Abstract
Krüppel-like factor 4 (Klf4) is a transcription factor and functions in regulating cell differentiation, cell growth, and cell cycle. Although Klf4 is expressed in lymphocytes, its function in lymphocytes is unknown. In this study, we report that the levels of Klf4 expression were low in pro-B cells and continuously increased in pre-B and in mature B cells. Upon activation, Klf4 was rapidly decreased in mature B cells after 2 h of activation. A modest decrease in numbers of pre-B cells in bone marrow and mature B cells in spleen was observed in Klf4-deficient mice. In the absence of Klf4, fewer B cells entered the S phase of the cell cycle and completed cell division in response to the engagement of BCR and/or CD40 in vitro. Furthermore, the delay in entering the cell cycle is associated with decreased expression of cyclin D2 in B cells that lack Klf4 expression. We then demonstrated that Klf4 directly bound to the promoter of cyclin D2 and regulated its expression. These findings demonstrate that Klf4 regulates B cell number and activation-induced B cell proliferation through directly acting on the promoter of cyclin D2.
Homeostasis of B cells is regulated by the processes of cell proliferation and death. Cell proliferation induced by engagement of the BCR and costimulatory receptors such as CD40 requires successful transition through different cell cycle phases and checkpoints (1). Upon receiving the activation signal, the decision of resting B cells to enter the cell cycle reflects a balance between growth-promoting and growth-inhibitory regulators (2). D-type cyclins are key regulators for the G1/S transition checkpoint and are primary targets for proliferation signals. Among the three members of D cyclins, cyclin D2 is the main D-type cyclin expressed in mature mouse B cells and plays an important role in cell proliferation after BCR-mediated proliferation (2-5). However, the transcriptional control of cyclin D2 expression in B cells is not completely understood.
Krüppel-like factor 4 (Klf4)3 is a zinc-finger transcription factor that regulates cell proliferation and differentiation (6-8). One unique feature of Klf4 is its function in cell cycle and growth depending on the surrounding context (9). Klf4 induces cell cycle arrest at the G1/S boundary in untransformed cells while acting as an oncogene by repressing p53 in transformed cells expressing RASV12 (7). In addition, Klf4 induces oncogenic transformation in the absence of functional cyclin D1 and the cyclin-dependent kinase inhibitor, p21WAF1/Cip1 (10). A recent report shows that Klf4 is expressed in several types of human B cell lymphoma/leukemia lineages (11), and overexpression of Klf4 can induce cell cycle arrest and apoptosis in the G1 phase of the cell cycle of transformed pro/pre-B cell lines. Pro/pre-B cells that overexpress Klf4 display increased expression of p21CIP1, and decreased expression of c-Myc and cyclin D2, indicating that Klf4 might regulate cell cycle in B cell malignancies. However, the physiological role of Klf4 in normal B cells has not yet been examined.
To determine the role of Klf4 in B cells, we generated mice deficient for Klf4 in B cells (bKlf4−/−) and studied their response to activation-induced proliferation. In this study, we report that bKlf4−/− mice had a modest reduction of pre-B cells in bone marrow and in mature B cells in spleen; B cells in the absence of Klf4 exhibited a defect in the BCR-mediated proliferation response. We demonstrate that this proliferation defect is due to the inability of cyclin D2 up-regulation in B cells from bKlf4−/− mice, and that Klf4 directly binds to the promoter and regulates cyclin D2 expression. These findings suggest that Klf4 plays an important role in regulating activation-induced B cell proliferation.
Materials and Methods
Generation of B cell-specific-Klf4-knockout mice
Klf4-loxP mice (12) were crossed with CD19-Cre transgenic mice (13) to generate bKlf4−/− mice in which the Klf4 allele is specifically deleted in B cells. The deletion of the Klf4 gene in B cells was confirmed by Southern blotting and by RT-PCR. The breeding was conducted between bKlf4+/− mice and genotyping was done by PCR around 4 wk of age. bKlf4−/− and bKlf4+/+ littermate mice were used in the experiments at 8–16 wk of age. Maintenance and experiments of mice were in accordance with the National Institutes of Health policies for animal care and use.
Isolation and stimulation of B cells
Pro- and pre-B cells were isolated from bone marrow by cell sorting (Mo-Flo) based on the expressions of CD43+/CD24−/B220+ for pro-B cells and CD43−/CD24+/B220+ for pre-B cells. Spleen B cells were isolated by a negative depletion method using a B cell isolation kit (Miltenyi Biotec) according to the manufacturer’s instruction. The purity of these isolated cells was >95%. Freshly isolated splenic B cells were resuspended in RPMI 1640 medium containing 10% FBS, 2 mM l-glutamine, 50 μM 2-ME, and 10 U/ml penicillin/10 μg/ml streptomycin (Invitrogen Life Technologies), and 24 μg/ml AffiniPure goat anti-mouse IgM μ-chain specific (Jackson ImmunoResearch Laboratories) alone or in combination with 5 μg/ml anti-mouse CD40 (clone 1C10; eBioscience) or with LPS (10 μg/ml; Sigma-Aldrich) were added. Stimulated B cells were harvested at specified time for mRNA, protein, and other analyses.
Flow cytometry analysis
Single-cell suspensions of bone marrow cells and splenocytes were incubated with different combinations of the following Abs against: CD4-FITC, CD8-PE, CD19-TC, CD24-FITC, CD43-PE, B220-Tri-color, IgM-PE (Invitrogen Life Technologies). Data were collected on the FACSCalibur and analyzed using CellQuest Pro software (BD Biosciences).
Cell cycle analysis
Cell cycle analysis was conducted using the propidium iodide staining kit (BD Biosciences) according to the manufacturer’s instruction. The data were collected using a FACSCalibur and analyzed using MultiCycle software (Phoenix Flow Systems).
[3H]Thymidine incorporation assays
Freshly isolated B cells were plated in triplicate at 2 × 105 cells/well in 96-well plates and stimulated with 24 μg/ml goat anti-mouse IgM, μ-chain specific (Jackson ImmunoResearch Laboratories) alone or in combination with 5 μg/ml anti-mouse CD40 or LPS for 48 h and then 1 μCi of [3H]thymidine was added to each well. Cells were harvested 20 h later and [3H]thymidine incorporation was measured.
CFSE assay
Freshly isolated B cells were labeled with 2.5 μM CFSE (Invitrogen Life Technologies) at 37°C for 10 min and washed twice with RPMI 1640 containing 10% FCS. After stimulation for 2 and 3 days, the CFSE profiles were evaluated by flow cytometry and the proliferation index (PI) was calculated using MODFIT software (Verity Software House).
Quantitative RT-PCR
The procedure of quantitative RT-PCR was previously described (14). Primers were designed using PRISM Primer Express 2.0 (Applied Biosystems) and made by Integrated DNA Technologies. Primers sequences are Klf4: 5′-GGTGCAGCTTGCAGCAGTAA-3′ and 5′-AAAGTCTAGGTCCAGGAGGTCGTT-3′; CcnD2: 5′-GCGTGCAGAAGGACATCCA-3′ and 5′-CCTCACAGACCTCTAGCATCCA-3′; CcnD3: 5′-ACGACTTCCTGGCCTTGATTC-3′ and 5′-CAAAGCCTGCCGGTCACT-3′; CdknA1 (p21): 5′-GGTGGGCCCGGAACAT-3′ and 5′-GCGCTTGGAGTGATAGAAATCTG-3′; c-Myc: 5′-GGGCCAGCCCTGAGCCCCTAGTG-3′ and 5′-ATGGAGATGAGCCCGACTCCGACC-3′; L32: 5′-ACCA GTCAGACCGATATGTGAAAA-3′ and 5′-CGCACCCTGTTGTCAATGC-3′; β-actin: 5′-AGGTCATCACTATTGGCAACGA-3′ and 5′-AGGA TTCCATACCCAAGAAGGAA-3′. The real-time PCR was conducted in 96-well plates in a 25 μl total volume using SYBR green PCR mix kit (Bio-Rad) under the manufacturer’s condition. The specificity of the PCR product was confirmed by gel (2% agarose) electrophoresis. Each RT-PCR was repeated twice and the mean was used in the figures. The threshold cycle values obtained from each reaction were normalized to the average of ribosomal protein L32 and β-actin at each correlating time point.
Western blot
Standard Western blots were performed using the following Abs: cyclin D2 (1:200; Abcam), cyclin D3 (1:2,000; Cell Signaling Technology), p21 (1:500; BD Biosciences), and β-actin (1:10,000; Sigma-Aldrich) as a loading control, followed by HRP-linked anti-mouse IgG (1:5,000; GE Healthcare). Signals were detected using the ECL detection system (GE Healthcare) according to the manufacturer’s instructions. Five different samples were analyzed for each Ab. The quantitation was conducted by collecting images (FluorChem; Alpha Innotech) and calculated using UN-SCAN-IT software (Silk Scientific). The intensity values were normalized to β-actin and the mean intensity values were presented.
Chromatin immunoprecipitation (ChIP) assay
The procedure of the ChIP assay was previously described (14). In brief, 2 million freshly isolated spleen B cells from C57BL/6 mice were incubated with 1% formaldehyde at 37°C for 10 min and washing twice with cold PBS containing protease inhibitors (Complete; Roche). The cells were lysed with lysis buffer, and the chromosomal DNA was then sheared with sonication (Sonic Dismembrator Model 100; Fisher Scientific). To eliminate nonspecific binding, Dynabeads protein G was first incubated with cell lysate and washed, then cell lysates were separated into three parts: 1) input, 2) immunoprecipitate with 2 μg of monoclonal anti-Klf4 (ABNOVA), and 3) immunoprecipitate with an isotype-matched control IgG. DNA from these three fractions were isolated and used for real-time PCR. Three pairs of primers were used: CcnD2_p2 (−233 to −372 bp): 5′-AAGCCTCCGAAGTTAGAGAGCAC-3′ and 5′-CTCCCCATCCAGCCCCGC-3′; CcnD2_p3 (−501 to −638 bp): 5′-AAGCACCCCTTTCTCCAACATC-3′ and 5′-GCAACCTCGCCAAACCAGG-3′; and CcnD2_p4 (−766 to −922 bp): 5′-AGCCAAACCTAAACCCTCCCTCTC-3′ and 5′-ACGCAGGAAAAACCCGCTTC-3′. The data were derived from two independent experiments and presented as the percentage of input value.
Statistical analysis
The Student t test was used to analyze the statistical differences between bklf4+/+ and bklf4−/− mice, and p values <0.05 were considered significant.
Results
Klf4 expression is regulated in B cells during development and activation
To examine the expression of Klf4 in B cells, we isolated pro-B cells to pre-B cells from bone marrow and mature B cells from spleen and measured levels of Klf4 mRNA by real-time quantitative RT-PCR. We found that levels of Klf4 expression were low in pro-B cells, increased in pre-B cells, and reached highest in mature B cells (Fig. 1A). Compared with pro-B cells, mature B cells expressed at least seven times more Klf4. To further examine Klf4 expression after activation, we stimulated mature B cells from spleen with anti-IgM (anti-μ-specific Ab) in vitro. We found that Klf4 levels rapidly down-regulate after 2 h of stimulation and reached the lowest level after 48 h stimulation (Fig. 1B). These findings suggest that expression of Klf4 increases with B cell maturation and decreases after activation.
FIGURE 1.
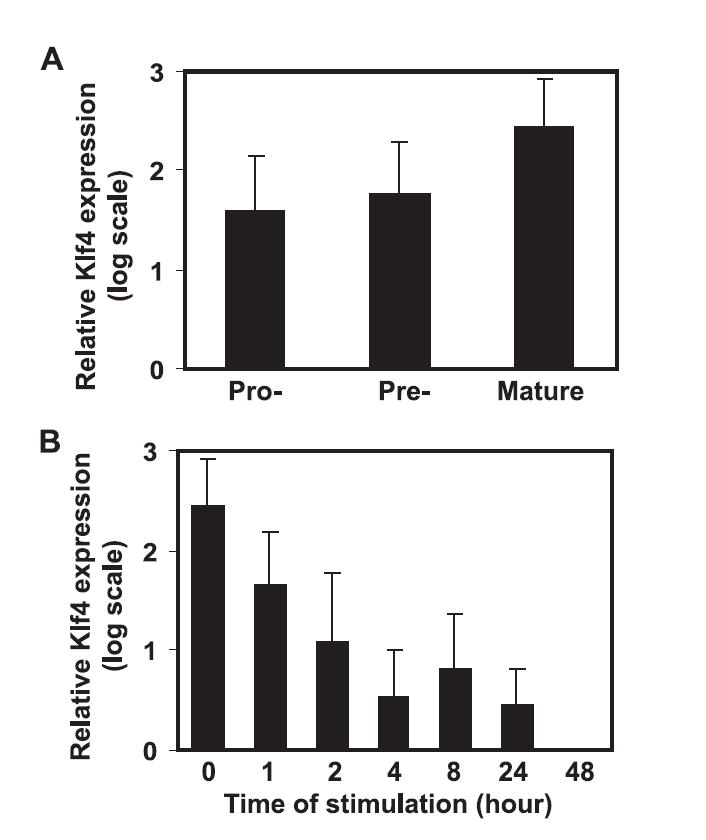
Klf4 expresses in pro-, pre-, and mature B cells. A, Expression of Klf4 during B cell development. Pro- and pre-B cells were isolated from bone marrow; mature B cells were isolated from spleen. B, Kinetic expression of Klf4 in mature B cells after stimulation with anti-IgM. The levels of Klf4, as measured by real-time quantitative RT-PCR and normalized to the mean of β-actin and L32, rapidly decrease after 2-h stimulation to undetectable at 48 h poststimulation. Data are presented as the mean and SEM of four independent experiments.
bKlf4−/− mice have a modest decrease in percentage/number of pre-B cells in bone marrow and of mature B cells in spleen
To further evaluate the function of Klf4, we generated mice that deleted the Klf4 gene specifically in B cells by crossing Klf4-loxP mice (12) with CD19-Cre transgenic mice (13) B cells isolated from the bKlf4−/− mice deleted the Klf4 gene only in B cells, but not in other cells as confirmed by Southern blot and PCR (Fig. 2, A and B). In addition, we demonstrated the loss of Klf4 at the mRNA level in B cells from bKlf4−/− mice by real-time RT-PCR (Fig. 2C). The bKlf4−/− mice were fertile and had no apparent abnormalities. Analysis of B cells in bone marrow from bKlf4−/− mice showed a modest increase of pro-B cells but it did not reach statistical significance (Fig. 3A, Table I). In contrast, a modest and statistical significant decrease of the percentage of pre-B cells (11% reduction, p = 0.001) was observed in bKlf4−/− mice (Fig. 3A, Table I). In spleen, a similar modest and statistical significant decrease in percentage (9% reduction, p < 0.05) and in numbers (25% reduction, p < 0.05) of mature B cells was observed (Fig. 3, B and C, Table I). The percentage of B1 B cells in the peritoneal cavity was not changed between bKlf4−/− mice and their controls (Table I). Together, these findings suggest that Klf4 may participate in the regulation of B cell development and homeostasis.
FIGURE 2.
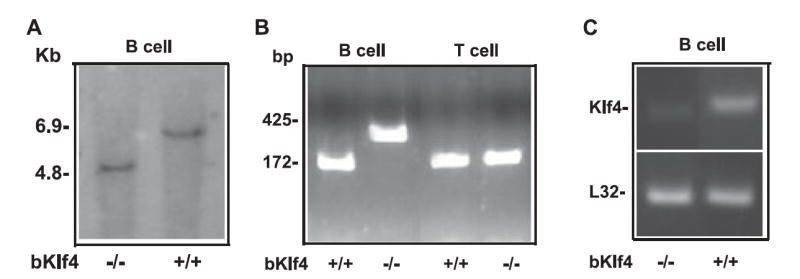
The Klf4 gene is deleted in B cells of bKlf4−/− mice. A, Deletion of Klf4 exon2 and exon3 in B cells from bKlf4−/− mice. A 4.8-kb band represents deletion of the Klf4 gene in B cells from bKlf4−/− mice and a 6.9-kb band represents the wild-type Klf4 gene in B cells from bKlf4+/+ mice by Southern blot hybridization. B, Specific deletion of Klf4 in B cells but not T cells from bKlf4−/− mice. Klf4 gene deletion resulted in a 450-bp band only in B cells from bKlf4−/− mice but not in B cells from bKlf4+/+ mice by PCR. In contrast, a 175-bp band can be detected in B and T cells from bKlf4+/+ mice and T cells from bKlf4−/− mice by PCR. C, Loss of Klf4 mRNA expression in B cells from spleen of bKlf4−/− mice by RT-PCR. L32 is a ribosomal protein that served as a control.
FIGURE 3.
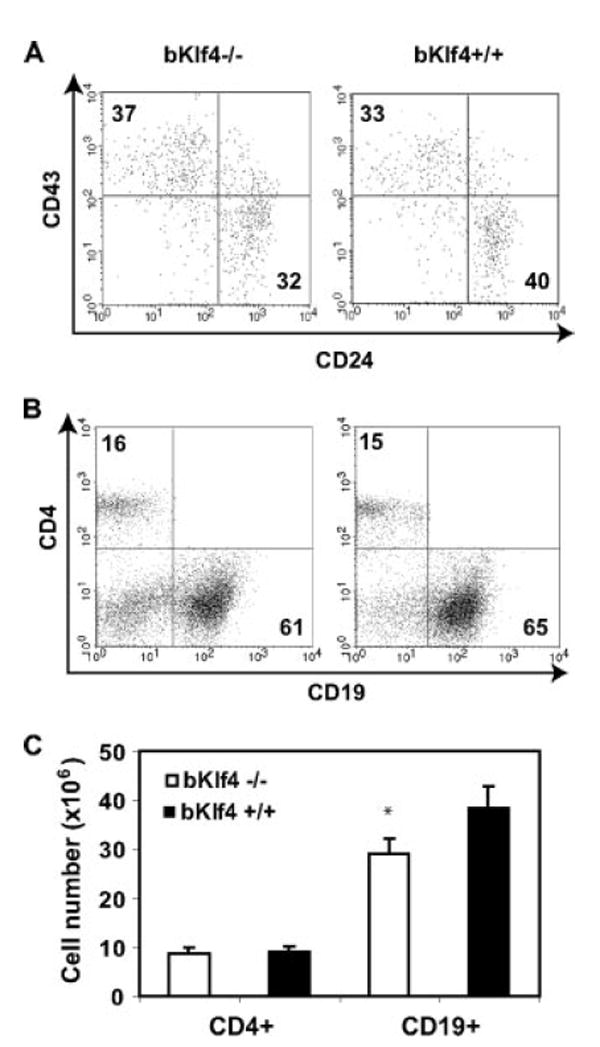
Modest loss of pre-B cells and mature B cells is observed in bKlf4−/− mice. A, Decrease of pre-B, but not pro-B, cells in bone marrow of bKlf4−/− mice. A representative dot plot of CD43 and CD24 staining of B220+ bone marrow cells from bKlf4−/− and bKlf4+/+ mice. The numbers indicate the mean percentage of cells. B, Decrease of mature B cells in spleen of bKlf4−/− mice. A representative dot plot of CD19 and CD4 staining of splenocytes from bKlf4−/− and bKlf4+/+ mice. C, Reduction of number (25%) of mature B cells but not CD4 T cells in spleen in bKlf4−/− mice. Data were presented as mean ± SEM (n = 12); *, p < 0.05.
Table I.
B cell subsets in bKlf4−/− mice
| Organs | Cell Types | Phenotype | bKlf4−/− | bKlf4+/+ | Significance (p)a |
|---|---|---|---|---|---|
| Bone marrow | B cell | B220+ | 26.4 ± 2.8 | 25.4 ± 2.8 | 0.405 |
| Pro-B cell | B220+CD43+ | 37.4 ± 1.9 | 33.2 ± 1.8 | 0.058 | |
| Pre-B cell | B220+CD24+ | 31.8 ± 1.7 | 40.2 ± 1.4 | 0.001 | |
| Spleen | Mature B cell | CD19+ | 60.7 ± 1.9 | 64.7 ± 1.7 | 0.030 |
| Peritoneum | B1 B cell | B220+CD5+ | 25.7 ± 2.5 | 25.1 ± 4.4 | 0.448 |
Bone marrow, n = 12; Spleen, n = 16; Peritoneum, n = 8.
Klf4 involves activation-induced proliferation
To determine whether Klf4 regulates B cell proliferation, we compared DNA synthesis and cell division of B cells between bKlf4−/− and bKlf4+/+ mice in response to anti-IgM alone, or anti-IgM plus anti-CD40, or LPS stimulation in vitro. [3H]Thymidine incorporation was used for evaluating DNA synthesis; B cells from bKlf4−/− mice showed significantly reduced [3H]thymidine uptake compared with B cells from bKlf4+/+ mice under stimulation conditions (p < 0.05 for both anti-IgM alone, n = 10 and for anti-IgM/anti-CD40, n = 5) but not under LPS stimulation (p = 0.11, n = 3) (Fig. 4A). CFSE-tracking dye was used to examine activation-induced cell division by comparing PI. PI is defined by the total number of cells after culture divided by the computed number of original parent cells, and was significantly lower in B cells from bKlf4−/− than from bKlf4+/+ mice (p < 0.05 for anti-IgM alone, n = 9 and p < 0.01 for anti-IgM/anti-CD40, n = 14) (Fig. 4B). Together, these findings suggest that Klf4 is involved in BCR-mediated, activation-induced B cell proliferation.
FIGURE 4.
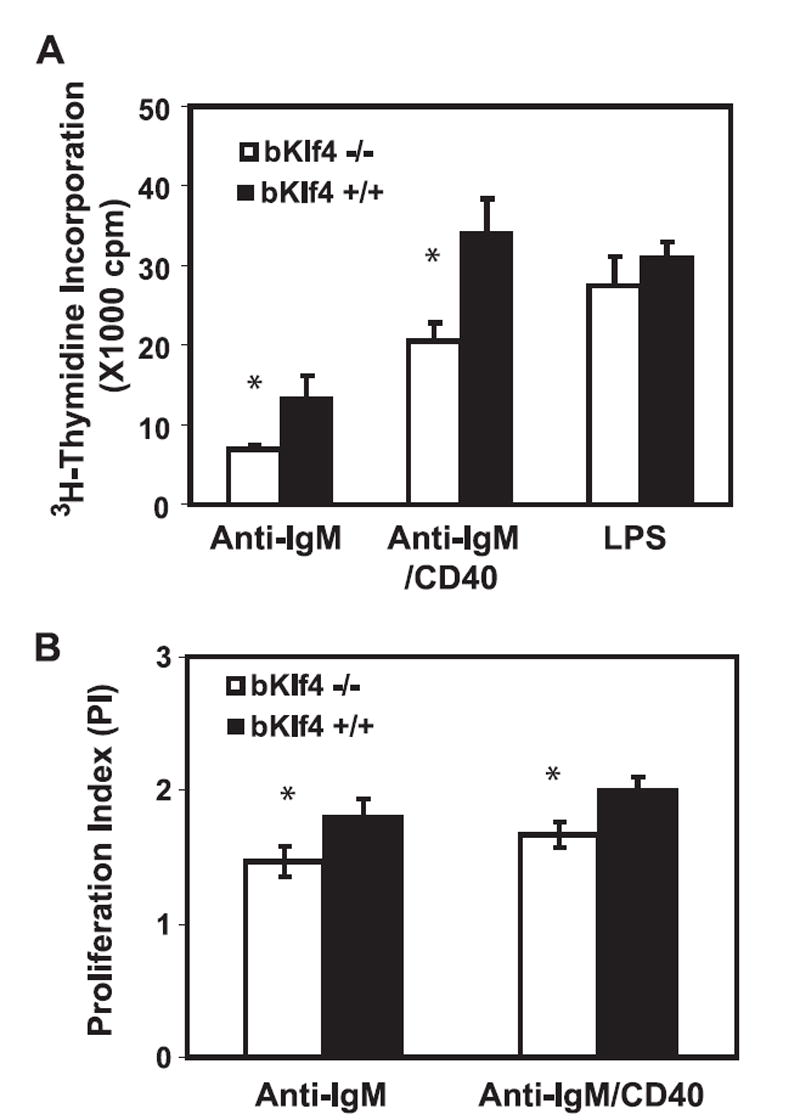
Klf4 involves in activation-induced B cell proliferation. A, Stimulation induced DNA synthesis of B cells from bKlf4−/− and bKlf4+/+ mice. B cells were isolated from spleen and stimulated in vitro with anti-IgM alone or anti-IgM plus anti-CD40 (anti-IgM/CD40) Abs or LPS (10 μg/ml) for 48 h and added [3H]thymidine for 20 h. B cells from bKlf4−/− show a significant reduction of [3H]thymidine uptake compared with control under stimulation conditions: anti-IgM alone (p < 0.05, n = 10) or anti-IgM/CD40 (p < 0.05, n = 5) but not under LPS (p = 0.11, n = 3). B, CFSE-tracking dye measuring proliferation of B cells from bKlf4−/− and bKlf4+/+ mice. Low PI of B cells from bKlf4−/− mice compared with bKlf4+/+ mice under both stimulation conditions (p < 0.05). Anti-IgM, n = 9; anti-IgM/CD40, n = 14.
Klf4 is required for the progression from the G1 to the S phase of B cell cycle
To further investigate the precise defects in activation-induced proliferation in bKlf4−/− B cells, the cell cycle status of B cells after activation was compared between bKlf4−/− and bKlf4+/+ mice. More B cells remained in G1 phase and fewer B cells in S phase from bKlf4−/− mice than from bKlf4+/+ mice after 48 h of anti-IgM (Fig. 5) and after anti-IgM/anti-CD40 stimulations (data not shown). This finding suggests that Klf4 regulates cell cycle progression from G1 to S phase in B cells and the absence of Klf4 results in partial blockade of B cell progression from G1 to S phase.
FIGURE 5.
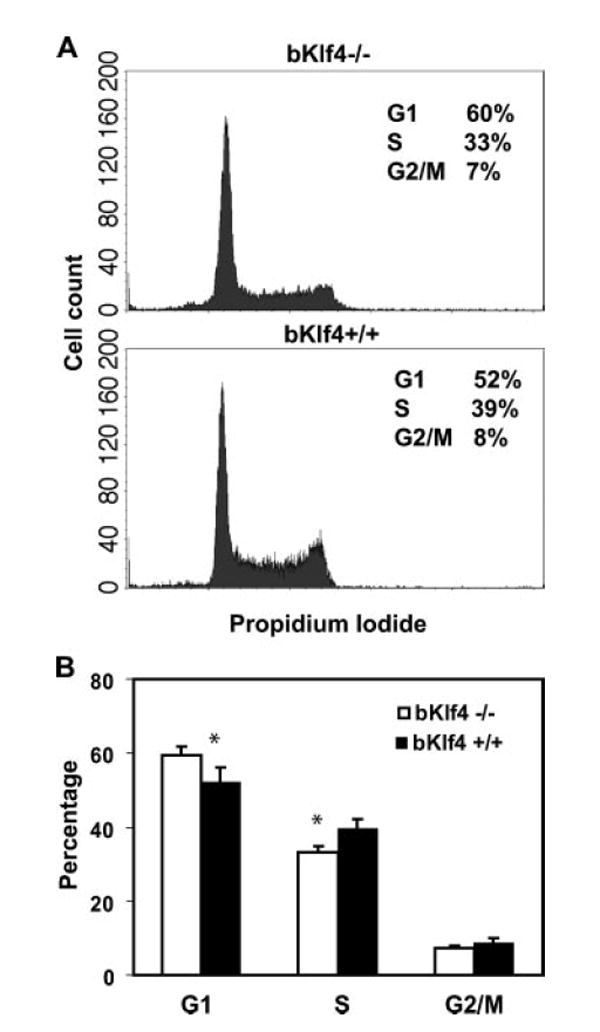
Klf4 regulates the G1 to S progression of cell cycle. A, Representative histograms of propidium iodide staining profile of spleen B cells from bKlf4−/− and bKlf4+/+ mice stimulated with anti-IgM/CD40 for 48 h were presented. B, More B cells from bKlf4−/− mice remain in G1 phase than those from bKlf4+/+ mice at 48 h after stimulation with anti-IgM/CD40 (p < 0.05, n = 8). Data were represented as mean ± SEM.
Klf4 is required for cyclin D2 induction in B cell activation-induced cell cycle
To understand the mechanisms underlying the defects of G1 to S phase progression, we examined the expression levels of four cell cycle and proliferation-related genes (p21WAF1/Cip1, cyclin D2, cyclin D3, and c-Myc). At the mRNA level, p21WAF1/Cip1 and cyclin D2 were expressed lower in freshly isolated and stimulated B cells from bKlf4−/− mice than from the control mice but cyclin D3 and c-Myc were expressed at similar levels (Fig. 6A). At the protein level, cyclin D2 was lower in B cells at resting and after 24-h stimulation from bKlf4−/− mice than from control mice but was similar after 48-h stimulation between bKlf4−/− and the control mice (Fig. 6B). The protein levels of cyclin D3 and p21 were similar in resting and activated B cells from bKlf4+/+ and bKlf4−/− mice (Fig. 6B). The low expression of cyclin D2 at resting B cells and the slow up-regulation in 24-h stimulated B cells from bKlf4−/− mice may explain the declined proliferation due to the defects of G1- to S-phase transition.
FIGURE 6.
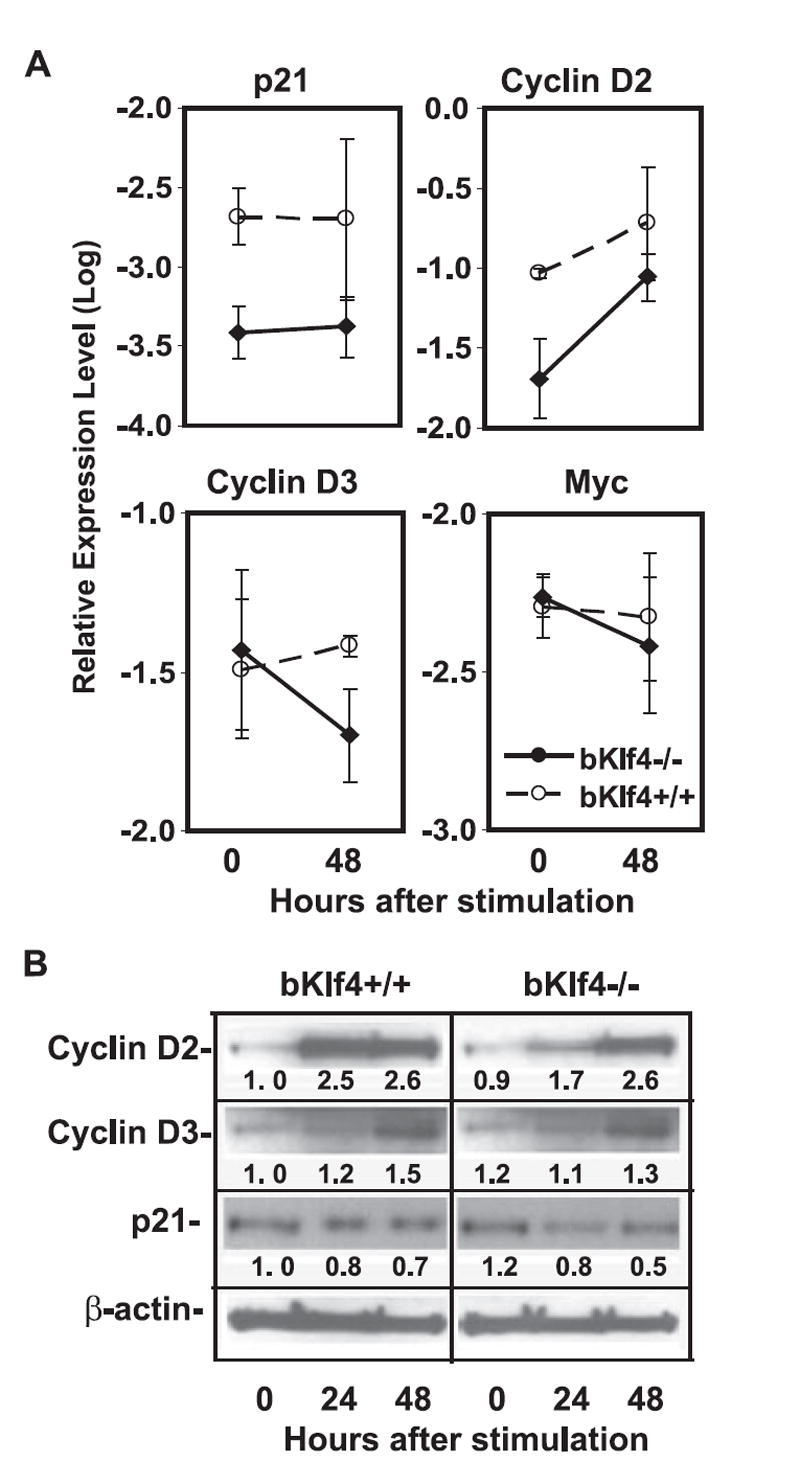
Klf4 regulates cyclin D2 expression. A, Expression of cell cycle and proliferation-related genes. Lower mRNA levels of cyclin D2 and p21 in B cells from bKlf4−/− than from bKlf4+/+ mice (p < 0.05, n = 4) but no significant differences in mRNA levels of cyclin D3 and c-myc in B cells between bKlf4−/− and bKlf4+/+ mice. B, Decreased expression of cyclin D2 protein in B cells from bKlf4−/− mice but no obvious differences of p21 and cyclin D3 proteins between B cells from bKlf4−/− and bKlf4+/+ mice after anti-IgM stimulation. A representative image of one set of samples from five independent experiments was shown. The mean intensity values of each band were presented.
Klf4 directly regulates cyclin D2 expression in B cells
To determine whether Klf4 directly regulates cyclin D2 expression, we applied ChIP with a mAb against Klf4. Based on the consensus Klf4-binding sequence (CACCC) (15), there were three potential sites located at the two regions (−336 to −340 bp and −622 to −626/−631 to −635 bp) (16) (Fig. 7A). Three pairs of primers were designed to cover these two putative-binding sites and one no-binding site as a control. We found that Klf4-binding sites had higher levels of binding (PCR region −233 to −372 bp and −501 to −638 bp) compared with anti-Klf4 Ab and a control Ab. There was no difference in the region containing no Klf4-binding site (region −776 to −922 bp) between anti-Klf4 Ab and control (Fig. 7B). These findings demonstrate that Klf4 directly binds to the promoter of cyclin D2 and regulated its expression.
FIGURE 7.
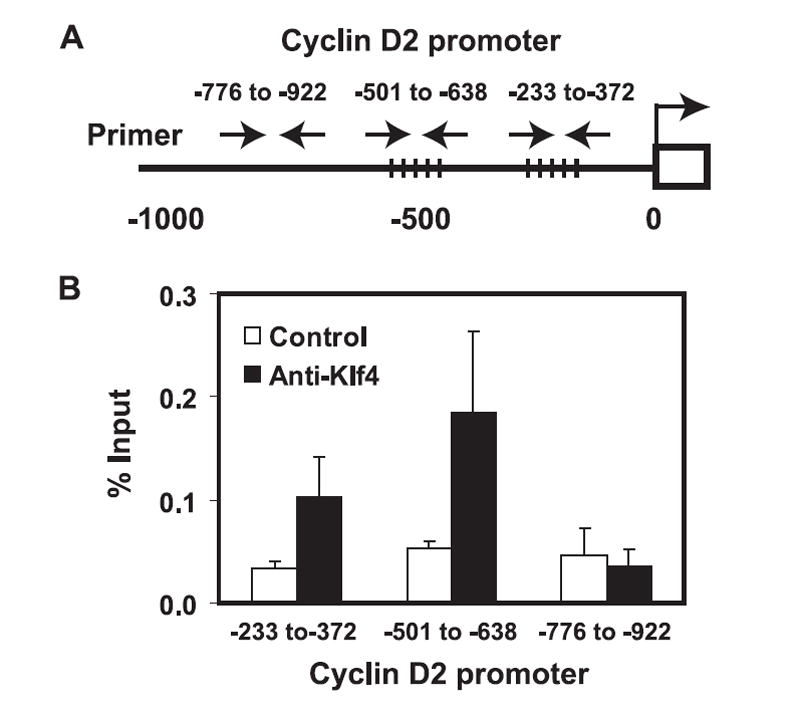
Klf4 directly binds to the promoter of the cyclin D2 gene. A, A diagram of the cyclin D2 gene promoter. The dotted lines cross the promoter region indicate the putative Klf4-binding sites. The arrows indicate the primers used for PCR. B, Klf4 bound specifically to the promoter of cyclin D2. ChIP assay and quantitative PCR were conducted with an anti-Klf4 mAb and an isotype control mAb. Three different regions of the promoter were examined: two putative-binding sites (−336 to −340 and region −626 to −635) showed 2.4- and 2.6-fold higher precipitated DNA by anti-Klf4 Ab than by the control Ab, but no difference in precipitated DNA between two Abs in the region that has no Klf4-putative binding site (−776 to −922). Data were represented as mean ± SEM (n = 3).
Discussion
In this report, we analyzed the function of transcription factor Klf4 in B cells. We found that the levels of Klf4 increased with B cell development and reached peak at resting mature B cells. In the absence of Klf4, we found that a modest but significant decrease in the percentage of pre-B cells in bone marrow and in the number of mature B cells in spleen of bKlf4−/− mice. Furthermore, mature B cells from bKlf4−/− mice had significant slow proliferation in response to BCR stimulations than those from wild-type mice. This proliferation defect was manifested by the partial blockade at the G1 to S cell cycle phase junction, which is in part due to the failure of up-regulation of cyclin D2 in B cells lacking Klf4. Finally, we demonstrated that Klf4 directly bound to the promoter of cyclin D2 and facilitated its expression. Together, these findings demonstrate that Klf4 plays an important role in B cell development and in activation-induced B cell proliferation.
Although the mechanism is not completely understood, the number of B cells in mice is tightly maintained (17). We found a significant reduction of pre-B cells in bone marrow and mature B cells in spleen in the absence of Klf4 but there was no decrease of pro-B cells in bone marrow. As the CD19-cre-mediated deletion occurs during pro-B cell to pre-B cell stages and Klf4 was expressed in relatively low levels in pro-B cells, it remains to be determined whether Klf4 plays any significant role in the pro-B cells. It is interesting to note that cyclin D2 has a role in regulation of B cell numbers (18, 19). In cyclin D2-deficient mice, there was a reduction of B-1 B cells and B cell progenitors but no obvious change in the number of spleen B cells. In this study, we show a decrease of pre-B cells and mature B cells, but not pro-B cells nor B-1 B cells, in bKlf4−/− mice. Although a reduced expression of cyclin D2 was found in resting and 24-h stimulated B cells of bKlf4−/− mice, the decrease of different types of B cells between bKlf4−/− mice and cyclin D2-deficient mice suggests that Klf4 may regulate a broad range of genes including cyclin D2 that may be involved in control of B cell numbers. Interestingly, we did not find a compensatory increase of cyclin D3 in B cells from bKlf4−/− mice or a decrease in proliferation in response to LPS. Together, these findings suggest that Klf4 regulates cyclin D2 but not cyclin D3 through a BCR-mediated activation pathway. Thus, it is necessary to identify additional Klf4 target genes that are involved in regulation of B cell number.
Activation-induced cell proliferation is a multiple-step process that contains many checkpoints. In the absence of Klf4, we found reduced DNA synthesis measured by [3H]thymidine uptake along with an increase in G1-phase cells and a reduction in the number of cells that underwent cell divisions after in vitro stimulation. Together, these findings suggest that Klf4 acts as a positive regulator in BCR-mediated B cell proliferation. Furthermore, the levels of cyclin D2, but not p21, were decreased in the absence of Klf4. Inability of up-regulation of cyclin D2 provides a partial explanation for the reduced cell proliferation in B cells that lack Klf4. Interestingly, overexpression of Klf4 in transformed pro-/pre-B cell lines causes cell cycle arrest of these transformed pro-/pre-B cell lines (11). This suggests that the consequence of Klf4 and its target gene expressions is dependent on the physiological makeup of the cells. As demonstrated in tumor cells, Klf4 can be either a tumor suppressor or an oncogene depending on the cellular conditions or “context” (20). Thus, understanding the availability and function of Klf4 target genes in B cells will help to elucidate the role of Klf4 in regulation of cell cycle and growth of B cells.
Acknowledgments
We thank Drs. Patricia Gearhart for critically reading this manuscript, Julie Segre for help and support of analyzing mice, and Monchou Fann, Lynn Heltemes-Harris, Gary Collins, and Ana Lustig for expert technical assistance. We also thank Robert Wersto, Christa Morris, and Joe Chrest from the National Institute on Aging Flow Cytometry Core Laboratory for cell sorting and cell cycle analysis.
Footnotes
This work was supported by the Intramural Research Program of the National Institute on Aging, National Institutes of Health.
Abbreviations used in this paper: Klf4, Krüppel-like factor 4; PI, proliferation index; ChIP, chromatin immunoprecipitation.
Disclosures The authors have no financial conflict of interest.
References
- 1.Banchereau J, Bazan F, Blanchard D, Briere F, Galizzi JP, Van Kooten C, Liu YJ, Rousset F, Saeland S. The CD40 antigen and its ligand. Annu Rev Immunol. 1994;12:881–922. doi: 10.1146/annurev.iy.12.040194.004313. [DOI] [PubMed] [Google Scholar]
- 2.Piatelli MJ, Tanguay D, Rothstein TL, Chiles TC. Cell cycle control mechanisms in B-1 and B-2 lymphoid subsets. Immunol Res. 2003;27:31–52. doi: 10.1385/IR:27:1:31. [DOI] [PubMed] [Google Scholar]
- 3.Chiles TC. Regulation and function of cyclin D2 in B lymphocyte subsets. J Immunol. 2004;173:2901–2907. doi: 10.4049/jimmunol.173.5.2901. [DOI] [PubMed] [Google Scholar]
- 4.Mohamedali A, Soeiro I, Lea NC, Glassford J, Banerji L, Mufti GJ, Lam EW, Thomas NS. Cyclin D2 controls B cell progenitor numbers. J Leukocyte Biol. 2003;74:1139–1143. doi: 10.1189/jlb.0803363. [DOI] [PubMed] [Google Scholar]
- 5.Solvason N, Wu WW, Parry D, Mahony D, Lam EW, Glassford J, Klaus GG, Sicinski P, Weinberg R, Liu YJ, et al. Cyclin D2 is essential for BCR-mediated proliferation and CD5 B cell development. Int Immunol. 2000;12:631–638. doi: 10.1093/intimm/12.5.631. [DOI] [PubMed] [Google Scholar]
- 6.Dai X, Segre JA. Transcriptional control of epidermal specification and differentiation. Curr Opin Genet Dev. 2004;14:485–491. doi: 10.1016/j.gde.2004.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen X, Johns DC, Geiman DE, Marban E, Dang DT, Hamlin G, Sun R, Yang VW. Kruppel-like factor 4 (gut-enriched Kruppel-like factor) inhibits cell proliferation by blocking G1/S progression of the cell cycle. J Biol Chem. 2001;276:30423–30428. doi: 10.1074/jbc.M101194200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X, Whitney EM, Gao SY, Yang VW. Transcriptional profiling of Kruppel-like factor 4 reveals a function in cell cycle regulation and epithelial differentiation. J Mol Biol. 2003;326:665–677. doi: 10.1016/S0022-2836(02)01449-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rowland BD, Peeper DS. KLF4, p21 and context-dependent opposing forces in cancer. Nat Rev Cancer. 2006;6:11–23. doi: 10.1038/nrc1780. [DOI] [PubMed] [Google Scholar]
- 10.Rowland BD, Bernards R, Peeper DS. The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat Cell Biol. 2005;7:1074–1082. doi: 10.1038/ncb1314. [DOI] [PubMed] [Google Scholar]
- 11.Kharas MG, Yusuf I, Scarfone VM, Yang VW, Segre JA, Huettner CS, Fruman DA. KLF4 suppresses transformation of pre-B cells by ABL oncogenes. Blood. 2007;109:747–755. doi: 10.1182/blood-2006-03-011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katz JP, Perreault N, Goldstein BG, Lee CS, Labosky PA, Yang VW, Kaestner KH. The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development. 2002;129:2619–2628. doi: 10.1242/dev.129.11.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rickert RC, Roes J, Rajewsky K. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 1997;25:1317–1318. doi: 10.1093/nar/25.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fann M, Godlove JM, Catalfamo M, Wood WH, III, Chrest FJ, Chun N, Granger L, Wersto R, Madara K, Becker K, et al. Histone acetylation is associated with differential gene expression in the rapid and robust memory CD8+ T cell response. Blood. 2006;108:3363–3370. doi: 10.1182/blood-2006-02-005520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dang DT, Pevsner J, Yang VW. The biology of the mammalian Kruppel-like family of transcription factors. Int J Biochem Cell Biol. 2000;32:1103–1121. doi: 10.1016/s1357-2725(00)00059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shields JM, Yang VW. Identification of the DNA sequence that interacts with the gut-enriched Kruppel-like factor. Nucleic Acids Res. 1998;26:796–802. doi: 10.1093/nar/26.3.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaudin E, Rosado M, Agenes F, McLean A, Freitas AA. B-cell homeostasis, competition, resources, and positive selection by self-antigens. Immunol Rev. 2004;197:102–115. doi: 10.1111/j.0105-2896.2004.0095.x. [DOI] [PubMed] [Google Scholar]
- 18.Mohamedali A, Soeiro I, Lea NC, Glassford J, Banerji L, Mufti GJ, Lam EW, Thomas NS. Cyclin D2 controls B cell progenitor numbers. J Leukocyte Biol. 2003;74:1139–1143. doi: 10.1189/jlb.0803363. [DOI] [PubMed] [Google Scholar]
- 19.Solvason N, Wu WW, Parry D, Mahony D, Lam EW, Glassford J, Klaus GG, Sicinski P, Weinberg R, Liu YJ, et al. Cyclin D2 is essential for BCR-mediated proliferation and CD5 B cell development. Int Immunol. 2000;12:631–638. doi: 10.1093/intimm/12.5.631. [DOI] [PubMed] [Google Scholar]
- 20.Rowland BD, Bernards R, Peeper DS. The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat Cell Biol. 2005;7:1074–1082. doi: 10.1038/ncb1314. [DOI] [PubMed] [Google Scholar]


