Abstract
Gap junction channels may be comprised of either connexin or pannexin proteins (innexins and pannexins). Membrane topologies of both families are similar, but sequence similarity is lacking. Recently, connexin-like sequences have been identified in mammalian and zebrafish genomes that have only four conserved cysteines in the extracellular domains (Cx23), a feature of the pannexins. Phylogenetic analyses of the non-canonical “C4” connexins reveal that these sequences are indeed connexins. Functional assays reveal that the Cx23 gap junctions are capable of sharing neurobiotin, and further, that Cx23 connexins form hemichannels in vitro.
Introduction
Gap junctions permit the direct exchange of small molecules (≤ 1200 Da) among neighboring cells, and form when dozens to thousands of channels aggregate in the plasma membrane. A single gap junction channel is comprised of two hexameric hemichannels, or connexons, contributed by adjacent cells. Two gene families, the connexins and pannexin proteins (i.e. innexins and pannexins), appear to be capable of building gap junction channels. The connexins are a large gene family (21 human, 20 in mouse, ref 1) found in both vertebrate and invertebrate chordates (2), and not in other invertebrates. Pannexins/innexins were initially identified in flies and worms and, while forming similar structures, lack sequence similarity with the connexins. Initially, these sequences were termed innexins, for invertebrate connexins. More recently, sequences related to the innexins were found in vertebrate genomes, and it was suggested that the innexins and their counterparts in chordates should be named pannexins (pan, from Latin for all, 3, 4, 5). We utilize this convention here (5). The pannexin family is moderately sized in D. melanogaster (8 pannexins), large in C. elegans (25 pannexins), and relatively small in mammals (3 in human and mouse) (6). It is not clear why such a large number of genes are required for the seemingly simple function of direct cell-cell communication. Since the connexins and pannexins likely evolved independently (7), it appears that both families have grown/are continuing to grow in parallel, perhaps providing opportunities for functional diversity and/or increased specialization. Identification of the ancestral forms for each family would likely contribute significant insights into the solution of local cellular communication.
Both the connexins and pannexins exhibit similar membrane topology (Figure 1A). The amino and carboxy termini are found in the cytoplasm, providing two extracellular loops (connecting transmembrane domains 1/2 and 3/4) and one intracellular loop (connecting transmembrane domains 2/3). An important feature of gap junction proteins is the presence of multiple cysteine residues in both extracellular domains. For example, a defining feature of the connexins is the presence of three invariant cysteines per loop (8), while the pannexins have only two cysteine residues per loop (reviewed in 6, 9). Disulfide bonds among these cysteines are thought to provide structural stability and to facilitate connexon-connexon docking at the plasma membrane (10).
Figure 1.
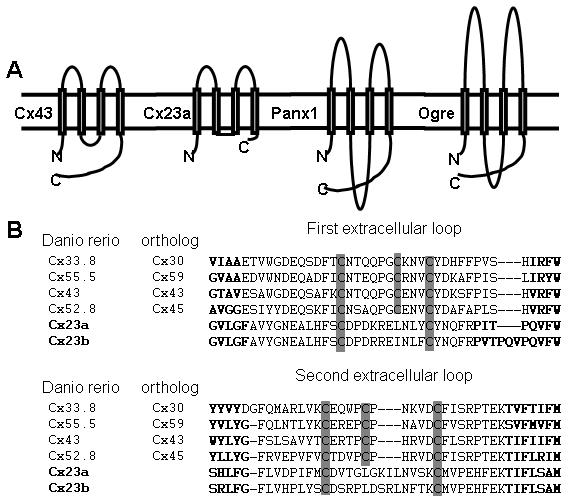
Comparison of predicted membrane topology of C4-connexins with other gap junction proteins. (A) Representative members of each family are shown: zebrafish Cx43 for the connexins, zebrafish Cx23a for the C4-connexins, zebrafish Panx1 for the chordate pannexins, and Drosophila Ogre for the protostomal pannexins. (B) Alignment of the extracellular domains of zebrafish connexins, Cx23a and Cx23b. Transmembrane domains are in bold and conserved cysteines are highlighted in grey. We previously found that the extracellular cysteines in the zebrafish connexins are conserved with cysteines in the mammalian connexins (11).
Recently, we identified two zebrafish sequences exhibiting significant sequence similarity with the connexins, but containing only two cysteine residues per extracellular loop as seen in the pannexins (11). Similar sequences were previously identified in the human and mouse genomes (1), suggesting evolutionary retention and diversification of a functional protein. These non-canonical connexins have inspired considerable speculation regarding function, but have not been evaluated for gene expression, gap junctional activity or hemichannel activity. In this report we examine the zebrafish C4-connexins (Cx23a and Cx23b). We find that these sequences form part of a monophyletic group that includes the mammalian C4 connexins (mouse and human Cx23) and that this group represents the closest known relative of the current connexin proteins. In heterologous assays, we find that untagged Cx23a is capable of forming channels that can transfer the small tracer molecule neurobiotin, but not the larger propidium iodide. Furthermore, Cx23a is capable of forming hemichannels. Together, this indicates that Cx23 can function as a connexin, albeit with only four extracellular cysteines.
Materials and Methods
Plasmid construction
cDNA was prepared from Trizol-treated embryos (Gibco) or fins followed by reverse transcription with an oligo dT12-15 primer. Oligos to amplify cx23a (NM_001013546 ) contained BglII (F-GCATTAGATCTTATGTCATTAAATTACATCAAAAAC) or EcoRI restriction sites (R- AGCTAAGAATTCCAGTCATTGGCTGTAAATAGCC). Oligos to attempt to amplify cx23b (ENSDART00000056324) included F- (GCATTAGATCTTATGTCCTTAAACTACATCAAGAAC) R1-(GGCATTGAATTCCCTGATGTGTTAAATACCCCAGCCT) and R2-(TGTTAAATACCCCAGCCTTCTGC). Amplified cx23a product (Advantage taq polymerase, Clontech) was cloned into pGEM-T and sequenced to confirm the absence of PCR-generated errors. BglII/EcoRI double digests were performed to subclone the cx23a gene sequence into pEGFP-N1 and pIRES2.
Sequence alignment and phylogenetic analysis
Because Cx23 proteins exhibit a defining characteristic of pannexins rather than connexins, i.e., two cysteines per extra-cellular loop, we conducted a broad scale phylogenetic analysis that included connexin sequences (zebrafish, human, mouse, Ciona), protostome pannexin sequences (fly, worm, hydra), chordate pannexin sequences (zebrafish, human, mouse, Ciona), and C4-connexins (zebrafish, human, and mouse). Accession numbers for all sequences utilized here may be found in supplementary material (Table S1). Amino acid sequences were aligned using ClustalX and NXS files were generated and imported into PAUP* (12).
A parsimony reconstruction was generated in PAUP* for the full alignment using a modified version of the PROTPARS executable in PHYLIP (13), where gaps were treated as missing data. A heuristic search based on 10 random addition sequences was conducted; TBR branch swapping was in effect. Bootstrap values for the parsimony tree are based on 1000 replicates using “fast” stepwise-addition. Trees are presented as unrooted.
In situ hybridization
Probes for cx23 were generated by digesting the pGEM-T construct with BglII for antisense and EcoRI for sense, followed by in vitro transcription (T7 or Sp6 RNA polymerase, respectively), NTPs, and digoxigenin-labeled UTP (Roche). Tissue was fixed overnight with 4% paraformaldehyde in PBS and stored in 100% methanol at -20°C. Gradual aqueous washes were completed in methanol/PBST. Tissue was treated with 5 μg/ml proteinase K (5 min for embryos; 45 min for fins) and re-fixed for 20 min. Prehybridization (50 % formamide, 5X SSC, 10 mM citric acid, 0.1 % Tween20) occurred for 1 hour at 65°C, and hybridization in the presence of digoxigenin-labeled antisense probes was completed overnight. Gradual washes into 0.2X SSC were followed by gradual washes into PBST. Anti-digoxigenin Fab fragments (pre-absorbed against zebrafish tissue) were used at 1:5000 overnight. Following extensive washes in PBST followed by three short washes in staining buffer (100 mM Tris, 9.5, 50 mM MgCl2, 100 mM NaCl, 0.1 % Tween20, pH 9.0). Tissue was next transferred to staining solution (staining buffer plus 0.22 mg/ml NBT and 0.175 mg/ml BCIP) for development.
Transfections and Dye coupling assays
Pairs of HeLa cells that had been transfected were microinjected using the Eppendorf FemtoJet microinjector and Eppendorf InjectMan® NI2 micromanipulator. One cell from each pair was injected with propidium iodide (PI at 1mg/mL, MW=668.4) or neurobiotin (2 % in PBS, MW= 322, Vector Labs). Propidium iodide was visualized in live cells. Cells injected with neurobiotin were fixed in 2 % paraformaldehyde in PBS for 30 minutes at room temperature, blocked using 2.5 % BSA; 6 % goat serum in PBS, and detected using 6.6 μg/ml streptavidin conjugated to Alexa-568 for one hour (Invitrogen). Cells were determined to have successfully transferred dye if the dye was detectable in the non-injected cell of a pair.
Hemichannel assays
Dye-uptake was examined under low extracellular Ca2+-conditions and mechanical stimulation in a modified hemichannel assay (14). In brief, HeLa cells were seeded at low density into 35 mm diameter dishes and transfected with respective cDNA constructs. After 48 hours, cells were chilled and washed several times in PBS. A 0.1 % sulpho-rhodamine dye-solution (Invitrogen, MW 558) containing 5 mM EGTA (pH7.4) was dripped from a height of 20 mm on the cells (3 times at 5 min intervals). Cells were rinsed repeatedly in PBS and images of stimulated cells were taken immediately. Cells expressing GFP or GFP-tagged connexins above a set threshold were counted in each image, and the number of GFP-positive cells that took up sulpho-rhodamine was determined. Dye-uptake ratios were determined for each image-area and cumulative ratios of 4 independent experiments were analyzed using ANOVA (single factor statistics). Differences in dye uptake were considered significant when p-values < 0.05 were obtained. Specificity of sulpho-rhodamine dye-uptake under the described conditions was verified in control experiments by failure to uptake fluorescent dextran (MW 10,000).
Results
Zebrafish Cx23 sequences are highly related to connexins
Given the results of Eastman et al. (11) showing that the “C4-connexins” exhibit significant sequence similarity with connexin proteins, but contain only two cysteine residues in each extracellular loop, we aimed to determine if the C4-connexins would emerge as connexins or pannexins when tested phylogenetically against both families. We began by comparing the predicted membrane topology of the zebrafish C4-connexins (Cx23a and Cx23b, see below) with representative members of the connexin and pannexin families (Figure 1). We find that the C4-connexins have a short intracellular loop and carboxy-terminus, which may suggest that they do not physically interact with a wide spectrum of cellular proteins. In contrast, the amino-terminus and extracellular loops are of similar length as other connexins, and the two cysteines in the C4-connexins align perfectly with the cysteines in the first and third positions of connexin proteins.
Our phylogenetic analysis included 110 amino acid sequences encompassing connexins, pannexins, and C4-connexins. Results reveal that the zebrafish C4-connexins are orthologous to the human and mouse Cx23 connexins, the four sequences forming a monophyletic group (Figure 2, inset). Indeed the zebrafish sequences are ∼75 % identical and ∼87 % similar to mammalian Cx23. Since both zebrafish C4-connexins are equally related to the mammalian Cx23 sequences, they will be referred to as Cx23a and Cx23b. Further, the predicted molecular weight of zebrafish Cx23a (23.92 kD) and Cx23b (23.74 kD) are similar to the human (23.76 kD) and mouse (23.83 kD) Cx23 sequences. Finally, all of the Cx23 sequences form a sister clade with the typical “C6” connexin sequences, indicating that these two groups share a most recent common ancestor (Figure 2 and Figure S1 in supplementary material for the expanded phylogenetic tree). Further evidence supporting the sister relationship of the C4-connexins and the connexins is the significant sequence similarity between them, ∼45 %. Sequence similarity between the C4-connexins and the pannexins was not found.
Figure 2.
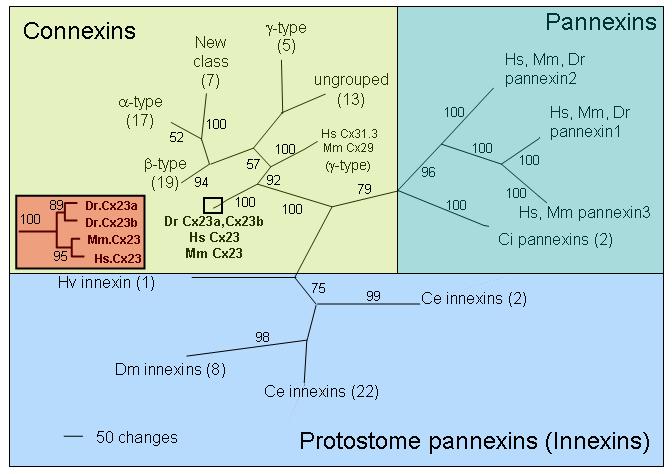
Phylogenetic analysis of connexins, pannexins, and C4-connexins. Parsimony reconstruction of 110 amino acid sequences representing the connexin, pannexin, and protostome pannexin (innexin) gene families (see Supplementary material for accession numbers, Table S1). Pannexin sequences include representatives of Hydra vulgaris (Hv), Ciona intestinalis (Ci), Caenorhabditis elegans (Ce), Drosophila melanogaster (Dm), Danio rerio (Dr), Homo sapiens (Hs), and Mus musculus (Mm). Connexin sequences include representatives of Ciona intestinalis (Ci), Danio rerio (Dr), Homo sapiens (Hs), and Mus musculus (Mm). Ciona connexins appear in the “ungrouped” connexin clade (see Eastman et al., 2006) along with representatives of the three vertebrate species. The remaining connexin groups include representatives of the three vertebrate species only. Numbers in parentheses indicate the number of sequences in each clade. Bootstrap values greater than 50% are reported. Inset: Expanded view, parsimony reconstruction of Cx23 sequences from Danio rerio (Dr), Homo sapiens (Hs), and Mus musculus (Mm).
cx23a is expressed in the embryonic lens
To provide evidence that cx23a and cx23b are functional genes and not pseudogenes, we monitored gene expression in zebrafish tissues. First, we attempted to amplify each gene from cDNA prepared from embryos at 24 hours post fertilization (hpf) and from regenerating fins. These tissues were tested because many genes are more highly expressed during early embryonic stages and/or during the process of rapid regenerative growth. Attempts to amplify cx23b were unsuccessful (data not shown). Further, the sequence for cx23b is not found in the zebrafish EST database suggesting the possibility that it is a pseudogene. Alternatively, cx23b may be expressed at low levels and/or at unsampled developmental timepoints. In contrast, it was possible to amplify cx23a from embryonic cDNA (but not from regenerating fin cDNA). Indeed, nine ESTs representing cx23a were found among embryonic cDNA libraries: day 0-3 (6), d3 (1), d5 larvae (1), or from early myoblasts (1). We next identified tissues that express cx23a by in situ hybridization. Expression is found in the lens in 24 hpf embryos (Figure 3). As a follow up, we searched the mammalian EST databases for CX23 sequences. Interestingly, we identified 6 mouse ESTs (5 from eye; 1 from head/neck), 1 cow EST from lens, 2 rabbit ESTs from eye, and 1 rhesus monkey EST from lens. This provides evidence that cx23-like genes are expressed in multiple mammalian species, and further, that expression is conserved (accession numbers provided in supplementary material, Table S2).
Figure 3.
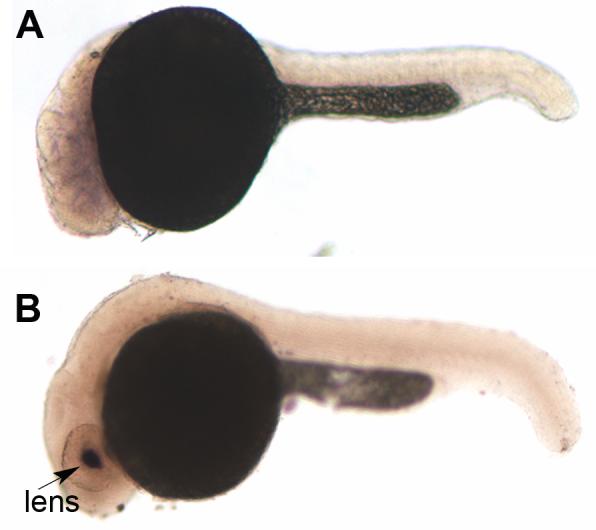
Expression of cx23a in zebrafish 24 hpf embryos. Anterior is to the left, dorsal is to the top. (A) In situ hybridization of representative wild-type embryos using a cx23a sense probe (negative control) and antisense probe (B) showing expression in the lens (arrow).
Cx23a forms gap junction channels
We next evaluated gap junction formation (i.e. the aggregation of multiple gap junction channels in the plasma membrane) by expressing an EGFP-tagged version of Cx23a (i.e. EGFP fusion to carboxy-terminus) in HeLa cells. Fluorescence of the Cx23a-EGFP fusion protein was compared with Cx43-EGFP, which forms readily identifiable gap junctions (Figure 4A and 15). In contrast, in Cx23a-EGFP expressing cells staining is observed in intracellular membranes and not in fluorescent puncta at the plasma membrane (Figure 4A).
Figure 4.
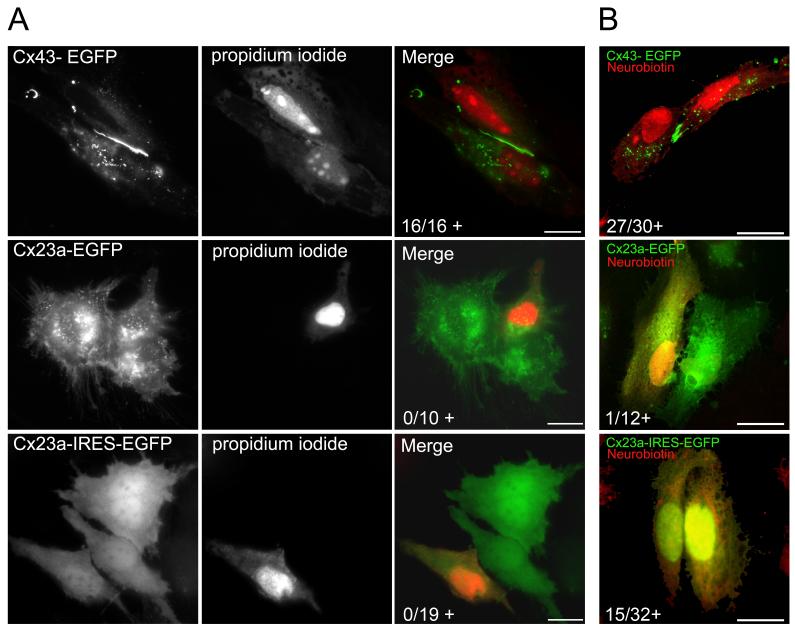
Cx23a gap junction channels do not transfer propidium iodide but can transfer neurobiotin. (A) Left panels show EGFP staining for Cx43-EGFP, Cx23a-EGFP, and Cx23-IRES-EGFP in HeLa cells. Middle panels show propidium iodide injections. Right panels show merge of propidium iodide and EGFP. (B) Merged images of neurobiotin staining and EGFP for Cx43-EGFP (top), Cx23a-EGFP (middle), and Cx23a-IRES-EGFP (bottom) in HeLa cells. The numbers in the merged photos indicate the frequency of positive dye transfer (+) in injected cell pairs. Scale bars are 20 μm.
The absence of obvious fluorescent puncta in the plasma membranes of adjacent cells suggests gap junction channels do not aggregate into typical areas of direct cell-cell communication, or that the EGFP-tag interferes with Cx23a trafficking or assembly. To test whether Cx23a forms functional cell-to-cell channels, we completed dye-coupling assays in HeLa cells transfected with Cx23a-EGFP. Since the EGFP tag may interfere with channel formation or function, we also completed dye coupling assays in cells transfected with Cx23a in the pIRES2 vector. This vector provides an internal ribosomal entry site between the multi-cloning site and the code for EGFP. Therefore, a single bicistronic mRNA is produced, but Cx23a and EGFP are translated as separate proteins.
Propidium iodide (668 MW) injections into zebrafish Cx43-EGFP transfected cells revealed robust dye coupling in all cell pairs (Figure 4A and reference 15). In contrast, cell pairs expressing either the Cx23a-EGFP construct or the untagged Cx23a did not transfer propidium iodide in any transfected cell pairs (Figure 4A). To test the possibility that Cx23a gap junction channels are capable of sharing smaller molecules, we repeated dye injections using the tracer molecule neurobiotin (322 MW). Again Cx43-EGFP channels were capable of dye coupling, and the Cx23a-EGFP was not capable of forming functional channels. Interestingly, transfection of the untagged Cx23a construct permitted transfer in about 47 % of the cell pairs examined (Figure 4B). This experiment reveals that untagged Cx23a is capable of forming functional gap junction channels, but with reduced efficiency, permitting visualization of coupling in only about half of transfected cell pairs. These data also indicate that the EGFP-tag inhibits Cx23a-channel trafficking, assembly or function.
Cx23a forms functional hemichannels
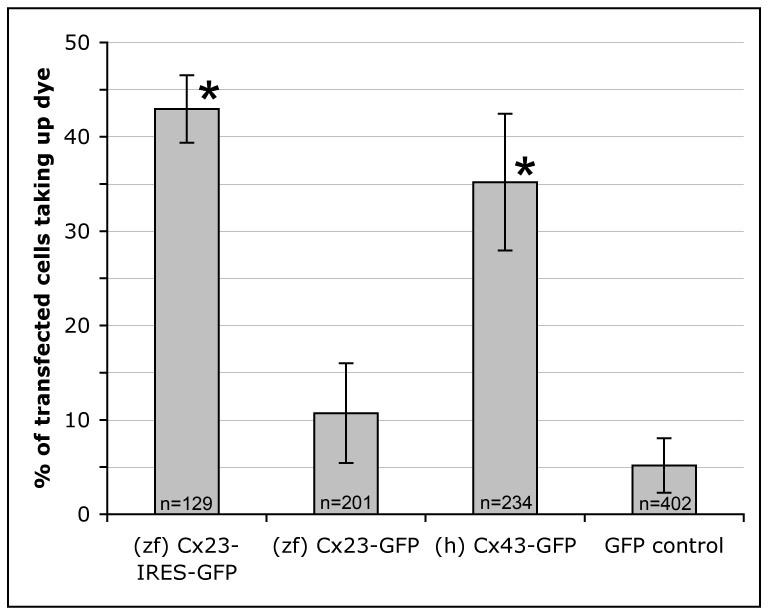
We next tested whether or not Cx23a is capable of forming hemichannels. HeLa cells were plated at low density and transfected with pEGFP-N1 expressing EGFP alone, human Cx43-EGFP (shown to exhibit hemichannel activity, ref 16), Cx23a-EGFP, or Cx23a-IRES2-EGFP. Note that zebrafish Cx43 does not exhibit hemichannel activity (15), and is therefore not the relevant positive control. Connexin hemichannels have been shown to take-up dye from the extracellular medium under conditions of low extracellular calcium and mechanical stimulation (14,17-21). Under these conditions and in the presence of sulforhodamine, we found significant hemichannel activity of both human Cx43-EGFP and untagged Cx23a when compared to cells transfected with EGFP alone (Figure 5). Cells transfected with Cx23a-EGFP did not exhibit hemichannel activity above background, consistent with the abolished dye transfer ability of EGFP-tagged Cx23 described above.
Figure 5.
Cx23 exhibits hemichannel activity. Cumulative dye-uptake efficiency (as a percentage of cells that took up dye) in 4 independent experiments. Bars represent standard deviation, and total number of transfected cells counted (n) is shown. Dye-uptake efficiency of untagged Cx23 and Cx43-EGFP was highly significant when compared with EGFP expressing control cells (p<0.0001, indicated by *). Hemichannel activity in Cx23-EGFP transfected cells was not statistically different from activity in cells transfected with EGFP alone (p>0.05).
Discussion
Here we describe the zebrafish C4-connexins, named Cx23a and Cx23b, which we find to be orthologous to previously identified C4-connexins in the human and mouse genomes (CX23, 1). While amino acid sequences for each of these genes reveal significant sequence similarity with typical connexins, they do not contain the requisite six cysteines in the two extracellular loops (8). It was therefore not clear if Cx23 would function as predicted for connexins, if they represent pseudogenes, or if they serve an undescribed function. This analysis represents the first evaluation of this as yet uncharacterized (and poorly understood) class of one such connexin-like protein in the zebrafish.
We find that the Cx23 sequences are indeed closely related to typical connexin proteins and that the four cysteines in the extracellular loops of the C4-connexins are conserved in both classes of molecules. The clade containing the Cx23 orthologs is most closely related to the connexins, thus sharing a most recent common ancestor with this diverse gene family. We provide evidence that cx23a is not a pseudogene: it is identified by 9 ESTs in zebrafish, may be amplified using RT-PCR, and is expressed in the lens by in situ hybridization. Indeed, mammalian CX23 is similarly expressed in the eye/lens based on the identification of ESTs from eye, lens, or head/neck cDNA libraries, suggesting the possibility that the function of the C4-connexins is both conserved and maintained in multiple species. Zebrafish Cx23a gap junction channels are not capable of transferring the large dye propidium iodide (MW 668), but are capable of sharing the smaller molecule neurobiotin (MW 322). However, the transfer of neurobiotin occurs only in about 47 % of transfected cell pairs, which may indicate that gap junction channel formation and/or function does not occur efficiently. In contrast, hemichannel activity appears to be as robust as the positive control (human Cx43-EGFP), and furthermore, is consistent with levels of positive hemichannel activity reported in other dye uptake studies (14).
Future studies will elucidate the primary function of Cx23a channels in vivo. For example, one possibility is that the Cx23 connexins serve as hemichannels in the lens, and may or may not be capable of forming gap junction channels when expressed at endogenous levels in a more complex tissue setting. Given the considerable diversification of the canonical connexin family relative to its sister group, Cx23, we further speculate that the additional extracellular loop cysteines represent a “key innovation” permitting the former to fill unoccupied and diverse cellular niches. If the additional cysteines indeed represent a key innovation leading to diversification, then the ancestral connexin, i.e., the most recent common ancestor of the connexin clade, may have had properties more similar to Cx23 than to the canonical connexin family.
Supplementary Material
Acknowledgements
The authors wish to thank Lynne Cassimeris for use of microinjection system and Jake Fugazzotto for care of zebrafish colony, preparation of plasmids, and in situ hybridization. The authors also thank Ross Johnson for helpful discussions and advice regarding the hemichannel assay. This work was funded from grants K22-DE014863 to MKI and GM55725 to MMF, in addition to the Bioengineering and Biosciences 2020 fund (MKI and MMF).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Sohl G, Willecke K. Gap junctions and the connexin protein family. Cardiovasc Res. 2004;62:228–32. doi: 10.1016/j.cardiores.2003.11.013. [DOI] [PubMed] [Google Scholar]
- [2].White TW, Wang H, Mui R, Litteral J, Brink PR. Cloning and functional expression of invertebrate connexins from Halocynthia pyriformis. FEBS Lett. 2004;577:42–8. doi: 10.1016/j.febslet.2004.09.071. [DOI] [PubMed] [Google Scholar]
- [3].Panchin Y, Kelmanson I, Matz M, Lukyanov K, Usman N, Lukyanov S. A ubiquitous family of putative gap junction molecules. Curr Biol. 2000;10:R473–4. doi: 10.1016/s0960-9822(00)00576-5. [DOI] [PubMed] [Google Scholar]
- [4].Baranova A, et al. The mammalian pannexin family is homologous to the invertebrate innexin gap junction proteins. Genomics. 2004;83:706–16. doi: 10.1016/j.ygeno.2003.09.025. [DOI] [PubMed] [Google Scholar]
- [5].Yen MR, Saier MH., Jr. Gap junctional proteins of animals: the innexin/pannexin superfamily. Prog Biophys Mol Biol. 2007;94:5–14. doi: 10.1016/j.pbiomolbio.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Phelan P. Innexins: members of an evolutionarily conserved family of gap-junction proteins. Biochim Biophys Acta. 2005;1711:225–45. doi: 10.1016/j.bbamem.2004.10.004. [DOI] [PubMed] [Google Scholar]
- [7].Alexopoulos H, et al. Evolution of gap junctions: the missing link? Curr Biol. 2004;14:R879–80. doi: 10.1016/j.cub.2004.09.067. [DOI] [PubMed] [Google Scholar]
- [8].Kumar NM, Gilula NB. Molecular biology and genetics of gap junction channels. Semin Cell Biol. 1992;3:3–16. doi: 10.1016/s1043-4682(10)80003-0. [DOI] [PubMed] [Google Scholar]
- [9].Panchin YV. Evolution of gap junction proteins--the pannexin alternative. J Exp Biol. 2005;208:1415–9. doi: 10.1242/jeb.01547. [DOI] [PubMed] [Google Scholar]
- [10].Foote CI, Zhou L, Zhu X, Nicholson BJ. The pattern of disulfide linkages in the extracellular loop regions of connexin 32 suggests a model for the docking interface of gap junctions. J Cell Biol. 1998;140:1187–97. doi: 10.1083/jcb.140.5.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Eastman SD, Chen TH, Falk MM, Mendelson TC, Iovine MK. Phylogenetic analysis of three complete gap junction gene families reveals lineage-specific duplications and highly supported gene classes. Genomics. 2006;87:265–74. doi: 10.1016/j.ygeno.2005.10.005. [DOI] [PubMed] [Google Scholar]
- [12].Swofford DL. Sinauer Associates. Sunderland, MA: 2002. PAUP*, Phylogenetic Analysis Using Parsimony (*and other methods) [Google Scholar]
- [13].Felsenstein J. PHYLIP Phylogeny Inference Package, Cladistics. 5ed. 1989. ^eds) [Google Scholar]
- [14].Li H, Liu TF, Lazrak A, Peracchia C, Goldberg GS, Lampe PD, Johnson RG. Properties and regulation of gap junctional hemichannels in the plasma membranes of cultured cells. J Cell Biol. 1996;134:1019–30. doi: 10.1083/jcb.134.4.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hoptak-Solga AD, Klein KA, Derosa AM, White TW, Iovine MK. Zebrafish short fin mutations in connexin43 lead to aberrant gap junctional intercellular communication. FEBS Lett. 2007;581:3297–302. doi: 10.1016/j.febslet.2007.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Contreras JE, Saez JC, Bukauskas FF, Bennett MV. Gating and regulation of connexin 43 (Cx43) hemichannels. Proc Natl Acad Sci U S A. 2003;100:11388–93. doi: 10.1073/pnas.1434298100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Pfahnl A, Dahl G. Gating of cx46 gap junction hemichannels by calcium and voltage. Pflugers Arch. 1999;437:345–53. doi: 10.1007/s004240050788. [DOI] [PubMed] [Google Scholar]
- [18].Quist AP, Rhee SK, Lin H, Lal R. Physiological role of gap-junctional hemichannels. Extracellular calcium-dependent isosmotic volume regulation. J Cell Biol. 2000;148:1063–74. doi: 10.1083/jcb.148.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Thimm J, Mechler A, Lin H, Rhee S, Lal R. Calcium-dependent open/closed conformations and interfacial energy maps of reconstituted hemichannels. J Biol Chem. 2005;280:10646–54. doi: 10.1074/jbc.M412749200. [DOI] [PubMed] [Google Scholar]
- [20].Trexler EB, Bennett MV, Bargiello TA, Verselis VK. Voltage gating and permeation in a gap junction hemichannel. Proc Natl Acad Sci U S A. 1996;93:5836–41. doi: 10.1073/pnas.93.12.5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bao L, Sachs F, Dahl G. Connexins are mechanosensitive. Am J Physiol Cell Physiol. 2004;287:C1389–95. doi: 10.1152/ajpcell.00220.2004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.