Abstract
Background
Dissemination of antimicrobial resistance genes has become an important public health and biodefense threat. Plasmids are important contributors to the rapid acquisition of antibiotic resistance by pathogenic bacteria.
Principal Findings
The nucleotide sequence of the Klebsiella pneumoniae multiresistance plasmid pMET1 comprises 41,723 bp and includes Tn1331.2, a transposon that carries the bla TEM-1 gene and a perfect duplication of a 3-kbp region including the aac(6′)-Ib, aadA1, and bla OXA-9 genes. The replication region of pMET1 has been identified. Replication is independent of DNA polymerase I, and the replication region is highly related to that of the cryptic Yersinia pestis 91001 plasmid pCRY. The potential partition region has the general organization known as the parFG locus. The self-transmissible pMET1 plasmid includes a type IV secretion system consisting of proteins that make up the mating pair formation complex (Mpf) and the DNA transfer (Dtr) system. The Mpf is highly related to those in the plasmid pCRY, the mobilizable high-pathogenicity island from E. coli ECOR31 (HPIECOR31), which has been proposed to be an integrative conjugative element (ICE) progenitor of high-pathogenicity islands in other Enterobacteriaceae including Yersinia species, and ICEKp1, an ICE found in a K. pneumoniae strain causing primary liver abscess. The Dtr MobB and MobC proteins are highly related to those of pCRY, but the endonuclease is related to that of plasmid pK245 and has no significant homology with the protein of similar function in pCRY. The region upstream of mobB includes the putative oriT and shares 90% identity with the same region in the HPIECOR31.
Conclusions
The comparative analyses of pMET1 with pCRY, HPIECOR31, and ICEKp1 show a very active rate of genetic exchanges between Enterobacteriaceae including Yersinia species, which represents a high public health and biodefense threat due to transfer of multiple resistance genes to pathogenic Yersinia strains.
Introduction
Plasmids are important contributors to the rapid acquisition of antibiotic resistance by pathogenic bacteria through their ability to acquire resistance genetic determinants and to transfer them among bacteria belonging to the same or different genera and species [1], [2]. These resistance genes are usually located inside transposable elements, integrons, or ISCRs (insertion sequence common regions), which facilitate their mobility at the molecular level and at least in the case of transposons could help expanding plasmid's host range [1], [3], [4], [5]. In the past few years, considerable efforts have been made to completely sequence resistance plasmids and understand their biological properties [1].
K. pneumoniae, an important opportunistic pathogen, is the causative agent of community-acquired infections and more frequently of infections in the urinary tract and soft tissue, pneumonia, septicemia, and meningitis in hospitalized patients [6]. Hospital outbreaks caused by multiresistant K. pneumoniae strains have been described throughout the world [7], [8]. It has been proposed that Crohn's disease as well as ankylosing spondylitis are linked to Klebsiella infection possibly through the mechanism of molecular mimicry [9]. K. pneumoniae has also been identified as the causative agent of liver abscess [10]. However, K. pneumoniae infections are especially dangerous in neonatal wards [6]. With the rise in the number of strains harboring resistance genes, usually plasmid-borne, the mortality of K. pneumoniae infections has increased and treatment has become more complicated [11], [12], [13]. A multiresistant K. pneumoniae isolate that caused high mortality in a neonatal ward harbors the self-transmissible plasmid pMET1, which carries the genetic determinants for resistance to several aminoglycosides and β-lactams [12]. In this paper we report the complete sequence and analysis of pMET1, and a comparative study with the Yersinia pestis cryptic plasmid pCRY, the mobilizable high-pathogenicity island found in Escherichia coli strain ECOR31 (HPIECOR31), an integrative conjugative element (ICE) that is a possible progenitor of high-pathogenicity islands in other Enterobacteriaceae including Yersinia species, and an ICE (ICEKp1) present in a clinical K. pneumoniae isolate. Our results indicate that pMET1 and pCRY are closely related and most possibly can be mobilized between Yersinia and other Enterobateriaceae, and be stably maintained in these bacteria. Furthermore the presence of elements of these plasmids in HPIECOR31 and at least another ICE is an indication of a very active genetic exchange between these bacteria, which represents a high public health and biodefense threat due to transfer of multiple resistance genes to pathogenic Yersinia strains.
Results and Discussion
The 41,723 bp multiresistance plasmid pMET1 (GenBank accession number EU383016) includes the replication region, a type 4 secretion system (T4SS), a partition system, and a transposable element that includes two genes coding for aminoglycoside modifying enzymes and two β-lactamases. Table 1 and Fig. 1 summarize the characteristics of pMET1 including the predicted functions of the ORFs on the basis of amino acid sequence comparison.
Table 1. Summary of ORFs found in pMET1.
| ORF‡ | Position | G+C Content (%) | Protein (aa) | Best Blast-X Hit | E value | Identity/similarity | Accession No. |
| 1 | 1,387–674 | 45 | 238 | Putative RepA protein [pCRY, Yersinia pestis biovar Microtus str. 91001] | 2e-119 | 89/95 | NP_995415 |
| 2 | 2,198–1,839 | 42.6 | 118 | Putative transcriptional regulator [pCRY, Yersinia pestis biovar Microtus str. 91001] | 2e-42 | 72/84 | NP_995417 |
| 3 | 2,451–2,906 | 48.7 | 151 | Transcription antiterminator [pCRY, Yersinia pestis biovar Microtus str. 91001] | 2e-83 | 99/100 | NP_995418 |
| 4 | 3,534–4,244 | 57.3 | 236 | Type IV secretory pathway VirB1 component [pCRY, Yersinia pestis biovar Microtus str. 91001] | 2e-132 | 99/99 | NP_995421 |
| 5 | 4,241–4,546 | 56.2 | 101 | Type IV secretory pathway VirB2 component [pCRY, Yersinia pestis biovar Microtus str. 91001] | 9e-31 | 98/99 | NP_995422 |
| 6 | 4,559–7,297 | 52.8 | 912 | Pilx3-4/VirB3-4-like protein [Escherichia coli] | 0.0 | 99/100 | AAP70302 |
| 7 | 7,316–8,026 | 49.2 | 236 | Type IV secretion system VirB5 component [pCRY, Yersinia pestis biovar Microtus str. 91001] | 2e-98 | 84/90 | NP_995424 |
| 8 | 8,275–9,351 | 46.4 | 358 | Type IV secretory pathway VirB6 component [pCRY, Yersinia pestis biovar Microtus str. 91001] | 1e-155 | 87/95 | NP_995426 |
| 9 † | 9443–9580 | 57.4 | 45 | Hypotetical protein[pCRY, Yersinia pestis biovar Microtus str. 91001] | 9e-13 | 91/91 | AE017044.1 |
| 10 | 9,620–10,255 | 55.4 | 211 | Type IV secretion system VirB8 component [pCRY, Yersinia pestis biovar Microtus str. 91001] | 4e-105 | 99/100 | NP_995427 |
| 11 | 10,252–11,160 | 56.9 | 302 | Type IV secretory pathway VirB9 component [pCRY, Yersinia pestis biovar Microtus str. 91001] | 1e-170 | 99/100 | NP_995428 |
| 12 | 11,201–12,475 | 57.4 | 424 | Type IV secretory pathway VirB10 component [pCRY, Yersinia pestis biovarMicrotus str. 91001] | 0.0 | 96/97 | NP_995429 |
| 13 | 12,465–13,490 | 47.6 | 341 | Type IV secretory pathway VirB11 component [pCRY, Yersinia pestis biovar Microtus str. 91001]. | 0.0 | 99/100 | NP_995430 |
| 14 | 13,487–13,885 | 46.5 | 132 | Hypothetical protein YP_pCRY17 [pCRY, Yersinia pestis biovar Microtus str. 91001] | 8e-73 | 98/100 | NP_995431 |
| 15 | 13,921–14,226 | 42.4 | 101 | Hypothetical protein YP_pCRY18 [pCRY, Yersinia pestis biovar Microtus str. 91001] | 3e-52 | 99/99 | NP_995432 |
| 16 | 14,251–14,565 | 40 | 104 | Putative dopa decarboxylase protein remnant [Yersinia pestis biovar Microtus str. 91001] | 8e-41 | 96/97 | NP_995433 |
| 17 | 15,331–17,082 | 39.8 | 583 | Putative mobilization MobB protein [pCRY, Yersinia pestis biovar Microtus str. 91001] | 0.0 | 99/99 | NP_995436 |
| 18 | 17,079 –17,846 | 48.7 | 255 | Putative mobilization protein MobC [pCRY, Yersinia pestis biovar Microtus str. 91001] | 9e-144 | 99/100 | NP_995437 |
| 19 | 17,949–18,749 | 54.2 | 168 | Putative endonuclease [pK245, Klebsiella pneumoniae] | 2e-54 | 62/76 | ABG56794 |
| 20 | 19,631–18,663 | 53.3 | 320 | Antirestriction protein ArdC [p29930, Yersinia enterocolitica] | 3e-152 | 86/92 | CAD58578. |
| 21 | 20,016–20,351 | 33.7 | 112 | Hypotetical protein AmetDRAFT_2972 [Alkaliphilus metalliredigenes QYMF] | 2e-14 | 39/86 | ZP_00799958 |
| 22 | 20,348–20,680 | 31.1 | 111 | Hypothetical protein AmetDRAFT_2972 [Alkaliphilus metalliredigenes QYMF] | 2e-05 | 43/60 | ZP_00799958 |
| 23 | 21,353–21,060 | 45.8 | 99 | Transcriptional regulador [Escherichia coli] | 5e-34 | 72/86 | CAH19146 |
| 24 | 21,706–21,356 | 47.8 | 117 | Hypothetical protein [Escherichia coli] | 1e-43 | 71/84 | CAH19145 |
| 25 | 22,100–22,717 | 45.8 | 205 | Plasmid partitioning protein ParF [p29930, Yersinia enterocolitica] | 3e-84 | 77/90 | CAD58556 |
| 26 | 22,772–23,011 | 47.3 | 79 | Partitioning protein ParG[p29930, Yersinia enterocolitica] | 9e-16 | 53/70 | CAD58557 |
| 27 | 24,886–23,630 | 46.8 | 417 | Hypothetical protein V12B01_09786 [Vibrio splendidus 12B01] | 6e-134 | 59/74 | ZP_00991171 |
| 28 | 26,905–24,890 | 46.6 | 675 | Hypothetical protein V12B01_09791 [Vibrio splendidus 12B01] | 0.0 | 73/84 | ZP_00991172 |
| 29 | 29,632–26,948 | 45.4 | 892 | Hypothetical protein V12B01_09796 [Vibrio splendidus 12B01] | 0.0 | 67/80 | ZP_00991173 |
| 30 | 30,731–29,871 | 50.3 | 286 | Beta-lactamase precursor (TEM-1)* [pJHCMW1, Tn1331, Klebsiella pneumoniae] | 2e-152 | 100/100 | NP_608310 |
| 31 | 32,270–31,431 | 50.3 | 279 | Beta-lactamase precursor * (Penicillinase) (Oxacillinase) (OXA-9) | 1e-157 | 100/100 | NP_608309 |
| 32 | 33,103–32,315 | 53.6 | 262 | Adenylyltranferase [pJHCMW1, Tn1331, Klebsiella pneumoniae]* | 6e-145 | 99/100 | NP_608308 |
| 33 | 33,778–33,173 | 54.1 | 201 | Aminoglycoside 6′-N-acetyltransferase type Ib (AAC(6′)-Ib) * [pJHCMW1, Tn1331, Klebsiella pneumoniae] | 1e-112 | 100/100 | NP_608307 |
| 34 | 35,317–34,478 | 50.3 | 279 | Beta-lactamase precursor * (Oxacillinase, OXA-9) [pJHCMW1, Tn1331, Klebsiella pneumoniae] | 1e-157 | 100/100 | NP_608309 |
| 35 | 36,150–35,362 | 53.6 | 262 | Adenylyltranferase [pJHCMW1, Tn1331, Klebsiella pneumoniae]* | 6e-145 | 99/100 | NP_608308 |
| 36 | 36,825–36,220 | 54.1 | 201 | Aminoglycoside 6′-N-acetyltransferase type Ib (AAC(6′)-Ib) * [pJHCMW1, Tn1331, Klebsiella pneumoniae] | 1e-112 | 100/100 | NP_608307 |
| 37 | 37,565–37,008 | 54.3 | 185 | Resolvase [pJHCMW1, Tn1331, Klebsiella pneumoniae] * | 2e-95 | 99/100 | NP_608306 |
| 38 | 37,694–40,732 | 51.8 | 1015 | Transposase [pJHCMW1, Tn1331, Klebsiella pneumoniae] * | 0.0 | 100/100 | NP_608305 |
| 39 | 41,223–40,942 | 52.7 | 93 | Predicted transcriptional regulator [pCRY, Yersinia pestis biovar Microtus str. 91001] | 7e-37 | 100/100 | NP_995443 |
| 40 | 41,157–41,516 | 52.1 | 120 | Hypothetical protein YP_pCRY30 [pCRY, Yersinia pestis biovar Microtus str. 91001] | 1e-50 | 97/98 | NP_995444 |
| 41 | 41,545–41,204 | 53 | 107 | Hypothetical protein plu0442 [Photorhabdus luminescens subsp.laumondii TTO1] | 2e-27 | 60/73 | NP_927795 |
Best hit selected according to genetic coherence rather than a minimal difference in score.
Done with tblastn
Including stop codon
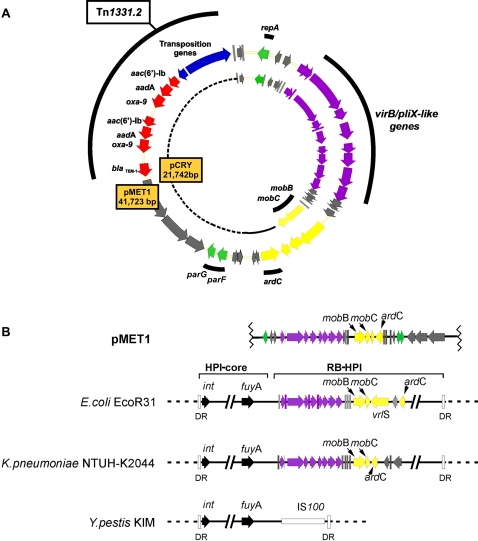
Figure 1. Genetic map of pMET1 and comparison to plasmid pCRY and chromosomal elements.
A. The genetic maps of pMET1 and pCRY are compared showing the homologous regions. The arrows indicate genes locations and orientation. Genes with different functions are shown with different colors and if the genes in the different structures shown are homologus they are represented with the same colors. Yellow: mobilization; green: replication and partition; red: antibiotic resistance; purple: virB/pilX-like; blue: transposition; grey: unknown. Since pCRY is smaller than pMET1, to represent it in circular form a dotted line was added to fill the gap. Solid line represents non-homologous DNA. B. Comparison of a pMET1 region with chromosomal HPIs or ICEs is shown using a linearized version of the plasmid. The HPIs shown are those from E. coli ECOR31 (HPIECOR31) [43], K. pneumoniae NTUH-K2044 (ICEKp1) [44], and Y. pestis KIM (HPIYp)[43]. The diagram shows the HP core regions, which are not at scale and are represented as in [43], and the RB-HPIs. The sequence described in this manuscript has been deposited in GenBank, accession number is EU383016.
The transposable element Tn1331.2
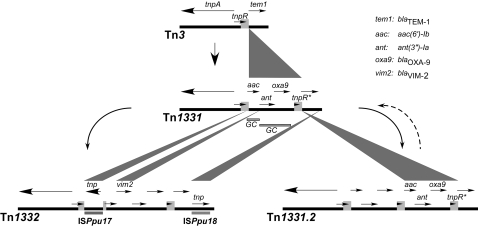
The plasmid pMET1 includes an 11,042-bp transposon, named Tn1331.2 [12] that is highly related to Tn1331 (see Fig. 2) [14], [15]. Tn1331 can be considered an evolutionary product of Tn3 by the addition of a 3,047-bp DNA region, which has the structure of the variable portion of the integrons, flanked by 520-bp direct repeats (Fig. 2). This region includes three resistance genes with a structure consisting of aac(6′)-Ib-attC-aadA1-bla OXA-9 -attC, potentially defining two gene cassettes including aac(6′)-Ib-attC and aadA1-bla OXA-9 -attC [14], [16], [17]. As a result of the presence of these three genes and bla TEM, cells harboring Tn1331 are resistant to several aminoglycosides and β-lactams. Possible mechanisms for the generation of Tn1331 have been discussed previously [16], [18]. Tn1331.2 has a perfect duplication of the 3,047-bp DNA region and as a consequence a 520-bp including a C-terminal fragment of tnpR is found as three direct repeats flanking the potential gene cassettes (Fig. 2). As it is the case for Tn1331 [14], the Tn1331.2 tnpA differs from the Tn3 version of the gene in the 9-nucleotide repeats that in Tn3 code for the amino acid sequence GFHGFH. Tn1331.2 includes only one copy of this 9-bp fragment coding for the amino acid sequence GFH.
Figure 2. Genetic organization of transposons Tn3, Tn1331, Tn1332, and Tn1331.2.
The horizontal arrows indicate the location and direction of transcription of the genes. The genes are named only once for the sake of clarity. The light gray blocks show the 520-bp region direct repeats. Note that in Tn1332 one of the direct repeats is interrupted by an insertion of ISPpu17. The upper vertices of the dark gray triangles show points of insertion of DNA fragments that generate a new transposable element. The length of the base of each triangle defines the sizes of such DNA insertions. The curved dashed arrow to the right indicates that the transition Tn1331.2 to Tn1331 by deletion of one of the 3,047-bp occurred in the laboratory but it has not been confirmed to occur in nature. Stippled bar below Tn1331 indicates potential gene cassettes (GC).
The duplication of the 3,047-bp DNA fragment including the resistance genes could be a strategy to increase the gene dosage when the transposon is present in a low copy number plasmid. Mechanisms of gene duplication when they are flanked by direct repeats have been described before [19], [20], [21]. Although we have observed a transition from Tn1331.2 to Tn1331 by the loss of one of the 3,047-bp repeats in the laboratory, we do not know if this is a process that happens in nature (dashed arrow, Fig. 2). While for many years no evolutionary derivatives of Tn1331 other than the duplication to render Tn1331.2 had been isolated, there was a recent report of such a phenomenon. The transposon Tn1332, isolated from Pseudomonas putida, can be considered as Tn1331 with the insertion of three DNA fragments that include the metallo-ß-lactamase gene bla VIM-2, and the insertion sequences ISPpu17 and ISPpu18, respectively (Fig. 2) [22].
Replication and partition
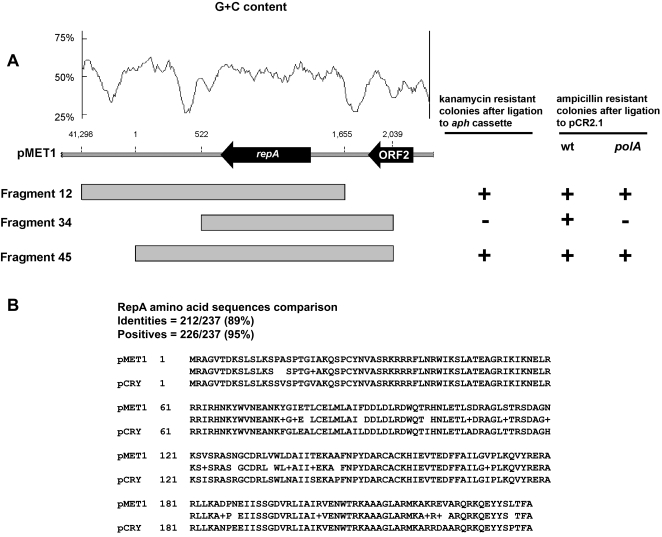
Blastn analysis revealed a pMET1 region of high homology to the Y. pestis 91001 plasmid pCRY putative replication region [23] (schematically shown in Fig. 1A). The characteristics of this region include two AT-rich DNA segments flanking ORF1, named RepA, whose amino acid sequence showed 89% identity and 95% similarity to the pCRY RepA putative protein (Fig. 3A and B). Lower but still significant similarities were found to other replication proteins from plasmids isolated from Buchnera aphidicola [24], [25] and Y. enterocolitica plasmid pYVe227 [26]. To functionally identify the replication region of pMET1, three fragments 12, 34, and 45 (coordinates 41298-1655, 522-2039, and 1-2039 respectively)(Fig. 3A) were ligated to the kanamycin resistance fragment and used to transform E. coli TOP10. Kanamycin resistant colonies were obtained only when the recombinant clones included the fragments 12 or 45 (Fig. 3A). These results were confirmed by cloning all three fragments using pCR2.1 as vector, and attempting to transform the DNA polymerase I deficient E. coli C2110. While all three recombinant plasmids replicated in a wild-type E. coli strain, only recombinant clones including fragment 12 or 45 could be stably maintained in E. coli C2110. The recombinant clone harboring the fragment 34 was unable to replicate in the absence of DNA polymerase I (Fig. 3A). Therefore the results of both experiments defined a replication region encompassing nucleotides 1 to 1655, which includes two essential features: one complete ORF, named repA, and the AT-rich sequence between nucleotides 1 and 522 (Fig. 3A). This latter region likely includes the ori locus. These two features are conserved in the pCRY replication region: the AT rich sequence is conserved as the G+C content profile is almost identical to pMET1, and repA shows 95% similarity and 89% identity to pMET1 RepA when translated (Fig. 3B). On the other hand, the overall identity at the nucleotide level between pMET1 and pCRY for this locus is 89%. This high relatedness between the pMET1 and pCRY replication regions strongly suggests that pMET1 can be replicated in Yersinia species.
Figure 3. Replication region of pMET1.
A. The bar shows a genetic map of the pMET1 replication region and the GC content plot generated using a window size of 100 bp on top. Recombinant clones were obtained by inserting the indicated fragments into pCR2.1 or ligated to the pUC4K aph cassette. The ability to be maintained in E. coli C2110 (a polA mutant) of the recombinant plasmids made using pCR2.1 as vector is indicated to the right by a + or − sign. The ability to generate kanamycin resistant colonies in E. coli TOP10 of the indicated fragments when ligated to the aph cassette from pUC4K is also represented by a + or − sign. B. BLASTP comparison of the amino acid sequences of the putative RepA proteins from pMET1 and pCRY.
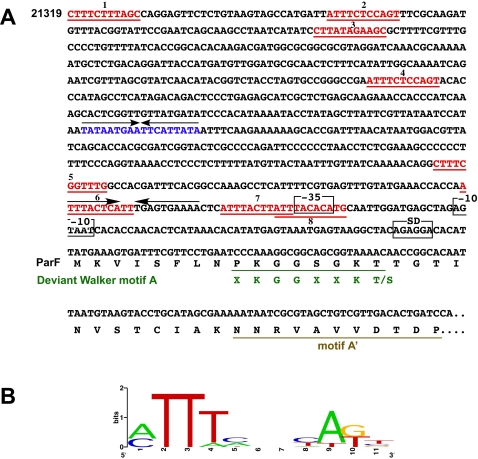
Plasmid pMET1 includes a putative partition region that has the general organization of that of plasmid pTP228, also known as the parFG locus [27]. The pMET1 partition region encompasses nucleotides 22,100 to 23,011 and includes two ORFs (ORF25 and ORF26, Table 1) encoding two putative proteins with strong homologies to ParF and ParG. The ParF homolog is a 205-amino acids protein that has shown significant homology with numerous proteins that belong to the type Ib subgroup of the ParA superfamily that is related to the MinD subgroup of cell division proteins [28], [29], [30]. These proteins are Walker-type ATPases that include a deviant motif A, which contains a conserved ‘signature’ lysine (XKGGXXKT) [29], [31], [32]. This motif as well as motif A' [29] are shown in Fig. 4A. The maximum pMET1 ParF identities and similarities were found with those encoded by the cryptic Y. enterocolitica plasmid p29930 [33] (90% similarity and 77% identity), the multidrug resistance plasmid pTP228 from Salmonella enterica subsp. Enterica serovar Newport [27] (87% similarity and 72% identity), and the Erwinia amylovora Ea88 plasmid pEA29 [34] (85% similarity and 73% identity).
Figure 4. Genetic structures located upstream of parF and parG.
A. The direct repeats within the pMET1 putative parH-like locus are shown in red. The diagram also shows the −35 and −10 sequences, as well as the inverted repeats (arrows). The inverted repeat within the putative parH locus is shown in blue. The beginning of the ParF amino acid sequence including the deviant Walker motif A and motif A' are shown. B. Logo plot [60], [61] of a multiple alignment of the direct repeats shown in red.
Unlike with ParF, the pMET1 ParG has significant homologues only in p29930 (70% similarity and 53% identity), pTP228 (71% similarity and 58% identity), and pEA29 (68% similarity and 51% identity), generally known as ParB-like proteins. Studies on the pTP228 ParG protein showed that it acts as a dimer, may interact in a sequence-specific manner with a direct repeats-containing DNA region located upstream of parF known as parH, stimulates ATP hydrolysis by ParF, and modulates polymerization of ParF [35]. In addition, the ParG dimer binds an operator and acts as an autoregulator of the parFG operon [31]. Structures of parB-like proteins alone or in complex with DNA have been determined [36], [37], [38]. Other plasmids that have ParF homologues include proteins that are functional analogs but heterologs to ParG. The nucleoprotein complex has been recently called “segrosome” and is essential to ensure proper partition of the plasmid molecules [31]. Analysis of the pMET1 region upstream of the parF and parG genes showed an organization similar to that found in pTP228, which includes the parH locus and a putative operator (Fig. 4A)[31], [39]. Interestingly, although the ParG proteins from pTP228 and pMET1 share homology, the repeats found upstream of parF in pMET1 are unrelated to those in pTP228. This region includes eight imperfect direct repeats that stretch over the promoter region. A graphical representation (sequence logo) of the multiple alignment of these repeats is shown in Fig. 4B. A region that includes some or all of these direct repeats could play the role of parH in pMET1 partition. Fig. 4A also shows the putative promoter, the −35 sequence is located within two partially overlapping direct repeats and upstream of this structure the repeat number 6 is part of an inverted repeat. One or more of these structures could function as operators, binding of ParG to one or both of these two structures may result in inhibition of transcription of parF and parG. Within the putative parH region there is also a perfect inverted repeat (Fig. 4A) unrelated to the putative ParG binding sites. The function of this structure is unknown.
Conjugation
Type 4 secretion systems (T4SSs) encoded by plasmids are involved in bacterial conjugation and transport of effectors to eukaryotic cells during infection [40]. Bacterial conjugation plays an important role in dissemination of antibiotic resistance genes at the cellular level [41], [42]. Previous studies indicated that pMET1 is self-transmissible [12], analysis of its nucleotide sequence shows that the transfer region of pMET1 lies within a ca. 16-kbp DNA segment, the ATG of the first gene, virB1, occurs at coordinate 3534 and the ATG of the last gene, ardC (which is transcribed opposite to all the rest) at coordinate 19,631. A block of predicted coding products (named VirB1–VirB11) are highly homologous to proteins that make up the mating pair formation complex (Mpf) in a large number of plasmids and other DNA elements with the highest identity with those in the Y. pestis 91001 plasmid pCRY [23], the high pathogenicity island HPIECOR31 [43], and ICEKp1, an ICE found in the primary liver abscess causing K. pneumoniae NTUH-K2044 [44] (see Fig 1B, Table 1, and Table 2). The putative functions of these proteins are discussed in several reviews [40], [42], [45]. In all four cases the virB3 and virB4 genes are fused. An homolog of published VirB7 proteins is absent in all four strucutures. However, a 45-amino acids hypothetical protein (ORF9) is present between virB6 and virB8 in all four elements. This high conservation and the synteny suggest that this ORF might play a role equivalent to that of VirB7. The pMET1 vir-like genes are followed by ORFs of unknown function and the predicted DNA transfer (Dtr) group of genes that potentially code for MobB, MobC, and an endonuclease. While the pMET1 MobB and MobC showed 99% identity to those in the Y. pestis plasmid pCRY, the endonuclease showed highest identity with that of the K. pneumoniae plasmid pK245, which carries genes coding for resistance to quinolones and extended-spectrum β-lactamases [46], and the cryptic Y. enterocolitica plasmid p29930 [33]. Interestingly, no significant homology was found with the nuclease encoded by pCRY. Further analysis of the pMET1 mobilization region showed a structure oriT-mobB-mobC, similar to those in other elements such as the mobilizable HPIECOR31 [43] or the plasmid CloDF13 [47]. The pMET1 region upstream of mobB has 90% identity with that of the HPIECOR31. As it is the case in this element, there are two nic (or oriT) sites in pMET1 due to their location inside inverted repeat 2 (IR2). It is then possible that either strand of pMET1 can be transferred to the recipient cell, an unusual phenomenon [48]. Furthermore, studies on the plasmid R1162, which also possesses two oriTs, suggested that transfer can be initiated at one of them and terminated at the other with the inverted repeat playing a role in termination [49].
Table 2. Comparison of amino acid sequences of VirB/PliX-like proteins.
| pCRY AE017044.1 | ICEECOR31 AY233333.1 | ICEKp1 AB298504.1 | |
| VirB1 | 99/99 | 63/77 | 63/77 |
| VirB2 | 98/99 | 76/85 | 76/85 |
| VirB3-4-like | 97/98 | 99/100 | 98/99 |
| VirB5 | 84/90 | 83/98 | 84/89 |
| VirB6 | 87/95 | 87/92 | 95/96 |
| ORF9 | 97/97 | 100/100 | 97/100 |
| VirB8 | 99/100 | 98/100 | 99/99 |
| VirB9 | 98/100 | 99/99 | 98/99 |
| VirB10 | 96/97 | 95/96 | 95/95 |
| VirB11 | 99/100 | 99/100 | 97/99 |
Values indicate identity/similarity obtained by blastx comparison of each pMET1 ORF and the corresponding hypothetical protein from pCRY, ICEECOR31 and ICEKp1.
Downstream of the endonuclease gene, there is an ORF that is transcribed in the opposite direction to all the genes described above with homology to the p29930 ardC [33], which codes for a putative antirestriction protein. The pSA ArdC protein has been studied in some detail. It has been shown to bind ssDNA and to protect single-stranded but not double-stranded plasmid DNA in vitro against the activity of HhaI, an enzyme that cleaves both single- and double-stranded DNA. It was proposed that ArdC is transported to the recipient cell with the transferred single-stranded DNA and protects the incoming single-stranded DNA from the host endonucleases [50].
Comparative analysis of pMET1 and the Y. pestis pCRY plasmid
pCRY is a 21,742-bp cryptic plasmid recently isolated from Y. pestis strain 91001 [23]. Comparative analysis of pMET1 and pCRY DNA sequences revealed highly similar backbones (see Fig. 1A). These analyses showed that the ca. 18-kb fragment including the replication region and the T4SS involved in conjugation of pMET1 shares high sequence homology as well as gene synteny with that of pCRY. These results suggest that they have a common ancestor and it would not be surprising if other plasmids with the same backbone but carrying other mobile genetic elements including resistance genes were found in other Enterobacteriaceae isolates. In the particular case of Y. pestis, our findings, taken together with the recent description of other multiresistance plasmids of Yersinia that share their backbone with plasmids of Salmonella [51], and the finding of a common ancestry between the Y. pestis pMT1/pFra plasmid and a cryptic plasmid from a clinical isolate of S. enterica serovar Typhi [52] indicate that there must be an active transfer of plasmids between Yersinia and other Enterobacteriaceae by direct contact or through other intermediate bacteria resulting in a high risk of dissemination of antibiotic resistance genes to pathogenic Y. pestis strains.
Comparative analysis of pMET1 and the integrative and conjugative element of E. coli ECOR31
The pMET1 T4SS is related to those found in the ICEKp1 and HPIECOR31 (Fig. 1B). A study on HPIECOR31 showed that it shares common structures, such as the genes for synthesis of the siderophore yersiniabactin, with other high-pathogenicity islands found in Enterobacteriaceae including Yersinia species; but differs in the presence of a DNA region within an end of the element, known as RB-HPIECOR31 (right border-high pathogenicity island ECOR31) (see Fig. 1B, ICEs from E. coli ECOR31 and Y. pestis KIM) [43]. This region includes a pMET1-related T4SS, which made HPIECOR31 the first pathogenicity island to be found harboring a complete set of conjugative plasmid genes [43]. Furthermore, the complete HPIECOR31 can be excised from the chromosome, circularized, and transferred indicating that it is an integrative conjugative element (ICE) [43], [53]. These properties taken together with the structure of high-pathogenicity islands found in Yersinia species and other Enterobacteriaceae led to the suggestion that HPIECOR31 is their progenitor [43]. It is possible that this is also the origin of the T4SS found in pCRY and pMET1, or conversely it has been captured from a plasmid related to pCRY or pMET1. These findings demonstrate the high rate of genetic exchanges that occurs between Yersinia species and other Enterobacteriaceae. These considerations also speak in favor of a high risk of acquisition of antibiotic resistance or virulence traits by pathogenic Yersinia species.
Materials and Methods
Bacterial strains and plasmids
Plasmid pMET1 was originally isolated from K. pneumoniae FC1, a strain isolated in a pediatric unit in Mendoza, Argentina (Hospital Luis C. Lagomaggiore) [12]. Plasmid pMET1 DNA was introduced into Escherichia coli XL1Blue (recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacI q ZM15 Tn10]) (Stratagene) by transformation for purification and further manipulations. Recombinant clones including fragments of pMET1 were hosted in E. coli XL1Blue or E. coli TOP10 F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (StrR) endA1 nupG (Invitrogen). E. coli C2110 polA gyrA Rifr [54] was used as host to isolate the pMET1 replication region. Recombinant clones were generated using pUC18 [55], pCR2.1 (Invitrogen), or pSMART (Lucigen) as a cloning vectors. Plasmid pUC4K [56] was the source of the kanamycin resistance fragment (aph gene).
Bacterial growth medium and general procedures
Growth of bacteria was in Lennox L broth (1% tryptone, 0.5% yeast extract, 0.5% NaCl). Plasmid DNA was prepared using the Qiagen plasmid mini kit (Qiagen, Inc.). Recombinant clones were sequenced using BigDye (ABI) and DYEnamic ET (Amersham) chemistries on an ABI Prism (model 310 or 3100) instrument and the appropriate commercial or custom-designed primers. The complete circular nucleotide sequence of pMET1 has been deposited in the GenBank sequence library and assigned the accession number EU383016. Sequences were examined and assembled with Sequencher 4.7 software (Gene Codes Corp.). DNA and protein sequence analyses were performed using the CLUSTAL W program (Pôle Bio-Informatique Lyonnais server [http://pbil.ibcp.fr/cgi-bin/npsa_automat.plpagenpsa_clustalw.html]) [57], the Promoter prediction code (http://www.fruitfly.org/seq_tools/promoter.html) [58], the Basic local alignment search tool (BLAST) (http://www.ncbi.nlm.nih.gov/BLAST/) [59], and Artemis (http://www.sanger.ac.uk/Software/Artemis/). The sequence logo was generated using the Weblogo web based application (http://weblogo.berkeley.edu/) [60], [61].
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was supported by Public Health Service grant 2R15AI047115 (to M.E.T.) from the National Institutes of Health and National Science Foundation grant 0420479 (to L.A.A.). J.N. and D.B. were supported in part by LA Basin Minority Health and Health Disparities International Research Training Program (MHIRT) 5T37MD001368-09 (National Center on Minority Health and Health Disparities). T.T. was the recipient of a Howell-CSUPERB Scholar Award.
References
- 1.Taylor D, Gibreel A, Lawley T, Tracz D. Antibiotic resistance plasmids. In: Funnell B, Phillips G, editors. Plasmid biology. Washington DC: ASM Press; 2004. pp. 473–491. [Google Scholar]
- 2.Cohen SN. Bacterial plasmids: their extraordinary contribution to molecular genetics. Gene. 1993;135:67–76. doi: 10.1016/0378-1119(93)90050-d. [DOI] [PubMed] [Google Scholar]
- 3.Bui D, Ramiscal J, Trigueros S, Newmark JS, Do A, et al. Differences in resolution of mwr-containing plasmid dimers mediated by the Klebsiella pneumoniae and Escherichia coli XerC recombinases: potential implications in dissemination of antibiotic resistance genes. J Bacteriol. 2006;188:2812–2820. doi: 10.1128/JB.188.8.2812-2820.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fluit AC, Schmitz FJ. Resistance integrons and super-integrons. Clin Microbiol Infect. 2004;10:272–288. doi: 10.1111/j.1198-743X.2004.00858.x. [DOI] [PubMed] [Google Scholar]
- 5.Walsh TR. Combinatorial genetic evolution of multiresistance. Curr Opin Microbiol. 2006;9:476–482. doi: 10.1016/j.mib.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 6.Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 1998;11:589–603. doi: 10.1128/cmr.11.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reish O, Ashkenazi S, Naor N, Samra Z, Merlob P. An outbreak of multiresistant Klebsiella in a neonatal intensive care unit. J Hosp Infect. 1993;25:287–291. doi: 10.1016/0195-6701(93)90115-g. [DOI] [PubMed] [Google Scholar]
- 8.van't Veen A, van der Zee A, Nelson J, Speelberg B, Kluytmans JA, et al. Outbreak of infection with a multiresistant Klebsiella pneumoniae strain associated with contaminated roll boards in operating rooms. J Clin Microbiol. 2005;43:4961–4967. doi: 10.1128/JCM.43.10.4961-4967.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ebringer A, Rashid T, Tiwana H, Wilson C. A possible link between Crohn's disease and ankylosing spondylitis via Klebsiella infections. Clin Rheumatol. 2007;26:289–297. doi: 10.1007/s10067-006-0391-2. [DOI] [PubMed] [Google Scholar]
- 10.Lederman ER, Crum NF. Pyogenic liver abscess with a focus on Klebsiella pneumoniae as a primary pathogen: an emerging disease with unique clinical characteristics. Am J Gastroenterol. 2005;100:322–331. doi: 10.1111/j.1572-0241.2005.40310.x. [DOI] [PubMed] [Google Scholar]
- 11.Gupta A. Hospital-acquired infections in the neonatal intensive care unit–Klebsiella pneumoniae. Semin Perinatol. 2002;26:340–345. doi: 10.1053/sper.2002.36267. [DOI] [PubMed] [Google Scholar]
- 12.Tolmasky ME, Chamorro RM, Crosa JH, Marini PM. Transposon-mediated amikacin resistance in Klebsiella pneumoniae. Antimicrob Agents Chemother. 1988;32:1416–1420. doi: 10.1128/aac.32.9.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woloj M, Tolmasky ME, Roberts MC, Crosa JH. Plasmid-encoded amikacin resistance in multiresistant strains of Klebsiella pneumoniae isolated from neonates with meningitis. Antimicrob Agents Chemother. 1986;29:315–319. doi: 10.1128/aac.29.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sarno R, McGillivary G, Sherratt DJ, Actis LA, Tolmasky ME. Complete nucleotide sequence of Klebsiella pneumoniae multiresistance plasmid pJHCMW1. Antimicrob Agents Chemother. 2002;46:3422–3427. doi: 10.1128/AAC.46.11.3422-3427.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tolmasky ME, Crosa JH. Tn1331, a novel multiresistance transposon encoding resistance to amikacin and ampicillin in Klebsiella pneumoniae. Antimicrob Agents Chemother. 1987;31:1955–1960. doi: 10.1128/aac.31.12.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tolmasky ME. Sequencing and expression of aadA, bla, and tnpR from the multiresistance transposon Tn1331. Plasmid. 1990;24:218–226. doi: 10.1016/0147-619x(90)90005-w. [DOI] [PubMed] [Google Scholar]
- 17.Tolmasky ME, Crosa JH. Genetic organization of antibiotic resistance genes (aac(6′)-Ib, aadA, and oxa9) in the multiresistance transposon Tn1331. Plasmid. 1993;29:31–40. doi: 10.1006/plas.1993.1004. [DOI] [PubMed] [Google Scholar]
- 18.Tolmasky ME. Aminoglycoside-modifying enzymes: characteristics, localization, and dissemination. In: Bonomo R, Tolmasky ME, editors. Enzyme-mediated resistance to antibiotics: mechanisms, dissemination, and prospects for inhibition. Washington, D.C.: ASM Press; 2007. pp. 35–52. [Google Scholar]
- 19.Peterson BC, Rownd RH. Homologous sequences other than insertion elements can serve as recombination sites in plasmid drug resistance gene amplification. J Bacteriol. 1983;156:177–185. doi: 10.1128/jb.156.1.177-185.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peterson BC, Rownd RH. Recombination sites in plasmid drug resistance gene amplification. J Bacteriol. 1985;164:1359–1361. doi: 10.1128/jb.164.3.1359-1361.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peterson BC, Rownd RH. Drug resistance gene amplification of plasmid NR1 derivatives with various amounts of resistance determinant DNA. J Bacteriol. 1985;161:1042–1048. doi: 10.1128/jb.161.3.1042-1048.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poirel L, Cabanne L, Collet L, Nordmann P. Class II transposon-borne structure harboring metallo-beta-lactamase gene blaVIM-2 in Pseudomonas putida. Antimicrob Agents Chemother. 2006;50:2889–2891. doi: 10.1128/AAC.00398-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song Y, Tong Z, Wang J, Wang L, Guo Z, et al. Complete genome sequence of Yersinia pestis strain 91001, an isolate avirulent to humans. DNA Res. 2004;11:179–197. doi: 10.1093/dnares/11.3.179. [DOI] [PubMed] [Google Scholar]
- 24.van Ham RC, Moya A, Latorre A. Putative evolutionary origin of plasmids carrying the genes involved in leucine biosynthesis in Buchnera aphidicola (endosymbiont of aphids). J Bacteriol. 1997;179:4768–4877. doi: 10.1128/jb.179.15.4768-4777.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perez-Brocal V, Gil R, Ramos S, Lamelas A, Postigo M, et al. A small microbial genome: the end of a long symbiotic relationship? Science. 2006;314:312–313. doi: 10.1126/science.1130441. [DOI] [PubMed] [Google Scholar]
- 26.Iriarte M, Lambermont I, Kerbourch C, Cornelis G. Yersinia enterocolitica plasmid pYVe227, complete sequence. GenBank NC_002120 2006 [Google Scholar]
- 27.Barilla D, Hayes F. Architecture of the ParF-ParG protein complex involved in prokaryotic DNA segregation. Mol Microbiol. 2003;49:487–499. doi: 10.1046/j.1365-2958.2003.03564.x. [DOI] [PubMed] [Google Scholar]
- 28.Moller-Jensen J, Jensen RB, Gerdes K. Plasmid and chromosome segregation in prokaryotes. Trends Microbiol. 2000;8:313–320. doi: 10.1016/s0966-842x(00)01787-x. [DOI] [PubMed] [Google Scholar]
- 29.Bignell C, Thomas CM. The bacterial ParA-ParB partitioning proteins. J Biotechnol. 2001;91:1–34. doi: 10.1016/s0168-1656(01)00293-0. [DOI] [PubMed] [Google Scholar]
- 30.Hayes F. The partition system of multidrug resistance plasmid TP228 includes a novel protein that epitomizes an evolutionarily distinct subgroup of the ParA superfamily. Mol Microbiol. 2000;37:528–541. doi: 10.1046/j.1365-2958.2000.02030.x. [DOI] [PubMed] [Google Scholar]
- 31.Hayes F, Barilla D. The bacterial segrosome: a dynamic nucleoprotein machine for DNA trafficking and segregation. Nat Rev Microbiol. 2006;4:133–143. doi: 10.1038/nrmicro1342. [DOI] [PubMed] [Google Scholar]
- 32.Lutkenhaus J, Sundaramoorthy M. MinD and role of the deviant Walker A motif, dimerization and membrane binding in oscillation. Mol Microbiol. 2003;48:295–303. doi: 10.1046/j.1365-2958.2003.03427.x. [DOI] [PubMed] [Google Scholar]
- 33.Strauch E, Goelz G, Knabner D, Konietzny A, Lanka E, et al. A cryptic plasmid of Yersinia enterocolitica encodes a conjugative transfer system related to the regions of CloDF13 Mob and IncX Pil. Microbiology. 2003;149:2829–2845. doi: 10.1099/mic.0.26418-0. [DOI] [PubMed] [Google Scholar]
- 34.McGhee GC, Jones AL. Complete nucleotide sequence of ubiquitous plasmid pEA29 from Erwinia amylovora strain Ea88: gene organization and intraspecies variation. Appl Environ Microbiol. 2000;66:4897–4907. doi: 10.1128/aem.66.11.4897-4907.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barilla D, Rosenberg MF, Nobbmann U, Hayes F. Bacterial DNA segregation dynamics mediated by the polymerizing protein ParF. Embo J. 2005;24:1453–1464. doi: 10.1038/sj.emboj.7600619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weihofen WA, Cicek A, Pratto F, Alonso JC, Saenger W. Structures of omega repressors bound to direct and inverted DNA repeats explain modulation of transcription. Nucleic Acids Res. 2006;34:1450–1458. doi: 10.1093/nar/gkl015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schumacher MA, Glover TC, Brzoska AJ, Jensen SO, Dunham TD, et al. Segrosome structure revealed by a complex of ParR with centromere DNA. Nature. 2007;450:1268–1271. doi: 10.1038/nature06392. [DOI] [PubMed] [Google Scholar]
- 38.Golovanov AP, Barilla D, Golovanova M, Hayes F, Lian LY. ParG, a protein required for active partition of bacterial plasmids, has a dimeric ribbon-helix-helix structure. Mol Microbiol. 2003;50:1141–1153. doi: 10.1046/j.1365-2958.2003.03750.x. [DOI] [PubMed] [Google Scholar]
- 39.Barilla D, Carmelo E, Hayes F. The tail of the ParG DNA segregation protein remodels ParF polymers and enhances ATP hydrolysis via an arginine finger-like motif. Proc Natl Acad Sci U S A. 2007;104:1811–1816. doi: 10.1073/pnas.0607216104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding Z, Atmakuri K, Christie PJ. The outs and ins of bacterial type IV secretion substrates. Trends Microbiol. 2003;11:527–535. doi: 10.1016/j.tim.2003.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rice L. Conjugative transposons. In: Bonomo R, Tolmasky ME, editors. Enzyme-mediated resistance to antibiotics Mechanisms, dissemination, and prospects for inhibition. Washington, DC: ASM Press; 2007. pp. 271–284. [Google Scholar]
- 42.Waters V. The dissemination of antibiotic resistance by bacterial conjugation. In: Bonomo R, Tolmasky ME, editors. Enzyme-mediated rersistance to antibiotics Mechanisms, dissemination, and prospects for inhibition. Washington, DC: ASM Press; 2007. pp. 285–312. [Google Scholar]
- 43.Schubert S, Dufke S, Sorsa J, Heesemann J. A novel integrative and conjugative element (ICE) of Escherichia coli: the putative progenitor of the Yersinia high-pathogenicity island. Mol Microbiol. 2004;51:837–848. doi: 10.1046/j.1365-2958.2003.03870.x. [DOI] [PubMed] [Google Scholar]
- 44.Lin TL, Lee CZ, Hsieh PF, Tsai SF, Wang JT. Characterization of integrative and conjugative element ICEKp1-associated genomic heterogeneity in a Klebsiella pneumoniae strain isolated from a primary liver abscess. J Bacteriol. 2008;190:515–526. doi: 10.1128/JB.01219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schroder G, Lanka E. The mating pair formation system of conjugative plasmids-A versatile secretion machinery for transfer of proteins and DNA. Plasmid. 2005;54:1–25. doi: 10.1016/j.plasmid.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 46.Chen YT, Shu HY, Li LH, Liao TL, Wu KM, et al. Complete nucleotide sequence of pK245, a 98-kilobase plasmid conferring quinolone resistance and extended-spectrum-beta-lactamase activity in a clinical Klebsiella pneumoniae isolate. Antimicrob Agents Chemother. 2006;50:3861–3866. doi: 10.1128/AAC.00456-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nuñez B, de la Cruz F. Two atypical mobilization proteins are involved in plasmid CloDF13 relaxation. Mol Microbiol. 2001;39:1088–1099. doi: 10.1046/j.1365-2958.2001.02308.x. [DOI] [PubMed] [Google Scholar]
- 48.Avila P, Nuñez B, de la Cruz F. Plasmid R6K contains two functional oriTs which can assemble simultaneously in relaxosomes in vivo. J Mol Biol. 1996;261:135–143. doi: 10.1006/jmbi.1996.0447. [DOI] [PubMed] [Google Scholar]
- 49.Bhattacharjee M, Rao X, Meyer R. Role of the origin of transfer in termination of strand transfer during bacterial conjugation. J Bacteriol. 1992;74:6659–6665. doi: 10.1128/jb.174.20.6659-6665.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Belogurov AA, Delver EP, Agafonova OV, Belogurova NG, Lee LY, et al. Antirestriction protein Ard (Type C) encoded by IncW plasmid pSa has a high similarity to the “protein transport” domain of TraC1 primase of promiscuous plasmid RP4. J Mol Biol. 2000;296:969–977. doi: 10.1006/jmbi.1999.3493. [DOI] [PubMed] [Google Scholar]
- 51.Welch TJ, Fricke WF, McDermott PF, White DG, Rosso ML, et al. Multiple antimicrobial resistance in plague: an emerging public health risk. PLoS ONE. 2007;2:e309. doi: 10.1371/journal.pone.0000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prentice MB, James KD, Parkhill J, Baker SG, Stevens K, et al. Yersinia pestis pFra shows biovar-specific differences and recent common ancestry with a Salmonella enterica serovar Typhi plasmid. J Bacteriol. 2001;183:2586–2594. doi: 10.1128/JB.183.8.2586-2594.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burrus V, Pavlovic G, Decaris B, Guedon G. Conjugative transposons: the tip of the iceberg. Mol Microbiol. 2002;46:601–610. doi: 10.1046/j.1365-2958.2002.03191.x. [DOI] [PubMed] [Google Scholar]
- 54.Leong SA, Ditta GS, Helinski DR. Heme biosynthesis in Rhizobium. Identification of a cloned gene coding for delta-aminolevulinic acid synthetase from Rhizobium meliloti. J Biol Chem. 1982;257:8724–8730. [PubMed] [Google Scholar]
- 55.Norrander J, Kempe T, Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983;26:101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- 56.Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 57.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reese MG. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput Chem. 2001;26:51–56. doi: 10.1016/s0097-8485(01)00099-7. [DOI] [PubMed] [Google Scholar]
- 59.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 60.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]