Abstract
The Caenorhabditis elegans heterochronic gene lin-28 regulates developmental timing in the nematode trunk. We report the dynamic expression patterns of Lin-28 homologues in mouse and chick embryos. Whole mount in situ hybridization revealed specific and intriguing expression patterns of Lin-28 in the developing mouse and chick limb bud. Mouse Lin-28 expression was detected in both the fore-limb and hindlimb at E9.5, but disappeared from the forelimb at E10.5, and finally from the forelimb and hindlimb at E11.5. Chicken Lin-28, which was first detected in the limb primordium at stage 15/16, was also downregulated as the stage proceeded. The amino acid sequences of mouse and chicken Lin-28 genes are highly conserved and the similar expression patterns of Lin-28 during limb development in mouse and chicken suggest that this heterochronic gene is also conserved during vertebrate limb development.
Keywords: Heterochronic gene, Whole mount in situ hybridization, Mouse, Chick, Limb bud, Lin-28, MicroRNAs, Developmental timing, Dynamic expression, Embryo
1. Results and discussion
Embryonic development is tightly regulated by spatio-temporal gene expression; however, systems regulating developmental timing are largely unknown. A series of mutagenesis studies using the nematode Caenorhabditis elegans as a model have identified a cluster of genes, the so-called “heterochronic genes,” which is expressed at the specific period of time during embryogenesis and play a critical role in determining the relative timing of developmental events (Ambros and Horvitz, 1984; Lee et al., 1993). Among these heterochronic genes, lin-28 is involved in specific developmental events, observed in the second of the nematode’s four larval stages (Moss et al., 1997). It should be also noted that lin-28 is regulated by lin-4, one of the heterochronic genes that encodes a microRNA (Moss et al., 1997). These studies demonstrated the critical role of non-coding RNA in development through posttranscriptional control.
Lin-28 is well conserved among various species and encodes an approximately 25 kDa protein with RNA-binding motifs and a “cold shock” domain (CSD) (Moss et al., 1997). Human LIN-28 is expressed in embryonic stem cells (Richards et al., 2004) and mouse Lin-28 protein is also detected in various tissues in embryos by immunohistochemistry (Yang and Moss, 2003). Importantly, the expression of Lin-28 is down-regulated or becomes restricted to a limited number of tissues as development proceeds. For instance, Lin-28 protein is expressed in developing bronchial epithelium and disappears in adult bronchial tissue. In addition, it is downregulated in the crypt/villus cells of the adult intestine. These results suggest that the function of Lin-28 as a heterochronic gene in C. elegans is conserved in mammals.
To further examine the spatio-temporal expression patterns of Lin-28 in developing embryos, we performed whole mount in situ hybridization using mouse and chicken embryos at different developmental stages.
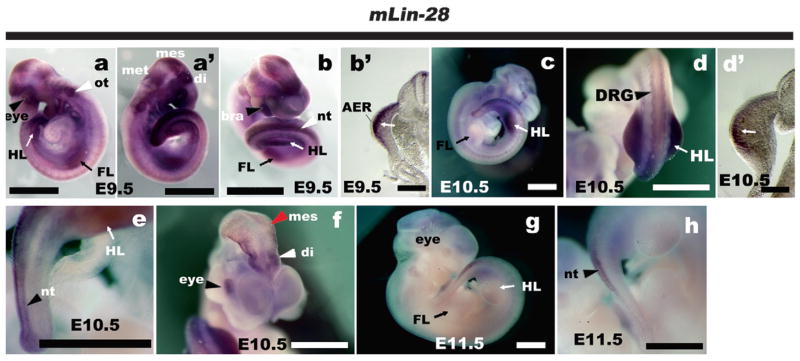
Although Lin-28 was ubiquitously expressed throughout the mouse embryo at E9.5 (Fig. 1a, a′), strong expression was observed in the most distal mesenchyme adjacent to the apical ectodermal ridge (AER) of both forelimb and hindlimb buds (Fig. 1b, b′, white arrow). Interestingly, at E10.5, the expression of Lin-28 in forelimb almost disappeared, and expression was restricted to the hindlimb and more caudal trunk at the hindlimb level (Fig. 1c–e). In a transverse section of the hindlimb, Lin-28 was expressed in the distal mesenchyme underneath the AER (Fig. 1d′) as well as E9.5. We also detected the specific expression of Lin-28 in the neural tube (Fig. 1e), diencephalon, mesencephalon (Fig. 1f) and eye primordium (Fig. 1f) in E10.5. At E11.5, expression was detected in the neural tube (Fig. 1g and h) and in the eyes (Fig. 1g), but disappeared from the hindlimb (Fig. 1g and h).
Fig. 1.
The expression pattern of Lin-28 in the developing mouse embryo (a–b′) Lin-28 is expressed ubiquitously at E9.5, with strong expression in forelimb and hindlimb buds, eyes, and otic vesicles (a), diencephalon, mesencephalon, and metencephalon (a′), branchial arch and neural tube (b). (b′) Transverse section of the hindlimb level of b. (c–f) At E10.5, expression is reduced in the forelimb (c) but maintained in the hindlimb (d, d′ (transverse section of d)), dorsal root ganglia (d, white arrowhead) and neural tube (e, black arrowhead). The eye, diencephalon, and mesencephalon are also Lin-28 positive at this stage (f). (g and h) At E11.5, Lin-28 expression mostly disappeared from forelimbs and hindlimbs (slightly detectable in the hindlimb). The eye and apical edge of the neural tube are still stained. All panels show anterior to the top. Bars, 200 μm (b′, d′), 1 mm (a, a′, b, c, d, e, f, g, h). Abbreviations: FL, forelimb; HL, hindlimb; ot, otic vesicle; di, diencephalon; mes, mesencephalon; met, metencephalon; DRG, dorsal root ganglia; nt, neural tube; AER, apical ectodermal ridge.
The expression of Lin-28 in the developing limb bud is particularly intriguing since it is differentially expressed in the forelimb and the hindlimb. Moreover, only a few genes are known to be expressed in a forelimb- or hindlimb-specific manner. For instance, the T-box transcription factor (Tbx) gene, Tbx5, is specifically expressed in the forelimb while Tbx4, the paired-like homeodomain transcription factor gene, Pitx1, and homeobox genes Hoxc9, c10, and c11, are expressed only in the hindlimb. To compare the dynamic expression pattern of Lin-28 (Fig. 2E) with other forelimb- or hindlimb-specific genes, we also performed whole mount in situ hybridization to detect Tbx5 (Fig. 2A), Tbx4 (Fig. 2B), Pitx1 (Fig. 2C) and Hoxc10 (Fig. 2D) transcripts. It is clear that the expression of Lin-28 dramatically changes as development proceeds, unlike other forelimb- or hindlimb-specific genes, which are persistent in either the forelimb or hindlimb throughout these stages.
Fig. 2.

Dynamic Lin-28 mRNA expression in the mouse limb bud whole mount in situ hybridization to detect Tbx5 (A), Tbx4 (B), Pitx1 (C), Hoxc10 (D), and Lin-28 (E) mRNA was performed using mouse E9.5, 10.5, 11.5 embryos. Dorsal views of the forelimb (FL) and hindlimb (HL) are shown with the developmental stage. The asterisk indicates expression in the limb bud. (A) Tbx5 expression is restricted to the forelimb. (B) In contrast, Tbx4 mRNA is detected predominantly in the hindlimb, but not in the forelimb. Pitx1 (C) and Hoxc10 (D) expression are also detected only in the hindlimb. Lin-28 expression is first detected in the both forelimb and hindlimb (E9.5); however, the expression diminished in the forelimb (E10.5), then hindlimb (E11.5). In each panel, a limb is viewed dorsally with anterior to the top and posterior to the bottom. A schematic diagram of the limb expression pattern is illustrated on the right side. Bars, 500 μm.
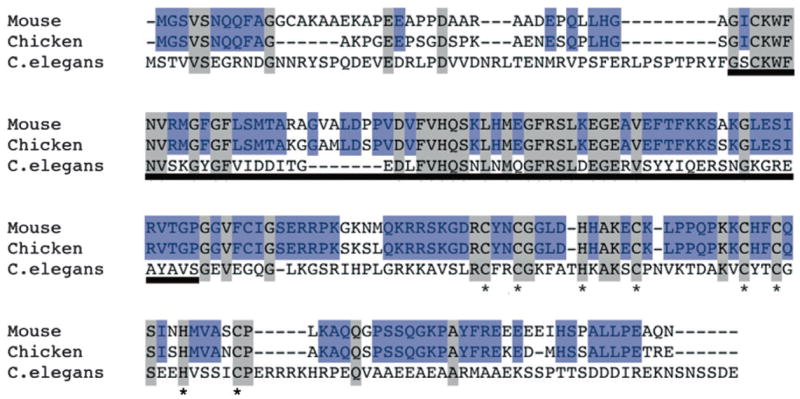
To examine whether this dynamic expression pattern of Lin-28 is conserved among vertebrate embryos, we next performed whole mount in situ hybridization using chicken embryos (Fig. 3). As shown in Table 1, amino acid sequence alignment of chicken Lin-28 and mouse Lin-28 (blue boxes) shows that the two proteins are highly homologous. In addition, the CSD (underline) and retroviral-type CCHC-zinc finger motif (asterisk) are highly conserved among chicken, mice and C. elegans Lin-28 proteins (Moss and Tang, 2003).
Fig. 3.
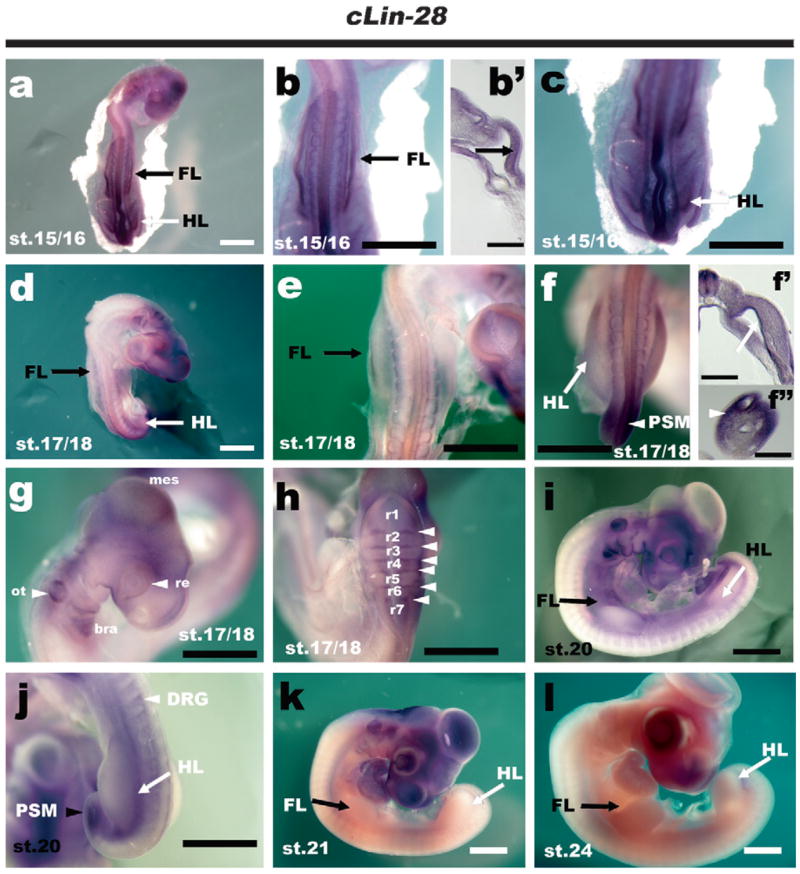
The expression patterns of chick Lin-28 at various stages (a–c) Chicken Lin-28 was expressed ubiquitously in stage 15/16 embryos. Strong expression was also detected in the presumptive limb primordium (b, c; enlargement of a, b′; transverse section of b). (d–h) Lin-28 mRNA slightly disappeared from the rostral trunk and forelimb (e, f; enlargement of d), but was still detectable in the hindlimb (f, white arrow, f′; transverse section of f) and PSM (f, white arrowhead, f″; section of white arrowhead of f). At this stage, expression was also observed in the otic vesicle (g, black arrow head), retina (g, white arrowhead), branchial arches, mesencephalon and rhombomere boundaries (h, white arrowhead), (i, j) At stage 20, strong expression of Lin-28 disappeared from the body, but faint expression was seen in the PSM (j, black arrowhead), DRG (j, white arrowhead) and posterior region of the hindlimb (white arrow), (k, i) Lin-28 expression in the trunk, forelimb and hindlimb could not be detected at stage 21 and stage 24. Bars, 200 μm (b′, f′, f″), 1 mm (a–f, g–l). Abbreviations: FL, forelimb; HL, hindlimb; PSM, presomitic mesoderm; mes, mesencephalon; ot, otic vesicle; bra, branchial arch; re, retina; r1–7, rhombomere 1–7; DRG, dorsal root ganglia.
Table 1.
Amino acid sequence alignments of mouse, chicken, and C. elegans Lin-28 homologs
|
Analysis was performed using the Clustal W method. Homologous residues are indicated by a gray shadow. The black bar indicates the “cold-shock” DNA-binding domain. The asterisk indicates a pair of CCHC zinc finger motifs. Note that mouse and chicken Lin-28 amino sequences are highly conserved (gray and blue). Accession numbers: mouse [Mus muscles] Lin-28 (209aa): NP_665832, chicken [Gallus gallus] Lin-28 (202aa): NP_001026944, C. elegans [Caenorhabditis elegans] Lin-28 (227 aa): AAC47476.
At stage 15/16, Lin-28 was expressed ubiquitously (Fig. 3a) in chicken embryos, with strong expression in the presumptive forelimb (Fig. 3b, b′ arrow) and hindlimb (Fig. 3C arrow) regions, a pattern similar to that observed in E9.5 mouse embryos (cf. Fig. 1a, a′). Although this ubiquitous expression was diminished from the rostral body (Fig. 3d) and forelimb (Fig. 3e) at around stage 17/18, caudal expression including hindlimb expression was still detectable (Fig. 3f). At this stage, Lin-28 was still weakly expressed in the hindlimb mesenchyme (Fig. 3f′ arrow) and strongly expressed in the presomitic mesoderm (Fig. 3f, f″ arrowhead), mesencephalon, retina, branchial arch, otic vesicle (Fig. 3g), and rhombomere boundaries (Fig. 3h, white arrowhead). At stage 20, Lin-28 expression in the forelimb and hindlimb disappeared (Fig. 3i and j), but was still detectable in the dorsal root ganglia (Fig. 3j). At stage 21 (Fig. 3k) and stage 24 (Fig. 3l), Lin-28 was not detected in the limb bud by in situ hybridization.
Yang and Moss (2003) reported Lin-28 protein localization as detected by immunohistochemistry in tissue sections of the mouse (Yang and Moss 2003). They revealed that Lin-28 protein is strongly localized in the epithelia of many tissues during early developmental stages and in muscles during late developmental stages. Additionally, they showed that Lin-28 is initially widely expressed at high levels in diverse tissues, but becomes down-regulated in many tissues as differentiation proceeds (Yang and Moss 2003; Polesskaya et al., 2007). A recent in vitro study showed that in skeletal myoblasts and in the embryonic carcinoma cell line P19, Lin-28 binds to polysomes where it increases the efficiency of IGF-2 protein synthesis, which is crucial for growth and differentiation of muscle tissue (Polesskaya et al., 2007).
Yang and Moss revealed that, overall, Lin-28 protein is localized in the epithelia during early development whereas we detected, at least in the limb bud, the Lin-28 mRNA is expressed in the distal mesenchyme at E9.5 and 10.5 (Fig. 1b′ and d′). One possible explanation for the discrepancy between mRNA expression observed in the current study and the protein localization described by Yang and Moss could be that Lin-28 is regulated post-transcriptionally by microRNAs such as lin-4 and let-7 (Moss et al., 1997; Reinhart et al., 2000; Moss and Tang, 2003). In this regard, it should be noted that another C. elegans heterochronic gene, Lin-41, which also contains complementary binding sites for lin-4 and let-7 in its 3′UTR like Lin-28, is highly expressed in the mouse limb bud at a time when the upstream microRNA gene let-7 is downregulated (Schulman et al., 2005; Mansfield et al., 2004). Schulman et al. (2005) also demonstrated by Northern blot analysis that Lin-41 mRNA is downregulated as development proceeds, while the upstream gene, miR-125 (the mammalian lin-4 ortholog), is upregulated (Schulman et al., 2005). To date, there is no functional data suggesting that a molecular network of heterochronic genes, including Lin-28, let-7 and Lin-41, is conserved in vertebrates; however, the dynamic expression of these genes in developing mouse limb buds may indicate a potential role for these genes in limb development.
Vertebrate paired-limb outgrowth is dependent on interactions between mesenchymal and epithelial cells mediated mainly by fibroblast growth factor (FGF) signaling. Cells in the lateral plate mesoderm at the prospective limb field express Fgf10, which induces Fgf8 in the adjacent epithelial tissue. The Fgf8-expressing epithelium develops into the thickened structure lying along the dorso-ventral boundary (AER) required for normal limb outgrowth (Ohuchi et al., 1997).
Gene misexpression experiments demonstrate that the genes Tbx5 and Tbx4 play a role in establishing the distinct morphological identity of the chick limb bud (Rodriguez-Esteban et al., 1999; Takeuchi et al., 1999). In mice, however, it has been suggested that these two T-box genes initiate limb bud development by inducing Fgf10 in the mesenchyme, but are not involved in establishing limb identity (Naiche and Papaioannou, 2003; Minguillon et al., 2005; Naiche and Papaioannou, 2007). The molecular mechanisms of forelimb and hindlimb specification are not completely understood. Given the dynamic expression pattern of Lin-28, progressively disappearing from the forelimb and then the hindlimb according to the stage of development, it would be interesting to examine the role of Lin-28 in limb initiation.
In this study, we demonstrated that mouse and chicken homologues of the nematode heterochronic gene lin-28 exhibit a dynamic expression pattern in developing mouse and chicken limb buds during embryogenesis. Further experiments are needed to determine the potential role of Lin-28 in limb development, such as determining the timing of forelimb and hindlimb bud initiation and differentiation.
2. Material and methods
2.1. Sequence analysis
Amino acid sequence alignments of mouse, chicken, and C. elegans Lin-28 homologues were analyzed using the Clustal W program. The accession numbers used were as follows: NP_665832 (mouse), NP_001026944 (chicken) and AAC47476 (C. elegans).
2.2. Preparation of non-RI RNA probe
The mouse Lin-28, Hoxc10, Tbx4, Pitx1 cDNA clones were kindly provided by RIKEN. The mouse Tbx5 cDNA was cloned by RT-PCR using the following primers: F-ATTGAGAACAACCCCTTCGC, R-CT CCACTCTGGCACCATGC.
Chicken Lin-28 (NCBI Accession No. NM_001031774) cDNA was synthesized by RT-PCR using total RNA extracted from a stage 14 whole embryo and E5.5 head. After a PCR amplification, the cDNA fragment was cloned into the pDrive cloning vector (QIAGEN). This plasmid was linearized with a restriction enzyme and used as a template for in vitro transcription with the following primers: (providing an 845 bp fragment including the 3′UTR and entire ORF of Lin28) F- GTCGGGATGGGGTCTGTTTC, R-GAGCTGTGCCTCCCTGCTC. Another primer set was used to confirm proper mRNA expression (providing a 669 bp fragment including the 3′UTR and partial ORF): F-AGAGCAAGAGCCTGCAGAAAC, R-TGAGCTCCACTGAGCCATG.
Digoxigenin (DIG) labeled antisense riboprobes were transcribed by incubating the primer sets and templates with DIG-RNA Labeling Mix (Roche) with T7/Sp6 RNA polymerase (Roche).
2.3. Animals
Pregnant mice (ICR strain) were purchased from a commercial source. Embryos (E9.5–11.5) were dissected from these mice into cold PBS.
Fertilized chicken eggs were purchased from a local farm and incubated at 37.5 °C in a humid atmosphere until the embryos reached the stage needed. Embryo staging was performed according to Hamburger and Hamilton (1951). After washing the embryos several times in cold PBS containing 0.1% Tween 20 (PBT), their amniotic membranes were removed. Embryos were collected in 4% paraformaldehyde (PFA) in PBT and fixed overnight with gentle shaking at 4 °C. Embryos were dehydrated with graduated methanol (MeOH) in a PBT series and stocked until needed at −30 °C in 100% MeOH.
2.4. Whole-mount in situ hybridization
Whole-mount in situ hybridization procedures were performed according to Wilkinson (1992) with some modifications.
Briefly, after rehydration with a graduated MetOH/PBT series, embryos were bleached with 6% H2O2 in PBT for 15 min and treated with 10 μg/ml Protease K (Roche) for 8–24 min at room temperature (RT) depending on the stage, stopped with 0.2% glycine and the embryos were then re-fixed with 4% PFA, 0.2% glutaraldehyde in PBT for 20 min at RT. Pre-hybridization (in 5× SSC (pH 4.5), 50% formamide, 1% SDS, 50 μg/ml yeast tRNA (Roche), and 50 μg/ml heparin (Nakarai-tesque)) was performed at 70 °C for 1 h, then a DIG-RNA probe (about 500 ng/ml) was added and hybridized for over 14 h at 70 °C.
Subsequently, embryos were subjected to a series of post-hybridization washes in wash buffers containing formamide, SSC, SDS, and 0.05% CHAPS. After blocking with 10% sheep serum (TRACE) in TBS containing 0.1% Tween 20 (TBST) for 1 h at RT, embryos were incubated overnight with anti-DIG-AP Fab fragments antibody (Roche) and 1% sheep serum in TBST at 4 °C. After a series of washes with TBST, embryos were equilibrated with NTMT (5 M NaCl, 1 M Tris-HCl (pH 9.5), 1 M MgCl2, and 0.1% Tween 20).
Color reaction was performed at 4 °C or RT with nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate (NBT/BCIP) (Roche), then embryos were stored in 4% PFA in PBT at 4 °C. Vibratome sections (100 μm thickness) were obtained using The Vibratome 1500 Sectioning System (Ted Pella, Inc.).
Stained samples were photographed with a SZX12 microscope and DP70 CCD camera system (Olympus).
Acknowledgments
We thank Dr. Hayashizaki for providing the FANTOM cDNA library. We also thank Akane Nagai, Tomohiro Hikata, Hironobu Naito, Takamasa Suzuki, and Noritsugu Fukuda for technical assistance and critical discussion, and all members of the Asahara laboratory for helpful discussion. This study was supported by Grants from NIH (AR50631), JST SORST, Grant ID05–24 from the National Institute of Biomedical Innovation, Genome Network Project (MEXT) and DECODE Grants-in-Aid for Scientific Research (MEXT) (H.A.)
Footnotes
Publisher's Disclaimer: This article was published in an Elsevier journal. The attached copy is furnished to the author for non-commercial research and education use, including for instruction at the author's institution, sharing with colleagues and providing to institution administration. Other uses, including reproduction and distribution, or selling or licensing copies, or posting to personal, institutional or third party websites are prohibited. In most cases authors are permitted to post their version of the article (e.g. in Word or Tex form) to their personal website or institutional repository. Authors requiring further information regarding Elsevier's archiving and manuscript policies are encouraged to visit: http://www.elsevier.com/copyright
References
- Ambros V, Horvitz HR. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226:409–416. doi: 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. J Morph. 1951;88:49–92. [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans hetero-chronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Mansfield JH, Harfe BD, Nissen R, Obenauer J, Srineel J, Chaudhuri A, Farzan-Kashani R, Zuker M, Pasquinelli AE, Ruvkun G, Sharp PA, Tabin CJ, McManus MT. MicroRNA-responsive ‘sensor’ transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat Genet. 2004;36:1079–1083. doi: 10.1038/ng1421. [DOI] [PubMed] [Google Scholar]
- Minguillon C, Del Buono J, Logan MP. Tbx5 and Tbx4 are not sufficient to determine limb-specific morphologies but have common roles in initiating limb outgrowth. Dev Cell. 2005;8:75–84. doi: 10.1016/j.devcel.2004.11.013. [DOI] [PubMed] [Google Scholar]
- Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- Moss EG, Tang L. Conservation of the heterochronic regulator Lin-28, its developmental expression and microRNA complementary sites. Dev Biol. 2003;258:432–442. doi: 10.1016/s0012-1606(03)00126-x. [DOI] [PubMed] [Google Scholar]
- Naiche LA, Papaioannou VE. Loss of Tbx4 blocks hindlimb development and affects vascularization and fusion of the allantois. Development. 2003;130:2681–2693. doi: 10.1242/dev.00504. [DOI] [PubMed] [Google Scholar]
- Naiche LA, Papaioannou VE. Tbx4 is not required for hindlimb identity or post-bud hindlimb outgrowth. Development. 2007;134:93–103. doi: 10.1242/dev.02712. [DOI] [PubMed] [Google Scholar]
- Ohuchi H, Nakagawa T, Yamamoto A, Araga A, Ohata T, Ishimaru Y, Yoshioka H, Kuwana T, Nohno T, Yamasaki M, Itoh N, Noji S. The mesenchymal factor, FGF10, initiates and maintains the outgrowth of the chick limb bud through interaction with FGF8, an apical ectodermal factor. Development. 1997;124:2235–2244. doi: 10.1242/dev.124.11.2235. [DOI] [PubMed] [Google Scholar]
- Polesskaya A, Cuvellier S, Naguibneva I, Duquet A, Moss EG, Harel-Bellan A. Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes Dev. 2007;21:1125–1138. doi: 10.1101/gad.415007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Richards M, Tan SP, Tan JH, Chan WK, Bongso A. The transcriptome profile of human embryonic stem cells as defined by SAGE. Stem Cells. 2004;22:51–64. doi: 10.1634/stemcells.22-1-51. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Esteban C, Tsukui T, Yonei S, Magallon J, Tamura K, Izpisua Belmonte JC. The T-box genes Tbx4 and Tbx5 regulate limb outgrowth and identity. Nature. 1999;398:814–818. doi: 10.1038/19769. [DOI] [PubMed] [Google Scholar]
- Schulman BR, Esquela-Kerscher A, Slack FJ. Reciprocal expression of lin-41 and the microRNAs let-7 and mir-125 during mouse embryogenesis. Dev Dyn. 2005;234:1046–1054. doi: 10.1002/dvdy.20599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi JK, Koshiba-Takeuchi K, Matsumoto K, Vogel-Hopker A, Naitoh-Matsuo M, Ogura K, Takahashi N, Yasuda K, Ogura T. Tbx5 and Tbx4 genes determine the wing/leg identity of limb buds. Nature. 1999;398:810–814. doi: 10.1038/19762. [DOI] [PubMed] [Google Scholar]
- Wilkinson DG. Whole mount in situ hybridization of vertebrate embryos. In: Wilkinson DG, editor. In situ Hybridization: A Practical Approach. IRL Press; Oxford: 1992. pp. 75–83. [Google Scholar]
- Yang DH, Moss EG. Temporally regulated expression of Lin-28 in diverse tissues of the developing mouse. Gene Expr Patterns. 2003;3:719–726. doi: 10.1016/s1567-133x(03)00140-6. [DOI] [PubMed] [Google Scholar]