Abstract
Sequence analysis using the Promoser program predicted two promoter like regions for rat mtGPAT: a distal promoter ∼30 kb upstream and a proximal promoter near the first translational codon. Rat liver cells transfected with pGL3-basic vector containing the distal and proximal promoter resulted in 10.8 and 4.8 fold increase in the luciferase activity, respectively. Results of electromobility shift assay and chromatin immunoprecipitation suggested binding of transcription factors to the distal and proximal promoter regions. 5′RACE PCR showed two transcripts with different transcriptional start sites. When transfected rat liver cells were starved and refed, there was about 2.7 fold increase in the luciferase activity with cells transfected with the distal promoter while the proximal promoter showed no change. Thus, the two promoters could be functionally distinguished. Taken together, we conclude that there are two promoters for rat mtGPAT gene and that the transcriptional regulation is mediated through the distal promoter.
Keywords: mitochondria; glycerophosphate acyltransferase; promoter; RACE-PCR, luciferase, ChIP
Introduction
Glycerol-3-phosphate acyltransferase (GPAT) converts sn-glycerol-3-phosphate to 1-acyl-sn-glycerol-3-phosphate, the first step in the biosynthetic pathway of glycerolipids. There are two major isoforms of this enzyme - one located on the mitochondrial outer membrane (mtGPAT) and the other on endoplasmic reticulum (microsomal; msGPAT). The mtGPAT is involved in the asymmetric distribution of fatty acids found in cellular glycerophospholipids. The asymmetric distribution of fatty acids plays an important role in maintaining structure and function of phospholipids present in cellular membranes (1-4) and also in modulation of cell signal transduction or in cell malignancy (2). MtGPAT is an enzyme that may switch the fate of fatty acids from oxidation to synthesis (1, 2, 3). The acyltransferase is highly induced upon fasting and refeeding high carbohydrate low fat diet to rodents (5, 6, 7). It has been implicated as one of the enzymes that can play a role in obesity and diabetes (5, 6, 7). The mtGPAT cDNA has been cloned (1, 2, 8, 9) and the protein purified from rat liver (10, 11). The mtGPAT is regulated not only transcriptionally (5, 6) but also by covalent modification such as by phosphorylation by casein kinase II (12, 13) and AMP-activated kinase (14). It is expected that understanding the regulation of this enzyme may help us in preventing diseases like obesity, diabetes, and atherosclerotic heart disease (5).
The location and sequence of mouse liver mtGPAT promoter was determined by Sul and her colleagues (15). It was subsequently reported that rat mtGPAT 5′ UTR was shorter than mouse 5′ UTR and that there was an internal deletion of 167 nucleotides in rat 5′ UTR (8). Therefore, it was important to locate and characterize the rat mtGPAT promoter. By analyzing the rat genome, we have found a sequence that is not only similar to mouse liver mtGPAT promoter and is highly conserved in rat, mouse, and human, but also is ∼30 kb nucleotides away from the first translational codon.
Since rat upstream sequence that showed homology with mouse liver mtGPAT promoter is ∼30 kb nucleotides away, we also decided to examine the sequence near the first translational codon. Here, we present the evidence for the presence and function of rat mtGPAT distal promoter and proximal promoter-like regulatory region close to the first initiation codon and also the difference in their response to fasting and refeeding.
Materials and Methods
In-silico analysis and amplification of rat liver mtGPAT promoter-like regions
Promoser program (16) was used to predict promoter-like regions which were further analyzed by Genomatrix Promoter inspector (19) and DNA manipulation program (einverted). Rat liver DNA was isolated and amplified by PCR using Taq polymerase (Qiagen). These promoter regions were submitted to GenBank and have the accession numbers: AY693775 (distal) and AY695517 (proximal). The proximal and distal regions were amplified by PCR by using the following primers:
- For distal region:
- F primer: AAACAGTTGCTAGCGCGTACCTG and
- R Primer: TCTGCAGACCATGGTTCATGGGG also shown in Fig. 1A.
- Two regions were amplified for the proximal region:
- Proximal 1050: F primer: AACGCGGTACCCCCTGTTGTGTGG and
- R primer: AAAAAACCGATCGCCTCATTCGTGTG
- Proximal 500: F primer: AAAAAAGGTACCAGTTATTAACAAAGGTG
and same reverse primer that was used for the proximal 1050 as shown above (cf. Fig. 1B).
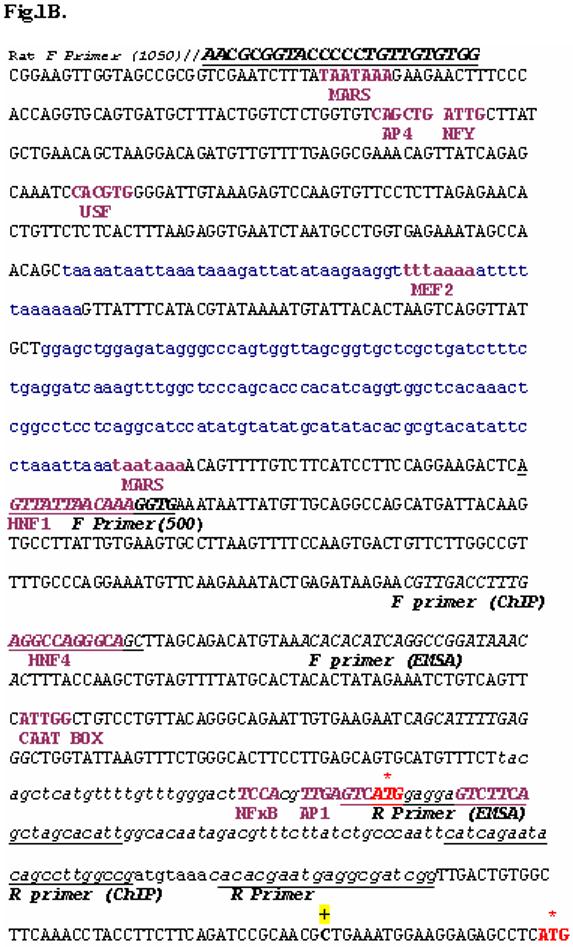
Fig.1.
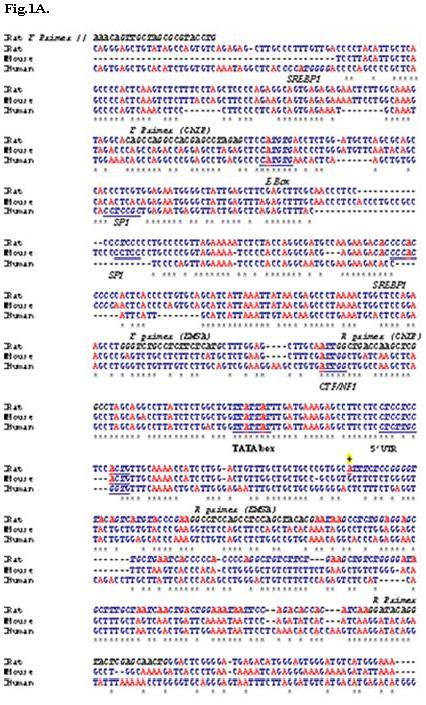
A. Alignment of rat and human mtGPAT upstream region (rat chromosome 1, human chromosome 10) with mouse mtGPAT promoter (chromosome 19) using Clustalw program and characterization of Distal promoter TATA box, E box, inverted CCAAT box, and SREBP1 are shown in bold italics underlined letters. The 5′ UTR in rat is shown in bold and italics. The primers designed for amplifying the promoter region, ChIP and EMSA are in black italicized and labeled. The sign ‘*’ designates identical nucleotides in all three species. The TSS1 predicted by RACE is denoted as ‘+’.
B. Proximal promoter sequence: The exon containing first translational codon is shown as bold and underlined. The first and second initiation codons of rat mtGPAT are shown by an asterisk ‘*’ and in red. Before 1st ATG the underlined sequence is part of 5′ UTR (8). The TSS2 predicted by RACE PCR is denoted as ‘+’. The forward and reverse primers and primers used for amplifying the promoter region, EMSA and ChIP are in upper case, italicized and underlined. The transcription factor binding sites are shown as bold letters. The sequences shown in lower case letters and underlined represent the repeat sequence present in the promoter. MARS = Matrix attachment regions.
Plasmid constructs
The promoter regions: distal and proximal regions were cloned at KpnI and NheI sites of promoterless mammalian expression vector pGL3-basic (Promega) that contains firefly luciferase as the reporter gene. The plasmid pRLTK (Promega) containing renilla luciferase reporter gene was used as an internal control for normalizing firefly luciferase activity (17).
Cell culture, transient transfection and luciferase assay
Rat liver cells (CRL-1442, ATCC) were maintained in Eagle’s minimum essential medium (EMEM) supplemented with 10%v/v fetal bovine serum and 50 U/ml penicillin-streptomycin (Gibco) (complete medium). The rat liver cells at 70% confluency were cotransfected with distal or proximal promoter containing plasmid and renilla luciferase plasmid (internal control). For the negative control, cells were cotransfected with promoterless plasmid and renilla luciferase plasmid (internal control). The transfections were performed using Lipofectamine reagent (Invitrogen). The cells were lysed 24 hrs after transfection with passive lysis buffer (Promega) and same amount of protein was used for the assay. The luciferase activity was measured using dual luciferase kit (Promega) by Berthold multiplate reader LB140. The concentration of total protein in cell lysate was determined using a Coomassie protein assay reagent kit (BioRad). Data (mean ± S.D.) were collected from triplicate assays of three independent transfections for each set.
Gel mobility shift assay
To examine the binding of transcription factors to both distal and proximal regions we amplified ∼200 bp size of each region by PCR.
- The primers designed to amplify the distal promoter region (Fig.1A):
- F Primer: AAAAAAGGTACCTGCCTCTTCTCATG
- R Primer: CGTGTAGCTAGCGAGGCTGGAGG
- Primers used for the proximal promoter region (Fig 1B):
- F Primer: ACACATCAGGTACCGGATAAACAC (positions -330 to -354)
- R Primer: AATGTGCTAGCTGAAGACTCCTCCATGAC
Both of these PCR products (2 pmoles/μl; 2 μl) were labeled by [γ-32P] ATP using T4 polynucleotide kinase. After labeling, they were passed through G-50 sephadex columns to remove unincorporated nucleotides and primers. The reaction mixtures containing 2μl HeLa nuclear extract (2.4 μg/μl), 2 μl of 5X binding buffer and nuclease-fee water to make total volume 9 μl, provided in the kit by Promega, were incubated at 4°C for 10 min. After adding 1 μl of labeled DNA, the samples were further incubated for 20 min at 4°C. For competition assay, 2 fold and 4 fold excess of cold oligonucleotides were incubated with HeLa nuclear extract before adding 1 μl of labeled DNA. Finally, 1 μl of gel loading 10X buffer was added to these reaction mixtures that were run on 5% nondenaturing polyacrylamide gel at 250 V. The gel was dried for 1 hr by BIO-RAD gel dryer-model 583 and scanned by phosphoimager (Packard Cyclone). The negative controls containing only labeled DNA and only HeLa nuclear extract were also run on the same gel.
Chromatin immunoprecipitation assays
ChIP analyses were performed using an assay kit (Upstate Biotechnology) according to the manufacturer’s protocol. Rat liver cells were grown to 80-90% confluency and treated with formaldehyde at a final concentration of 1% and incubated at 37°C for 10 min. Cells were washed with PBS containing protease inhibitors (1mM phenylmethylsulfonyl fluoride PMSF, aprotinin (1μg/ml) and pepstatin A (1μg/ml)) scraped and collected by centrifugation. Cells were resuspended in SDS lysis buffer (Upstate Biotechnology) and incubated at 4°C for 10 min and sonicated for five cycles of 10s each at setting 4 in a Vibra Cell. The entire process was perfomed at 4°C. The lysate was centrifuged at 13,000 rpm for 10 min at 4°C. The supernatant fluid was then diluted 10 fold with ChIP dilution buffer (Upstate Biotechnology). The cell supernatant fraction was pre-cleared with Salmon Sperm DNA/Protein A agarose-50% slurry for 30 min at 4°C. An aliquot (about 20 μl) was removed to use as an input control. Immunoprecipitation was performed with antibodies against ChREBP (Novus Biologicals) and NFκB (Santa Cruz Biotechnology) overnight at 4°C. For a negative control, no-antibody immunoprecipitation was performed by incubating the supernatant fraction with 50% slurry of Salmon Sperm DNA/Protein A agarose for 1 hr at 4°C with rotation. Following immunoprecipitation, samples were treated with 50% slurry of Salmon sperm DNA/Protein A Agarose and incubated for 1 hr. Agarose beads were collected by gentle centrifugation (1000 rpm at 4°C) and subsequently washed with Low Salt Immune Complex Wash Buffer (Upstate Biotechnology), High Salt Immune Complex Wash Buffer (Upstate Biotechnology), LiCl Immune Complex Wash Buffer (Upstate Biotechnology) and twice with 1X TE. Agarose beads were extracted twice with 1% SDS-0.1M NaHCO3 and the eluent was heated with 5M NaCl at 65°C for 4 hr to reverse the crosslink. Chromatin associated protein was digested with proteinase K and samples were extracted with phenol/CHCl3 followed by precipitation with ethanol. The pellet was resuspended in nuclease free water and subjected to PCR. The PCR product was run on a 2% agarose gel.
- The primers designed to amplify distal promoter region (Fig.1A):
- Forward primer: 5′CAGCCAGGCCACGAGCCTAGAG 3′
- Reverse primer: 5′GCTAGGCCGAGCTTGGTCAGCC3′
- Primers used for proximal promoter region amplification (Fig 1B):
- Forward primer:5′CGTTGACCTTTGAGGCCAGGGCAGC3′
- Reverse primer: 5′ CGGCCAAGGCTGTATTCTGATG 3′
- Primers used for beta-actin:
- Forward primer: 5′TTGCTGACAGGATGCAGAAGGAG3′
- Reverse primer: 5′TGAGGCCAGGATAGAGCCACCAATCC3′
Amplification of 5′ cDNA ends
Total RNA was isolated from rat liver cells (Qiagen’s RNeasy minikit) and used for 5′RACE using total RNA 5′-RACE System for Rapid Amplification of cDNA Ends kit (Version 2.0; Invitrogen) according to the manufacturer’s protocol. The first strand was synthesized using gene specific primer of rat mtGPAT cDNA (GSP1, 5′GCTTTAGTAACACCCAGCCAGTCAGCC3′). Homopolymeric (C) tails were added to the 3′ end of the first strand cDNA using dCTP and terminal deoxynucleotidyl transferase. First round of PCR was performed for amplification of the target first strand cDNA using Abridged Anchor Primer (5′GGCCACGCGTCGACTAGTACGGGIIGGGII GGGIIG-3′, where I is inosine) and GSP1. The amplified products were subjected to a second round of PCR with Abridged Universal Anchor Primer (5′GGCCACGCGTCGACTAGTAC3′) and the nested specific primer NGSP1 (5′GGAGACTGTAGCAACCATTTCCTGGAGG3′). Both GSP1 and NGSP1 were designed from exon 6 of mtGPAT gene as shown in Fig.5B. The amplified DNA was run on 1.5% agarose gel. Two amplified bands were excised from the gel and the DNA was isolated following gel extraction (Qiagen’s minelute gel extraction kit) and subsequently cloned into TOPO TA cloning vector (Invitrogen) and sequenced by Genewiz.
Fig.5. Analysis of 5′-RACE-PCR products subjected to 1.5% agarose gel electrophoresis.
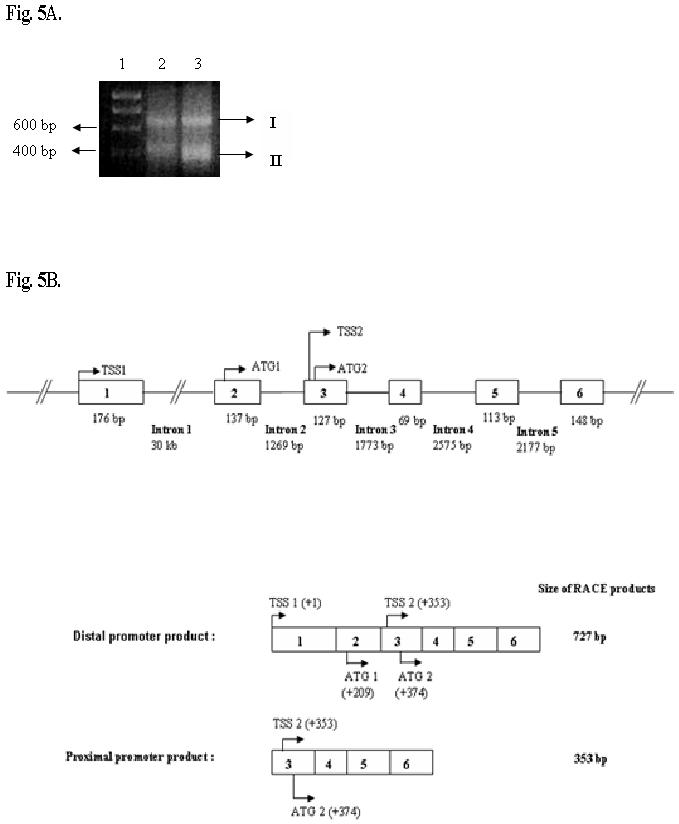
A. Lane 1, DNA marker containing Hyperladder I (Bioline) with arrows indicating the positions of the 400 bp and 600 bp markers. Lane 2, second round of PCR performed with NGSP1 for 30 cycles. Lane 3, second round of PCR performed for 35 cycles. Both the bands were excised and sequenced. For both lanes 2 and 3 arrows with I and II indicate amplified bands.
B. Schematic of the two transcripts obtained from the two promoter regions and the arrangement of the exons and introns. Boxes with numbers represent exons and their sizes are mentioned. The nested primer (NGSP1) used for 5′RACE was from exon 6 at position +727. The two transcriptional and translational start sites with their respective positions relative to TSS1 (+1) are shown.
Nutritional effect on rat mtGPAT promoters
The rat liver cells at 70% confluency were cotransfected with vector containing either distal promoter or proximal promoter and with pRLTK (internal control). Vector control was performed with cells cotransfected with promoterless vector, pGL3-basic and pRLTK. After 24 hrs of transfection, EMEM medium was replaced by glucose, serum and pyruvate-free medium. The cells were then grown for 24 hrs, 48 hrs and 72 hrs in the starvation medium after which the medium was replaced by complete medium containing 5.5mM glucose, 1mM pyruvate and 10%v/v FBS, and the cells were allowed to grow for 24 hrs and then lysed. The luciferase activity was measured as described above.
Isolation of Mitochondria
Mitochondria were isolated from rat liver cell culture starved for 24 hrs, 48 hrs or 72 hrs and refed for 24 hrs as mentioned above, by differential centrifugation at 600xg and 11000xg. Intactnes of isolated mitochondrial membrane was determined by measuring the cytochrome c-oxidase activity (∼95%). Protein estimation of mtGPAT was done using Bradford assay. Mitochondrial GPAT specific activity was determined in the presence of N-Ethylmaleimide (NEM, 5mM).
Immunodetection of mtGPAT using IM1GPAT antibody
Whole mitochondria (5 μg) isolated from above was run on 10% SDS-PAGE and the proteins were transferred to a nitrocellulose membrane at a constant voltage of 80V for 90 minutes at 4°C. Immunodetection was performed using ECL’s advanced Western blotting kit according to their protocol using IM1GAT antibody according to Onorato T. M. et al. (13). The membrane was stripped following ECL’s advanced Western blotting kit protocol and reprobed for β-actin (Sigma Aldrich).
Results
Identification of rat mtGPAT promoters
By aligning rat mtGPAT cDNA (8) with rat genomic sequence available at NCBI database, we found that the rat genomic sequence upstream of 5′UTR on chromosome 1 has 99% similarity with mouse mtGPAT promoter (15). Initially, we aligned rat mtGPAT cDNA (2, 8) with rat genomic sequence from NCBI. We found 21 exons and 20 introns in this gene (Table 1). We also analyzed the exons and introns of mouse and human mtGPAT cDNA. The results are included in Table 1. This table suggests that mtGPAT is spliced differently in rat, mouse, and human. Promoser program predicted two promoter-like regions - proximal region near the first ATG and a distal region ∼ 30 kb upstream on chromosome 1 similar to mouse mtGPAT (Fig. 1A and 1B). Promoter inspector analysis showed different transcription factor binding sites on distal and proximal promoter regions (Fig. 1A and 1B).
Table 1. Putative exons and introns of mtGPAT gene in rat, mouse, and human.
The exons and introns were manually counted after aligning each mtGPAT cDNA with the respective genomic sequence. Also, exon prediction program SIM 4 (18) was used to reinforce the results
| Source of mtGPAT gene | Location (Chromosome) | Exons | Introns | Number of amino acids |
|---|---|---|---|---|
| Rat (AF021348) | 1 | 21 | 20 | 828 |
| Mouse (NM_008149.2) | 19 | 25 | 24 | 827 |
| Human (NM_020918.2) | 10 | 22 | 21 | 828 |
Luciferase activity due to the distal and proximal promoter regions
Based on the Promoser’s prediction, we amplified the proximal and distal sequences using the primers as shown in Fig. 1A and 1B. The constructs made for luciferase assay were as follows: (a) plasmid containing the 1050 bp of the proximal promoter region upstream of luciferase, (b) plasmid containing the 500 bp of the proximal promoter region upstream of luciferase (500 bp does not contain the repeat sequence as shown in Fig. 1B) and (c) plasmid containing 932 bp of the distal promoter region upstream of luciferase gene as shown in Fig 2. The 500 bp proximal region gave about 4.8-fold increase in luciferase activity 24 hrs after transfection (Fig. 2, “Proximal 500”) while the 1050 bp region of proximal promoter (Fig.2, “Proximal 1050) gave about 2.5 fold increase in luciferase activity as compared to promoterless vector. The vector with the distal promoter gave about 10.8-fold increase in luciferase activity as compared to the promoterless vector. These results suggest that both distal and proximal sequences can function as promoters.
Fig.2. Luciferase assay with the mtGPAT proximal and distal promoter-like regions cloned in pGL3-basic vector.
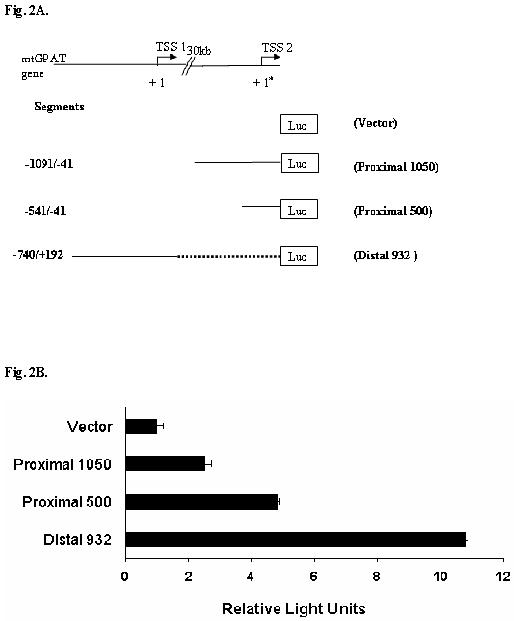
A. Schematic presentation of the promoter constructs used for luciferase assay. TSS1 is represented as +1 and TSS2 is represented as +1* and there is a 30 kb gap between the promoter regions. Solid line denotes the actual promoter sequence used. The distal promoter region and the two proximal promoter regions (1050 and 500) relative to TSS1 (+1) and TSS2 (+1*) respectively are shown. B. The figure shows the average of three independent transfection experiments (firefly luciferase normalized by renilla luciferase) in rat liver cells. Each experiment was done in triplicates. Luciferase assay was done 24 hrs after transfection. The results are the average of three independent transfections performed in triplicate ± S.E.
Transcription factors binding to proximal and distal promoter regions
To determine if the two promoter regions bind transcription factors, we took the following two approaches. In the first set of experiments, we used EMSA (Fig. 3). We amplified by PCR, a portion of (∼200 bp) each of the distal and proximal regions (see methods and Fig.1A and Fig.1B). After labeling with [γ-32P] ATP, these labeled fragments were incubated with HeLa nuclear extract (5μg) and were run on a non-denaturing polyacrylamide gel. One major complex was seen for both promoters: distal promoter, Fig. 3, lane 2, proximal promoter, Fig. 3, lane 6. In both cases, the complex was reduced by increasing amounts of unlabeled probe, Fig.3 lanes 3 & 4 for distal and lane 7 & 8 for proximal region. The complex was not competed away by using unlabeled mtGPAT cDNA as a probe. Similarly, no shift was observed with either distal or proximal regions when they were incubated with BSA (5μg) (data not shown). These tests demonstrate the specificity of DNA-protein complex. The results show that these two regions have the ability to bind with transcription factors and hence have the potential to act as promoters.
Fig.3. Gel Mobility Shift assay for distal and proximal promoter.
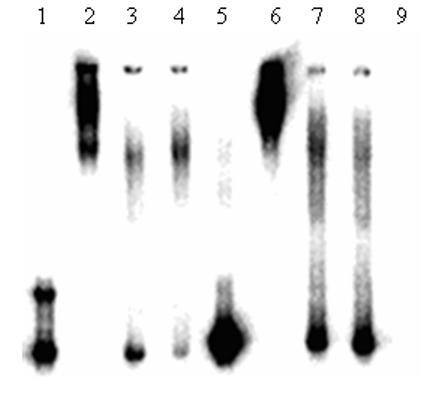
Lane 1 = Distal promoter, Lane 2 = Distal promoter incubated with HeLa nuclear extract, Lane 3 = Distal promoter with HeLa nuclear extract + 4 fold cold oligonucleotide, Lane 4 = Distal promoter with HeLa nuclear extract + 2 fold cold oligonucleotide, Lane 5 = Proximal promoter, Lane 6 = Proximal promoter incubated with HeLa nuclear extract, Lane 7 = Proximal promoter with HeLa nuclear extract + 2 fold cold oligonucleotide, Lane 8 = Proximal promoter with HeLa nuclear extract + 4 fold cold oligonucleotide and Lane 9 = HeLa nuclear extract.
To determine if the promoter regions can bind specific transcription factors we performed the second set of experiments. We performed ChIP assays with carbohydrate response element binding protein (ChREBP) binding for the distal promoter and NFκB binding for the proximal promoter regions. The primers for ChIP assays used for distal and proximal regions as shown in Fig1A and 1B. No antibody treatment was performed as a negative control (-IP). The results are documented in Fig. 4. Immunoprecipitated bands were observed only when the ChIP was done in the presence of the respective antibodies, not in the absence. PCR primers specific for rat beta actin detected actin only in input DNA and not in any other sample DNA. These results suggest that distal promoter and proximal promoters are capable of recruiting ChREBP and NFκB transcription factors, respectively.
Fig.4. Chromatin immunoprecipitation of distal and proximal promoter regions.

Binding of NFκB and ChREBP to proximal and distal promoter regions. Input DNA or DNA immunoprecipitated by NFκB and ChREBP antibodies were amplified for 30 cycles in 25 μl of reaction using pairs of the primers indicated in materials and method. -IP: No immunoprecipiatation; In: input DNA; IP: immunoprecipitated DNA.
Analysis of the transcription start sites of mtGPAT by 5′RACE PCR
In an effort to determine if the two promoter like regions lead to the production of transcripts with different transcriptional start sites we performed 5′RACE PCR of cDNA obtained from rat liver cells. Analysis of the products revealed two bands about 400 bp and 700 bp length as shown in Fig 4A. Subcloning of the RACE products into pCR-TOPO vector and sequencing of the inserts from the two clones showed the 400 bp and 700 bp species that had different transcriptional start sites. The 700 bp clone’s transcriptional start site begins from the 5′UTR region of the distal promoter as shown in Fig.1A while the 400 bp transcriptional start site lies between the first and second initiation codons as shown in Fig.4B. These results are in keeping with the idea that the two promoters are responsible for synthesizing two transcripts that differ in their 5′UTR regions.
Nutritional regulation by distal and proximal promoters
Sul and her associates reported 30 fold increase in the amount of mtGPAT mRNA in the liver of mice when the animals were starved for 48 hrs and refed a fat free, carbohydrate rich diet (38). To determine if both the distal and proximal promoters of mtGPAT respond to fasting and refeeding, we performed a similar experiment but using rat liver cells. The cells, cotransfected with either proximal (500 bp) or distal promoter (932 bp) cloned into pGL3- basic vector and pRLTK vector (internal control), were starved for 24 hrs, 48 hrs or 72 hrs and then refed with complete medium for 24 hrs. Cells were lysed and luciferase assay was performed. Cells, transfected with the distal promoter, were starved for 24 hrs, 48 hrs and 72 hrs and refed had about 2, 2.7 and 1.9 - fold increase in luciferase activity, respectively when compared to control cells that were transfected with the distal promoter and grown in complete medium (Fig. 6A). In the case of proximal promoter, the luciferase activity remained almost the same for all the samples at about 1.3 fold compared to control cells which were transfected with proximal promoter and grown in complete medium (Fig. 6A). Similarly, when rat liver cells were starved and refed, mtGPAT activity (Fig. 6B) and protein (Fig. 6C) also increased for the cells starved for 1 and 2 days before refeeding, and then decreased for the 3 day starved cells.
Fig.6. Nutritional effect on luciferase activity of distal and proximal promoters, on mtGPAT specific activity and mtGPAT protein level.
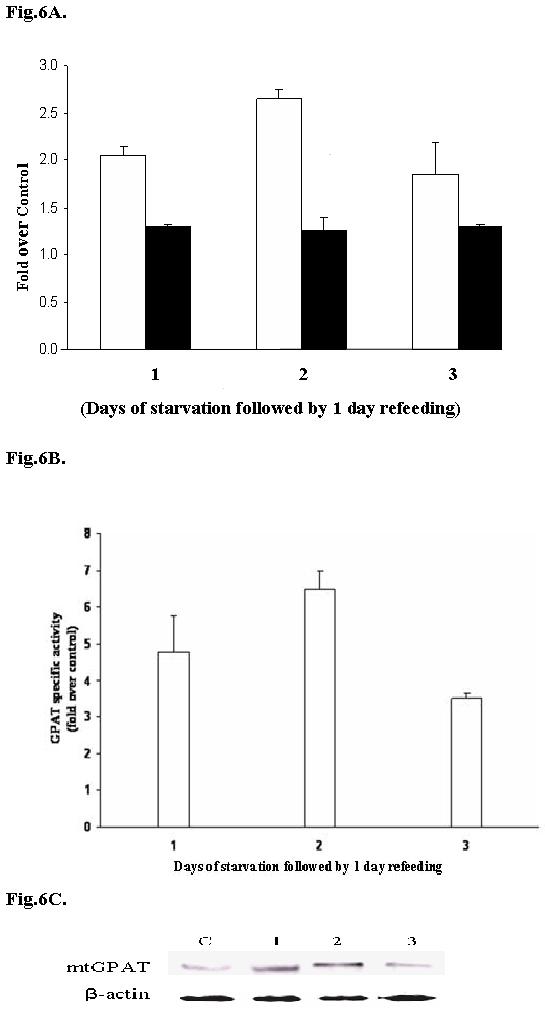
A. Distal promoter (932bp) or proximal promoter (500bp) were cotransfected separately into rat liver cells with pRLTK vector. Cells were starved for 24 hrs, 48 hrs or 78 hrs and then refed with complete medium for 24 hrs and luciferase assay was performed. Control shows cells that were cotransfected with the respective vectors and pRLTK and normally grown (no starvation). White bars represent values obtained with distal promoter and black bars represent values obtained with proximal promoter. The results are the averages of three independent experiments performed in triplicate ± S.E. B. GPAT specific activity (nM/min/mg) was measured for mitochondria isolated from control cells (normally grown cells; without starvation and refeeding) and starved and refed rat liver cell culture as described in materials and methods. Results are represented as fold over control cells. C. Whole mitochondria (5μg) was subjected to 10% SDSPAGE and immunoprecipitated with IM1GAT antibody. C : control cells,1: 24 hrs starvation followed by 24 hrs refeeding, 2 : 48 hrs starvation followed by 24 hrs refeeding, 3 : 72 hrs starvation followed by 24 hrs refeeding. Equal amount of loading was confirmed by stripping the membrane and reprobing with β-actin.
Discussion
A number of new information has emerged from this work. First, rat mtGPAT has two promoters: distal and proximal. Second, the distal promoter is ∼30 kb upstream of the first initiation codon. Third, there are two transcripts synthesized - one from each promoter. Finally, only the distal promoter, not the proximal promoter, responds to starvation and refeeding in rat liver cells.
The promoter inspector analysis shows the presence of hepatocyte nuclear factor (HNF-1 and HNF-4), NFκB, NFY binding reverse CAAT box (32), activating protein binding site (AP1 and AP4), myocyte enhancer factor (MEF2), upstream stimulatory factor (USF), and matrix attachment regions (MARS) in the proximal promoter and SREBP, ChREBP, SP1 and CTF1 binding sites in distal promoter (Fig. 1A and 1B). The presence of MARS sequence shows that the proximal region may have a significant role in transcription of mtGPAT gene. The MARS sequences link chromatin loops to protein scaffold known as the nuclear matrix and they have been found in repeat sequences (27). They are the nucleation site for unwinding DNA as seen in human 5′ beta-interferon gene and immunoglobulin heavy chain gene enhancer (28). Distal promoter region has ChREBP binding site which directly promotes lipogenic enzymes like ACC, FAS and LPK gene transcription (31). The 30 kb intron has three inverted loops that might be responsible in bringing the distal promoter region close to the first initiation codon thus enabling the transcription of the distal promoter product to occur. Inverted repeats have been shown to be involved in transcription, replication, and gene amplification (21).
Comparison of 5′UTR regions of mouse mtGPAT and rat mtGPAT revealed that there was a deletion of 167 nucleotides in rat making the rat 5′UTR region shorter than that of the mouse (8). This 167 nucleotide sequence is actually present in the rat genomic sequence. So, its absence in the rat mtGPAT cDNA suggests that this length of 5′UTR is spliced out in the rat. If so, the splicing of 5′ UTR in rat mtGPAT may generate more than one type of transcripts. This consideration suggested the presence of another promoter (22-26). So, we examined the genomic region just upstream of first initiation codon. The luciferase assay with transfected rat liver cells shows about 4.8-fold increase in luciferase activity with the proximal 500 bp (Fig. 2) and a 2.5-fold increase with the proximal 1050 bp (Fig. 2) as compared to the cells transfected with the promoterless vector. The distal promoter’s luciferase activity is about 10.8 fold meaning that the promoter activity is more in case of the distal promoter. Highest promoter activity could be localized to the 500 bp region of the proximal promoter which contains consensus binding sites for transcription factors. The promoter activity of outside this region (1050 bp) is significantly lower. Portions of the 1050 bp region of proximal promoter contain repeat sequences (Fig. 1B) that might cause a reduction in transcription as seen with luciferase assay (Fig. 2). The general view about multiple promoters is that when a single gene is transcribed from multiple promoters, an organism gains additional flexibility in the control of expression of that gene (35). In the case of the α-amylase gene, there are two promoters: weak downstream promoter which is active in liver while a strong upstream promoter is active in the parotid gland. This difference in the two promoters account for almost 100-fold difference in expression levels of the gene in the two above mentioned tissues (36). This could be the case with rat mtGPAT gene where the two promoters function differentially in a tissue specific way or nutritionally or hormonally.
In the preliminary EMSA (Fig. 3), 200 bp probes were used for both distal and proximal promoters to check for binding of a number of transcription factors in the probe region. The binding of ChREBP and NFκB to distal and proximal regions was further confirmed by ChIP assays (Fig. 4). This result shows that these factors bind specifically to the predicted sites and hence are major determinants of activity of the promoters.
The two transcripts of rat mtGPAT that we obtained after RACE PCR show the presence of alternative exons. The longer transcript has 176 bp from the first exon that is ∼30 kb upstream of the second exon while the proximal promoter transcript starts from exon 3 (Fig. 4B). This situation is very similar to that of ACC gene that has three promoters and three different transcripts which are nutritionally and hormonally regulated (25).
It has been shown that ChREBP is expressed at high levels in the liver, adipose, kidney and small intestine (32,33). Distal promoter region has consensus binding sites for key transcription factors like SREBP and ChREBP which are responsible for the transcriptional regulation of lipogenic enzymes. It has been shown that distal promoter can recruit ChREBP (Fig. 4) implying that this promoter could be responsible for the transcriptional regulation of mtGPAT (54).
From the combined data of computer analysis (Fig. 1A and Fig. 1B), luciferase assay (Fig. 2), EMSA (fig. 3), ChIP assay (Fig. 4) and 5′RACE PCR (Fig. 5), we conclude that mtGPAT has two promoters (distal and proximal) that are responsible for generating two transcripts.
It is known that activities of critical lipogenic enzymes like mtGPAT and FAS are tightly controlled nutritionally and hormonally (20). So, when cells were starved and refed, luciferase activity regulated by the distal promoter almost tripled although that by the proximal promoter remained unchanged (Fig. 6). The results suggest that the distal but not the proximal promoter is the inducible one. Thus, the two promoters can be functionally differentiated.
It is interesting to note that, like GPAT, human ACC gene has more than one promoter (32). Both enzymes are positively affected by starvation and refeeding (20) and also by insulin and EGF (30) and negatively by glucagon and adrenaline (29, 34). Our preliminary work with EGF shows that EGF increases luciferase expression by two fold in rat liver cells transfected with rat mtGPAT distal promoter (data not shown). A question arises why both ACC and GPAT, the first step in fatty acid and glycerolipid synthesis, respectively, need to be similarly regulated in the overall synthesis of the glycerolipids? Probably, without GPAT being regulated similar to ACC, the fatty acids produced may be used either for synthesis of glycerolipids or for β-oxidation.
Since promoters are tissue specific and function differently at different developmental levels (29), the two regulatory regions might be functioning differently in other cell lines. Our results suggest that distal promoter and not the proximal promoter is the one that is inducible by nutritional regulation. The elucidation of the function of these transcription factors for these promoters and the role of ∼30 kb intron are expected to help in further understanding the mechanism of transcription and regulation of the mtGPAT gene.
Acknowledgements
This research was supported by the National Institutes of Health Grant GM-57643 and by Clare Boothe Luce fellowship (KKA). We thank Dr. Issar Smith for his helpful comments and Ms. Pamela Gaddi for helping with portions of this work. This work formed a portion of a doctoral thesis submitted by KKA to the faculty of Biological Sciences, St. John’s University.
The abbreviations used are
- mtGPAT
mitochondrial glycerol-3-phosphate acyltransferase
- ChREBP
carbohydrate response element binding protein
- S14
Spot 14
- 5′UTR
5′ untranslated region
- LPK
L pyruvate kinase
- ACC
acetyl-CoA carboxylase
- FAS
fatty acid synthase
- EGF
epidermal growth factor
- HNF-1 and HNF-4
hepatocyte nuclear factor
- AP1 and AP4
activating protein
- MEF2
myocyte enhancer factor
- USF
upstream stimulatory factor
- MARS
matrix attachment regions
- ChIP
chromatin immunoprecipitation
- EMSA
electromobility shift assay
- NFκB
nuclear factor kappa B
- ChREBP
carbohydrate response element binding protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Balija VS, Chakraborty TR, Nikonov AV, Morimoto T, Haldar D. Identification of two transmembrane regions and a cytosolic domain of rat mitochondrial glycerophosphate acyltransferase. J. Biol. Chem. 2000;275:31668–31673. doi: 10.1074/jbc.M002963200. [DOI] [PubMed] [Google Scholar]
- [2].Nikonov AV, Morimoto T, Haldar D. Recent Res. In: Pandalai SG, editor. Developments Lipids Res. Part II. Vol. 2. Transworld Research Network; Trivandarum, India: 1998. pp. 207–222. [Google Scholar]
- [3].Onorato TM, Chakraborty S, Balija VS, Haldar D. Mitochondrial Glycerol-3-Phosphate Acyltransferase. In: Haldar D, Das SK, editors. LIPIDS: Glycerolipid Metabolizing Enzymes. Research Signpost; Trivandrum: 2002. [Google Scholar]
- [4].Hesler CB, Carroll MA, Haldar D. The topography of glycerophosphate acyltransferase in the transverse plane of the mitochondrial outer membrane. J. Biol. Chem. 1985;260:7452–7456. [PubMed] [Google Scholar]
- [5].Coleman RA, Lewin TM, Muoio DM. Physiological and nutritional regulation of enzymes of triacylglycerol synthesis. Annu. Rev. Nutr. 2000;20:77–103. doi: 10.1146/annurev.nutr.20.1.77. [DOI] [PubMed] [Google Scholar]
- [6].Coleman RA, Douglas PL. Enzymes of triacylglycerol synthesis and their regulation. Prog. Lipid Res. 2004;43:134–176. doi: 10.1016/s0163-7827(03)00051-1. [DOI] [PubMed] [Google Scholar]
- [7].Thuresson ER. Inhibition of glycerol-3-phosphate acyltransferase as a potential treatment for insulin resistance and type 2 diabetes. Curr. Opin. Investig. Drug. 2004;5:411–418. [PubMed] [Google Scholar]
- [8].Bhat BG, Wang P, Ji-Hyeon Kim, Black TM, Lewin TM, Fiedorek FT, Jr., Coleman RA. Rat sn-glycerol-3-phosphate acyltransferase: molecular cloning and characterization of the cDNA and expressed protein. Biochim. Biophys. Acta. 1999;1439:415–423. doi: 10.1016/s1388-1981(99)00103-1. [DOI] [PubMed] [Google Scholar]
- [9].Shin DH, Paulauskis JD, Moustaid N, Sul HS. Transcriptional regulation of p90 with sequence homology to Escherichia coli glycerol-3-phosphate acyltransferase. J. Biol. Chem. 1991;266:23834–23839. [PubMed] [Google Scholar]
- [10].Vancura A, Haldar D. Purification and characterization of glycerophosphate acyltransferase from rat liver mitochondria. J. Biol. Chem. 1994;269:27209–27215. [PubMed] [Google Scholar]
- [11].Haldar D, Vancura A. Glycerophosphate acyltransferase from liver. Methods Enzymol. 1992;209:64–72. doi: 10.1016/0076-6879(92)09008-q. [DOI] [PubMed] [Google Scholar]
- [12].Onorato TM, Haldar D. Casein kinase II stimulates rat liver mitochondrial glycerophosphate acyltransferase activity. Biochem. Biophys. Res. Commun. 2002;296:1091–1096. doi: 10.1016/s0006-291x(02)02064-8. [DOI] [PubMed] [Google Scholar]
- [13].Onorato TM, Chakraborty S, Haldar D. Phosphorylation of Rat Liver Mitochondrial Glycerol-3-phosphate Acyltransferase by Casein Kinase 2. J. Biol Chem. 2005;280:19527–19534. doi: 10.1074/jbc.M410422200. [DOI] [PubMed] [Google Scholar]
- [14].Muoio DM, Seefeld K, Witters LA, Coleman RA. AMP-activated kinase reciprocally regulates triacylglycerol synthesis and fatty acid oxidation in liver and muscle: evidence that sn-glycerol-3-phosphate acyltransferase is a novel target. Biochem. J. 1999;338:783–791. [PMC free article] [PubMed] [Google Scholar]
- [15].Jerkins AA, Robert LW, Lee S, Sul HS. Characterization of the Murine Mitochondrial Glycerol-3-phosphate Acyltransferase Promoter. J. Biol. Chem. 1995;270:1416–421. doi: 10.1074/jbc.270.3.1416. [DOI] [PubMed] [Google Scholar]
- [16].Halees AS, Leyfer D, Zhiping W. PromoSer: a large-scale mammalian promoter and transcription start site identification service. Nucl. Acids Res. 2003;13:3554–3559. doi: 10.1093/nar/gkg549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sherf BA, Navarroi SL, Hannah RR, Wood KV. Dual-Luciferase™ Reporter Assay: An Advanced Co-Reporter Technology Integrating Firefly and Renilla Luciferase Assays. Promega Notes Magazine. 1996;57:2–7. [Google Scholar]
- [18].Florea L, Hartzell G, Zhang Z, Rubin GM, Miller W. A Computer Program for Aligning a cDNA Sequence with a Genomic DNA Sequence. Genome Res. 1998;8:967–974. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sul HS, Wang D. Nutritional and hormonal regulation of enzymes in fat synthesis: studies of fatty acid synthase and mitochondrial glycerol-3-phosphate acyltransferase gene transcription. Annu. Rev. Nutr. 1998;18:331–51. doi: 10.1146/annurev.nutr.18.1.331. [DOI] [PubMed] [Google Scholar]
- [21].Lin C-T, Lin W-H, Lyu YL, Whang-Peng J. Inverted repeats as genetic elements for promoting DNA inverted duplication: implications in gene amplification. Nucl. Acids Res. 2001;29:3529–3538. doi: 10.1093/nar/29.17.3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Landry JR, Mager DL, Wilhelm BT. Complex controls: the role of alternative promoters in mammalian genomes. Trends in Genetics. 2003;19:640–648. doi: 10.1016/j.tig.2003.09.014. [DOI] [PubMed] [Google Scholar]
- [23].Trinklein ND, Aldred SJK, Saldanha AJ, Myers RM. A Computer Program for Aligning a cDNA Sequence with a Genomic DNA Sequence. Genome Res. 2003;13:308–312. [Google Scholar]
- [24].Zavolan M, Van Nimwegen E, Gaasterland T. Splice Variation in Mouse Full-Length cDNAs Identified by Mapping to the Mouse Genome. Genome Res. 2002;12:1377–1385. doi: 10.1101/gr.191702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Jianqiang M, Subrahmanyam SC, Salih JW. Human acetyl-CoA carboxylase 1 gene: Presence of three promoters and heterogeneity at the 5′-untranslated mRNA region. Proc. Natl. Acad. Sci. USA. 2003;100:7515–7520. doi: 10.1073/pnas.1332670100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ayoubi TA, Van De Ven WJ. Regulation of gene expression by alternative promoters. The FASEB J. 1996;10:453–460. [PubMed] [Google Scholar]
- [27].Bode J, Kohwi Y, Dickinson L, John JT, Klehr D, Mielke C, Kohwi-Shigematsu T. Biological significance of unwinding capability of nuclear matrix-associating DNAs. Science. 1992;255:195–197. doi: 10.1126/science.1553545. [DOI] [PubMed] [Google Scholar]
- [28].Cockerill PN. Nuclear matrix attachment occurs in several regions of the IgH locus. Nucl. Acids Res. 1990;18:2643–8. doi: 10.1093/nar/18.9.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Munday MR. Regulation of mammalian acetyl-CoA carboxylase. Biochem Soc Trans. 2002;30:1059–64. doi: 10.1042/bst0301059. [DOI] [PubMed] [Google Scholar]
- [30].Wang D, Sul HS. Upstream Stimulatory Factor Binding to the E-box at -65 is Required for Insulin Regulation of the Fatty Acid Synthase Promoter. J. Biol. Chem. 1997;272:26367–74. doi: 10.1074/jbc.272.42.26367. [DOI] [PubMed] [Google Scholar]
- [31].Ishii S, Iizuka K, Miller BC, Uyeda K. Carbohydrate response element binding protein directly promotes lipogenic enzyme gene transcription. Proc. Natl. Acad. Sci. USA. 2004;101:15597–602. doi: 10.1073/pnas.0405238101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Koo S-H, Towle HC. Glucose Regulation of Mouse S14 Gene Expression in Hepatocytes. Involvement of a Novel Transcription Factor Complex. J. Biol. Chem. 2000;275:5200–5207. doi: 10.1074/jbc.275.7.5200. [DOI] [PubMed] [Google Scholar]
- [33].Iizuka K, Bruick RK, Liang G, Horton JD, Uyeda K. From the Cover: Deficiency of carbohydrate response element-binding protein (ChREBP) reduces lipogenesis as well as glycolysis. Proc. Natl. Acad. Sci. USA. 2004;101:7281–7286. doi: 10.1073/pnas.0401516101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Haystead TA, Campbell DG, Hardie DG. Analysis of sites phosphorylated on acetyl-CoA carboxylase in response to insulin in isolated adipocytes. Comparison with sites phosphorylated by casein kinase-2 and the calmodulin-dependent multiprotein kinase. Eur. J. Biochem. 1988;175:347–354. doi: 10.1111/j.1432-1033.1988.tb14203.x. [DOI] [PubMed] [Google Scholar]
- [35].Schihler U, Sierra F. Alternative promoters in developmental gene expression. Annu. Rev. Gene. 1987;21:237–257. doi: 10.1146/annurev.ge.21.120187.001321. [DOI] [PubMed] [Google Scholar]
- [36].Sehibler J, Hagenhuehle U, Wcllauer PK, Pittet AC. Two promoters of different strengths control the transcription of the mouse alpha amylase gene 4,ny-I& in the parotid gland and the liver. Cell. 1983;33:501–508. doi: 10.1016/0092-8674(83)90431-2. [DOI] [PubMed] [Google Scholar]


