Abstract
Background
Clustering is a popular data exploration technique widely used in microarray data analysis. Most conventional clustering algorithms, however, generate only one set of clusters independent of the biological context of the analysis. This is often inadequate to explore data from different biological perspectives and gain new insights. We propose a new clustering model that can generate multiple versions of different clusters from a single dataset, each of which highlights a different aspect of the given dataset.
Results
By applying our SigCalc algorithm to three yeast Saccharomyces cerevisiae datasets we show two results. First, we show that different sets of clusters can be generated from the same dataset using different sets of landmark genes. Each set of clusters groups genes differently and reveals new biological associations between genes that were not apparent from clustering the original microarray expression data. Second, we show that many of these new found biological associations are common across datasets. These results also provide strong evidence of a link between the choice of landmark genes and the new biological associations found in gene clusters.
Conclusion
We have used the SigCalc algorithm to project the microarray data onto a completely new subspace whose co-ordinates are genes (called landmark genes), known to belong to a Biological Process. The projected space is not a true vector space in mathematical terms. However, we use the term subspace to refer to one of virtually infinite numbers of projected spaces that our proposed method can produce. By changing the biological process and thus the landmark genes, we can change this subspace. We have shown how clustering on this subspace reveals new, biologically meaningful clusters which were not evident in the clusters generated by conventional methods. The R scripts (source code) are freely available under the GPL license. The source code is available [see Additional File 1] as additional material, and the latest version can be obtained at http://www4.ncsu.edu/~pchopra/landmarks.html. The code is under active development to incorporate new clustering methods and analysis.
Background
Microarrays have enabled scientists to monitor the activities of thousands of genes simultaneously. Clustering methods provide a useful technique for exploratory analysis of microarray data since they group genes with similar expression patterns together. It is believed that genes that display similar expression patterns are often involved in similar functions. Various clustering techniques have been proposed [1,2]. Some of the popular techniques for clustering genes employ k-means [3], hierarchical clustering [4], self-organizing maps [5] or some of their variants. Although clustering is a data exploration tool, there is a shortage of clustering algorithms that enable the exploration of a dataset from multiple different biological perspectives. Most of these conventional clustering algorithms generate only one set of clusters, thus forcing a very restricted view of gene associations. They leave little room for data exploration and re-interpretation of existing data. It would be difficult to interpret the complex biological regulatory mechanisms and genetic interactions from this restrictive interpretation of microarray expression data. In this paper we show that biologically meaningful gene clusters can be developed with our gene signature algorithm SigCalc. Our algorithm uses elements of subspace projection, along with existing knowledge on gene associations to come up with multiple new cluster sets. We show that each of these new cluster sets reveal biological associations that were not apparent from clustering the original gene expression data. The proposed method is fundamentally different from the conventional subspace clustering methods in that it projects the original expression data into a different information space where genes are described in relative terms against a chosen subset of genes called landmarks.
Random Projection
Random projection is one of the dimensionality reduction techniques that is useful for eliminating features that may be irrelevant. The high dimensionality data is projected onto a smaller random subspace. Random projections and subspaces have been extensively used in data mining. They have been used to reduce dimensionality and search for similarity in clustering [6,7] and for information retrieval [8]. Some of the application areas include classification [9], image processing [10], and other machine learning topics [11,12]. The key difference between our method and other random projection methods is that we project our data onto a known set of genes that are functionally related, whereas in other methods, random points are chosen for the subspace.
Subspace Clustering
Subspace clustering or biclustering [13,14], has been a popular method for analyzing microarray datasets. The main idea of subspace clustering is to find a subset of genes and a subset of conditions under which these genes exhibit a similar trend. The major differences between the subspace clustering and the method proposed in this paper are: (1) The subspace clusters are static; whereas, our framework provides a tool for users to choose landmark genes, and then to analyze the dataset based on these landmark genes. (2) Unlike the subspace clusters, the clusters generated from our method using the same landmarks are comparable across different datasets.
Semi-supervised Clustering
Semi-supervised clustering [15-17] uses existing domain knowledge to guide the clustering process. One popular method is constraint based clustering, where pairwise constraints (i.e 'must-link' and 'cannot-link' pairs) guide the clustering. The objective function of the underlying clustering algorithm is modified to accomodate these constraints. Our method differs from this clustering method as it does not constrain all the landmark genes to belong to one cluster. In our biological context, it is not unusual for genes to have more than one function.
Gene Ontology
Gene Ontology (GO) is a collection of controlled vocabularies that describe the biology of a gene product [18]. It consists of approximately 20,000 terms arranged in three independent ontologies: Biological Process, Cellular Component, and Molecular function, each represented by a directed acyclic graph (DAG). Gene Ontology has proven to be very important for secondary analysis of microarray expression data [19], and a wide range of tools have been developed to aid in this analysis. A comprehensive analysis of the available tools is given by Khatri [20]. Some of the prominent ones are ontoTools [21], GOminer [22], and GOstat [23].
In this paper we use the Biological Process ontology. A Biological Process (BP) is defined as "A phenomenon marked by changes that lead to a particular result, mediated by one or more gene products". As of 2006, there were approximately 10,000 GO terms associated with Biological Process [24]. We use Gene Ontology to provide external validation for the clusters. We use statistical significance tests to determine if the genes in a cluster belong to a specific Biological Process. A biologically meaningful cluster would consist of many genes that are annotated to a specific GO term.
Results and Discussion
Results
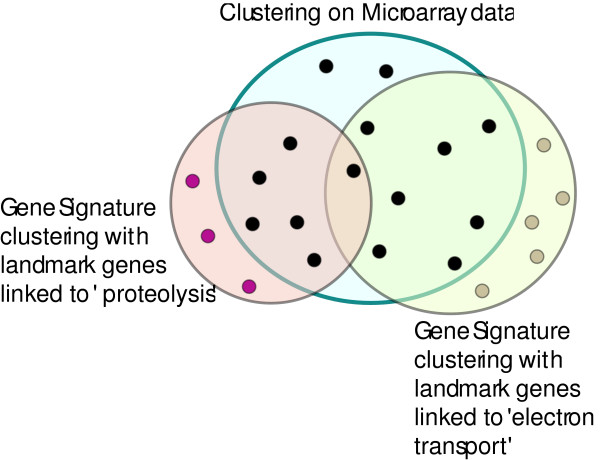
In the gene signature model, genes are points in a projected subspace whose coordinates are the landmark genes. The gene signature consists of relative distance to these landmark genes. So, by changing the landmark genes, a different perspective of the subspace can be obtained. Even using the same clustering algorithm, we can get different sets of clusters by changing this subspace. We repeated gene signature clustering for several biological processes (i.e., we used several different sets of landmark genes). The details for the overlapping GO terms and the unique GO terms, using different biological processes as landmarks for the Spellman dataset are shown in Table 1 (see Additional File 2 for DeRisi dataset). We analyzed genes in some of the clusters that produced the unique GO terms. These genes, annotated to the same GO term, clustered together when gene signatures were used, but did not cluster together when the original microarray data was used. Some of these genes are shown in Figures 1 and 2.
Table 1.
Details of overlaps between significant GO terms found by original clustering of microarray data, and those found by using gene signature clustering for the Spellman dataset.
| Biological process used for landmark genes | Number of Landmark Genes | Number of Original GO terms | Number of Overlapping GO terms | Number of Unique GO terms |
| proteolysis | 51 | 182 | 120 | 41 |
| electron transport | 20 | 182 | 126 | 44 |
| regulation of transcription | 100 | 182 | 126 | 41 |
| protein biosynthesis | 194 | 182 | 101 | 20 |
| carbohydrate metabolism | 121 | 182 | 142 | 58 |
| signal transduction | 52 | 182 | 121 | 53 |
| ubiquitin-dependent protein catabolism | 40 | 182 | 129 | 61 |
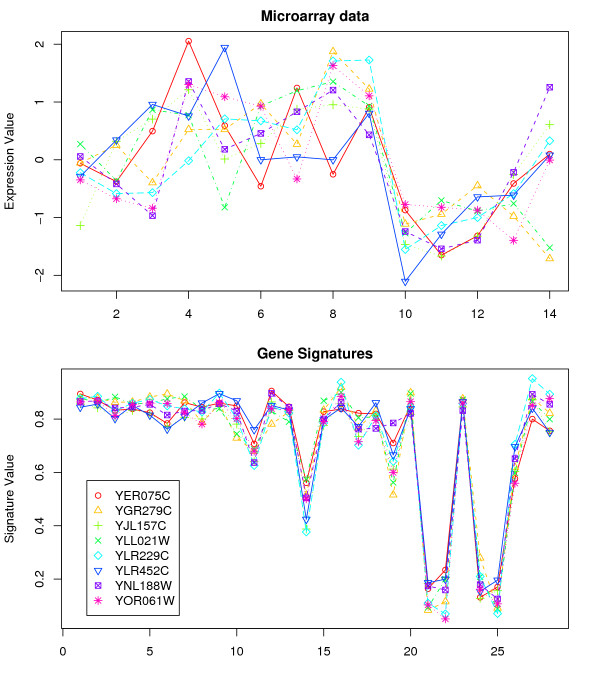
Figure 1.
Comparison of microarray expression data with gene signatures for genes that clustered together using gene signatures. Gasch dataset: Genes associated with multi-organism process (GO:0051704) were clustered together.
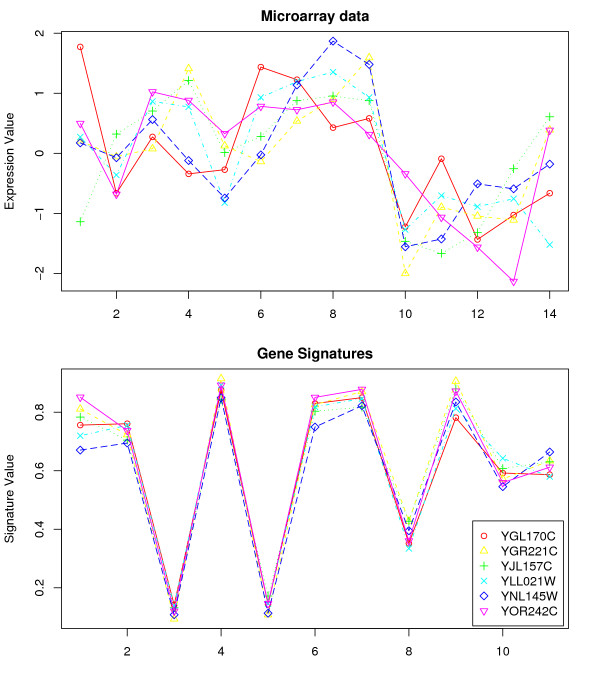
Figure 2.
Comparison of microarray expression data with gene signatures for genes that clustered together using gene signatures. Gasch dataset: Genes associated with reproduction (GO:0000003) were clustered together.
As illustrated, the gene expression patterns do not appear to be highly correlated, while the gene signatures show a strong correlation. For example, in the Gasch dataset, eight genes all relating to the GO term multi-organism process (GO:0051704), were in one cluster when gene signatures (with electron transport as landmark) were used. These genes did not cluster together with the original microarray data. Similarly, the six genes YGL170C, YGR221C, YJL157C, YLL021W, YNL145W and YOR242C, associated with reproduction (GO:0000003), only clustered together when gene signatures (with protein ubiquitination) were used. Although the biological significance between the landmark genes and the new GO term discovered is not immediately clear in this case, there might be some inherent relationships between them that are worth further investigation. Nonetheless, there were many other GO terms discovered using signatures (but not with the original expression data) whose associations with signature terms are much clearer. Some of these terms are investigated in detail in the discussion section.
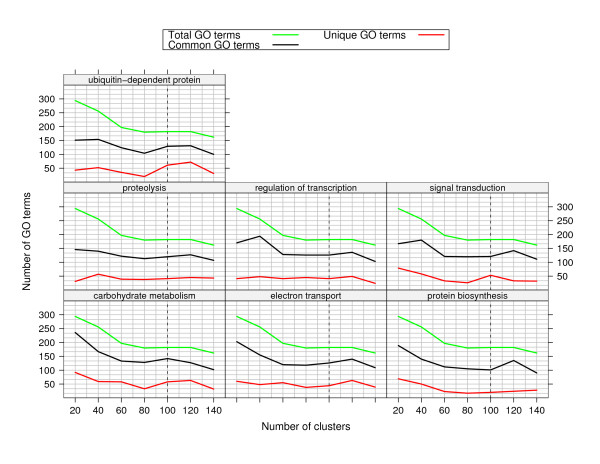
In order to test the effect of the number of clusters on the number of unique GO terms discovered for each landmark, we performed an experiment varying the numbers of clusters from 20 to 140. The results are shown in Figure 3. These indicate that there are a substantial number of unique GO terms for each set of landmark genes, that are largely independent of the number of clusters.
Figure 3.
Number of GO terms for varying number of clusters. For each landmark, a number of unique GO terms are found irrespective of the number of clusters.
Next, we compared the unique GO terms from two datasets for different landmark genes. Table 2 shows details of this comparison for three datasets taken two at a time. For example, the first and second column indicate the number of unique GO terms found for the Spellman and the Gasch datasets. The third column indicates the number of unique GO terms that were common between the Spellman and the Gasch datasets, and the p-value associated with this. In effect, this indicates the number of significant GO terms found in both datasets, by clustering of gene signatures, that were not found in the original clustering of either of the two datasets. Similarly, Table 3 shows the comparison when SOM was used for clustering. As can be seen from the tables, both the clustering algorithms produced a substantial number of unique GO terms that were common across datasets.
Table 2.
Common Unique GO Terms between datasets (taken two at a time), using Tight Clustering algorithm.
| Spellman-Gasch (2038 genes) Unique GO terms | Gasch-DeRisi (2474 genes) Unique GO terms | Spellman-DeRisi (1408 genes) Unique GO terms | |||||||
| Biological process used to get landmark genes | Spellman | Gasch | Common (p-value) | Gasch | DeRisi | Common (p-value) | Spellman | DeRisi | Common (p-value) |
| proteolysis | 28 | 89 | 12 (7.7 × 10-6) | 117 | 80 | 27 (1.2 × 10-7) | 32 | 17 | 3 (3.4 × 10-2) |
| electron transport | 28 | 89 | 9 (1.3 × 10-3) | 121 | 125 | 28 (6.5 × 10-4) | 47 | 59 | 15 (1.6 × 10-6) |
| regulation of transcription | 23 | 57 | 5 (1.3 × 10-2) | 83 | 76 | 20 (1.6 × 10-6) | 31 | 24 | 5 (2.8 × 10-3) |
| protein biosynthesis | 32 | 85 | 7 (2.6 × 10-2) | 101 | 83 | 20 (1.4 × 10-4) | 21 | 53 | 1 (3.2 × 10-1) |
| carbohydrate metabolism | 22 | 72 | 7 (1.4 × 10-2) | 97 | 81 | 16 (3.6 × 10-3) | 28 | 33 | 1 (3.6 × 10-1) |
| signal transduction | 43 | 68 | 23 (1.0 × 10-15) | 76 | 98 | 22 (1.6 × 10-6) | 44 | 28 | 8 (1.6 × 10-4) |
| protein folding | 29 | 72 | 10 (6.3 × 10-5) | 110 | 81 | 31 (4.9 × 10-11) | 24 | 32 | 4 (1.8 × 10-2) |
| intracellular protein transport | 38 | 79 | 9 (5.1 × 10-3) | 137 | 83 | 25 (7.6 × 10-5) | 33 | 43 | 7 (2.4 × 10-3) |
| lipid metabolism | 43 | 73 | 17 (1.3 × 10-8) | 97 | 85 | 27 (6.4 × 10-9) | 32 | 27 | 4 (2.6 × 10-2) |
| ribosome biogenesis | 66 | 94 | 22 (4.4 × 10-7) | 111 | 124 | 22 (1.2 × 10-2) | 55 | 30 | 9 (2.4 × 10-4) |
Table 3.
Common Unique GO Terms between datasets (taken two at a time), using SOM algorithm.
| Spellman-Gasch (2038 genes) Unique GO terms | Gasch-DeRisi (2474 genes) Unique GO terms | Spellman-DeRisi (1408 genes) Unique GO terms | |||||||
| Biological process used to get landmark genes | Spellman | Gasch | Common (p-value) | Gasch | DeRisi | Common (p-value) | Spellman | DeRisi | Common (p-value) |
| proteolysis | 28 | 90 | 1 (1.3 × 10-1) | 90 | 56 | 17 (4.4 × 10-6) | 29 | 49 | 8 (4.6 × 10-4) |
| electron transport | 55 | 76 | 23 (1.1 × 10-11) | 97 | 52 | 17 (4.13 × 10-6) | 36 | 57 | 3 (2.4 × 10-1) |
| regulation of transcription | 39 | 79 | 15 (5.0 × 10-7) | 69 | 45 | 8 (9.8 × 10-3) | 32 | 40 | 1 (2.9 × 10-1) |
| protein biosynthesis | 64 | 71 | 19 (2.07 × 10-7) | 77 | 69 | 10 (2.8 × 10-2) | 20 | 72 | 2 (2.9 × 10-1) |
| carbohydrate metabolism | 37 | 73 | 11 (1.3 × 10-4) | 76 | 60 | 13 (4.2 × 10-4) | 42 | 45 | 2 (2.5 × 10-1) |
| signal transduction | 74 | 74 | 14 (2.5 × 10-3) | 92 | 61 | 10 (3.7 × 10-2) | 45 | 36 | 6 (1.9 × 10-2) |
| protein folding | 47 | 71 | 5 (1.7 × 10-1) | 77 | 44 | 5 (1.4 × 10-1) | 39 | 36 | 4 (9.6 × 10-2) |
| intracellular protein transport | 41 | 98 | 16 (3.5 × 10-6) | 113 | 51 | 18 (6.0 × 10-6) | 42 | 46 | 6 (1.4 × 10-2) |
| lipid metabolism | 47 | 83 | 9 (2.3 × 10-2) | 84 | 64 | 12 (5.5 × 10-3) | 73 | 37 | 0 (1.3 × 10-2) |
| ribosome biogenesis | 40 | 71 | 19 (1.6 × 10-11) | 99 | 77 | 9 (1.4 × 10-1) | 27 | 55 | 0 (9.7 × 10-2) |
We also compared our gene signature model against a base line approach built using a k-nn classifier. We used ten fold cross validation to impute functional annotations using k-nn and clusters obtained from our model. For all the landmarks tested, our approach produced a higher classification accuracy than the k-nn based approach, irrespective of 'k' (see Additional File 2).
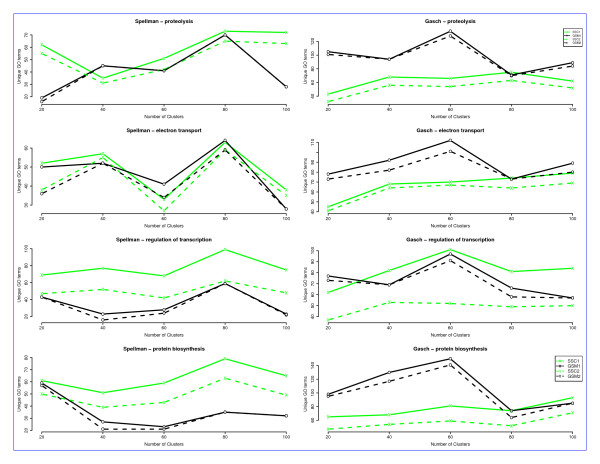
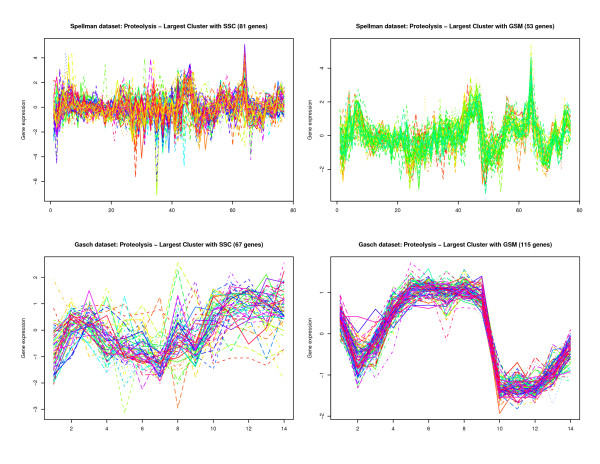
Finally, in order to validate the effectiveness of our approach, we compared our model, using tight clustering with gene signatures (GSM), to an existing semi-supervized clustering (SSC) model. For the SSC, the landmark genes were considered as 'must-link' constraints. All the landmark genes were thus clustered together in one cluster using the SSC. We then compared our model to the SSC by comparing the number of unique GO terms found for each set of landmark genes. We used the Spellman and the Gasch datasets for these experiments. The results of this comparison are shown in Figure 4. These indicate that in general, our model does better for the Gasch dataset while the SSC model does better for the Spellman dataset. The two models may be able to exploit different aspects of the underlying gene expression data. Even for the same set of landmark genes, one model may do better in one dataset than in the other. This is exemplified in the case of 'protein biosynthesis' where the SSC model does better in the Spellman dataset, whereas our model does better in the Gasch dataset (Figure 4). One difference between the two models is that the SSC forces the landmark genes in one cluster. This could lead to a large, less compact cluster, especially in cases where there are a large number of landmark genes with varied expression patterns. For example, for the Spellman dataset (Figure 5), the gene expression pattern of the landmark genes correlates well and the SSC model performs better, whereas for the Gasch dataset (Figure 5) the gene expression pattern of the landmark genes does not correlate well, and the GSM performs better.
Figure 4.
Comparison of unique GO terms found using gene signatures versus those found using semi-supervized clustering (SSC) for the Spellman and Gasch datasets. For the semi-supervized clustering (SSC), the landmark genes were considered as 'must-link' constraints. SSC1 denotes the number of unique GO terms found by using landmark genes as constraints in SSC. GSM1 denotes the number of unique GO terms found by using the gene signature model. SSC2 denotes the number of unique GO terms found for SSC if we remove the largest cluster (containing all the landmark genes) from analysis. GSM2 denotes the number of unique GO terms found using the gene signature model if we remove the largest cluster from analysis. The results for other landmarks are shown in Figure 3 in Additional File 2.
Figure 5.
Comparison of gene expression patterns in the largest cluster of semi-supervized clustering (SSC) versus the gene signature model (GSM) for the Gasch dataset using landmark genes associated with 'proteolysis'.
Discussion
These results indicate that clusters using gene signatures have biological significance, and that many of these gene associations are not found using clustering on the original microarray expression datasets. Each set of landmark genes carries the potential of defining its own set of clusters from the same dataset. To study this more closely, we examined several pairs of biological processes, i.e., the biological process that was used for selecting the landmark genes and its corresponding common unique GO terms found across datasets. For the Spellman and Gasch datasets, we analyze two of these biological processes (proteolysis and electron transport) and some of their common unique GO terms. These are listed in Table 4.
Table 4.
Some examples of biological processes used to select landmark genes and the common unique GO terms found across the Spellman and Gasch datasets
| Biological Process | Common Unique GO terms |
| Proteolysis | transcription |
| transcription, DNA-dependent | |
| phosphorylation | |
| energy reserve metabolism | |
| microtubule-based process | |
| sporulation | |
| sporulation (sensu Fungi) | |
| cellular lipid metabolism | |
| regulation of transcription | |
| reproductive sporulation | |
| ribosomal large subunit export from nucleus | |
| regulation of transcription, DNA-dependent | |
| Electron Transport | oxidative phosphorylation |
| ATP synthesis coupled electron transport | |
| ATP synthesis coupled electron transport (sensu Eukaryota) | |
| cellular respiration | |
| DNA strand elongation | |
| phosphorylation | |
| phosphorus metabolism | |
| phosphate metabolism | |
| aerobic respiration |
Proteolysis and Transcription
The connection between proteolysis and transcription has been well established. Proteolysis has been known to regulate transcription [25,26]. Interaction between the two processes is important for gene control and signaling pathways [27], and for the regulation of the cell cycle [28].
Proteolysis and Phosphorylation
The two processes interweave and interact with each other resulting in chromosome replication and segregation in budding yeast [29]. The two processes have also been linked to the Cdc28 protein kinase complex and other proteins involved in the budding yeast [30]. Recently it was reported that the human homolog of Mcm10 (a protein in yeast involved in DNA replication) is also regulated by proteolysis and phosphorylation during the cell cycle [31]. One article explores how the signaling molecule Hedgehog prevents the proteolyis (by phosphorylation) of Cubitus interruptus (Ci-155) transcriptional activator [32] and another touches on how phosphorylation-induced proteolysis eliminates unwanted by-products of protein kinases [33].
Electron Transport and Oxidative Phosphorylation
The relationship between these two processes has been studied across organisms. The inhibitory effects of Salicylic Acid on both the mitochondrial functions were presented in [34]. Salicylic acid inhibited mitochondrial electron transport which in turn inhibits oxidative phosphorylation. A recent article has studied the neurological diseases in humans and found that they may be caused by a defective electron transport system and its effect on oxidative phosphorylation [35]. Many other papers have also studied the relationship between these processes [36-39].
Electron Transport and ATP Synthesis
The relationship between these two processes has also been well studied. Allakhverdiev [40] studied the role of these two interlinked processes on photodamage and repair in Synechocystis. Electron transport is also tightly coupled to ATP synthesis in chloroplasts [41]. The effect of the two processes on the frequencies and harmonics of yeast Saccharomyces cerevisiae were studied in [42]. Faxen [43] and Belevich [44] study the mechanics of the intermediate steps between Electron transport and energy requiring processes like ATP synthesis.
We chose the Biological Process Ontology to select the landmark genes. Nevertheless, other sources that list genes belonging to a particular process or function can also be used. The biologist should also be able to define their own set of landmark genes and use these as the co-ordinates for projection.
We showed that clustering on gene signatures using different sets of landmark genes creates new sets of clusters that are different from the clusters obtained from the original microarray data. Genes in these new clusters reveal biological insights that were not present in the clustering of the original microarray data. We also showed that the new clusters are associated with biological terms that have some ties with the genes used for landmark selection.
Conclusion
We have used the SigCalc algorithm to project the microarray data onto a subspace whose co-ordinates are genes (called landmark genes), known to belong to a Biological Process. By changing the biological process and thus the landmark genes, we can change this subspace. We have shown how clustering on this subspace reveals new, biologically meaningful clusters which were not evident in the clusters generated by conventional methods. Each unique choice of a biological process would result in a unique subspace and a new set of clusters, enabling biologists to have more than one interpretation of the dataset. We have used three datasets to show that many of these unique GO terms are common across datasets. We have compared our model to an existing model, semi-supervized clustering, and shown that it compares favorably to existing models exploiting some prior knowledge of the data. We have done a literature survey and find strong evidence to support a link between the biological process used to select the landmark genes and the newly found unique GO terms that are common across the datasets.
Methods
Datasets
We use three yeast Saccharomyces cerevisiae datasets in our experiments. First, we use the cell cycle dataset of Spellman [45] available in R [46], comprising of 5624 genes and 77 samples. Second, we use the diauxic shift dataset of DeRisi [47] comprising of 6066 genes and 7 samples, and third the heat shock dataset of Gasch [48] comprising of 6097 genes and 14 samples. We applied a filter based on variation in gene expression, to focus our computations on informative genes across the samples. We selected genes that had a standard deviation greater than 0.35, and selected only those genes that were annotated in the biological process ontology of GO. The reduced datasets had 2288 genes for Spellman, 2794 genes for DeRisi and 4508 genes for Gasch. We then normalized them to a mean of zero and a standard deviation of one.
SigCalc
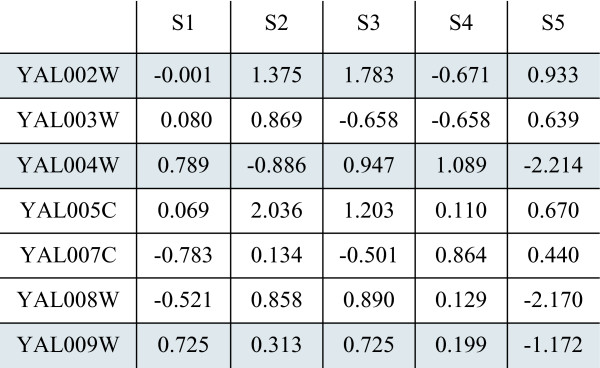
We introduced the concept of gene signatures in our previous work [49] where it was used as a basis for biological data integration. We formally define the signature calculation algorithm, SigCalc, in this subsection. Let M represent the microarray table consisting of n genes and m samples. SigCalc takes as input a microarray table M and a biological process. Using Gene Ontology, we find all the GO terms associated with the chosen biological process, and then find all genes associated with these GO terms. These genes are called landmark genes. For example, in yeast, the biological process "Protein Folding" is associated with several genes: YHR189W, YCR024C, YMR097C, etc. The algorithm for calculating the gene signatures, given a biological process, is shown in Algorithm 1 [see Appendix]. The SigCalc algorithm would convert a microarray data matrix (Figure 6) into a gene signature matrix (Figure 7). SigCalc projects the data onto a subspace, in which each coordinate corresponds to a landmark gene. The projected genes are represented as points in a multi-dimensional subspace. If two genes are close to each other in this projected subspace, then these two genes may show similar expression patterns relative to the landmark genes. By varying the set of landmark genes, we are able to vary this subspace.
Figure 6.
Microarray expression data matrix. The selected landmark genes are highlighted.
Figure 7.
Gene signatures derived from microarray data using SigCalc. Gene signature matrix, where each row represents a gene signature.
The SigCalc algorithm uses a distance function, dist, to measure the similarity between two gene vectors in microarray M. A variety of distance metrics such as Euclidean and cosine distances, or some other variants can be used. In our experiments we used the pearson correlation, a popular similarity metric [50] to arrive at the distance. Given two gene vectors and , the pearson correlation is given by:
To calculate our gene signatures, we define our correlation distance function as:
The correlation distance thus ranges from zero to one. A distance of zero indicates perfect positive correlation, and a distance of one indicates perfect negative correlation. A value of 0.5 would indicate no correlation between the gene vectors. Given a set of landmark genes k and a microarray M containing n genes and m samples, the SigCalc algorithm will return an n × k matrix, where each row represents a gene signature, as shown in Figure 7.
Clustering algorithms used
We chose two popular algorithms, tight clustering that is based on k-means clustering and self organizing maps (SOM) [5] to validate our Gene Signature model. The Tight Clustering algorithm [51] is a re-sampling based algorithm, that uses k-means clustering, to return genes that are clustered together consistently upon resampling. Re-sampling based methods have been found to return consistent clusters [52,53]. The Tight Clustering algorithm forms clusters that are stable and tight, and excludes genes from clusters that are 'noisy' and only serve to dilute the cluster. It has been widely used in microarray data clustering [54-57]. SOM is another clustering algorithm we used in our experiments. We use the R [46] implementation of SOM.
Cluster validation
Cluster results can be validated using external or internal criteria. External criteria are preferred because they provide a source to validate the clusters independent of the underlying datasets. We use the Gene Ontology to provide this external validation. Gene Ontology validates clustering results by comparing the genes in the clusters to genes known to be associated with specific biological functions. A "good" cluster will have a statistically significant over-representation of genes belonging to a specific biological process, as represented by a GO term. Our approach shows how the choice of landmark genes results in different sets of clusters, and that each set of clusters is associated with different sets of biological processes (GO terms).
Significant GO terms from clustering microarray data
We partition the microarray data M (n genes × m samples) into N clusters (N = 100 for results presented). We evaluate the biological significance of each cluster as follows: For a set of genes in a cluster, we evaluate if there are any GO terms that are over-represented than would be expected by chance. We evaluate the probability of a set of genes in a cluster being associated with the same GO term by using the hypergeometric distribution of the genes in the cluster. The probability of a cluster of size S containing x genes belonging to a particular GO term, given that the reference dataset of N genes has a total of A genes belonging to that particular GO term is:
where X is a random variable representing the number of genes in a cluster, that are associated with a particular GO term [58]. A cluster is considered to contain a significant GO term only if it has more than two genes associated with a specific GO term, and has a p-value less than 0.01. We used the GOstat package [23] for the hypergeometric test to find the set of statistically significant GO terms.
The set of significant GO terms for the original microarray clusters is the union of the significant GO terms for all of the clusters. This set of GO terms will be called the Original GO terms, as shown in Figure 8.
Figure 8.
Significant GO terms in microarray data. The dots indicate Significant GO terms found by performing clustering on microarray data (i.e., original GO terms).
Significant GO terms for a dataset using gene signatures
We build the gene signature matrix, for a selected biological process, by using SigCalc as given in Algorithm 1 [see Appendix]. Next, we partition the n × k gene signature matrix into N clusters (N = 100, i.e., the same number of clusters that were used for clustering the original microarray data). All other parameters for the clustering algorithm were kept the same as were used to cluster the original microarray data, as described in the previous section. This clustering of Gene signatures will be termed as Gene Signature Clustering. The set of significant GO terms from the clusters is derived using the hypergeometric distribution in the same way as described in the previous section. This set of significant GO terms obtained by clustering gene signatures, associated with a set of landmark genes, will be called landmark GO terms, as shown in Figure 9. The set of significant GO terms that are present in both the landmark GO terms and the original GO terms are called overlapping GO terms, and the set of significant GO terms that are present in the landmark GO terms but not in the original GO terms are called unique GO terms.
Figure 9.
Significant GO terms in microarray data and in gene signatures. Shows a comparison of Significant GO terms found by clustering gene signatures (i.e., landmark GO terms) with the original GO terms.
Unique GO terms common across datasets
Next, we determined if there were unique GO terms that were common across datasets. To ensure that the two datasets were comparable, we selected only those genes that were common to both datasets. For example, when comparing the Spellman (2288 genes) and Gasch (4508 genes) datasets, there were 2038 genes that were common to both datasets. So for this comparison, the Spellman dataset comprised 2038 genes × 77 samples, and the Gasch dataset comprised 2038 genes × 14 samples. This also ensured that, for a biological process, the same set of genes would be picked as landmarks for both datasets. For each dataset, we found the unique GO terms for a set of landmark genes, and then compared the two sets to determine which unique GO terms were common across datasets.
Source code availability and requirements
Project name: Landmark gene-guided clustering.
Project home page: http://www4.ncsu.edu/~pchopra/landmarks.html
Operating system: Windows
Programming languages: R (download at http://cran.r-project.org/). All R packages for Gene Ontology can be downlaoded at Bioconductor http://www.bioconductor.org/.
Licence: The R source code is freely available under the GPL license. The source code can be obtained as as additional material (see Additional file 1). This source code is provided only for academic use. By using the code, the user agrees to cite the main paper if results obtained from this code are used in the manuscript.
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
PC, JK and JY contributed to the algorithm design, implementation and systematic analysis of the framework. HC, HK and ML contributed to the biological validation of the findings obtained using the framework. All authors read and approved the final manuscript.
Appendix
Input: Microarray table M (n genes × m samples), and a Biological Process in Gene Ontology (GO), X.
Output: Set of gene signatures S = (g1), ..., (gn).
List all the genes linked to X in Gene Ontology. This set of k genes are the landmarks and will be represented by L = {l1, ..., lk}.
foreach gene gi in M do
foreach gene lj in L do
dj ← dist(, )
end
(gi) ← [d1,d2, ..., dk]
end
Algorithm 1: SigCalc: Signature Computation Algorithm.
Supplementary Material
R source code file
Supplementary Material for 'Microarray data mining using landmark gene-guided clustering'
Acknowledgments
Acknowledgements
This work was supported in part by Microsoft Bioinformatics Grant and Korea University Grant. The authors would like to thank Dr. Steffen Heber, Dr. Min-Kyu Oh and the two anonymous reviewers for their valuable suggestions.
Contributor Information
Pankaj Chopra, Email: pchopra@ncsu.edu.
Jaewoo Kang, Email: kangj@korea.ac.kr.
Jiong Yang, Email: jiong.yang@case.edu.
HyungJun Cho, Email: hj4cho@korea.ac.kr.
Heenam Stanley Kim, Email: hstanleykim@korea.ac.kr.
Min-Goo Lee, Email: mingoolee@korea.ac.kr.
References
- Jiang D, Tang C, Zhang A. Cluster Analysis for Gene Expression Data: A Survey. IEEE Transactions on Knowledge and Data Engineering. 2004;16:1370–1386. [Google Scholar]
- Handl J, Knowles J, Kell DB. Computational cluster validation in post-genomic data analysis. Bioinformatics. 2005;21:3201–3212. doi: 10.1093/bioinformatics/bti517. [DOI] [PubMed] [Google Scholar]
- Tavazoie S, Hughes JD, Campbell MJ, Cho RJ, Church GM. Systematic determination of genetic network architecture. Nat Genet. 1999;22:281–285. doi: 10.1038/10343. http://dx.doi.org/10.1038/10343 [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. PNAS. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Lander ES, Golub TR. Interpreting patterns of gene expression with self-organizing maps: Methods and application to hematopoietic differentiation. PNAS. 1999;96:2907–2912. doi: 10.1073/pnas.96.6.2907. http://www.pnas.org/cgi/content/abstract/96/6/2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons L, Haque E, Liu H. Subspace clustering for high dimensional data: a review. SIGKDD Explor Newsl. 2004;6:90–105. [Google Scholar]
- Fern X, Brodley C. Random Projection for High Dimensional Data Clustering: A Cluster Ensemble Approach. The Twentieth International Conference on Machine Learning (ICML-2003) 2003.
- Papadimitriou CH, Raghavan P, Tamaki H, Vempala S. Latent semantic indexing: a probabilistic analysis. J Comput Syst Sci. 2000;61:217–235. [Google Scholar]
- Deegalla S, Bostrom H. Reducing High-Dimensional Data by Principal Component Analysis vs. Random Projection for Nearest Neighbor Classification. icmla. 2006. pp. 245–250.
- Bingham E, Mannila H. Random projection in dimensionality reduction: applications to image and text data. Knowledge Discovery and Data Mining. 2001. pp. 245–250.
- Dasgupta S. Experiments with Random Projection. UAI '00: Proceedings of the 16th Conference on Uncertainty in Artificial Intelligence. 2000. pp. 143–151.
- Fradkin D, Madigan D. Experiments with Random Projections for Machine Learning. SIGKDD2003. 2003.
- Cheng Y, Church GM. Biclustering of Expression Data. Eighth International Conference on Intelligent Systems for Molecular Biology. 2000. pp. 93–103. [PubMed]
- Zhao L, Zaki MJ. Proceedings of the 2005 ACM SIGMOD international conference on Management of data. New York, NY, USA: ACM Press; 2005. TRICLUSTER: an effective algorithm for mining coherent clusters in 3D microarray data; pp. 694–705. [Google Scholar]
- Basu S, Banerjee A, Mooney RJ. Active Semi-Supervision for Pairwise Constrained Clustering. 2004. pp. 333–344.
- Bilenko M, Basu S, Mooney RJ. ICML '04: Proceedings of the twenty-first international conference on Machine learning. New York, NY, USA: ACM; 2004. Integrating constraints and metric learning in semi-supervised clustering; p. 11. [Google Scholar]
- Wagsta K, Cardie C, Rogers S, Schroedl S. Constrained K-means Clustering with Background Knowledge. Proceedings of 18th International Conference on Machine Learning (ICML-01) 2001. pp. 577–584.
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wang W, Yang J. A framework for ontology-driven subspace clustering. KDD '04: Proceedings of the tenth ACM SIGKDD international conference on Knowledge discovery and data mining. 2004. pp. 623–628.
- Khatri P, Draghici S. Ontological analysis of gene expression data: current tools, limitations, and open problems. Bioinformatics. 2005;21:3587–3595. doi: 10.1093/bioinformatics/bti565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draghici S, Khatri P, Bhavsar P, Shah A, Krawetz SA, Tainsky MA. Onto-Tools, the toolkit of the modern biologist: Onto-Express, Onto-Compare, Onto-Design and Onto-Translate. Nucl Acids Res. 2003;31:3775–3781. doi: 10.1093/nar/gkg624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeberg BR, Feng W, Wang G, Wang MD, Fojo AT, Sunshine M, Narasimhan S, Kane DW, Reinhold WC, Lababidi S, Bussey KJ, Riss J, Barrett JC, Weinstein JN. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 2003;4 doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beissbarth T, Speed TP. GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- Consortium GO. The Gene Ontology (GO) project in 2006. Nucl Acids Res. 2006;34:D322–326. doi: 10.1093/nar/gkj021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzouki N, Camier S, Ruet A, Moenne A, Sentenac A. Selective proteolysis defines two DNA binding domains in yeast transcription factor. Nature. 1986;323:176–178. doi: 10.1038/323176a0. [DOI] [PubMed] [Google Scholar]
- Wang X, Sato R, Brown MS, Hua X, Goldstein JL. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- Tansey W. Transcriptional regulation: RUPture in the ER. Nat Cell Biol. 2000;2:175–177. doi: 10.1038/35036420. [DOI] [PubMed] [Google Scholar]
- Cross F, Levine K. Regulation of the yeast cell cycle by transcription and proteolysis of cyclin-dependent kinase regulators. Kidney International. 1999;56:1185–1186. [Google Scholar]
- Deshaies RJ. Phosphorylation and proteolysis: partners in the regulation of cell division in budding yeast. Curr Op Gen and Development. 1997;7:7–16. doi: 10.1016/s0959-437x(97)80103-7. [DOI] [PubMed] [Google Scholar]
- Tyers M, Tokiwa G, Nash R, Futcher B. The Cln3-Cdc28 kinase complex of S. cerevisiae is regulated by proteolysis and phosphorylation. EMBO J. 1992;11:1773–1784. doi: 10.1002/j.1460-2075.1992.tb05229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi M, Yatagai F, Hanaoka F. Cell cycle-dependent proteolysis and phosphorylation of human Mcm10. J Biol Chem. 2001;276:M107190200. doi: 10.1074/jbc.M107190200. [DOI] [PubMed] [Google Scholar]
- Price MA, Kalderon D. Proteolysis of the Hedgehog Signaling Effector Cubitus interruptus Requires Phosphorylation by Glycogen Synthase Kinase 3 and Casein Kinase 1. Cell. 2002;108:823–835. doi: 10.1016/s0092-8674(02)00664-5. [DOI] [PubMed] [Google Scholar]
- Elion EA, Qi M, Chen W. SIGNAL TRANSDUCTION: Signaling Specificity in Yeast. Science. 2005;307:687–688. doi: 10.1126/science.1109500. [DOI] [PubMed] [Google Scholar]
- Xie Z, Chen Z. Salicylic Acid Induces Rapid Inhibition of Mitochondrial Electron Transport and Oxidative Phosphorylation in Tobacco Cells. Plant Physiol. 1999;120:217–226. doi: 10.1104/pp.120.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussbaum RL. Mining yeast in silico unearths a golden nugget for mitochondrial biology. J Clin Invest. 2005;115:2689–2691. doi: 10.1172/JCI26625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattoon JR, Sherman F. Reconstitution of Phosphorylating Electron Transport in Mitochondria from a Cytochrome c-deficient Yeast Mutant. J Biol Chem. 1966;241:4330–4338. [PubMed] [Google Scholar]
- Wakiyama S, Ogura Y. Oxidative phosphorylation and the electron transport system of castor bean mitochondria. Plant Cell Physiol. 1970;11:835–848. [Google Scholar]
- Van Verseveld HW, Stouthamer AH. Electron-transport chain and coupled oxidative phosphorylation in methanol-grown Paracoccus denitrificans. J Arch Microbiology. 1978;118:13–20. doi: 10.1007/BF00406068. [DOI] [PubMed] [Google Scholar]
- Hatefi Y. The Mitochondrial Electron Transport and Oxidative Phosphorylation System. Annu Rev Biochem. 1985;54:1015–1069. doi: 10.1146/annurev.bi.54.070185.005055. [DOI] [PubMed] [Google Scholar]
- Allakhverdiev SI, Nishiyama Y, Takahashi S, Miyairi S, Suzuki I, Murata N. Systematic Analysis of the Relation of Electron Transport and ATP Synthesis to the Photodamage and Repair of Photosystem II in Synechocystis. Plant Physiol. 2005;137:263–273. doi: 10.1104/pp.104.054478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JF. Photosynthesis of ATPElectrons, Proton Pumps, Rotors, and Poise. Cell. 2002;110:273–276. doi: 10.1016/s0092-8674(02)00870-x. [DOI] [PubMed] [Google Scholar]
- Miller J, Nawarathna D, Vajrala V, Gardner J, Widger W. Electromagnetic probes of molecular motors in the electron transport chains of mitochondria and chloroplasts. 2005.
- Faxen K, Gilderson G, Adelroth P, Brzezinski P. A mechanistic principle for proton pumping by cytochrome c oxidase. Nature. 2005;437:286–289. doi: 10.1038/nature03921. [DOI] [PubMed] [Google Scholar]
- Belevich I, Verkhovsky MI, Wikstrm M. Proton-coupled electron transfer drives the proton pump of cytochrome c oxidase. Nature. 2006;440:829–832. doi: 10.1038/nature04619. [DOI] [PubMed] [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive Identification of Cell Cycle-regulated Genes of the Yeast Saccharomyces cerevisiae by Microarray Hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria; 2006. [ISBN 3-900051-07-0] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO. Exploring the Metabolic and Genetic Control of Gene Expression on a Genomic Scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic Expression Programs in the Response of Yeast Cells to Environmental Changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Yang J, Xu W, Chopra P. Integrating Heterogeneous Microarray Data Sources Using Correlation Signatures. In: Ludäscher B, Raschid L, editor. DILS, Volume 3615 of Lecture Notes in Computer Science. Springer; 2005. pp. 105–120. [Google Scholar]
- D'Haeseleer P. How does gene expression clustering work? Nature Biotechnology. 2005;23:1499–1501. doi: 10.1038/nbt1205-1499. [DOI] [PubMed] [Google Scholar]
- Tseng GC, Wong WH. Tight Clustering: A Resampling-Based Approach for Identifying Stable and Tight Patterns in Data. Biometrics. 2005;61:10–16. doi: 10.1111/j.0006-341X.2005.031032.x. [DOI] [PubMed] [Google Scholar]
- Yeung K, Medvedovic M, Bumgarner R. Clustering gene-expression data with repeated measurements. Genome Biology. 2003;4:R34. doi: 10.1186/gb-2003-4-5-r34. http://genomebiology.com/2003/4/5/R34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison DB, Cui X, Page GP, Sabripour M. Microarray data analysis: from disarray to consolidation and consensus. Nature Reviews Genetics. 2006;7:406–406. doi: 10.1038/nrg1749. [DOI] [PubMed] [Google Scholar]
- Zhou XJ, Kao MCJ, Huang H, Wong A, Nunez-Iglesias J, Primig M, Aparicio OM, Finch CE, Morgan TE, Wong WH. Functional annotation and network reconstruction through cross-platform integration of microarray data. Nature Biotechnology. 2005;23:238–243. doi: 10.1038/nbt1058. [DOI] [PubMed] [Google Scholar]
- Huang D, Wei P, Pan W. Combining Gene Annotations and Gene Expression Data in Model-Based Clustering: Weighted Method. OMICS: A Journal of Integrative Biology. 2006;10:28. doi: 10.1089/omi.2006.10.28. [DOI] [PubMed] [Google Scholar]
- Kabbarah O, Mallon MA, Pfeifer JD, Goodfellow PJ. Transcriptional profiling endometrial carcinomas microdissected from DES-treated mice identifies changes in gene expression associated with estrogenic tumor promotion. International Journal of Cancer. 2006;119:1843–1849. doi: 10.1002/ijc.22063. [DOI] [PubMed] [Google Scholar]
- Casati P, Stapleton AE, Blum JE, Walbot V. Genome-wide analysis of high-altitude maize and gene knockdown stocks implicates chromatin remodeling proteins in response to UV-B. The Plant Journal. 2006;46:613–627. doi: 10.1111/j.1365-313X.2006.02721.x. [DOI] [PubMed] [Google Scholar]
- Draghici S, Khatri P, Martins RP, Ostermeier GC, Krawetz SA. Global functional profiling of gene expression. Genomics. 2003;81:98–104. doi: 10.1016/s0888-7543(02)00021-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
R source code file
Supplementary Material for 'Microarray data mining using landmark gene-guided clustering'