hutson and Mortimore, 1982; Volarevic et al., 2000). Immediately before re-feeding, each mouse was given an intraperitoneal injection of 32P-orthophosphate (30 μCi per gram body weight). Livers were snap frozen after re-feeding. Total liver RNA (1–10 μg per lane) was separated by formaldehyde agarose gel electrophoresis, transferred to a nitrocellulose membrane and exposed to X-ray film (Biomax, Kodak). Density tracings were analyzed using NIH Image software (Scion). The membrane was stained with Methylene Blue to confirm equal RNA loading.
Polysome profiles
Livers from sex-matched adult littermates were homogenized in ice-cold hypotonic lysis buffer (1.5 mM KCl, 2.5 mM MgCl2, 5 mM Tris-HCl pH 7.4, 1% Na deoxycholate, 1% Triton X-100) using 1 ml buffer per 10 mg liver. After centrifugation for 15 minutes at 2500 g, heparin (1 mg/ml) and cycloheximide (100 μg/ml) were added, and 700 μl of the supernatants were applied to 13 ml 7–47% (w/v) sucrose gradients. Samples were centrifuged in a Beckman SW-41 rotor at 288,000 g for 2 hours at 4°C. Gradients were collected using a Biocomp fractionator with UV photometer.
Murine embryonic fibroblasts
Murine embryonic fibroblast (MEF) cultures were generated at E13.5 from co-isogenic C57BLKS Bst/+ and +/+ embryos and from (CAST/Ei × C57BLKS Bst/+) F2 embryos as described (Robertson, 1987) and tested at equivalent early passages (<P8). Experiments were performed in triplicate unless otherwise noted.
To evaluate protein synthesis, MEFs were plated at a density of 5×104 cells per 35 mm dish. After 24 hours, cells were incubated for 2 hours in 2 ml fresh media containing 10 μCi [3H]leucine, washed with ice-cold phosphate buffered saline (PBS), and treated with 5% trichloroacetic acid (TCA) for at least 2 hours at 4°C. [3H]leucine incorporation was measured after lysis in 0.5 N NaOH, 0.5% sodium dodecyl sulphate (SDS) and was normalized to mitochondrial dehydrogenase activity in parallel cultures, measured using a water soluble tetrazolium (WST) assay (Roche).
To measure cell-doubling time, parallel cultures were plated in complete media at a density of 5×104 cells per 35 mm dish. Starting 36 hours after plating, adherent cells were harvested at 12-hour intervals and counted using a Coulter Z2 counter. The growth rate was determined between 36 and 60 hours after plating.
Bivariate cell cycle analysis was performed on asynchronous cultures using a BrdU Flow Kit (BD Pharmingen). Cultures were labeled with 10 μM BrdU for 75 minutes, stained with an FITC-conjugated anti-BrdU antibody (new DNA synthesis) and 7-AAD (total DNA content), and sorted using a Coulter Elite ESP Fluorescence Cell Sorter. The fraction of cells in G1, S and G2/M is directly related to the duration of each phase in the cell cycle (Gray et al., 1990). Flow cytometry data were analyzed using WinMDI v.2.8 software.
To evaluate S-phase entry after serum starvation, parallel MEF cultures were plated in complete media at 5×104 cells per 35 mm dish. After plating, cells were washed twice with serum-free media and re-fed Dulbecco’s modified Eagle’s medium (DMEM) containing 0.5% fetal bovine serum (FBS) for 48 hours. Cultures were released from serum starvation (Herrera et al., 1996) by the addition of complete DMEM containing 15% FBS. DNA synthesis was measured periodically by adding 0.8 μCi [3H]thymidine per dish for 1 hour. These cultures were washed twice with ice-cold PBS and treated with 5% trichloroacetic acid (TCA) for at least 3 hours at 4°C. The cells were washed, lysed in 0.5 N NaOH, 0.5% SDS and counted for [3H]thymidine incorporation. This experiment was repeated using MEFs derived from three separate litters.
Chimera analysis
E3.5 blastocysts were harvested as described (Robertson, 1987) from 24- to 30-day-old C57BLKS +/+ females that had been superovulated and mated to C57BLKS Bst/+ males. Blastocysts were microinjected with exactly five or ten ROSA26 embryonic stem (ES) cells, which were between passage 15 and 18 (kindly provided by Liz Robertson and Phil Soriano). After reimplantation into pseudopregnant (C57BL/6 × DBA) F2 females, chimeras were analyzed at E12.5 or after birth.
The percent agouti coat color was estimated independently by our lab and the University of Michigan transgenic core at the time of weaning. Rpl24 genotypes were determined afterward from tail DNA by allele-specific PCR. To further evaluate the extent of chimerism, adult tissues were fixed at 4°C for 2–3 hours in 2% paraformaldehyde, 0.2% glutaraldehyde 0.1 M NaPO4 (pH 7.3), 0.02% NP40, 0.02% Na deoxycholate; cryopreserved through 10–30% sucrose in PBS with 5 mM MgCl2; and embedded in optimal cutting temperature (OCT) medium (TissueTek). Cryosections (10 μm) were stained for β-galactosidase in 100 mM NaPO4 (pH 7.3), 3 mM K3Fe(CN)6, 3 mM K4Fe(CN)6, 1.3 mM MgCl2, 0.1% X-gal for 7 hours at 37°C, counterstained with neutral red, dehydrated and mounted in Permount.
Genomic DNA was extracted from E12.5 embryos, amplified by PCR, and tested using an XbaI polymorphism in the βh1 locus that distinguishes 129/SvEv (ROSA26) and C57BLKS alleles (Fiering et al., 1995). The relative abundance of alleles was determined using a Phosphorimager.
Results
Dysmorphic features and reduced somatic growth in Bst/+ mice
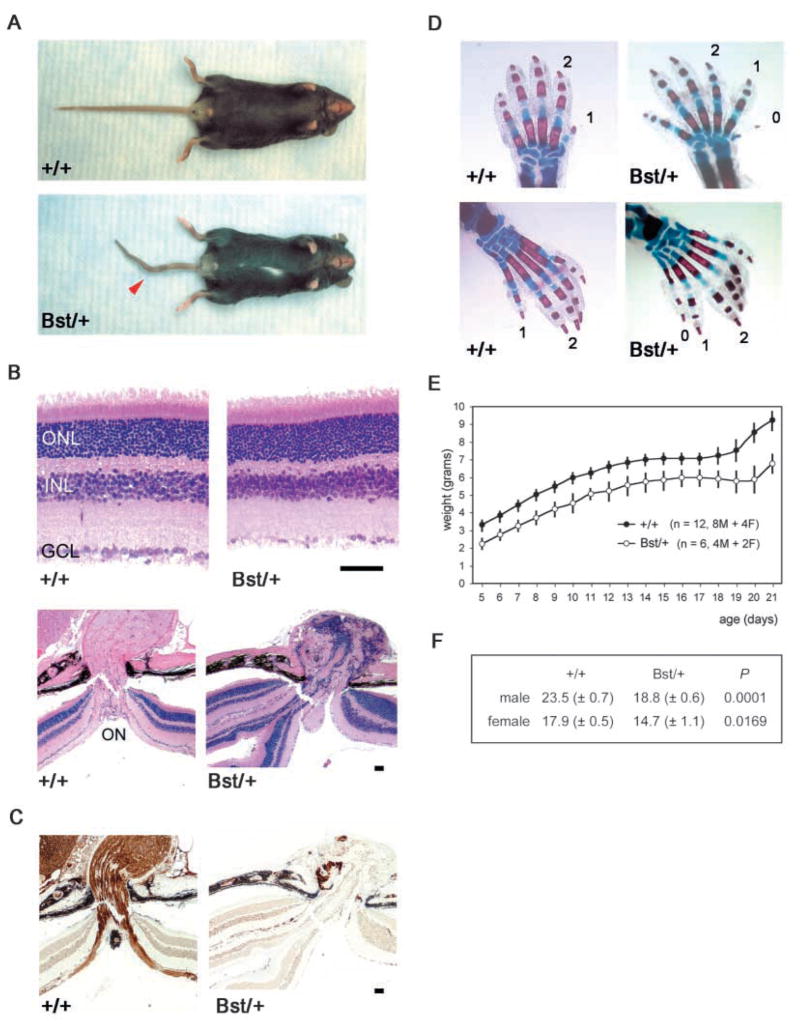
Bst/+ mice have a pleiotropic phenotype that includes a white ventral midline spot, white hind feet and a kinked tail (Fig. 1A). Bst homozygotes die before E9.5 (data not shown). Abnormal retinal histogenesis in Bst/+ mice causes a 30–100% reduction in the number of retinal ganglion cells (RGCs), with some laminar disorganization, hypoplastic or absent optic nerves, and occasional optic nerve colobomas (Fig. 1B,C) (Rice et al., 1997). In addition to tail kinks, there are other skeletal abnormalities. Bst/+ mice frequently have a triphalangeal first digit and an extra preaxial digit (Fig. 1D). They also have six lumbar vertebrae, compared with the five in co-isogenic C57BLKS controls (data not shown). Skeletal stains show fewer caudal vertebrae in Bst/+ mice and fused or wedge-shaped hemivertebrae at the sites of tail kinks (data not shown).
Fig. 1.
C57BLKS Bst/+ phenotypes. (A) Kinked tail (arrowhead), white hind feet and belly spot. (B) Hematoxylin and eosin staining of +/+ and Bst/+ retinal sections. (C) Neurofilament (NF160) immunostaining of adjacent sections at the optic nerve head, marking ganglion and horizontal cells. (D) Skeletal stain of newborn fore limbs (upper) and hind limbs (lower), showing preaxial polydactyly (0) and triphalangy of the first digit (1) in Bst/+ limbs. (E) Postnatal growth curves of littermate +/+ and Bst/+ pups. (F) Weights (grams±s.e.m.) of adult +/+ and Bst/+ littermates, compared using a two-tailed unpaired t-test (n=44 total). Scale bars in B,C: 50 μm. ON, optic nerve head; ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer.
Bst/+ mice appear smaller than wild-type littermates. To determine the onset and magnitude of this size difference, we compared growth curves and adult weights of co-isogenic cohorts. The Bst/+ mice were 20% smaller than wild-type littermates through the first 3 postnatal weeks and remained so into adulthood (Fig. 1E,F).
Molecular identification of Bst
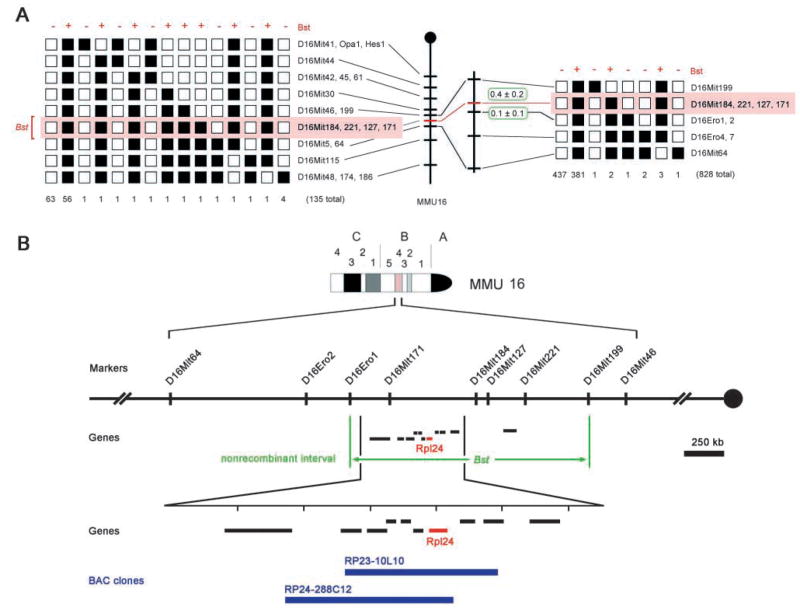
Bst arose spontaneously in the inbred C57BLKS strain (Southard and Eicher, 1977) and was mapped to a 10 cM interval on mouse chromosome 16 (Rice et al., 1995). To further localize Bst, we performed an intersubspecific backcross between (C57BLKS Bst/+ × CAST/Ei) F1 and C57BLKS+/+ mice. N2 progeny were scored for Bst pigmentation and tail phenotypes and genotyped with informative microsatellite DNA markers. Among 828 N2 progeny, 386 were Bst/+. This segregation ratio (386:442) is less than 1:1 (P=0.092), consistent with decreased viability of Bst/+ mice. Bst maps within a 0.5 cM interval, delimited by D16Mit199 and D16Ero1 (Fig. 2A). Our data formally exclude Opa1, the mouse homolog of the human optic nerve atrophy gene (MIM605290), and the Hes1 basic helix-loop-helix transcription factor, which has roles in retinal and somite development (Saga and Takeda, 2001; Tomita et al., 1996). Both genes lie more than 5cM centromeric to Bst (Fig. 2A).
Fig. 2.
Positional cloning of Bst. (A) Recombination data localizing Bst to a 0.5 cM interval on mouse chromosome 16 (shaded in red) and excluding Opa1 and Hes1. Bst and microsatellite alleles transmitted by F1 parents are indicated together with the number of N2 progeny in each class. The 135 N2 mice typed with the extended marker set (left) are included in the total (right). □, CAST/Ei; ■, C57BLKS. (B) Physical map of critical Bst region derived from Ensembl mouse genome database (3.35 Mb, v. 4.1.1). The map shows informative microsatellite markers, candidate transcription units and relevant BAC clones (blue). The nonrecombinant interval maps within cytogenetic band B4, spans 1.5 Mb (green arrows) and includes Rpl24 (red).
The non-recombinant interval spans 1.5 Mb (Fig. 2B) and contains at least 18 known or predicted genes (Ensembl mouse and UC Santa Cruz human databases, April 2002). Eleven are expressed in E13.5 head and E15.5 eye (data not shown) and were evaluated as Bst candidates by comparing genomic DNA sequences from C57BLKS Bst/+ and +/+ mice. These studies revealed only a single change, within Rpl24 (Fig. 3A,B).
Rpl24 encodes a protein component of the large (60S) ribosomal subunit. L24 is a highly basic protein of 157 amino acids and is conserved among eukaryotes and archaebacteria. Its position near the polypeptide exit and elongation factor binding sites (Ban et al., 2000), its contacts with the 40S subunit (Spahn et al., 2001) and the properties of L24 mutant yeast (Dresios et al., 2000) suggest that L24 helps catalyze the peptidyl transferase reaction. Rpl24 spans six exons, extends over 5.2 kb and contains a characteristic 5′ terminal oligopyrimidine (TOP) initiator element (Hariharan et al., 1989). More than 20 processed Rpl24 pseudogenes are interspersed throughout the genome (data not shown), similar to other riboprotein genes (Zhang et al., 2002). However, only one of these loci, within the Bst critical interval, contains introns and is therefore the bona fide Rpl24 gene.
Rpl24 branchpoint mutation alters mRNA splicing
Bst destroys the first intron splice branchpoint of Rpl24, which matches the consensus sequence (UACUAAC) exactly. The deletion removes four nucleotides, including the critical adenosine (underlined) that reacts to form the lariat intermediate (Fig. 3A,B). No other obvious branchpoints are present within this 108 bp intron. Using an allele-specific PCR assay, we determined that the deletion in Rpl24 is unique to the Bst mice (Fig. 3C). This branchpoint is intact in the parental C57BLKS strain, highly related C57 strains, different Mus species (M. spretus, M. castaneus and M. molossinus) and rat Rpl24 (accession no. X78443).
To determine how Bst affects Rpl24 splicing, we performed RT-PCR experiments (Fig. 3D). Using primers in exons 1 and 5, we amplified a novel 500 bp product from Bst/+ RNA, which reflects inclusion of intron 1. No other products were detected, including a 320 bp product that would result if exon 2 were skipped. This result was confirmed by direct PCR sequencing and a second RT-PCR using primers in intron 1 and exon 5 (Fig. 3D). Rpl24 translation begins in the first exon (Fig. 3A). Retention of intron 1 would cause premature termination of the L24 polypeptide after the first two codons. The first intron contains two other stop codons and the next AUG codon (M91) is in the fourth exon.
Activation of a cryptic branchpoint could restore proper splicing (Ruskin et al., 1985). Because Bst homozygotes are not available, we evaluated Rpl24 splicing in interspecies hybrid mice, derived from a (C57BLKS Bst/+ × CAST/Ei) F1 × SPRET/Ei cross, by a PCR strategy, using a polymorphic AluI restriction site in exon 4 and an upstream primer spanning the exon 1–2 junction. This allowed us to resolve and compare the amount of correctly spliced Rpl24 transcripts from wild-type and Bst alleles (Fig. 3E–G). These results show that 20–25% of Bst transcripts are correctly spliced. Bst is therefore hypomorphic. Inbred C57BLKS Bst heterozygotes and homozygotes thus have approximately 60% and 20% of normal Rpl24 transcript levels, respectively.
Rpl24 BAC and cDNA transgenes correct Bst
Like the Drosophila Minutes, Bst has general effects on somatic growth and specific effects on different tissues (Fig. 1). To confirm that the Rpl24 mutation is solely responsible for the pleiotropic Bst phenotypes, we created transgenic mouse lines expressing wild-type Rpl24 from a 181 kb mouse BAC (RP23-10L10) or a human cDNA expression cassette (Fig. 4A,D). Four BAC transgenic lines were bred to C57BLKS Bst/+ mice (Fig. 4C). Founders Tg321 and Tg326 have intact transgenes that completely correct the Bst phenotypes (Fig. 4B,C). Founders Tg898 and Tg901 have partial BAC transgenes lacking Rpl24 and these fail to correct the Bst phenotype. We generated cDNA transgenics using human RPL24 and the ubiquitously active murine ROSA26 promoter (Zambrowicz et al., 1997). Human and mouse L24 proteins are identical. We injected co-isogenic C57BLKS Bst/+ and +/+ eggs with the R26-huL24 construct (Fig. 4D) and recovered seven transgenic founders. Three of the four Bst/+ founders were indistinguishable from wild-type littermates (data not shown), proving that RPL24 alone is sufficient to correct the Bst phenotype and that Rpl24 is Bst. Taken together, our data show Bst acts via a partial loss-of-function mechanism and should be considered a mouse Minute.
Bst impairs ribosome biogenesis
Ribosome biogenesis requires highly coordinated synthesis of ribosomal proteins and rRNAs (Warner, 1999). These are assembled in a tightly ordered process involving more than 200 accessory proteins and small nucleolar ribonucleoproteins (Fatica and Tollervey, 2002). Decreased availability of individual components can impair these events (Saveanu et al., 2001; Volarevic et al., 2000; Zhao et al., 2003).
Although large and small subunits assemble independently, transcription and processing of the 45S rRNA precursor into 18S and 28S rRNAs couples formation of both subunits (Fatica and Tollervey, 2002). As such, rRNA processing is a sensitive indicator of ribosome biogenesis. We therefore examined rRNA synthesis in a pulse labeling experiment. Sex-matched co-isogenic C57BLKS Bst/+ and wild-type littermates were fasted for 48 hours, injected with 32P-orthophosphate and re-fed for 2 hours. Profiles of unlabeled (steady state) rRNA were similar after re-feeding (Fig. 5A, Methylene Blue), but the amount of newly processed rRNA in the Bst/+ mouse was dramatically decreased (Fig. 5A, autoradiogram). The 28S rRNA was reduced to a greater extent than the 18S rRNA, consistent with a defect in large subunit assembly (Fig. 5A, densitometry). In yeast, mutations specific to the 60S subunit can affect 18S rRNA processing indirectly, most probably through the exosome (Zanchin et al., 1997). This nuclease complex degrades improperly spliced mRNAs, excess rRNA precursors and unused rRNA intermediates, and is required for rRNA processing (Allmang et al., 2000; Butler, 2002; Mitrovich and Anderson, 2000). Likewise, ribosome subunits are stable once assembled and most unincorporated riboproteins are rapidly eliminated (Warner, 1977). Bst is therefore unlikely to affect the abundance of L24 at steady state. The labeling of the 34S and 45S pre-rRNAs was similar in both mice (Fig. 5A, autoradiogram), suggesting that the decrease in 28S rRNA is due to increased turnover rather than a specific bottleneck in rRNA processing. This cellular response is consistent with a dynamic shortage of L24 polypeptide.
Fig. 5.
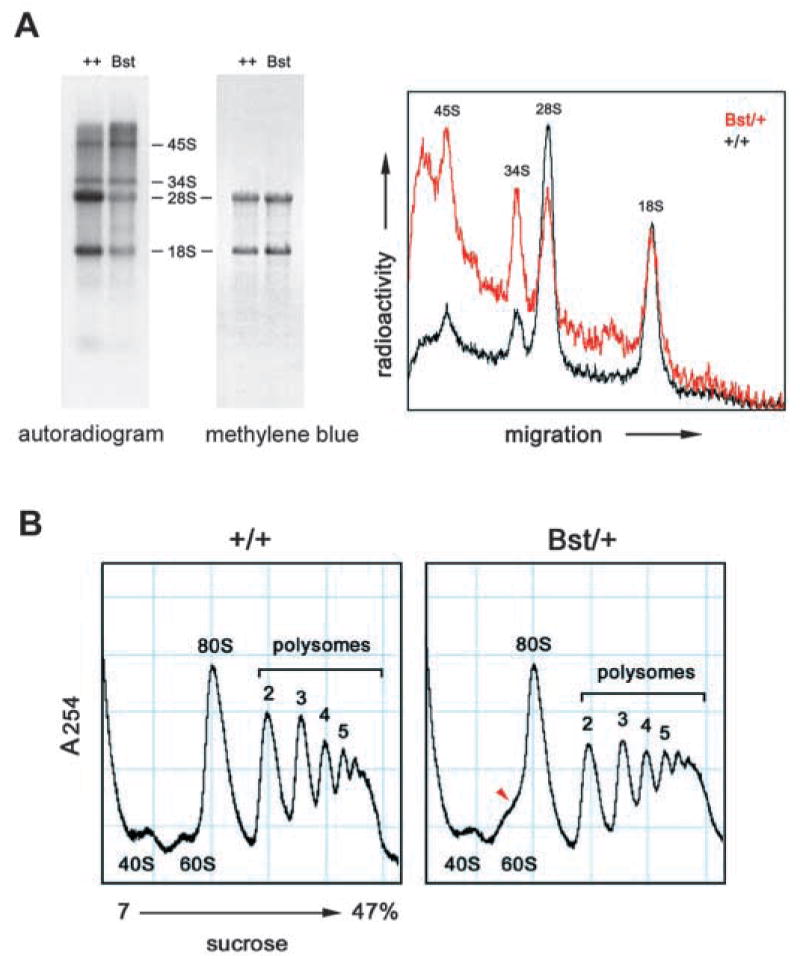
Bst impairs ribosome biogenesis. (A) Bst/+ causes a decrease in processed rRNAs. C57BLKS Bst/+ and +/+ littermates were fasted for 48 hours and re-fed for 2 hours. Tissue RNAs were radiolabeled by an intraperitoneal 32P injection at the start of the re-feeding period. (left) Autoradiogram of pulse-labeled liver RNA (5 μg/lane). Mature 18S and 28S rRNAs and 34S and 45S precursors are indicated. (center) Methylene Blue stain of the same filter, showing total RNA. (right) Density tracing of autoradiogram showing that Bst has a greater effect on 28S rRNA. The Bst/+ profile is scaled twofold to equalize 18S rRNA levels. The relative peak areas measured for 45S, 34S, 28S and 18S rRNAs are 0.9, 1.1, 5.9 and 4.2 (+/+) and 0.9, 1.1, 1.5 and 1.8 (Bst/+), respectively. (B) Sucrose gradients of liver homogenates from normally fed +/+ and Bst/+ adult littermates. UV absorbance peaks corresponding to ribosomal subunits and polysomes are labeled. A prominent shoulder is present between the 60S and 80S subunits (arrowhead).
There were no changes in the ratio of small to large subunits or in the polyribosome fraction of Bst/+ liver homogenates (Fig. 5C). However, a prominent shoulder between the 60S subunit and 80S monosome peaks was consistently observed in sucrose gradients from Bst/+ mice, compatible with an alteration in 60S subunit structure. This change was observed under fed, fasted and re-fed conditions (data not shown).
Bst/+ fibroblasts have altered properties
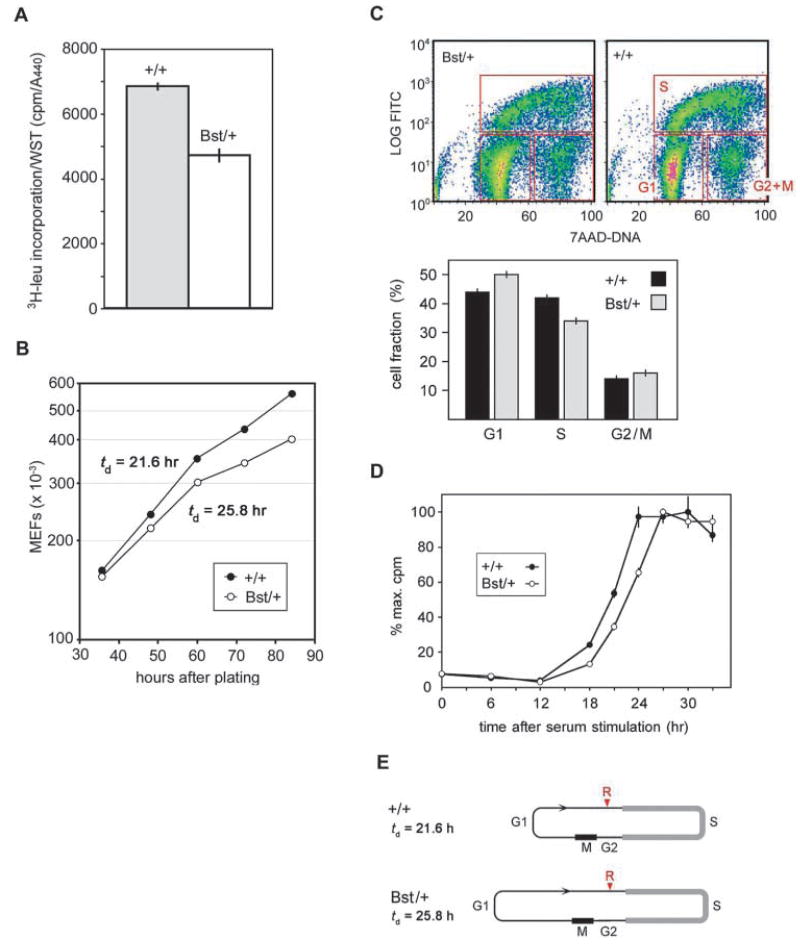
In spite of its apparent tissue specificity, L24 deficiency should affect every cell in the Bst/+ mouse, in the same way that cell growth is universally slower in Minute flies (Simpson and Morata, 1981). To test this hypothesis, we examined cell size, protein synthesis, proliferation rates and cycle kinetics in exponentially growing mouse embryonic fibroblast (MEF) cultures. We found no difference in size between Bst/+ and wild-type MEFs by flow cytometry (forward light scatter, data not shown). This is consistent with recent studies in Drosophila showing no size difference between Minute and wild-type imaginal disc cells (Montagne et al., 1999). We examined overall protein synthesis in MEFs by [3H]leucine incorporation, normalized to cell viability (Fig. 6A). Bst/+ cultures showed a 31.2±5.8% reduction in the rate of protein synthesis, similar to the 30% reduction described for M(3)i55/+ Drosophila Minute embryos (Boring et al., 1989).
Fig. 6.
Cellular effects of the Bst mutation. (A) Protein synthesis ([3H]leu incorporation) is decreased in Bst/+ MEFs (P<0.001, two-tailed unpaired t-test). (B) Growth rates of subconfluent +/+ and Bst/+ MEF cultures (cell counts), showing decreased doubling time of Bst/+ cells (P<0.001, two-tailed unpaired t-test). (C) Bivariate cell cycle analysis. Cells were sorted by total DNA content (7-AAD fluorescence) and BrdU incorporation (FITC-conjugated antibody). Density plots represent combined data from two experiments (20,000 cells each). The histogram shows the proportion of cells in G1, S and G2/M sectors (red boxes); bars indicate the range for two experiments. The distribution of cells in G1, S and G2/M differ significantly between Bst/+ and +/+ (P<0.001, χ2=233 for 2×3 contingency table). (D) S-phase entry of Bst/+ MEFs is delayed following 48 hour serum starvation. [3H]thymidine incorporation was normalized for each genotype to the highest level measured during the experiment. The panel shows one of three independent experiments performed. Each point shows the average of three cultures; bars indicate standard deviation. (E) Model for increased cell cycle length, based on bivariate FACS analysis and doubling times. Assuming the length of S phase is constant, Bst/+ MEFs remain significantly longer in G1 before reaching the restriction point (R).
Bst/+ MEFs proliferate significantly more slowly than wild-type cultures. The population doubling time (Td) is 25.8±0.4 hours for Bst/+ MEFs compared with 21.6±0.2 hours for wild-type MEFs (Fig. 6B). To determine what part of the cell cycle is elongated, we performed bivariate flow cytometry on asynchronous MEF cultures using BrdU incorporation and DNA content as sorting parameters. The ratio of G1 to S-phase cells was 1.45 (±0.05) for Bst/+ MEFs and 1.04 (±0.01) for control MEFs (Fig. 6C). This suggests that Bst causes actively growing cells to remain longer in G1 phase. A small increase in the G2/M fraction was also observed. If the absolute length of S phase is constant (9.1 hours) and the cell cycle is increased by 20% overall (Fig. 6B), then G1 phase is increased by 40% in Bst/+ MEFs (Fig. 6E).
To further investigate the prolongation of G1 by Bst, we compared the time required for Bst/+ MEF cultures to enter S phase after serum stimulation. Parallel cultures were arrested in G1 by serum starvation, released by the addition of serum-rich media, and assayed periodically for DNA synthesis by [3H]thymidine incorporation. We observed a 2.4±0.5 hour increase in the time required to reach half-maximal incorporation in Bst/+ cultures compared with wild type (Fig. 6D). These results confirm the increased length of G1 in Bst/+ MEFs and are consistent with classical studies in which the protein synthesis inhibitor cycloheximide was shown to specifically lengthen G1 phase following serum stimulation (Brooks, 1977).
Wild-type cells outcompete Bst/+ cells during development
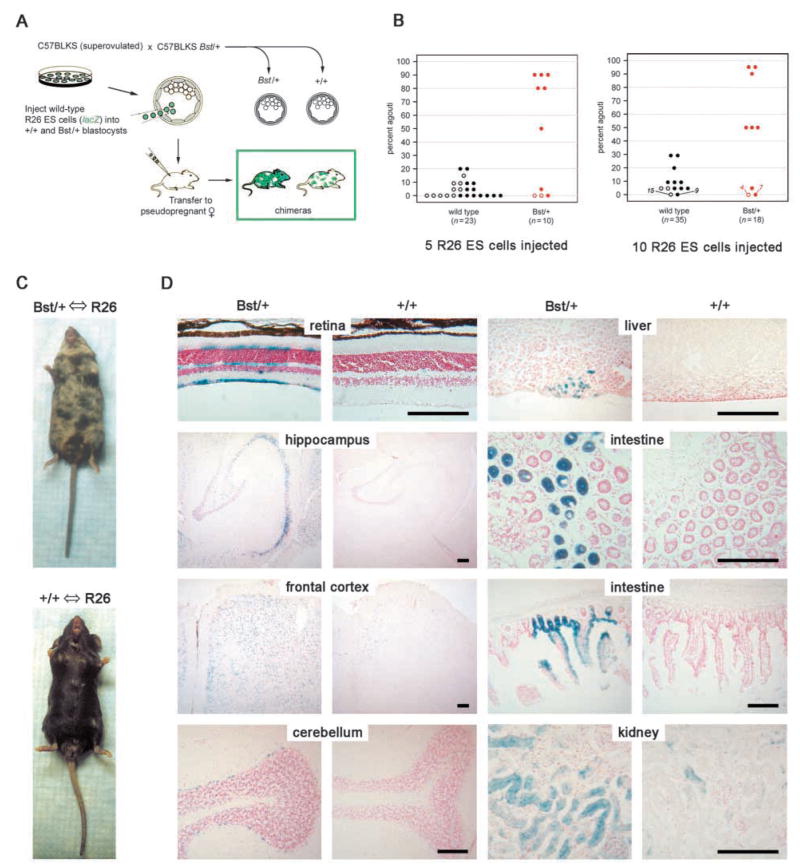
Among the most striking features of Drosophila Minutes is the competition that occurs between M/+ and wild-type cells in somatic mosaics (Garcia-Bellido et al., 1973; Morata and Ripoll, 1975; Simpson, 1979). The increased doubling time of Bst/+ MEFs suggests that a similar competition effect may occur between mutant and wild-type cells in developing mice. To test this prediction, we generated chimeras by injecting exactly five or ten wild-type ROSA26 (R26) embryonic stem (ES) cells into a/a (non-agouti) C57BLKS Bst/+ and +/+ E3.5 blastocysts (Fig. 7A). The R26 ES cells are derived from the A/A (agouti) 129/SvEv strain and contain a ubiquitously expressed lacZ enhancer trap (Zambrowicz et al., 1997). In every experiment, we found that a greater fraction of mice derived from Bst/+ blastocysts were detectably chimeric (⩾5% agouti coloration). These chimeras had a greater agouti coat content (Fig. 7B,C) and more extensive lacZ staining throughout the body (Fig. 7D). Some tissues, such as brain, have a greater R26 contribution than others, such as liver, regardless of the recipient blastocyst genotype. This is most likely due to strain differences between C57 and the 129 ES cells. A similar strain effect was described in neural tissues of interspecific aggregation chimeras (Goldowitz, 1989).
Fig. 7.
Bst causes a growth disadvantage in vivo. (A) Chimera analysis protocol. Exactly five or ten R26 ES cells were injected into mutant and control blastocysts. (B) Coat color comparison of adult chimeras. Rpl24 genotypes were determined by allele-specific PCR. C57BLKS and R26 contributions were determined by the percent black (a/a) and agouti (A/A) coloration, respectively. The R26 contribution was significantly greater in chimeras derived from Bst/+ blastocysts, in experiments where five (left) or ten (right) ES cells were injected (P<0.01, Mann-Whitney nonparametric rank sum tests). Open and closed symbols represent female and male chimeras, respectively. (C) Typical Bst/+ and +/+ chimeras showing 50% and 5% agouti coats, respectively (ten ES cell injection). (D) β-Galactosidase staining of cryopreserved tissues from chimeras in (C). The increased R26 contribution (lacZ positive) in Bst/+ chimeras is evident in all tissues examined. Scale bars: 150 μm.
To assess competition during embryogenesis, we determined the relative amount of C57 single and 129 diffuse βh1 globin alleles in total E12.5 genomic DNA. The fractional R26 contribution was 0.43±0.06 for +/+ chimeras and 0.63±0.12 for Bst/+ chimeras injected with 10 ES cells (P=0.18, n=11). While these values suggest that some differential growth has occurred by E12.5, most of the competition appears to take place between late gestation and weaning. About half of embryos and adults had no detectable ES cell contribution (Fig. 7B and data not shown), in keeping with the efficiency of chimera recovery reported elsewhere (Gossler et al., 1986). These combined results show that Bst/+ cells have a significant growth and survival disadvantage in the presence of wild-type cells, and this extends throughout the animal.
Discussion
The Rpl24 mutation causes a kinetic defect in ribosome biogenesis. Bst/+ mice have features remarkably similar to Drosophila Minutes. Our chimera studies show that the cell competition first described in Drosophila also occurs in mice. In Minute fly larvae, wild-type somatic clones outcompete surrounding M/+ cells, filling and so defining entire compartments of the wing, while respecting developmental boundaries (Garcia-Bellido et al., 1973; Morata and Ripoll, 1975; Simpson, 1979). Conversely, M/+ clones arising in wild-type flies are selectively eliminated, by a TGFβ signaling mechanism that recognizes cells with slower proliferation rates (Moreno et al., 2002). This active process occurs in some, but not all, cell types and serves to maximize tissue fitness (Abrams, 2002). Future studies are needed to show whether cell competition in mice is an active process similar to that in the fly wing.
If wild-type cells outcompete Bst/+ cells in the germline as expected, our results may have practical implications for generating high level ES cell chimeras, as a useful alternative to tetraploid recombination methods (Nagy et al., 1993; Wang et al., 1997). Bst/+ blastocysts may represent ideal hosts, as the introduced ES cells should readily populate the embryo.
Paucity of mammalian riboprotein mutations
Riboprotein mutations appear to be rare in mammals, in contrast to bacteria, yeast and Drosophila. The paucity of mutations can be explained if heterozygous phenotypes are too mild, too severe, heterogeneous or otherwise poorly ascertained. The fact that Bst is hypomorphic suggests that a threshold level of gene expression might be required for survival. Null mutations may be lethal or subvital. Likewise, ribosomes may tolerate mutations in few of their components. L24 does not exist in eubacteria and is one of seven dispensable riboproteins in yeast (Baronas-Lowell and Warner, 1990). Yeast lacking L24 have an increased doubling time and a reduced peptidyl transferase rate (Baronas-Lowell and Warner, 1990; Dresios et al., 2000). These observations suggest that L24 enhances translation efficiency but does not have a unique structural role. Indeed, L24 is added to the 60S subunit relatively late, most probably during export through the nuclear pore or in perinuclear cytoplasm (Harnpicharnchai et al., 2001; Saveanu et al., 2001). A special case of redundancy might also exist for L24. An evolutionarily conserved paralog, Rlp24 (ribosomal-like protein 24), associates with pre-60S complexes (Saveanu et al., 2001). Mouse Rlp24 and L24 are 31% identical and equally similar to the archaean L24 homolog. It has been proposed that Rlp24 plays an essential role in ribosome biogenesis and that Rlp24 and L24 sequentially occupy the same docking site on the 60S subunit, in the nucleus and cytoplasm, respectively (Saveanu et al., 2001). If Rlp24 can reach the cytoplasm, it might in principle partially substitute for L24 in the mature ribosome. Alternatively, a large number of unrecognized riboprotein mutations may exist in mice as they do in Drosophila, with semidominant spotting, tail defects and small size as core phenotypes, and homozygous lethality (Hrabe de Angelis and Balling, 1998; Morgan, 1950; Tease and Fisher, 1993).
Bst phenotypes
Since Rpl24 is essential for protein translation, Bst homozygotes are likely to die at or before the blastocyst stage, when the pool of maternal ribosomes is exhausted (Copp, 1995), or slightly later, owing to low-level Rpl24 expression from the Bst allele (Fig. 4E-G). In a normal mouse blastocyst, ≤20% of the 2.5×108 ribosomes originate from the oocyte (Piko and Clegg, 1982) and ribosomal proteins account for 8% of total protein synthesis (LaMarca and Wassarman, 1979). By contrast, 0-nu Xenopus mutants lacking rRNA genes can survive to the tadpole stage using maternal ribosomes only (Brown and Gurdon, 1964).
The specific defects in Bst/+ mice (Fig. 1) are difficult to reconcile with a global reduction in protein synthesis. Developing retinas, somites and melanocytes may be hypersensitive because mRNAs critical for these tissues are translated at or near a required threshold. Indeed, different mRNAs are translated with widely varying rates of initiation and elongation (Dever, 2002; Lodish, 1976). However, Bst phenotypes may be more easily explained by tissue differences in (1) the extent of coupling between cell division and organogenesis, (2) levels of riboprotein gene expression or (3) extraribosomal functions.
The cell cycle alterations in Bst/+ mice (Fig. 6) provide the basis for the first model. In Drosophila, mutations in the promitotic dmyc gene are phenotypically similar to Minutes (Johnston et al., 1999). Mice with graded c-myc alleles are likewise small, have short tails and prolonged cell cycles (Trumpp et al., 2001). Bst phenotypes could arise in tissues in which the timecourse of organogenesis is relatively uncoupled from cell proliferation. During the life cycle of Minute flies, delayed eclosion allows for complete development. In mice, however, gestation cannot be prolonged; Bst/+ pups are born at the same time as wild-type littermates. Organogenesis must proceed within the time allotted to ensure viability and must be relatively uncoupled from cell proliferation. In principle, defects could result from situations in which development outpaces cell proliferation. Such a dyschronic mechanism may explain the phenotypes in the retina and vertebral column, where timing of developmental events is particularly critical.
In the case of the developing retina, a single progenitor population gives rise to seven major cell types in a characteristic order (Turner et al., 1990). The progenitors are thought to proceed through a sequence of competency states, each of which favors the differentiation of one or a few particular cell types (Livesey and Cepko, 2001). RGCs are the first-born retinal neurons in all vertebrate species (Altshuler et al., 1991). Although histogenesis can occur when cell division is blocked (Harris and Hartenstein, 1991), the timing of cell cycle exit greatly influences retinal development and progenitor cell fate (Dyer and Cepko, 2001). Furthermore, cell cycle length is significantly modulated during the normal course of retinal histogenesis (Alexiades and Cepko, 1996; Li et al., 2000). In the Bst/+ retina, there is no change in the fraction of cells undergoing proliferation or apoptosis, but the total number of neuroblasts is reduced by roughly 60% (E10.5) to 20% (E13.5), and the onset of terminal mitoses is delayed from E10.5 to E12.5 (Tang et al., 1999). These findings can now be considered in light of the general increase in Bst/+ cell-cycle length (Fig. 6). Assuming our MEF results apply in vivo, Bst is likely to have a disproportionately greater effect during the early stages of retinal histogenesis, when the cycle time is shorter (Alexiades and Cepko, 1996), protein synthetic demands are greatest, and RGCs are born (Livesey and Cepko, 2001). The subretinal neovascularization (Smith et al., 2000) is probably a secondary consequence of RGC deficiency, similar to that observed in Math5 mutant mice and humans with optic nerve aplasia (Brown et al., 2001; Lee et al., 1996; Brzezinski et al., 2003).
Timing is similarly critical during somite development. Somites are added iteratively to the caudal end of the neural tube, in response to periodic waves of gene expression (Saga and Takeda, 2001). This process is controlled by an intrinsic clock that is sensitive to cycloheximide (Hirata et al., 2002; Palmeirim et al., 1997). The vertebral defects in Bst/+ mice are more severe posteriorly, consistent with a progressive deterioration in periodicity and phase coherence of the intrinsic somite clock (Saga and Takeda, 2001). This pattern could occur if high levels of the protein synthesis were needed to propagate successive cycles of somitogenesis.
In the second model, pleiotropic Bst phenotypes may reflect tissue-specific differences in Rpl24 expression. To achieve unimolar stoichiometry, dispersed riboprotein genes are coordinately expressed via 5′TOP elements, which define transcriptional start sites and control translation of riboprotein mRNAs (Hariharan et al., 1989; Jefferies et al., 1997). In spite of this convergently evolved feature, 5′TOP sequences may vary in potency across tissues. Differential regulation of riboprotein mRNAs has been described in pluripotent embryonal carcinoma cells, various cancer cell lines and in RNA profiling studies (Blackshaw et al., 2001; Panda et al., 2002). If the developing retina, somites and melanoblasts express relatively low levels of Rpl24 mRNA, they may be more sensitive to quantitative reduction caused by the Bst mutation.
In the third model, Bst phenotypes may occur because L24 has unique extraribosomal functions, apart from protein translation (Wool, 1996). For example, the small subunit riboprotein S3 functions as an endonuclease during DNA repair, while S0 acts on the cell surface, as the primary laminin receptor (Ardini et al., 1998). This explanation would be supported if mice with mutations in other riboprotein genes have phenotypes unlike Bst. The high incidence of RPS19 mutations among DBA patients (Draptchinskaia et al., 1999) suggests that S19 has extraribosomal functions in erythropoiesis. Recent studies show that L24 from a variety of species interacts directly with the transactivator (TAV) protein of cauliflower mosaic virus to promote translation re-initiation on polycistronic mRNAs (Park et al., 2001). In principle, the unique Bst phenotypes could reflect tissue specificity of cellular mRNAs that utilize internal ribosome entry (Hinnebusch, 1997; Johannes et al., 1999; Morris and Geballe, 2000).
Implications for human disease
Human RPS19 and mouse Rpl24 mutations are both associated with decreased growth and skeletal defects. Curiously, triphalangeal first digits are frequent in Bst/+ mice and are the single most common extramedullary findings in DBA patients. This suggests a shared mechanism in which limb malformations arise through impairment of ribosome biogenesis. However, unlike DBA patients, Bst/+ mice appear hematologically normal in the absence of erythroid stress (data not shown). Conversely, RPS19 mutations do not cause optic nerve disease or obvious hypopigmentation. These findings might reflect differences between humans and mice, or specific roles for S19 and L24 in hematopoiesis and retinogenesis, respectively. Accordingly, if the retinal phenotype is unique to Bst/+ mice, then RPL24 is a good candidate for dominant human optic nerve disorders associated with small stature, including colobomas of the posterior segment (Onwochei et al., 2000) and septo-optic dysplasia (De Morsier syndrome, MIM182230). Affected children have small optic nerves and growth retardation, similar to Bst/+ mice. A few patients have mutations in the HESX1 homeobox gene (Dattani et al., 1998), but in most cases the cause is unknown. Growth hormone levels are often decreased, but pituitary anatomy and endocrine profiles are usually normal.
Riboprotein gene mutations may also contribute more broadly to common human malformations, such as the CHARGE (MIM214800) and VACTERL (MIM192350) syndromes, which involve a constellation of developmental defects but as yet have no clear genetic basis (Lalani et al., 2003). Eighty percent of CHARGE syndrome patients have retinal colobomas, anatomically equivalent to those in Bst/+ mice (Fig. 1B,C), and growth retardation as core diagnostic features (Russell-Eggitt et al., 1990; Tellier et al., 1998). Conversely, 11% of patients with ocular colobomas have diagnostic features of CHARGE syndrome (Chestler and France, 1988).
Acknowledgments
We thank Margaret Van Keuren, Elizabeth Hughes, Donalyn Scheuner, Mark Adams, Deanna Kulpa, Stephanie Schmoll, Bob Lyons, Dave Siemieniak, Sean Morrison, Ömer Yilmaz, Phil Andrews, John Strahler, Sinisa Volarevic, Joe Brzezinski and Sara Schulz for technical advice; Elizabeth Robertson and Philippe Soriano for providing ES cells; and David Ginsburg, Sally Camper, Dave Turner, John Moran, Miriam Meisler, Randy Kaufman and Steve Weiss for helpful discussions and critically reading the manuscript. E.R.O. was supported by MSTP and UM Organogenesis NIH training grants (T32 GM07863 and HD07505). This work was supported by NIH grants to T.G. (EY14259, EY12994 and GM067840) and the UM Diabetes Research and Training Center (DK20572).
Footnotes
Note added in proof
Amsterdam et al. (Amsterdam et al., 2004) recently reported that zebrafish with heterozygous ribosomal protein gene mutations have a high incidence of malignant nerve sheath tumors (11 out of 16 riboprotein gene insertions identified in a recessive lethal screen). However, tumor formation was not elevated in the L24 mutant line.
References
- Abrams JM. Competition and compensation: coupled to death in development and cancer. Cell. 2002;110:403–406. doi: 10.1016/s0092-8674(02)00904-2. [DOI] [PubMed] [Google Scholar]
- Alexiades MR, Cepko C. Quantitative analysis of proliferation and cell cycle length during development of the rat retina. Dev Dyn. 1996;205:293–307. doi: 10.1002/(SICI)1097-0177(199603)205:3<293::AID-AJA9>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Allmang C, Mitchell P, Petfalski E, Tollervey D. Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res. 2000;28:1684–1691. doi: 10.1093/nar/28.8.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler DM, Turner DL, Cepko CL. Specification of cell type in the vertebrate retina. In: Lam DMK, Shatz CJ, editors. Cell Lineage and Cell Fate in Visual System Development. Cambridge, MA: MIT Press; 1991. pp. 37–58. [Google Scholar]
- Amsterdam A, Sadler KC, Lai K, Farrington S, Bronson RT, Lees JA, Hopkins N. Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol. 2004;2:E139. doi: 10.1371/journal.pbio.0020139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardini E, Pesole G, Tagliabue E, Magnifico A, Castronovo V, Sobel ME, Colnaghi MI, Menard S. The 67-kDa laminin receptor originated from a ribosomal protein that acquired a dual function during evolution. Mol Biol Evol. 1998;15:1017–1025. doi: 10.1093/oxfordjournals.molbev.a026000. [DOI] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- Baronas-Lowell DM, Warner JR. Ribosomal protein L30 is dispensable in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:5235–5243. doi: 10.1128/mcb.10.10.5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackshaw S, Fraioli RE, Furukawa T, Cepko CL. Comprehensive analysis of photoreceptor gene expression and the identification of candidate retinal disease genes. Cell. 2001;107:579–589. doi: 10.1016/s0092-8674(01)00574-8. [DOI] [PubMed] [Google Scholar]
- Boring LF, Sinervo B, Schubiger G. Experimental phenocopy of a minute maternal-effect mutation alters blastoderm determination in embryos of Drosophila melanogaster. Dev Biol. 1989;132:343–354. doi: 10.1016/0012-1606(89)90231-5. [DOI] [PubMed] [Google Scholar]
- Brooks RF. Continuous protein synthesis is required to maintain the probability of entry into S phase. Cell. 1977;12:311–317. doi: 10.1016/0092-8674(77)90209-4. [DOI] [PubMed] [Google Scholar]
- Brown DD, Gurdon JB. Absence of ribosomal RNA synthesis in the anucleolate mutant of Xenopus laevis. Proc Natl Acad Sci USA. 1964;51:139–146. doi: 10.1073/pnas.51.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NL, Patel S, Brzezinski J, Glaser T. Math5 is required for retinal ganglion cell and optic nerve formation. Development. 2001;128:2497–2508. doi: 10.1242/dev.128.13.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzezinski JA, IV, Schulz SM, Crawford S, Wroblewski E, Brown NL, Glaser T. Math5 null mice have abnormal retinal and persistent hyaloid vasculatures. Dev Biol. 2003;259:553. [Google Scholar]
- Butler JS. The yin and yang of the exosome. Trends Cell Biol. 2002;12:90–96. doi: 10.1016/s0962-8924(01)02225-5. [DOI] [PubMed] [Google Scholar]
- Chestler RJ, France TD. Ocular findings in CHARGE syndrome. Six case reports and a review. Ophthalmology. 1988;95:1613–1619. doi: 10.1016/s0161-6420(88)32968-4. [DOI] [PubMed] [Google Scholar]
- Copp AJ. Death before birth: clues from gene knockouts and mutations. Trends Genet. 1995;11:87–93. doi: 10.1016/S0168-9525(00)89008-3. [DOI] [PubMed] [Google Scholar]
- Dattani MT, Martinez-Barbera JP, Thomas PQ, Brickman JM, Gupta R, Martensson IL, Toresson H, Fox M, Wales JK, Hindmarsh PC, et al. Mutations in the homeobox gene HESX1/Hesx1 associated with septo-optic dysplasia in human and mouse. Nat Genet. 1998;19:125–133. doi: 10.1038/477. [DOI] [PubMed] [Google Scholar]
- Dever TE. Gene-specific regulation by general translation factors. Cell. 2002;108:545–556. doi: 10.1016/s0092-8674(02)00642-6. [DOI] [PubMed] [Google Scholar]
- Dietrich W, Katz H, Lincoln SE, Shin HS, Friedman J, Dracopoli NC, Lander ES. A genetic map of the mouse suitable for typing intraspecific crosses. Genetics. 1992;131:423–447. doi: 10.1093/genetics/131.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draptchinskaia N, Gustavsson P, Andersson B, Pettersson M, Willig TN, Dianzani I, Ball S, Tchernia G, Klar J, Matsson H, et al. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat Genet. 1999;21:169–175. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]
- Dresios J, Derkatch IL, Liebman SW, Synetos D. Yeast ribosomal protein L24 affects the kinetics of protein synthesis and ribosomal protein L39 improves translational accuracy, while mutants lacking both remain viable. Biochemistry. 2000;39:7236–7244. doi: 10.1021/bi9925266. [DOI] [PubMed] [Google Scholar]
- Dyer MA, Cepko CL. Regulating proliferation during retinal development. Nat Rev Neurosci. 2001;2:333–342. doi: 10.1038/35072555. [DOI] [PubMed] [Google Scholar]
- Fatica A, Tollervey D. Making ribosomes. Curr Opin Cell Biol. 2002;14:313–318. doi: 10.1016/s0955-0674(02)00336-8. [DOI] [PubMed] [Google Scholar]
- Fiering S, Epner E, Robinson K, Zhuang Y, Telling A, Hu M, Martin DI, Enver T, Ley TJ, Groudine M. Targeted deletion of 5′HS2 of the murine beta-globin LCR reveals that it is not essential for proper regulation of the beta-globin locus. Genes Dev. 1995;9:2203–2213. doi: 10.1101/gad.9.18.2203. [DOI] [PubMed] [Google Scholar]
- Garcia-Bellido A, Ripoll P, Morata G. Developmental compartmentalisation of the wing disk of Drosophila. Nat New Biol. 1973;245:251–253. doi: 10.1038/newbio245251a0. [DOI] [PubMed] [Google Scholar]
- Goldowitz D. Cell allocation in mammalian CNS formation: evidence from murine interspecies aggregation chimeras. Neuron. 1989;3:705–713. doi: 10.1016/0896-6273(89)90239-0. [DOI] [PubMed] [Google Scholar]
- Gossler A, Doetschman T, Korn R, Serfling E, Kemler R. Transgenesis by means of blastocyst-derived embryonic stem cell lines. Proc Natl Acad Sci USA. 1986;83:9065–9069. doi: 10.1073/pnas.83.23.9065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JW, Dolbeare F, Pallavicini MG. Quantitative cell-cycle analysis. In: Melamed MR, Lindmo T, Mendelsohn ML, editors. Flow Cytometry and Sorting. New York: Wiley-Liss; 1990. pp. 445–467. [Google Scholar]
- Hariharan N, Kelley DE, Perry RP. Equipotent mouse ribosomal protein promoters have a similar architecture that includes internal sequence elements. Genes Dev. 1989;3:1789–1800. doi: 10.1101/gad.3.11.1789. [DOI] [PubMed] [Google Scholar]
- Harnpicharnchai P, Jakovljevic J, Horsey E, Miles T, Roman J, Rout M, Meagher D, Imai B, Guo Y, Brame CJ, et al. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol Cell. 2001;8:505–515. doi: 10.1016/s1097-2765(01)00344-6. [DOI] [PubMed] [Google Scholar]
- Harris WA, Hartenstein V. Neuronal determination without cell division in Xenopus embryos. Neuron. 1991;6:499–515. doi: 10.1016/0896-6273(91)90053-3. [DOI] [PubMed] [Google Scholar]
- Herrera RE, Sah VP, Williams BO, Makela TP, Weinberg RA, Jacks T. Altered cell cycle kinetics, gene expression, and G1 restriction point regulation in Rb-deficient fibroblasts. Mol Cell Biol. 1996;16:2402–2407. doi: 10.1128/mcb.16.5.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. Translational regulation of yeast GCN4. A window on factors that control initiator-trna binding to the ribosome. J Biol Chem. 1997;272:21661–21664. doi: 10.1074/jbc.272.35.21661. [DOI] [PubMed] [Google Scholar]
- Hirata H, Yoshiura S, Ohtsuka T, Bessho Y, Harada T, Yoshikawa K, Kageyama R. Oscillatory expression of the bHLH factor Hes1 regulated by a negative feedback loop. Science. 2002;298:840–843. doi: 10.1126/science.1074560. [DOI] [PubMed] [Google Scholar]
- Hrabe de Angelis M, Balling R. Large scale ENU screens in the mouse: genetics meets genomics. Mutat Res. 1998;400:25–32. doi: 10.1016/s0027-5107(98)00061-x. [DOI] [PubMed] [Google Scholar]
- Hutson NJ, Mortimore GE. Suppression of cytoplasmic protein uptake by lysosomes as the mechanism of protein regain in livers of starved-refed mice. J Biol Chem. 1982;257:9548–9554. [PubMed] [Google Scholar]
- Jefferies HB, Fumagalli S, Dennis PB, Reinhard C, Pearson RB, Thomas G. Rapamycin suppresses 5′TOP mRNA translation through inhibition of p70s6k. EMBO J. 1997;16:3693–3704. doi: 10.1093/emboj/16.12.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes G, Carter MS, Eisen MB, Brown PO, Sarnow P. Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. Proc Natl Acad Sci USA. 1999;96:13118–13123. doi: 10.1073/pnas.96.23.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston LA, Prober DA, Edgar BA, Eisenman RN, Gallant P. Drosophila myc regulates cellular growth during development. Cell. 1999;98:779–790. doi: 10.1016/s0092-8674(00)81512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalani SR, Stockton DW, Bacino C, Molinari LM, Glass NL, Fernbach SD, Towbin JA, Craigen WJ, Graham JM, Jr, Hefner MA, et al. Toward a genetic etiology of CHARGE syndrome: I. A systematic scan for submicroscopic deletions. Am J Med Genet. 2003;118A:260–266. doi: 10.1002/ajmg.a.20002. [DOI] [PubMed] [Google Scholar]
- LaMarca MJ, Wassarman PM. Program of early development in the mammal: changes in absolute rates of synthesis of ribosomal proteins during oogenesis and early embryogenesis in the mouse. Dev Biol. 1979;73:103–119. doi: 10.1016/0012-1606(79)90141-6. [DOI] [PubMed] [Google Scholar]
- Lambertsson A. The Minute genes in Drosophila and their molecular functions. Adv Genet. 1998;38:69–134. doi: 10.1016/s0065-2660(08)60142-x. [DOI] [PubMed] [Google Scholar]
- Lee BL, Bateman JB, Schwartz SD. Posterior segment neovascularization associated with optic nerve aplasia. Am J Ophthalmol. 1996;122:131–133. doi: 10.1016/s0002-9394(14)71982-8. [DOI] [PubMed] [Google Scholar]
- Li Z, Hu M, Ochocinska MJ, Joseph NM, Easter SS., Jr Modulation of cell proliferation in the embryonic retina of zebrafish (Danio rerio) Dev Dyn. 2000;219:391–401. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1063>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Livesey FJ, Cepko CL. Vertebrate neural cell-fate determination: lessons from the retina. Nat Rev Neurosci. 2001;2:109–118. doi: 10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- Lodish HF. Translational control of protein synthesis. Annu Rev Biochem. 1976;45:39–72. doi: 10.1146/annurev.bi.45.070176.000351. [DOI] [PubMed] [Google Scholar]
- McLeod MJ. Differential staining of cartilage and bone in whole mouse fetuses by alcian blue and alizarin red S. Teratology. 1980;22:299–301. doi: 10.1002/tera.1420220306. [DOI] [PubMed] [Google Scholar]
- Mitrovich QM, Anderson P. Unproductively spliced ribosomal protein mRNAs are natural targets of mRNA surveillance in C. elegans. Genes Dev. 2000;14:2173–2184. doi: 10.1101/gad.819900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagne J, Stewart MJ, Stocker H, Hafen E, Kozma SC, Thomas G. Drosophila S6 kinase: a regulator of cell size. Science. 1999;285:2126–2129. doi: 10.1126/science.285.5436.2126. [DOI] [PubMed] [Google Scholar]
- Morata G, Ripoll P. Minutes: mutants of Drosophila autonomously affecting cell division rate. Dev Biol. 1975;42:211–221. doi: 10.1016/0012-1606(75)90330-9. [DOI] [PubMed] [Google Scholar]
- Moreno E, Basler K, Morata G. Cells compete for decapentaplegic survival factor to prevent apoptosis in Drosophila wing development. Nature. 2002;416:755–759. doi: 10.1038/416755a. [DOI] [PubMed] [Google Scholar]
- Morgan WC. A new tail-short mutation in the mouse. J Hered. 1950;41:208–215. doi: 10.1093/oxfordjournals.jhered.a106131. [DOI] [PubMed] [Google Scholar]
- Morris DR, Geballe AP. Upstream open reading frames as regulators of mRNA translation. Mol Cell Biol. 2000;20:8635–8642. doi: 10.1128/mcb.20.23.8635-8642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onwochei BC, Simon JW, Bateman JB, Couture KC, Mir E. Ocular colobomata. Surv Ophthalmol. 2000;45:175–194. doi: 10.1016/s0039-6257(00)00151-x. [DOI] [PubMed] [Google Scholar]
- Palmeirim I, Henrique D, Ish-Horowicz D, Pourquie O. Avian hairy gene expression identifies a molecular clock linked to vertebrate segmentation and somitogenesis. Cell. 1997;91:639–648. doi: 10.1016/s0092-8674(00)80451-1. [DOI] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- Park HS, Himmelbach A, Browning KS, Hohn T, Ryabova LA. A plant viral “reinitiation” factor interacts with the host translational machinery. Cell. 2001;106:723–733. doi: 10.1016/s0092-8674(01)00487-1. [DOI] [PubMed] [Google Scholar]
- Piko L, Clegg KB. Quantitative changes in total RNA, total poly(A), and ribosomes in early mouse embryos. Dev Biol. 1982;89:362–378. doi: 10.1016/0012-1606(82)90325-6. [DOI] [PubMed] [Google Scholar]
- Rice DS, Tang Q, Williams RW, Harris BS, Davisson MT, Goldowitz D. Decreased retinal ganglion cell number and misdirected axon growth associated with fissure defects in Bst/+ mutant mice. Invest Ophthalmol Vis Sci. 1997;38:2112–2124. [PubMed] [Google Scholar]
- Rice DS, Williams RW, Ward-Bailey P, Johnson KR, Harris BS, Davisson MT, Goldowitz D. Mapping the Bst mutation on mouse chromosome 16: a model for human optic atrophy. Mamm Genome. 1995;6:546–548. doi: 10.1007/BF00356174. [DOI] [PubMed] [Google Scholar]
- Robertson EJ. Teratocarcinomas and Embryonic Stem Cells: a Practical Approach. Oxford: IRL Press; 1987. [Google Scholar]
- Ruskin B, Greene JM, Green MR. Cryptic branch point activation allows accurate in vitro splicing of human beta-globin intron mutants. Cell. 1985;41:833–844. doi: 10.1016/s0092-8674(85)80064-7. [DOI] [PubMed] [Google Scholar]
- Russell-Eggitt IM, Blake KD, Taylor DS, Wyse RK. The eye in the CHARGE association. Br J Ophthalmol. 1990;74:421–426. doi: 10.1136/bjo.74.7.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saga Y, Takeda H. The making of the somite: molecular events in vertebrate segmentation. Nat Rev Genet. 2001;2:835–845. doi: 10.1038/35098552. [DOI] [PubMed] [Google Scholar]
- Saveanu C, Bienvenu D, Namane A, Gleizes PE, Gas N, Jacquier A, Fromont-Racine M. Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps. EMBO J. 2001;20:6475–6484. doi: 10.1093/emboj/20.22.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J. The Minute reaction in the development of Drosophila melanogaster. Genetics. 1929;14:366–419. doi: 10.1093/genetics/14.4.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson P. Parameters of cell competition in the compartments of the wing disc of Drosophila. Dev Biol. 1979;69:182–193. doi: 10.1016/0012-1606(79)90284-7. [DOI] [PubMed] [Google Scholar]
- Simpson P, Morata G. Differential mitotic rates and patterns of growth in compartments in the Drosophila wing. Dev Biol. 1981;85:299–308. doi: 10.1016/0012-1606(81)90261-x. [DOI] [PubMed] [Google Scholar]
- Smith RS, John SW, Zabeleta A, Davisson MT, Hawes NL, Chang B. The Bst locus on mouse chromosome 16 is associated with age-related subretinal neovascularization. Proc Natl Acad Sci USA. 2000;97:2191–2195. doi: 10.1073/pnas.040531597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southard JL, Eicher EM. Belly spot and tail (Bst) Mouse News Lett. 1977;56:40. [Google Scholar]
- Spahn CM, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, Frank J. Structure of the 80S ribosome from Saccharomyces cerevisiae – tRNA-ribosome and subunit-subunit interactions. Cell. 2001;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- Stern C, Tokunaga C. On cell lethals in Drosophila. Proc Natl Acad Sci USA. 1971;68:329–331. doi: 10.1073/pnas.68.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q, Rice DS, Goldowitz D. Disrupted retinal development in the embryonic belly spot and tail mutant mouse. Dev Biol. 1999;207:239–255. doi: 10.1006/dbio.1998.9142. [DOI] [PubMed] [Google Scholar]
- Tease C, Fisher G. Tail kinks, small size and white spotting (Tks): a radiation-induced mutation on chromosome 9. Mouse Genome. 1993;91:137–138. [Google Scholar]
- Tellier AL, Cormier-Daire V, Abadie V, Amiel J, Sigaudy S, Bonnet D, de Lonlay-Debeney P, Morrisseau-Durand MP, Hubert P, Michel JL, et al. CHARGE syndrome: report of 47 cases and review. Am J Med Genet. 1998;76:402–409. doi: 10.1002/(sici)1096-8628(19980413)76:5<402::aid-ajmg7>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Tomita K, Ishibashi M, Nakahara K, Ang SL, Nakanishi S, Guillemot F, Kageyama R. Mammalian hairy and Enhancer of split homolog 1 regulates differentiation of retinal neurons and is essential for eye morphogenesis. Neuron. 1996;16:723–734. doi: 10.1016/s0896-6273(00)80093-8. [DOI] [PubMed] [Google Scholar]
- Trumpp A, Refaeli Y, Oskarsson T, Gasser S, Murphy M, Martin GR, Bishop JM. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature. 2001;414:768–773. doi: 10.1038/414768a. [DOI] [PubMed] [Google Scholar]
- Turner DL, Snyder EY, Cepko CL. Lineage-independent determination of cell type in the embryonic mouse retina. Neuron. 1990;4:833–845. doi: 10.1016/0896-6273(90)90136-4. [DOI] [PubMed] [Google Scholar]
- Uechi T, Tanaka T, Kenmochi N. A complete map of the human ribosomal protein genes: assignment of 80 genes to the cytogenetic map and implications for human disorders. Genomics. 2001;72:223–230. doi: 10.1006/geno.2000.6470. [DOI] [PubMed] [Google Scholar]
- Volarevic S, Stewart MJ, Ledermann B, Zilberman F, Terracciano L, Montini E, Grompe M, Kozma SC, Thomas G. Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science. 2000;288:2045–2047. doi: 10.1126/science.288.5473.2045. [DOI] [PubMed] [Google Scholar]
- Wang ZQ, Kiefer F, Urbanek P, Wagner EF. Generation of completely embryonic stem cell-derived mutant mice using tetraploid blastocyst injection. Mech Dev. 1997;62:137–145. doi: 10.1016/s0925-4773(97)00655-2. [DOI] [PubMed] [Google Scholar]
- Warner JR. In the absence of ribosomal RNA synthesis, the ribosomal proteins of HeLa cells are synthesized normally and degraded rapidly. J Mol Biol. 1977;115:315–333. doi: 10.1016/0022-2836(77)90157-7. [DOI] [PubMed] [Google Scholar]
- Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- Wool IG. Extraribosomal functions of ribosomal proteins. Trends Biochem Sci. 1996;21:164–165. [PubMed] [Google Scholar]
- Zambrowicz BP, Imamoto A, Fiering S, Herzenberg LA, Kerr WG, Soriano P. Disruption of overlapping transcripts in the ROSA beta geo 26 gene trap strain leads to widespread expression of beta-galactosidase in mouse embryos and hematopoietic cells. Proc Natl Acad Sci USA. 1997;94:3789–3794. doi: 10.1073/pnas.94.8.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanchin NI, Roberts P, DeSilva A, Sherman F, Goldfarb DS. Saccharomyces cerevisiae Nip7p is required for efficient 60S ribosome subunit biogenesis. Mol Cell Biol. 1997;17:5001–5015. doi: 10.1128/mcb.17.9.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Harrison P, Gerstein M. Identification and analysis of over 2000 ribosomal protein pseudogenes in the human genome. Genome Res. 2002;12:1466–1482. doi: 10.1101/gr.331902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Sohn JH, Warner JR. Autoregulation in the biosynthesis of ribosomes. Mol Cell Biol. 2003;23:699–707. doi: 10.1128/MCB.23.2.699-707.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]