Abstract
The human ATOH7 gene encodes a basic helix-loop-helix (bHLH) transcription factor that is highly similar to Drosophila Atonal within the conserved bHLH domain. The ATOH7 coding region is contained within a single exon. We mapped ATOH7 to Chromosome (Chr) 10q21.3–22.1, a region syntenic to the segment of mouse Chr 10 where Atoh7 (formerly Math5) is located. The evolutionary relationship between ATOH7 and other atonal homologs was investigated using parsimony analysis. A direct comparison of ATH5/7 and ATH1 protein subgroups to Atonal also revealed a nonrandom distribution of amino acid changes across the bHLH domain, which may be related to their separate visual and proprioceptive sensory functions. Among bHLH genes, ATOH7 is most closely related to Atoh7. This sequence conservation extends significantly beyond the coding region. We define blocks of strong homology in flanking human and mouse genomic DNA, which are likely to include cis regulatory elements. Because targeted deletion of Atoh7 causes optic nerve agenesis in mice, we propose ATOH7 as a candidate for human optic nerve aplasia and related clinical syndromes.
Early development of the mammalian retina proceeds through a series of cell fate decisions in which progenitor cells in the optic cup progressively exit mitosis, migrate to specific laminar positions, and terminally differentiate into one of seven basic neural or glial cell types. Defects in retinal cell determination or differentiation lead to specific malformations in humans, including Leber’s congenital amaurosis (Freund et al. 1998; Sohocki et al. 1998), cone-rod dystrophy (Freund et al. 1997; Sohocki et al. 1998; Swain et al. 1997), enhanced S-cone syndrome (Cepko 2000; Haider et al. 2000), and optic nerve aplasia or hypoplasia (Lee et al. 1996; Scott et al. 1997; Weiter et al. 1977). Although the genetic basis for retinal histogenesis is poorly understood, recent findings suggest that key proteins controlling some of these developmental processes are nuclear transcription factors (Brown et al. 2001; Freund et al. 1997; Kobayashi et al. 1999; Swain et al. 1997; Wang et al. 2001).
Basic helix-loop-helix (bHLH) transcription factors regulate multiple aspects of retinal neuron formation in vertebrates and invertebrates (Jan and Jan 1993; Vetter and Brown 2001; Cepko 1999). Proneural bHLH proteins contain basic DNA-binding and helix-loop-helix dimerization motifs and act to promote neuron formation. They are generally expressed by groups of equivalent neural progenitor cells, with each group ultimately giving rise to one or more neurons. A key proneural bHLH gene that controls photoreceptor development in Drosophila is atonal, named for its chordotonal stretch receptor mutant phenotype (Jarman et al. 1993). While several vertebrate atonal homologs (Atoh proteins) have been identified, most act only during peripheral or central nervous system development and are not expressed significantly during eye formation. In contrast, the ATH5/7 subclass (named for Xenopus Xath5 and mouse Atoh7) is highly expressed by retinal progenitors during the early stages of eye development in zebrafish, chick, frog, and mouse (Brown et al. 1998; Kanekar et al. 1997; Liu et al. 2001; Masai et al. 2000).
Mutations in the fly atonal, zebrafish Zath5, or mouse Atoh7 (Math5) genes cause agenesis of the initial neuron class in the diverse eye types of these organisms, which consist of R8 photo-receptors in Drosophila (Jarman et al. 1994, 1995) and retinal ganglion cells (RGCs) in vertebrates (Brown et al. 2001; Jarman et al. 1994, 1995; Kay et al. 2001; Wang et al. 2001). Because Drosophila photoreceptors are induced sequentially, by an iterative process that depends on previously formed photoreceptors, atonal mutant flies are nearly eyeless, whereas zebrafish Zath5 and mouse Atoh7 mutants have eyes but lack RGCs and optic nerves. These phenotypes are consistent with gain-of-function experiments in which ectopic expression of frog Xath5 or chicken Cath5 was shown to bias retinal progenitors toward an RGC fate (Kanekar et al. 1997; Liu et al. 2001).
We report the cloning, chromosome mapping, and phylogenetic analysis of human ATOH7. By comparing the flanking genomic DNA in humans and mice, we have identified multiple discrete segments of sequence homology. On the basis of these conserved features, we propose that ATOH7 is the human ortholog of Atoh7. Given the central role of Atoh7 (Math5) in RGC and optic nerve formation, ATOH7 mutations may underlie clinically important congenital malformations or degenerative diseases of the optic nerve.
Materials and methods
DNA library screening
A Sau3A-partial human genomic DNA library constructed in λFix (Stratagene) was screened using a radiolabeled full-length Atoh7 cDNA probe (Brown et al. 1998) at reduced stringency. Two positive clones (#3 and #6) were purified for further analysis.
DNA cloning and sequence analysis
Restriction maps of phage clones were prepared by using BamHI, EcoRI, SalI, HindIII, and XhoI, and Southern analysis. Human genomic DNA fragments hybridizing to the Atoh7 cDNA probe were subcloned in pGEM3Z and used as templates for automated sequencing. A 5.7-kb segment from the assembled sequence of phage clone #6 was deposited in GenBank (accession no. AF418922) and is largely identical to the sequence of an independent BAC clone (AC016395). We also identified a short 3′ EST for ATOH7 (H05728) and sequenced the corresponding 1.7-kb cDNA clone. The genomic sequence of mouse Atoh7 (accession no. AF418923) was obtained from a 6.5-kb EcoRI clone (Brown et al. 1998, 2001). Some sequence comparisons were also made to a Drosophila BAC clone containing the atonal gene (AC008094).
Nucleotide and amino acid sequence analysis was performed by using MacVector and AssemblyLIGN software programs (Oxford Molecular). Amino acid sequences were initially aligned with Clustal W (Thompson et al. 1994) and subsequently adjusted by eye. Two-way genomic DNA sequence comparisons were performed by using BLASTN 2.1.2 (Tatusova and Madden 1999) via the National Center for Biotechnology Information (NCBI) website (www.ncbi.nlm.nih.gov/blast/bl2seq/bl2.html). Scoring parameters for the noncoding regions were match, 1; mismatch, 2; gap open, 1; and gap extension, 2. Global human-mouse genomic sequence alignments were generated and displayed using the VISTA (VISualization Tool for Alignments) program (Dubchak et al. 2000; Mayor et al. 2000) via the Lawrence Berkeley Laboratory website (www.gsd.lbl.gov/vista) with a 100-bp window size and 70% nucleotide identity threshold for conserved regions. Human repetitive elements and CpG islands were identified by using RepeatMasker at ftp.genome.washington.edu/RM/RepeatMasker.html (AFA Smit and P Green, unpublished).
Fluorescence in situ hybridization (FISH)
Phytohemagglutinin-stimulated lymphocytes from whole human blood were cultured and harvested under standard conditions. Giemsa-trypsin banding (G-banding) and FISH were performed on metaphase spreads as previously described (Dagenais et al. 1999). Chromosomes were G-banded, photographed, and de-stained. After air-drying, slides were denatured and hybridized overnight with ATOH7 phage clone #6 DNA. This probe was labeled with biotin-14-dATP using the BioNick Translation Kit (GibcoBRL) and pre-annealed to human Cot-1 DNA (GibcoBRL). Signals were visualized by incubation with two layers of FITC-conjugated avidin-DCS and fluorescein-conjugated anti-avidin IgG (Vector Laboratories). Chromosomes were counterstained with propidium iodide (Vector Laboratories), analyzed with a Zeiss Axiophot epifluorescence microscope, and photographed with Kodak technical Pan and Royal Gold ASA 1000 films.
Phylogenetic analysis
Amino acid sequences from 26 bHLH domains, including human ATOH7, were aligned and subjected to parsimony analysis using PAUP* version 4.0b8 (Swofford 1998). The bHLH domains (taxa) each consist of 56 amino acids (characters), as designated by Ferré-D’Amaré et al. (1993). A full heuristic search for shortest trees was performed by using 100 random sequences of taxa addition, branch swapping via the TBR (tree-bisection and recombination) algorithm, and the PROTOPARS stepmatrix for amino acid substitutions. This matrix gives the minimum number of nucleotide substitutions needed to convert one amino acid into another on the basis of the genetic code used by nuclear genes of most eukaryotes (Felsenstein 1993). To assess support for the inferred phylogeny, the heuristic search was repeated 100 times using bootstrap data sets derived by sampling the matrix of aligned characters with replacement, and a majority rule consensus tree was generated from the bootstrap replicates (Swofford 1998; Swofford et al. 1996). Mash1 and Mash2 bHLH domains were used as outgroup taxa.
Accession numbers for the phylogenetic analysis are: Homo sapiens ATOH7 (AF418922); Mus musculus Atoh7 cDNA (AF071223); Gallus gallus Cath5 (AJ001178); Xenopus laevis Xath5a (U93170) and Xath5b (U93171); Danio rerio Zath5 (AB049457); Drosophila melanogaster Atonal (L36646), Cato (AF134869) and Amos (AF166113); Caenorhabditis elegans Lin32 cDNA (U15418) and genomic (U50199); Homo sapiens ATOH1 (U61148); Mus musculus Atoh1 (D43694); Gallus gallus Cath1 (U61149); Danio rerio Zath1 (AF024536); Mus musculus Atoh2 (D44480) and Atoh3 (AF036257); Xenopus laevis Xath3 (D85188); Mus musculus NeuroD (U28068) and NeuroD2 (U58471); Mus musculus Ngn1 (U67776), Ngn2 (AF303001) and Ngn3 (AF364300); Xenopus laevis Xngnr1a (U67778); Rattus norvegicus Mash1 (X53725) and Mash2 (X53724).
Results and Discussion
The mouse bHLH transcription factor Atoh7 (Math5) is expressed in the developing optic cup, beginning at embryonic day E11.5, in a temporal and spatial pattern that coincides with RGC formation (Brown et al. 1998). Atoh7 is required for development of RGC neurons, optic nerves and chiasmata (Brown et al. 2001; Wang et al. 2001). Congenital defects of the optic nerve, including aplasia, hypoplasia and coloboma (Hellstrom et al. 1999; Lee et al. 1996; Onwochei et al. 2000; Scott et al. 1997; Weiter et al. 1977), are among the most common causes of childhood blindness and severe visual impairment (DeCarlo and Nowakowski 1999; Steinkuller et al. 1999). RGCs are also the final target of pathogenesis in glaucoma. The human ATOH7 ortholog is thus a candidate gene for these disorders.
Isolation and molecular characterization of human ATOH7
Like other members of the atonal family, the mouse Atoh7 gene has no introns. We were therefore able to isolate the entire ATOH7 transcription unit as a single contiguous segment by screening a human genomic phage library with a mouse Atoh7 cDNA probe. Two positive clones were purified and analyzed in detail (Figure 1A). We subcloned and sequenced a 5.0 kb EcoRI fragment that hybridized to the cDNA probe, and identified a 456 bp open reading frame with strong homology to Atoh7. The adjoining 3′ UTR sequence also matched a human EST (H05728). The corresponding 1.7 kb clone contains an essentially full-length cDNA and its sequence matches the genomic phage clones and a bacterial artificial chromosome (BAC) identified as part of the human genome project (AC016395).
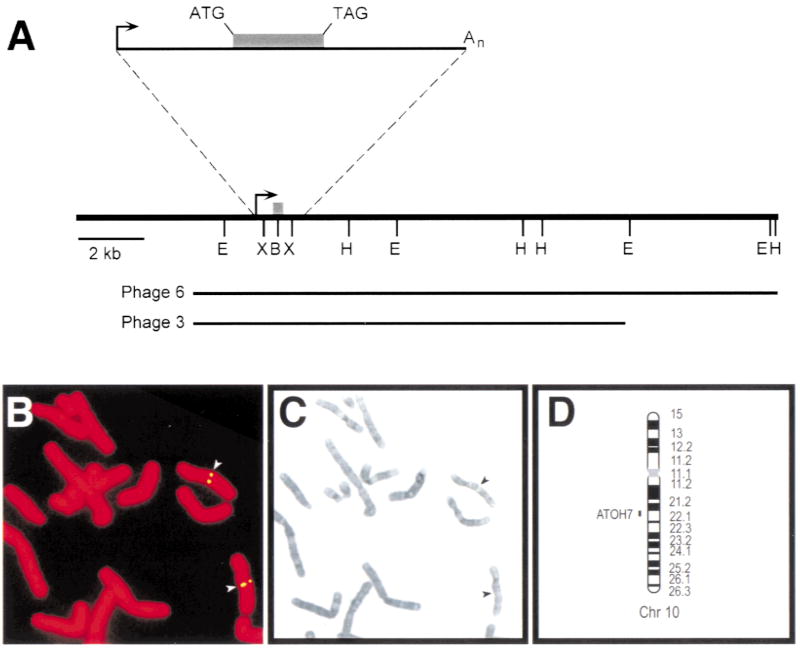
Fig. 1.
Map of ATOH7 and localization to human Chr 10q21.3–22.1 A. Restriction map derived from phage clones 3 and 6. The shaded box denotes the coding region, and the arrow indicates the start of transcription. Clone 6 was used for FISH analysis. The cDNA clone is represented by the solid line in the expanded view, and is coextensive with the genomic sequence. E, EcoRI; B, BamHI; H, HindIII; X, XhoI. B. Hybridization of biotin-labeled ATOH7 probe to propidium iodide-stained human female metaphase chromosomes. The signal was visualized indirectly with FITC. C. Giemsa-trypsin banding of the same metaphase spread. D. Ideogram showing the cytogenetic location of ATOH7 in 10q21.3–22.1.
ATOH7 maps to human chromosome 10q21–q22
Phage clone #6 was used as a FISH probe on normal human metaphase chromosome spreads (Fig. 1B,C). Fluorescent antibody and propidium iodide labeling, in combination with Giemsa-trypsin banding, showed that ATOH7 maps to the q21.3–q22.1 interval of Chr 10 (Fig. 1D). These cytogenetic data are consistent with the regional assignment of BAC clone AC016395 on human Chr 10 (McPherson et al. 2001) and genetic mapping of Atoh7 on mouse Chr 10 in an interspecific cross (Brown et al. 1998). At least 17 other markers have been identified in the HSA10-MMU10 syntenic group (Burmeister et al. 1998), including two cadherins that are implicated in neurosensory development. CDH23 (otocadherin) is strongly expressed in the retina and inner ear, and is mutated in humans with retinal degeneration and deafness (Usher syndrome, USH1D) and isolated autosomal recessive deafness (DFNB12), and in waltzer (v) mice (Bolz et al. 2001; Bork et al. 2001; Di Palma et al. 2001). PCDH15 (protocadherin) is mutated in Ames waltzer (av) mice and is a candidate for the USH1F locus (Alagramam et al. 2001; Zobeley et al. 1998). The pattern of CDH23 and PCDH15 expression within the retina, including the RGCs, has not been determined, but neither gene maps close to ATOH7. Chr 10q21 also contains a second atonal homolog, NEUROG3 (del Bosque-Plata et al. 2001), which encodes Neurogenin 3 and is distantly related to ATOH7 (Fig. 2B). ATOH7 and NEUROG3 (AF234829) are clearly distinct bHLH genes, separated by >1000 kb.
Fig. 2.
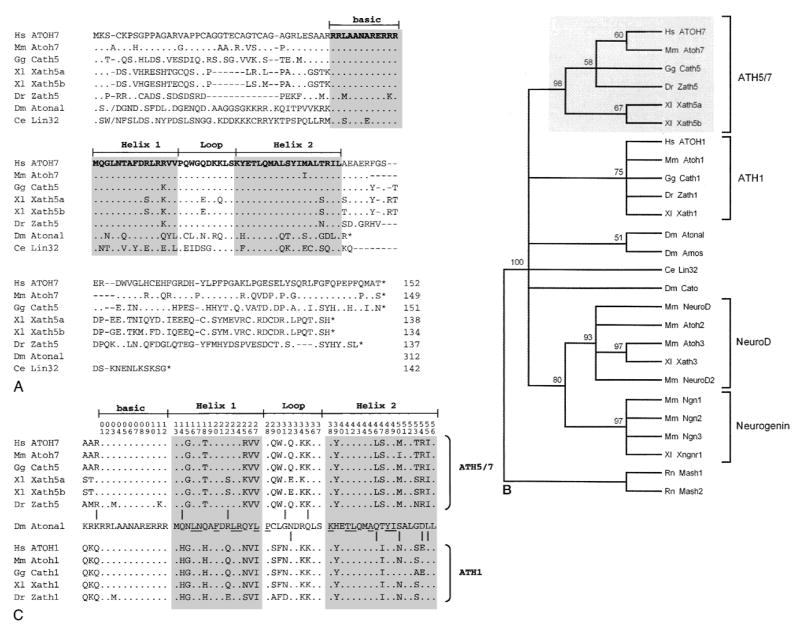
Amino acid alignment and evolutionary tree showing ATOH7 within the Atonal bHLH family. A. Optimal alignment of full-length bHLH proteins. Amino acid identities to ATOH7, sequence gaps, and stop codons are indicated by periods, dashes, and asterisks, respectively. The slashes (/) near the N-termini of Atonal and Lin32 mark the positions where 196 and 31 amino acids were omitted, respectively. Lin32 was recently found to extend further upstream from the bHLH domain (Portman and Emmons 2000). B. Bootstrap majority rule consensus tree of the Atonal family. The tree was generated by comparing 26 bHLH domains (56 characters) using maximum parsimony analysis, with Mash1 and Mash2 as outgroup taxa. Above each branchpoint, we show the proportion of bootstrap replications (out of 100) that support the group descending from that node. A value over 70 indicates very strong support (Hillis and Bull 1993). The Atonal-Amos group is weakly supported. This phylogram is completely congruent with the strict consensus tree generated by a heuristic search. C. Distribution of 10 informative amino acid sites across the bHLH domain, where proteins in the ATH5/7 (top) and ATH1 (bottom) groups differ from each other, but where at least one member of the ATH5/7 or ATH1 groups is identical to Atonal. Amino acid positions are numbered at the top. The periods indicate identity with Atonal. Underlined residues are predicted to form intra- or intermolecular helical contacts (Atchley et al. 2000; Ferré-D’Amaré et al. 1993). These informative changes are nonrandomly distributed (Mann-Whitney rank test, US = 29, P = 0.005).
In spite of its established role in RGC development, our mapping data also exclude ATOH7 from 10q24-qter, where trisomy has been associated with optic nerve aplasia (Pfeiffer et al. 1995). Apart from USH1D, no human genetic ocular disease has been mapped to the 10q21–q22 region.
Evolutionary comparisons within the Atonal family
To better understand the conservation between ATOH7 and other Atonal-related proteins, we aligned the full-length amino acid sequences of Atonal, C. elegans Lin32 and six vertebrate homologs (Fig. 2A). This comparison highlights the significant degree of amino acid identity within the bHLH domain. For example, the basic DNA-binding region, which largely determines target site specificity (Chien et al. 1996; Dang et al. 1992; Fisher and Goding 1992), is identical among human, mouse, chicken, frog, and fly proteins in the ATH5/7 group (Fig. 2A). However, outside of the bHLH domain, there are no segments of extended sequence homology. In fact, Atonal has a significantly longer N-terminal coding region than the other proteins, by >200 amino acids, and is predicted to truncate immediately C-terminal to the bHLH domain. These observations are consistent with the modular evolution proposed for bHLH proteins (Morgenstern and Atchley 1999) and suggest that functional constraints on the external amino acids are relatively limited.
To further delineate phylogenetic relationships among these proteins, we used maximum parsimony analysis to compare 24 bHLH domains in the Atonal class and two outgroups. This data matrix contains 56 characters, of which 34 are informative. Our initial heuristic search gave 12 shortest trees with 216 steps and consistency index (CI) 0.690. The strict consensus tree has three major clades: NeuroD/Neurogenin, ATH5/7 and ATH1. Atonal and Amos are grouped with ATH1 and Cato is grouped with NeuroD/Neurogenin; Lin 32 is basal. The bootstrap majority-rule consensus tree (Fig. 2B) generated from 100 replicate searches has a similar overall topology, but does resolve Atonal, Amos, Cato and Lin32 branches with respect to the ATH5/7, ATH1 and NeuroD/Neurogenin clades. As a further measure of support, we generated a congruent strict consensus tree using an exact branch-and-bound algorithm and a pruned data set, in which the NeuroD and Neurogenin clades were represented by single taxa (not shown).
Our analysis confirms the existence of four well-supported clades within the Atonal bHLH family, corresponding to the vertebrate ATH5/7, ATH1, NeuroD, and Neurogenin subfamilies (Brown et al. 1998; Ledent and Vervoort 2001), and shows that human ATOH7 is most closely related to mouse Atoh7 (Fig. 2B). There is additional strong support for a combined NeuroD-Neurogenin clade. The phylogenetic data do not resolve whether the ATH5/7 and ATH1 clades are sister groups or are equivalently divergent from Atonal. However, the hypothesis that ATH5/7 and ATH1 arose by gene duplication after divergence from the Atonal lineage is compatible with our analysis. This model is particularly appealing in view of the probable genome-wide duplications that occurred during early vertebrate evolution (Holland 1999a; Nadeau and Sankoff 1997; Ohno 1999) and the segregation of visual and proprioceptive functions demonstrated in mice between paralogous Atoh7 and Atoh1 genes, respectively (Bermingham et al. 1999, 2001; Brown et al. 2001; Hassan and Bellen 2000).
Although largely separated in vertebrates, these dual functions are mediated by the single atonal gene in Drosophila. Indeed, closer inspection of the ten informative residues that alternately resolve the ATH5/7 or ATH1 clade from Atonal shows that these amino acid changes are not uniformly distributed across the bHLH domain (Fig. 2C). This pattern is highly significant (P = 0.005). There is relatively greater similarity between the ATH5/7 subfamily and Atonal in the N-terminal half of the bHLH domain, including Helix 1, whereas there is greater similarity between the ATH1 subfamily and Atonal in the C-terminal half, including Helix 2. The affected residues do not directly participate in bHLH dimerization or intrahelical contacts (Atchley et al. 2000; Ferré-D’Amaré et al. 1993), but are predicted to be exposed and so may affect important heterologous protein-protein interactions. Although the primary functional divergence of ATH5/7 and ATH1 paralogs is most likely to have occurred through modular evolution of cis regulatory elements (Li and Noll 1994; Sun et al. 1998), it is also possible that Helices 1 and 2 have been differentially selected to interact with distinct but conserved proteins required for visual or proprioceptive development, similar to functional changes acquired during the evolution of Ubx proteins (Grenier and Carroll 2000). In both respects, the proposed partitioning of ancestral functions between ATH5/7 and ATH1 clades conforms to the duplication-degeneration-complementation (DDC) model for duplicate gene preservation (Force et al. 1999). In this context, the Atoh7 and Atoh1 genes may be regarded as semi-orthologs of atonal (Holland 1999b; Sharman 1999).
Noncoding sequences flanking ATOH7 and Atoh7 are highly conserved
As a first step to understand the cis regulation of ATOH7 transcription, we determined the sequence of genomic DNA surrounding the human (5.7 kb) and mouse (7.3 kb) coding regions. The human sequence was subsequently extended when public BAC sequence data became available. We identified CpG islands and repetitive elements, using the RepeatMasker search algorithm, and compared the human and mouse sequences using BLASTN2 (Tatusova and Madden 1999) and VISTA (Dubchak et al. 2000; Mayor et al. 2000) alignment programs. These analyses were performed on 6.5 kb of genomic DNA that extends 3 kb from the coding region on each side (Fig. 3A).
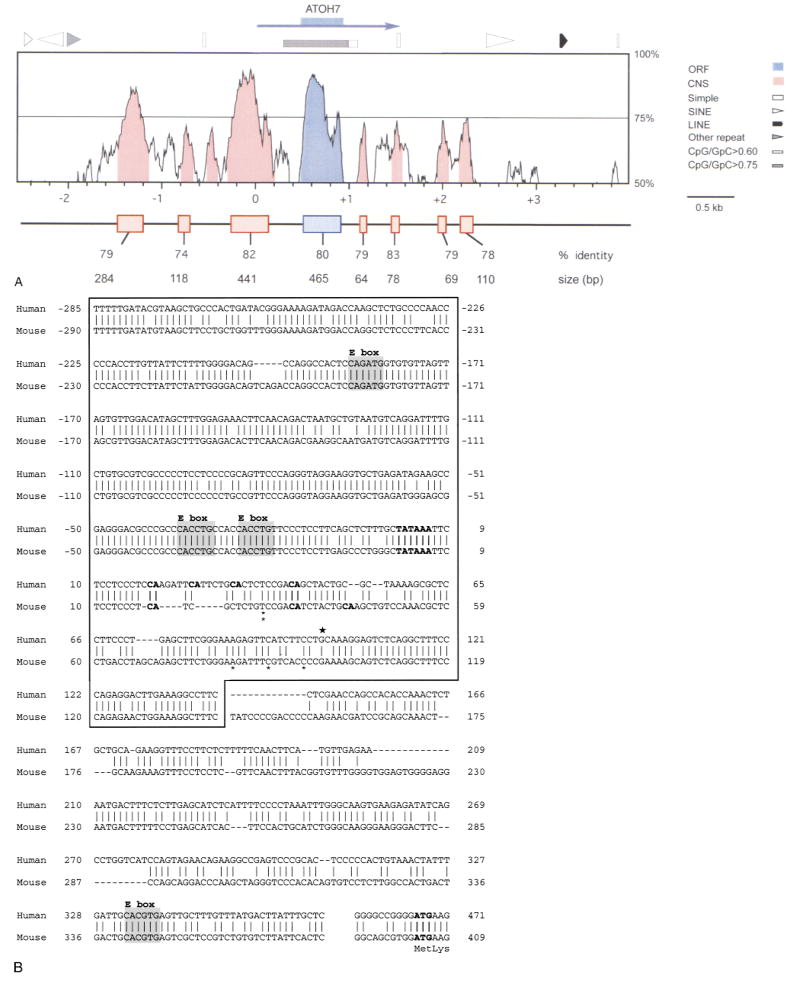
Fig. 3.
Extended DNA sequence similarity between human ATOH7 and mouse Atoh7. A. Distribution of human-mouse sequence homology for a DNA segment extending 3000 bp from each side of the ATOH7 open reading frame (ORF, blue box). The sequence is numbered in relation to the human transcription unit (blue arrow), starting with the conserved TATAAA box. The profile was created using VISTA and plots the percentage nucleotide identity (y-axis) calculated for a moving 100 bp window centered at each position of the aligned sequence (x-axis). Eight conserved nucleotide segments (CNS, shaded in red) were found in noncoding DNA within which all 100 bp windows are ≥70% identical. SINE, LINE, simple (low complexity) and other (MER) repetitive elements and CpG-rich regions in the human DNA are indicated above the graph. The boxes below the graph show highly conserved DNA segments revealed by pairwise BLAST2N analysis and the size and percentage identity of each alignment. Because of introduced gaps, the alignments are typically longer than the corresponding sequence in each species. In human DNA, the conserved segments extend from −1474 to −1193, −816 to −701, −285 to +142, +465 to +923 (coding), +1126 to +1189, +1438 to +1515, +1963 to +2031, and +2189 to +2297. B. Conservation of the promoter region. ATOH7 and Atoh7 sequences directly 5′ to the coding region are aligned. Vertical lines and dashes indicate nucleotide identity and gaps, respectively. The box encloses a 441-bp segment, revealed by BLASTN2, with 82% identity. The 5′ ends of the one human (★) and five mouse (*) cDNA clones (Brown et al. 1998) are shown above and below the aligned sequences, respectively. The TATAAA box, translational start site (ATG), and all potential transcription initiator (m7G cap) sites (CA) located 15–45 bp from the TATAAA box are indicated in bold type. The four conserved E box binding sites are shaded.
The ATOH7 coding region is extremely GC-rich (78%) and lies inside within a single large CpG island, where the ratio of CpG to GpC dinucleotides exceeds 0.75. Both alignment programs revealed seven segments of strong nucleotide identity (≥74%) outside the coding region (Fig. 3A). An additional conserved nucleotide segment (CNS) was found in the VISTA profile at the 70% identity level, but was below our threshold for detection with BLASTN2. The order and relative spacing of these segments has also been conserved (Fig. 3A). In view of the evolutionary distance between rodents and primates, these segments are likely to be functionally important. Several examples of noncoding DNA conservation have been described for homologous regions of mouse and human chromosomes (Hardison 2000). In some cases, functional tests have confirmed a role for these sequences in controlling gene expression (Heintz 2000; Loots et al. 2000). We therefore predict that the seven conserved DNA segments surrounding ATOH7 and Atoh7 likewise contain transcriptional regulatory elements. However, similar comparisons between these two mammalian genes and atonal (Sun et al. 1998) failed to uncover significant stretches of sequence conservation in flanking genomic DNA (data not shown).
The most striking nucleotide conservation is directly 5′ to the start of ATOH7 translation (Fig. 3B). This 441-bp segment is 82% identical between human and mouse genomic DNA and contains sequence motifs characteristic of eukaryotic promoters. It encompasses the 5′ ends of the human cDNA clone and five mouse cDNA clones (Brown et al. 1998) and contains a conserved TATAAA box. Several potential transcription initiator sites (weak consensus Y2CAY5) are located within 18–43 bp of the TATAAA box (Smale 1994), including one site that is identical to the optimal TdT initiator (Smale and Baltimore 1989). The longest two mouse cDNA clones start within a group of three potential initiator sites. The size of the human (Fig. 1A) and mouse cDNA clones is similar to the single 1.7-kb Atoh7 transcript detected by Northern analysis in E15.5 mouse eyes (data not shown). Together these observations strongly suggest that ATOH7 and Atoh7 use this conserved TATAAA box to initiate transcription.
The proximal 5′ segment is more highly conserved than the coding region itself (80%) and contains three E boxes (CANNTG), which are core DNA recognition sites for the bHLH domain. The relative spacing and sequence of the two E boxes closest to the transcription unit suggest that a pair of bHLH protein hetero- or homodimers may bind cooperatively to each of these sites to regulate ATOH7 and Atoh7 expression. In particular, the CACCTG sequence has been associated with binding of Group A bHLH proteins (Dang et al. 1992; Murre et al. 1989), including E47 and Atonal (Jarman et al. 1993). Our mouse knockout data, however, suggest that Atoh7 does not significantly autoregulate itself in retinal progenitor cells. In these experiments, a lacZ cassette inserted 18 codons downstream from the start of translation was expressed similarly in the retinas of Atoh7 −/− and +/− embryos (Brown et al. 2001). Although we cannot rule out the possibility of subtle autoregulation in some expression domains or differentiated cell types, Atoh7 appears to differ in this respect from Atoh1, which partially regulates its hindbrain expression via a 3′ enhancer (Helms et al. 2000), and atonal, which autoregulates its expression in intermediate clusters and R8 photoreceptors through a 5′ enhancer (Sun et al. 1998).
We have characterized ATOH7, a human homolog of atonal, and the distribution of phylogenetically conserved flanking sequences. Together, these elements significantly regulate the genesis of retinal ganglion cells, through which all visual and photo-sensory information is transmitted to the human brain. Our findings provide a foundation for mutation screening and investigation of upstream factors that control its spatial and temporal expression.
Acknowledgments
We thank Tom Glover for generous support and advice; Priscilla Tucker, Jim Lauderdale, David Ginsburg, and Ed Oliver for valuable discussions; Debra Gemza for technical assistance; and Priscilla Tucker and Anand Swaroop for critically reading the manuscript. This work was supported by a grant from the National Institutes of Health (EY11729) to T. Glaser.
Footnotes
References
- Alagramam KN, Murcia CL, Kwon HY, Pawlowski KS, Wright CG, et al. The mouse Ames waltzer hearing-loss mutant is caused by mutation of Pcdh15, a novel protocadherin gene. Nat Genet. 2001;27:99–102. doi: 10.1038/83837. [DOI] [PubMed] [Google Scholar]
- Atchley WR, Wollenberg KR, Fitch WM, Terhalle W, Dress AW. Correlations among amino acid sites in bHLH protein domains: an information theoretic analysis. Mol Biol Evol. 2000;17:164–178. doi: 10.1093/oxfordjournals.molbev.a026229. [DOI] [PubMed] [Google Scholar]
- Bermingham NA, Hassan BA, Price SD, Vollrath MA, Ben-Arie N, et al. Math1: an essential gene for the generation of inner ear hair cells. Science. 1999;284:1837–1841. doi: 10.1126/science.284.5421.1837. [DOI] [PubMed] [Google Scholar]
- Bermingham NA, Hassan BA, Wang VY, Fernandez M, Banfi S, et al. Proprioceptor pathway development is dependent on math1. Neuron. 2001;30:411–422. doi: 10.1016/s0896-6273(01)00305-1. [DOI] [PubMed] [Google Scholar]
- Bolz H, von Brederlow B, Ramirez A, Bryda EC, Kutsche K, et al. Mutation of CDH23, encoding a new member of the cadherin gene family, causes Usher syndrome type 1D. Nat Genet. 2001;27:108–112. doi: 10.1038/83667. [DOI] [PubMed] [Google Scholar]
- Bork JM, Peters LM, Riazuddin S, Bernstein SL, Ahmed ZM, et al. Usher syndrome 1D and nonsyndromic autosomal recessive deafness DFNB12 are caused by allelic mutations of the novel cadherin-like gene CDH23. Am J Hum Genet. 2001;68:26–37. doi: 10.1086/316954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NL, Kanekar S, Vetter ML, Tucker PK, Gemza DL, et al. Math5 encodes a murine basic helix-loop-helix transcription factor expressed during early stages of retinal neurogenesis. Development. 1998;125:4821–4833. doi: 10.1242/dev.125.23.4821. [DOI] [PubMed] [Google Scholar]
- Brown NL, Patel S, Brzezinski J, Glaser T. Math5 is required for retinal ganglion cell and optic nerve formation. Development. 2001;128:2497–2508. doi: 10.1242/dev.128.13.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmeister M, Bryda EC, Bureau JF, Noben-Trauth K. Encyclopedia of the mouse genome VII. Mouse chromosome 10. Mamm Genome. 1998;8:S200–S214. doi: 10.1007/s003359900655. [DOI] [PubMed] [Google Scholar]
- Cepko C. Giving in to the blues. Nat Genet. 2000;24:99–100. doi: 10.1038/72887. [DOI] [PubMed] [Google Scholar]
- Cepko CL. The roles of intrinsic and extrinsic cues and bHLH genes in the determination of cell fates. Curr Opin Neurobiol. 1999;9:37–46. doi: 10.1016/s0959-4388(99)80005-1. [DOI] [PubMed] [Google Scholar]
- Chien CT, Hsiao CD, Jan LY, Jan YN. Neuronal type information encoded in the basic-helix-loop-helix domain of proneural genes. Proc Natl Acad Sci USA. 1996;93:13239–13244. doi: 10.1073/pnas.93.23.13239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagenais SL, Guevara-Fujita M, Loechel R, Burgess AC, Miller DE, et al. The canine copper toxicosis locus is not syntenic with ATP7B or ATX1 and maps to a region showing homology to human 2p21. Mamm Genome. 1999;10:753–756. doi: 10.1007/s003359901085. [DOI] [PubMed] [Google Scholar]
- Dang CV, Dolde D, Gillison ML, Dato GJ. Discrimination between related DNA sites by a single amino acid residue of myc-related basic-helix-loop-helix proteins. Proc Natl Acad Sci USA. 1992;89:599–602. doi: 10.1073/pnas.89.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCarlo DK, Nowakowski R. Causes of visual impairment among students at the Alabama School for the Blind. J Am Optom Assoc. 1999;70:647–652. [PubMed] [Google Scholar]
- del Bosque-Plata L, Lin J, Horikawa Y, Schwarz PE, Cox NJ, et al. Mutations in the coding region of the neurogenin 3 gene (NEUROG3) are not a common cause of maturity-onset diabetes of the young in Japanese subjects. Diabetes. 2001;50:694–696. doi: 10.2337/diabetes.50.3.694. [DOI] [PubMed] [Google Scholar]
- Di Palma F, Holme RH, Bryda EC, Belyantseva IA, Pellegrino R, et al. Mutations in Cdh23, encoding a new type of cadherin, cause stereocilia disorganization in waltzer, the mouse model for Usher syndrome type 1D. Nat Genet. 2001;27:103–107. doi: 10.1038/83660. [DOI] [PubMed] [Google Scholar]
- Dubchak I, Brudno M, Loots GG, Mayor C, Pachter L, Rubin EM, et al. Active conservation of noncoding sequences revealed by 3-way species comparisons. Genome Res. 2000;10:1304–1306. doi: 10.1101/gr.142200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.5c. 1993. (Department of Genetics, University of Washington, Seattle: Distributed by the author) [Google Scholar]
- Ferré-D’Amaré AR, Prendergast GC, Ziff EB, Burley SK. Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature. 1993;363:38–45. doi: 10.1038/363038a0. [DOI] [PubMed] [Google Scholar]
- Fisher F, Goding CR. Single amino acid substitutions alter helix-loop-helix protein specificity for bases flanking the core CANNTG motif. EMBO J. 1992;11:4103–4109. doi: 10.1002/j.1460-2075.1992.tb05503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, et al. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund CL, Gregory-Evans CY, Furukawa T, Papaioannou M, Looser J, et al. Cone-rod dystrophy due to mutations in a novel photoreceptor-specific homeobox gene (CRX) essential for maintenance of the photoreceptor. Cell. 1997;91:543–553. doi: 10.1016/s0092-8674(00)80440-7. [DOI] [PubMed] [Google Scholar]
- Freund CL, Wang QL, Chen S, Muskat BL, Wiles CD, et al. De novo mutations in the CRX homeobox gene associated with Leber congenital amaurosis. Nat Genet. 1998;18:311–312. doi: 10.1038/ng0498-311. [DOI] [PubMed] [Google Scholar]
- Grenier JK, Carroll SB. Functional evolution of the Ultrabithorax protein. Proc Natl Acad Sci USA. 2000;97:704–709. doi: 10.1073/pnas.97.2.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haider NB, Jacobson SG, Cideciyan AV, Swiderski R, Streb LM, et al. Mutation of a nuclear receptor gene, NR2E3, causes enhanced S cone syndrome, a disorder of retinal cell fate. Nat Genet. 2000;24:127–131. doi: 10.1038/72777. [DOI] [PubMed] [Google Scholar]
- Hardison RC. Conserved noncoding sequences are reliable guides to regulatory elements. Trends Genet. 2000;16:369–372. doi: 10.1016/s0168-9525(00)02081-3. [DOI] [PubMed] [Google Scholar]
- Hassan BA, Bellen HJ. Doing the MATH: is the mouse a good model for fly development? Genes Dev. 2000;14:1852–1865. [PubMed] [Google Scholar]
- Heintz N. Analysis of mammalian central nervous system gene expression and function using bacterial artificial chromosome-mediated transgenesis. Hum Mol Genet. 2000;9:937–943. doi: 10.1093/hmg/9.6.937. [DOI] [PubMed] [Google Scholar]
- Hellstrom A, Wiklund LM, Svensson E. The clinical and morphologic spectrum of optic nerve hypoplasia. J Aapos. 1999;3:212–220. doi: 10.1016/s1091-8531(99)70005-4. [DOI] [PubMed] [Google Scholar]
- Helms AW, Abney AL, Ben-Arie N, Zoghbi HY, Johnson JE. Autoregulation and multiple enhancers control Math1 expression in the developing nervous system. Development. 2000;127:1185–1196. doi: 10.1242/dev.127.6.1185. [DOI] [PubMed] [Google Scholar]
- Hillis DM, Bull JJ. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol. 1993;42:182–192. [Google Scholar]
- Holland PW. Gene duplication: past, present and future. Semin Cell Dev Biol. 1999a;10:541–547. doi: 10.1006/scdb.1999.0335. [DOI] [PubMed] [Google Scholar]
- Holland PWH. The effect of gene duplication on homology. In: Bock GR, Cardew G, editors. Homology. New York: John Wiley and Sons; 1999b. pp. 226–236. [Google Scholar]
- Jan YN, Jan LY. HLH proteins, fly neurogenesis, and vertebrate neurogenesis. Cell. 1993;75:827–830. doi: 10.1016/0092-8674(93)90525-u. [DOI] [PubMed] [Google Scholar]
- Jarman AP, Grau Y, Jan LY, Jan YN. atonal is a proneural gene that directs chordotonal organ formation in the Drosophila peripheral nervous system. Cell. 1993;73:1307–1321. doi: 10.1016/0092-8674(93)90358-w. [DOI] [PubMed] [Google Scholar]
- Jarman AP, Grell EH, Ackerman L, Jan LY, Jan YN. atonal is the proneural gene for Drosophila photoreceptors. Nature. 1994;369:398–400. doi: 10.1038/369398a0. [DOI] [PubMed] [Google Scholar]
- Jarman AP, Sun Y, Jan LY, Jan YN. Role of the proneural gene, atonal, in formation of Drosophila chordotonal organs and photoreceptors. Development. 1995;121:2019–2030. doi: 10.1242/dev.121.7.2019. [DOI] [PubMed] [Google Scholar]
- Kanekar S, Perron M, Dorsky R, Harris WA, Jan LY, et al. Xath5 participates in a network of bHLH genes in the developing Xenopus retina. Neuron. 1997;19:981–994. doi: 10.1016/s0896-6273(00)80391-8. [DOI] [PubMed] [Google Scholar]
- Kay JN, Finger-Baier KC, Roeser T, Staub W, Baier H. Retinal ganglion cell determination requires lakritz, a zebrafish homolog of Drosophila atonal. Neuron. 2001;30:725–736. doi: 10.1016/s0896-6273(01)00312-9. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Takezawa SI, Hara K, Yu RT, Umesono Y, et al. Identification of a photoreceptor cell-specific nuclear receptor. Proc Natl Acad Sci USA. 1999;96:4814–4819. doi: 10.1073/pnas.96.9.4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledent V, Vervoort M. The basic helix-loop-helix protein family: comparative genomics and phylogenetic analysis. Genome Res. 2001;11:754–770. doi: 10.1101/gr.177001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BL, Bateman JB, Schwartz SD. Posterior segment neovascularization associated with optic nerve aplasia. Am J Ophthalmol. 1996;122:131–133. doi: 10.1016/s0002-9394(14)71982-8. [DOI] [PubMed] [Google Scholar]
- Li X, Noll M. Evolution of distinct developmental functions of three Drosophila genes by acquisition of different cis-regulatory regions. Nature. 1994;367:83–87. doi: 10.1038/367083a0. [DOI] [PubMed] [Google Scholar]
- Liu W, Mo Z, Xiang M. The Ath5 proneural genes function upstream of Brn3 POU domain transcription factor genes to promote retinal ganglion cell development. Proc Natl Acad Sci USA. 2001;98:1649–1654. doi: 10.1073/pnas.98.4.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loots GG, Locksley RM, Blankespoor CM, Wang ZE, Miller W, et al. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science. 2000;288:136–140. doi: 10.1126/science.288.5463.136. [DOI] [PubMed] [Google Scholar]
- Masai I, Stemple DL, Okamoto H, Wilson SW. Midline signals regulate retinal neurogenesis in zebrafish. Neuron. 2000;27:251–263. doi: 10.1016/s0896-6273(00)00034-9. [DOI] [PubMed] [Google Scholar]
- Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, et al. VISTA: Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 2000;16:1046–1047. doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
- McPherson JD, Marra M, Hillier L, Waterston RH, Chinwalla A, et al. A physical map of the human genome. Nature. 2001;409:934–941. doi: 10.1038/35057157. [DOI] [PubMed] [Google Scholar]
- Morgenstern B, Atchley WR. Evolution of bHLH transcription factors: modular evolution by domain shuffling? Mol Biol Evol. 1999;16:1654–1663. doi: 10.1093/oxfordjournals.molbev.a026079. [DOI] [PubMed] [Google Scholar]
- Murre C, McCaw PS, Vaessin H, Caudy M, Jan LY, et al. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- Nadeau JH, Sankoff D. Comparable rates of gene loss and functional divergence after genome duplications early in vertebrate evolution. Genetics. 1997;147:1259–1266. doi: 10.1093/genetics/147.3.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Gene duplication and the uniqueness of vertebrate genomes circa 1970–1999. Semin Cell Dev Biol. 1999;10:517–522. doi: 10.1006/scdb.1999.0332. [DOI] [PubMed] [Google Scholar]
- Onwochei BC, Simon JW, Bateman JB, Couture KC, Mir E. Ocular colobomata. Surv Ophthalmol. 2000;45:175–194. doi: 10.1016/s0039-6257(00)00151-x. [DOI] [PubMed] [Google Scholar]
- Pfeiffer RA, Junemann A, Lorenz B, Sieber E. Aplasia of the optic nerve in two cases of partial trisomy 10q24-ter. Clin Genet. 1995;48:183–187. doi: 10.1111/j.1399-0004.1995.tb04085.x. [DOI] [PubMed] [Google Scholar]
- Portman DS, Emmons SW. The basic helix-loop-helix transcription factors LIN-32 and HLH-2 function together in multiple steps of a C. elegans neuronal sublineage. Development. 2000;127:5415–5426. doi: 10.1242/dev.127.24.5415. [DOI] [PubMed] [Google Scholar]
- Scott IU, Warman R, Altman N. Bilateral aplasia of the optic nerves, chiasm, and tracts in an otherwise healthy infant. Am J Ophthalmol. 1997;124:409–410. doi: 10.1016/s0002-9394(14)70840-2. [DOI] [PubMed] [Google Scholar]
- Sharman AC. Some new terms for duplicated genes. Semin Cell Dev Biol. 1999;10:561–563. doi: 10.1006/scdb.1999.0338. [DOI] [PubMed] [Google Scholar]
- Smale ST. Core promoter architecture for eukaryotic protein-coding genes. In: Conaway RC, Conaway JW, editors. Transcription: Mechanisms and Regulation. New York: Raven Press, Ltd; 1994. pp. 63–81. [Google Scholar]
- Smale ST, Baltimore D. The “initiator” as a transcription control element. Cell. 1989;57:103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- Sohocki MM, Sullivan LS, Mintz-Hittner HA, Birch D, Heckenlively JR, et al. A range of clinical phenotypes associated with mutations in CRX, a photoreceptor transcription-factor gene. Am J Hum Genet. 1998;63:1307–1315. doi: 10.1086/302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinkuller PG, Du L, Gilbert C, Foster A, Collins ML, et al. Childhood blindness. J Aapos. 1999;3:26–32. doi: 10.1016/s1091-8531(99)70091-1. [DOI] [PubMed] [Google Scholar]
- Sun Y, Jan LY, Jan YN. Transcriptional regulation of atonal during development of the Drosophila peripheral nervous system. Development. 1998;125:3731–3740. doi: 10.1242/dev.125.18.3731. [DOI] [PubMed] [Google Scholar]
- Swain PK, Chen S, Wang QL, Affatigato LM, Coats CL, et al. Mutations in the cone-rod homeobox gene are associated with the cone-rod dystrophy photoreceptor degeneration. Neuron. 1997;19:1329–1336. doi: 10.1016/s0896-6273(00)80423-7. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP* Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4.0. Sunderland, MA: Sinauer Associates; 1998. [Google Scholar]
- Swofford DL, Olsen GJ, Waddell PJ, Hillis DM. Phylogenetic inference. In: Hillis DM, Moritz C, Mable BK, editors. Molecular Systematics. Sunderland, Mass: Sinauer Associates; 1996. pp. 407–509. [Google Scholar]
- Tatusova TA, Madden TL. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter MV, Brown NL. The role of basic helix-loop-helix genes in vertebrate retinogenesis. Semin Cell Dev Biol. 2001;12 doi: 10.1006/scdb.2001.0273. in press. [DOI] [PubMed] [Google Scholar]
- Wang SW, Kim BS, Ding K, Wang H, Sun D. Requirement for math5 in the development of retinal ganglion cells. Genes Dev. 2001;15:24–29. doi: 10.1101/gad.855301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiter JJ, McLean IW, Zimmerman LE. Aplasia of the optic nerve and disk. Am J Ophthalmol. 1977;83:569–576. doi: 10.1016/0002-9394(77)90569-4. [DOI] [PubMed] [Google Scholar]
- Zobeley E, Sufalko DK, Adkins S, Burmeister M. Fine genetic and comparative mapping of the deafness mutation Ames waltzer on mouse chromosome 10. Genomics. 1998;50:260–266. doi: 10.1006/geno.1998.5298. [DOI] [PubMed] [Google Scholar]