Abstract
The transcription factor Sp1 is ubiquitously expressed in different cells and thereby regulates the expression of genes involved in many cellular processes. This study reveals that Sp1 was phosphorylated during the mitotic stage in three epithelial tumor cell lines and one glioma cell line. By using different kinase inhibitors, we found that during mitosis in HeLa cells, the c-Jun NH2-terminal kinase (JNK) 1 was activated that was then required for the phosphorylation of Sp1. In addition, blockade of the Sp1 phosphorylation via inhibition JNK1 activity in mitosis resulted in the ubiquitination and degradation of Sp1. JNK1 phosphorylated Sp1 at Thr278/739. The Sp1 mutated at Thr278/739 was unstable during mitosis, possessing less transcriptional activity for the 12(S)-lipoxygenase expression and exhibiting a decreased cell growth rate compared with wild-type Sp1 in HeLa cells. In N-methyl-N-nitrosourea–induced mammary tumors, JNK1 activation provided a potential relevance with the accumulation of Sp1. Together, our results indicate that JNK1 activation is necessary to phosphorylate Sp1 and to shield Sp1 from the ubiquitin-dependent degradation pathway during mitosis in tumor cell lines.
INTRODUCTION
The transcription factor Sp1 is ubiquitously expressed in mammalian cells, and it is important in a variety of physiological processes, including cell cycle regulation, apoptosis, and differentiation (Firestone and Bjeldanes, 2003; Chu and Ferro, 2005; Wong et al., 2005; Deniaud et al., 2006). Sp1 binds specifically to the GC-rich promoter elements, via three C2H2-type zinc finger regions at the C terminus of Sp1, and it regulates the transcriptional activity of the target genes by using two major glutamine-rich transactivation domains localized, respectively, at the N terminus and the medial region (Suske, 1999; Bouwman and Philipsen, 2002). In addition, in Sp1, there are serine/threonine-rich sequences between the two transactivation domains that may be a target for posttranslational modification (Bouwman and Philipsen, 2002).
The transcriptional activity of a transcription factor is determined at least by three factors: transactivational activity, DNA binding affinity, and protein level. Previous studies on the regulation of Sp1 activities focused mostly on transactivational activity, thereby allowing study of its interaction with other proteins and its DNA binding affinity. However, one of the apparent key elements regulating the activity of Sp1 is via its stability, which certainly needs to be explored and established. Recent studies revealed that the DNA binding ability, transactivational activity, and protein stability of Sp1 might be influenced by its posttranslational modifications such as sumoylation, glycosylation, ubiquitination, acetylation, and phosphorylation (Han and Kudlow, 1997; Mortensen et al., 1997; Wells et al., 2001; Ryu et al., 2003; Abdelrahim and Safe, 2005; Chu and Ferro, 2005; Hung et al., 2006; Spengler and Brattain, 2006). For example, Sp1 is sumoylated at Lys16, which might repress the transactivational activity of p21WAF1/CIP1 (Spengler and Brattain, 2006), although the detailed mechanism is still unknown. Acetylation of Sp1 is related to its DNA binding affinity and its transactivational activity (Ryu et al., 2003; Hung et al., 2006). In addition, glycosylation and ubiquitination of Sp1 regulate its stability in a proteasome-dependent manner (Han and Kudlow, 1997; Wells et al., 2001; Abdelrahim and Safe, 2005). The phosphorylation of Sp1 has been widely studied, and the results showed that some kinases phosphorylate Sp1, which affects transactivation, but some other kinases phosphorylate Sp1, which affects its DNA binding affinity (Chu and Ferro, 2005). Serine or threonine residues could be phosphorylated by different kinases, including DNA-dependent protein kinase, ERK1/2, casein kinase II, and CDK2. Thus, DNA-dependent protein kinase was shown to associate with nuclear protein Tat of human immunodeficiency virus 1 and to phosphorylate Ser131 of Sp1, thus increasing the transcriptional activity of Sp1 through increasing DNA binding affinity (Chun et al., 1998). In addition, some growth factors, such as hepatocyte growth factor/scatter factor, can activate the pathways of phosphatidylinositol 3-kinase (PI3-kinase) and protein kinase C-ζ, which then phosphorylate Sp1 to increase the transcriptional activity of Sp1, but do not change the DNA binding affinity (Reisinger et al., 2003). Conversely, some kinases inhibit the transcriptional ability of Sp1. For example, during terminal differentiation of the liver, casein kinase II modifies Thr579 on Sp1 and down-regulates the DNA binding ability of Sp1 (Armstrong et al., 1997). Stimulation by different substances induces ERK1/2 to phosphorylate Sp1, which, in turn, has different effects on different genes. Estradiol-activated extracellular signal-regulated kinase (ERK)1/2 modifies Thr453/739 of Sp1, which induces Sp1 binding with vascular endothelial growth factor (VEGF) promoter, stimulating gene activation (Milanini-Mongiat et al., 2002). However, when cells are stimulated with platelet-derived growth factor (PDGF), Sp1 phosphorylation represses PDGF receptor-α promoter activity (Bonello and Khachigian, 2004). Recent studies also indicate that the cell cycle might regulate the phosphorylation states of Sp1. When cells enter the S phase of the cell cycle, Sp1 is phosphorylated by cyclin-dependent kinase 2 (CDK2) at Ser59, thus increasing the expression of dihydrofolate reductase, a key enzyme in the production of thymidine (Fojas de Borja et al., 2001). Whether Sp1 is also phosphorylated by other kinases during different phases of the cell cycle and why Sp1 must be modified as such are still not clear.
Previous studies also indicated the Sp1 could be maintained and divided into daughter cells during and after mitosis (He and Davie, 2006). However, how Sp1 is shielded from ubiquitin-dependent degradation during mitosis is an interesting question. The c-Jun NH2-terminal kinase (JNK) is a major member of the mitogen-activated protein kinase family, and it is known to respond to stress stimuli such as UV irradiation, heat shock, and reactive oxygen species (Nagata et al., 1997; Davis, 2000). JNK consists of three isoforms, including JNK1, JNK2, and JNK3. JNK1 and JNK2 are ubiquitously distributed, and JNK3 is found in neuronal tissue (Sabapathy et al., 2004). Different JNK isoforms may have evolved for different specific biological functions. JNK1, in the absence of JNK2, increases the expression of cyclin D1 to increase the proliferation rate. JNK1 has higher kinase activity for c-Jun than JNK2. JNK2 negatively affects cell proliferation and c-Jun kinase activity (Sabapathy et al., 2004). SP600125 inhibits both JNK1 and JNK2 kinase activities in human HMC-1 cells, and as a result, it attenuates cell proliferation associated with cell cycle arrest at the G1 phase (Wang et al., 2006). In addition, inactive JNK causes the degradation of c-Jun, ATF2, JunB, and p53, but activated JNK protects these proteins from ubiquitination (Fuchs et al., 1996, 1997, 1998). Thus, JNK enzymatic activity may be important both for the modulation of cell cycle progression and for the regulation of the stability of certain proteins. It was recently reported that the expression of bile acid-inducible DR5/tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-R2, an apoptosis-inducing membrane receptor for TRAIL, is controlled by the JNK pathway that targets transcription factor Sp1 (Higuchi et al., 2004). In turn, Sp1 drives the transcription from the −243/−137 region of the DR5/TRAIL-R2 promoter, which contains two Sp1 binding sites at −198/189 and −152/−143 (Higuchi et al., 2004). Interestingly, JNK1/2 inhibitor SP600125 reduces the formation of Sp1-DR5/TRAIL-R2 promoter DNA complex (Higuchi et al., 2004). Therefore, these studies indicate that JNK1/2 might interact with Sp1, consequently raising an interesting question of how exactly JNK1/2 affects the transcriptional activity of Sp1.
In the present study, we found that JNK1 was activated during mitosis and then caused most of the Sp1 to be the phosphorylated form. JNK1 phosphorylated Sp1 at Thr278 and Thr739, thereby increasing the Sp1 stability by repressing the Sp1 degradation in the proteasome-dependent pathway. In MNU-induced mammary tumors, we also found a high JNK1 activation and a high Sp1 accumulation. This study characterizes the stability of Sp1 and suggests a tumorigenic action of Sp1.
MATERIALS AND METHODS
Cell Culture and Transfection
Human cervical adenocarcinoma HeLa cells, breast carcinoma MDA-MB-231 cells, lung adenocarcinoma A549 cells, and rat glioma C6 cells were cultured in DMEM (Carlsbad, CA) containing 10% fetal bovine serum, 100 μg/ml streptomycin sulfate, and 100 U/ml penicillin G sodium at 37°C and 5% CO2. The 90% confluent cells were treated with 45 ng/ml nocodazole (Sigma-Aldrich, St. Louis, MO) for 16 h or cotreated with 10 μM different kinase inhibitors (Calbiochem, San Diego, CA, and Promega, Madison, WI) before nocodazole treatment. Transfection of HeLa cells with expression vectors was done using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol, with a slight modification. The cells were replated 24 h before transfection at an optimal cell density in 2 ml of fresh culture medium in a 3.5-cm dish. For transfection, 1 μl of Lipofectamine was incubated with 1 μg of different hemagglutinin (HA)-Sp1 or green fluorescent protein (GFP)-Sp1 mutant plasmids in 0.2 ml of Opti-MEM medium (Invitrogen) for 30 min at room temperature. The cells were transfected by changing the medium with 2 ml of Opti-MEM medium containing the plasmid and Lipofectamine, and then they were incubated at 37°C in 5% CO2 for 6 h. Next, the cells were incubated for an additional 36 h by using 2 ml of fresh medium. Luciferase assay (Promega) was performed using a luminometer (model LB9506; Berthold Technologies, Bad Wildbad, Germany), according to a method described previously (Liaw et al., 1998). Each transfection experiment was performed three times, and each sample in each experiment was prepared in duplicate.
Immunofluorescence and Confocal Microscopy
HeLa cells were seeded onto glass slides overnight and fixed with 4% paraformaldehyde (Sigma-Aldrich) in phosphate-buffered saline (PBS) at 4°C for 10 min. The cells were then rinsed with PBS three times and permeabilized with 1% Triton X-100 for 7 min. Next, the cells were pretreated with 1% bovine serum albumin (BSA) in PBS at 25°C for 60 min and incubated with rabbit anti-Sp1 polyclonal antibodies (Upstate Biotechnology, Waltham, MA) and mouse anti-lamin A/C monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA) at a dilution of 1:200 for 1 h and treated with fluorescein isothiocyanate (FITC)-conjugated donkey anti-mouse immunoglobulin G (IgG) polyclonal antibodies and cyanine (Cy)5-conjugated donkey anti-rabbit IgG polyclonal antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA) at a dilution of 1:250 for 1 h. Finally, the cells were washed with PBS, mounted in 90% glycerol containing 4,6-diamidino-2-phenylindole (DAPI; Invitrogen), and examined using a confocal laser scanning microscope (FluoView FV 1000; Olympus, Melville, NY).
Fluorescence-activated Cell Sorting (FACS) Analysis
HeLa cells were treated with nocodazole for 16 h, and the attached and rounded-up cells were collected separately. The cells were washed with ice-cold PBS and fixed in 4% paraformaldehyde at 4°C for 10 min. The cells were then permeabilized with 1% Triton X-100 for 5 min. After the cells had been treated with 10 μg/ml RNase A (QIAGEN, Valencia, CA) at 37°C for 20 min, they were stained with 50 μg/ml propidium iodide (Sigma-Aldrich) at room temperature for 5 min and analyzed using a flow cytometer (FACSCalibur; BD Biosciences, Mountain View, CA).
Western Blot Analysis
Total cell lysates were fractionated using SDS-polyacrylamide gel electrophoresis (PAGE) and transferred onto a polyvinylidene difluoride membrane (GE Healthcare, Baie d'Urfe, QC, Canada) by using a transfer apparatus according to the manufacturer's protocols (Bio-Rad, Hercules, CA). After incubation with 5% nonfat milk in Tris-buffered saline/Tween 20 (TBST; 10 mM Tris, pH 8.0, 150 mM NaCl, and 0.5% Tween 20) for 1 h, the membranes were incubated with anti-Sp1 (1:3000 dilution) (Upstate Biotechnology), anti-phospho (p)-JNK (Thr183/Thr185) (1:2000 dilution) (Cell signaling Technology, Beverly, MA), anti-cyclin B1 (1:3000 dilution), anti-JNK (1:1000 dilution), anti-cyclin E (1:1000 dilution), anti-glutathione transferase (GST) (1:1000 dilution), anti-histone H3 (1:1000 dilution), anti-Sp3 (1:3000 dilution) (Santa Cruz Biotechnology), anti-HA (1:2000 dilution) (Roche Diagnostics, Indianapolis, IN), anti-α-tubulin (1:5000 dilution), or anti-actin (1:5000 dilution) (Sigma-Aldrich) antibodies at room temperature for 2 h. The membranes were washed for 5 min three times and incubated with a 1:3000 dilution of horseradish peroxidase-conjugated anti-mouse, anti-rabbit, or anti-rat antibodies (Santa Cruz Biotechnology) at room temperature for 1 h. Blots were washed with TBST three times and developed using the ECL system (Pierce Chemical, Rockford, IL) according to the manufacturer's protocols.
In Vitro Calf Intestinal Alkaline Phosphatase (CIP) Assay
For in vitro dephosphorylation of Sp1, the mitotic cell extracts were combined with 10 U of alkaline phosphatase (New England Biolabs, Beverly, MA) containing 50 mM Tris, pH 7.9, 100 mM NaCl, 10 mM MgCl2, and 1 mM dithiothreitol in CIP buffer, and the mixture was incubated at 37 or 4°C for 1 h.
Immunoprecipitation
HeLa cells (1 × 107) were washed with PBS. Lysate was prepared using a radioimmune precipitation assay (RIPA) buffer [50 mM Tris, pH 7.8, 150 mM NaCl, 5 mM EDTA, 0.5% Triton X-100, 0.1% Nonidet P-40, and 10 μg/ml each of MG132, leupeptin, aprotinin, and 4-(2-aminoethyl) benzenesulfonyl fluoride]. The supernatant was added with anti-ubiquitin (1:250 dilution) antibodies (Santa Cruz Biotechnology) at 4°C for 1 h. Protein-A/G agarose beads (30 μl) were added to the lysate, and the mixture was incubated under shaking at 4°C for 1 h. The beads were collected using centrifugation and washed three times with RIPA buffer. Proteins binding to the beads were eluted by adding 30 μl of 2× electrophoresis sample buffer and analyzed using immunoblotting with anti-Sp1 antibodies.
In Vitro JNK1 Kinase Assay
For the in vitro phosphorylation analysis, full-length Sp1 protein was prepared from HeLa cells by using immunoprecipitation with anti-Sp1 antibodies, and the different GST-Sp1 fragments and the point mutations of Sp1 were purified from Escherichia coli BL21 (DE3). These different Sp1 proteins and active JNKα1/stress-activated protein kinase 1c (Upstate Biotechnology) were used to examine Sp1 phosphorylation in vitro. Each reaction (20 μl) contained 1 μg of purified Sp1, 0.015 U or different doses of active JNK1, 2 μCi of [γ-32P]ATP (GE Healthcare) or 1 mM ATP (New England Biolabs), and 2 μl of 10× kinase buffer containing 500 mM HEPES, pH 7.4, 10 mM MgCl2, 1 mM EGTA, and 1 mM dithiothreitol (DTT). The phosphorylation reactions were incubated at 30°C for 15 min. After the incubation, one-half of the reaction was added to 10 μl of 2× electrophoresis sample buffer, which was then heated to 95°C for 5 min. Proteins in the mixtures were immediately separated using SDS-PAGE.
Cell Synchronization
Mitotic cells were collected by incubating HeLa cells in complete medium with 45 ng/ml nocodazole at 37°C for 16 h, after which the mitotic cells were obtained by mechanical shake-off. Cells were then washed three times with PBS and replated in fresh medium. The released cells were then collected after different time intervals (12, 16, 20, 22, and 24 h) and lysed in RIPA buffer as described above. Equal amounts of proteins from these cell extracts were analyzed using immunoblotting. For another kind of cell synchronization, HeLa cells were blocked at the G1/S boundary with 2 mM thymidine (Sigma-Aldrich) for 14 h. The cells were then washed three times with PBS and incubated with fresh medium. And 12 h after release, the cells were replated onto 6-cm plates with 2 mM thymidine and reincubated for 14 h. Plates were then washed three times with PBS, and fresh medium was added. The time point, corresponding to the G1/S transition, was defined as 0 h. The cells were collected at different times and then analyzed using immunoblotting.
Expression of Plasmids
Plasmids pCMV HA-Sp1 and pEGFP-Sp1 both contains the cDNA of full-length Sp1 transcribed from the cytomegalovirus (CMV) immediate-early promoter (Hung et al., 2006). Plasmids pGEX6P-1-Sp1 (8-290), pGEX6P-1-Sp1 (8-618), and pGEX6P-1-Sp1 (619-785) were kindly provided by Dr. Wen-Chun Hung (National Sun Yat-Sen University, Kaohsiung, Taiwan), which express GST-tagged Sp1 containing the regions from amino acids 8-290, 8-618, and 619-785 in E. coli BL21(DE3). The amino acid sequence of Sp1 protein includes Ser59, Ser73, Thr117, Thr278, Thr355, Thr453, Thr503, Ser588, and Thr739 residues, which involve Ser/Thr prophosphorylation of consensus sites by JNK. These Ser/Thr residues were mutated to alanine or aspartic acid by using a polymerase chain reaction (PCR) mutagenesis method.
Purification of GST Fusion Proteins
To purify different GST-Sp1 fragments and the point mutations of Sp1, the E. coli BL21 (DE3) was cultured to mid-log phase in 200 ml of LB medium containing 50 mg/ml ampicillin. Isopropyl-1-thio-β-d-galactopyranoside (IPTG) (Sigma-Aldrich) was then added to the medium to a final concentration of 1 mM. Cells were harvested after IPTG treatment for 4 h, and then they were suspended in ice-cold buffer A [50 mM Tris, pH 8.0, 500 mM NaCl, 1 mM DTT, 1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride, and 1 mM leupeptin], and homogenized using sonication for 1 min. Cell lysate was then centrifuged at 12,000 rpm at 4°C for 10 min. The supernatant was incubated with 0.2 ml of glutathione-agarose beads (GE Healthcare) at 4°C for 1 h. The beads were washed five times with buffer A. GST fusion proteins were finally eluted from the beads by adding buffer A containing 20 mM glutathione (GE Healthcare). The eluted GST fusion proteins were dialyzed for more than 16 h against a dialysis buffer containing 50 mM Tris, pH 7.6, 100 mM NaCl, and 1 mM DTT. Dialyzed GST fusion proteins were stored at −80°C until use.
Reverse Transcription (RT)-PCR
Total RNA of cells was isolated with a TRIzol RNA extraction kit (Invitrogen), and 3 μg of RNA was subjected to RT-PCR with SuperScript II (Invitrogen). The primers used for PCR for HA-Sp1 were Sp1-forward, 5′-AGATGCCCAACCCCAAGC-3′, which specifically bound the nucleotide 1767-1784 region of Sp1 cDNA; and SP6 reverse, 5′-ATTTGGTGACACTATAGAA-3′, which specifically bound the SP6 region before the simian virus 40 poly-A of pCMV-HA vector. The primers for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were 5′-CCATCACCATCTTCCAGGAG-3′ and 5′-CCTGCTTCACCACCTTCTTG-3′. The PCR products were separated using 1% agarose gel electrophoresis and visualized with ethidium bromide staining.
Experimental Animals
Female Sprague-Dawley rats at 50 d of age were used and housed in group cages of two or three rats each in an air-conditioned vivarium with free access to food and water. Throughout the study, a 12-h light/dark cycle was maintained with lights on at 8 am. All procedures adhere to the Guidelines for Care and Use of Experimental Animals of the National Cheng-Kung University (Tainan, Taiwan). Rats were treated with an intraperitoneal injection of N-methyl-N-nitrosourea (MNU, 50 mg/kg body weight) (Sigma-Aldrich) dissolved in saline to initiate mammary tumorigenesis (Kotsopoulos et al., 2005). MNU has a half-life of <1 h under physiological conditions (Druckrey et al., 1967). Starting at 4 wk after MNU injection, the rats were palpated twice weekly for mammary tumors. When palpating a tumor, its location and date of detection were recorded. Rats were killed with CO2 inhalation either when tumors reached 300 mm3 or if the mice became moribund.
Cultured Rat Primary Glial Cells
Rat pups no more than 36 h old (postnatal day 1) were anesthetized on ice for several minutes until they were immobile and unresponsive to any peripheral stimulation. Animals were then decapitated, brains were removed, and cortex was dissected and digested in 7 ml of trypsin (10 U/ml) in PBS at 37°C for 30 min. After rinsing, the tissue was triturated and filtered through a nylon mesh filter (70 μm; Small Parts, Florida, MI). The cells were plated at a density of 2 × 105 cells/cm2 onto a plastic culture plate that was precoated with 50 μg/ml poly-ld-lysine (Sigma-Aldrich). Cells were maintained in the DMEM supplemented with 100 U/ml penicillin, 0.1 mg/ml streptomycin, 0.5 mM l-glutamine, and 10% FBS (Invitrogen) and kept at 37°C in a humidified atmosphere of 5% CO2 and 95% air.
Immunohistochemistry (IHC)
All immunohistochemical analyses were done on 5-μm, archival paraformaldehyde-fixed, paraffin-embedded rat mammary tumor or normal tissue sections. For IHC staining, after 2 h of heating at 60°C, the tissues were deparaffinized in xylene, rehydrated with graded alcohols, and rinsed in tap water. Slides were then pretreated for antigen retrieval with citrate buffer (10 mM citric acid, pH 6.0) for 15 min at 100°C in a microwave oven. After heating, slides were remained in citrate buffer for an additional 20 min at room temperature and then incubated in 0.3% hydrogen peroxide for 10 min to quench endogenous peroxidase activity. The EnVision + Dual Link System-HRP detection system (Dako North America, Carpinteria, CA) was used. After antigen retrieval, the primary antibody (mouse monoclonal anti-estrogen receptor; Cell Marque, Hot Springs, AR) or rabbit polyclonal anti-Sp1) at a dilution of 1:100 was applied, and the slides were left in humidity chambers at room temperature for 1 h. After a wash, peroxidase-labeled polymer conjugated to goat anti-mouse and goat anti-rabbit immunoglobulins (Dako North America) was applied for 1 h at room temperature. After all IHC procedures, the immunohistochemical reaction was revealed with 3′,3-diaminobenzidine chromagen solution (Dako North America) for 3 min, and the slides were thoroughly rinsed in distilled water. Sections were counterstained with Harris hematoxylin (Surgipath, Richmond, IL), dehydrated, and coverslipped with permanent mounting media.
RESULTS
The Localization of Sp1 during Cell Cycle Period Is Distinguishable
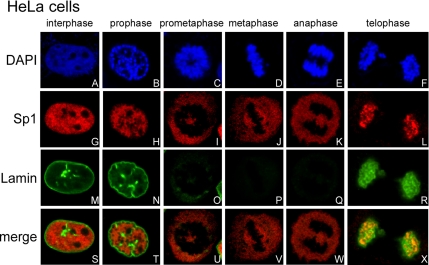
Although a previous study characterized the distribution of Sp1 during the cell cycle (He and Davie, 2006), we were interested in studying how Sp1 is able to occupy the same nuclear space as DNA in interphase cells but is devoid of DNA in metaphase cells. Therefore, we examined the relation between nuclear membrane disturbance and Sp1 distribution at different stages of the cell cycle (Figure 1). Chromatin and Sp1 were evenly distributed within the nuclear, and they were colocalized in interphase cells (Figure 1, A, G, and S). Lamin, a marker of the nuclear membrane, was also observed at different stages (Figure 1M). When cells entered the prophase, chromatin and Sp1 were both more condensed but were still colocalized (Figure 1, B, H, and T). At this stage, the nucleus was still integrated (Figure 1N). At the metaphase, the chromatin was even more condensed (Figure 1, C and D), but the Sp1 distributed throughout the cell because of the disappearance of the nuclear membrane (Figure 1, I, J, O, and P). At metaphase, chromatin and Sp1 were not colocalized (Figure 1, U and V). When the cell cycle entered anaphase, the nuclear membrane was absent (Figure 1Q), chromatin was divided (Figure 1E), and Sp1 was still not colocalized with the chromatin (Figure 1K). During telophase, the colocalization of chromatin, Sp1, and lamin A/C became evident (Figure 1, F, L, R, and X). As such, Sp1 was present in every different phase with its level well maintained at different stages, and its distribution correlated with the integrity of the nuclear membrane. Together, these findings suggest that Sp1 was stably reserved during mitosis, and there was different Sp1 localization between interphase and mitotic periods.
Figure 1.
Distribution of Sp1 during the cell cycle. HeLa cells were grown on coverslips in DMEM, fixed using 4% paraformaldehyde, double-labeled with rabbit anti-Sp1 antibodies (G–L) and mouse anti-lamin A/C antibodies (M–R), and then stained with secondary antibodies conjugated with FITC or Cy5, respectively. DNA was stained with DAPI (A–F). Finally, the cells were examined using a confocal laser scanning microscope.
Sp1 Is Highly Phosphorylated in the Mitotic Stage
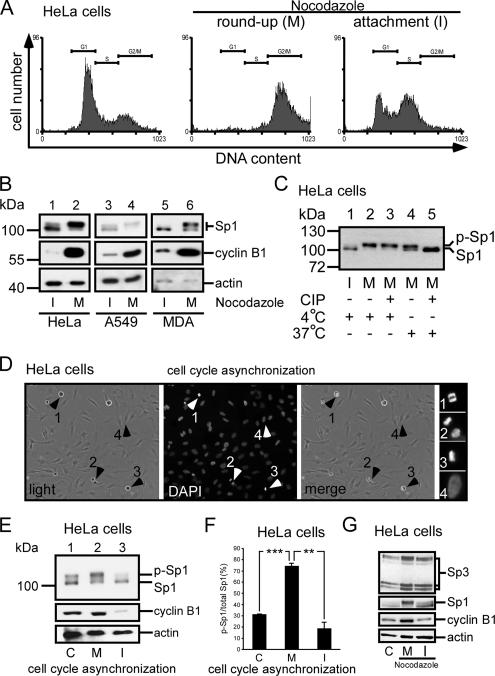
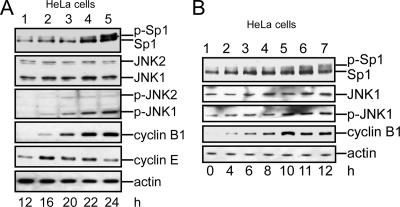
To study how the level of Sp1 is shielded from the ubiquitin-dependent degradation pathway, HeLa cells were divided into round-up cell (mitosis) and attachment cell (interphase) groups after 16 h of treatment with nocodazole (Tighe et al., 2001). The cell cycle stages were then studied using flow cytometry (Figure 2A). Our results confirmed that most of the round-up cells stayed in the G2/M stage, but the attachment cells mostly stayed in G1 and S phases. In the interphase, cyclin B, considered as a marker of mitosis, was expressed less in HeLa cells, A549 cells, and MDA-MB-231 cells (Figure 2B, lanes 1, 3, and 5). In Western blottings examining Sp1 at interphase, two bands (≈100 kDa) were found, and the bottom band was the major band (Figure 2B, lanes 1, 3, and 5). When the cells entered the mitotic stage, however, the major signal shifted to the top band (Figure 2B, lanes 2, 4, and 6). A CIP assay was done next to determine whether phosphorylation had caused this band-shift. The result revealed that the major band recognized by anti-Sp1 antibodies at the interphase had now shifted to the top band at the mitotic stage (Figure 2C, lanes 1 and 2). When cell lysate from mitotic cells was incubated with alkaline phosphatase at 37°C, the major signal shifted to the bottom band (Figure 2C, lane 5). To examine further whether Sp1 was highly phosphorylated during mitosis in normal growth cells, each group of interphase cells and mitotic cells was divided into two parts and then checked for the Sp1 phosphorylation level (Figure 2, D–F). Technically, we checked first the cell cycle stage by staining DNA with DAPI, and then we divided cells into round-up cells, which were mitotic, and attached cells, which were at interphase stage (Figure 2D). An equal number of cells from each group, i.e., mixed, interphase, or mitosis, was used for Sp1 immunoblotting to examine the Sp1 phosphorylation level. Data show that Sp1 was highly phosphorylated in mitotic cells (Figure 2, E and F). Sp3 is also a ubiquitous transcription factor closely related to Sp1. We then characterized the Sp3 pattern in interphase and mitosis period (Figure 2G), and the result revealed that there was no significant alternation in Sp3 pattern between interphase and mitosis in HeLa cells.
Figure 2.
Sp1 was highly phosphorylated during the mitotic stage. HeLa cells were treated with 45 ng/ml nocodazole for 16 h, and then they were divided into two parts: round-up and attachment cells. To check the stage of the cell cycle, samples were taken for FACS and Western blot analysis. (A) FACS results of paraformaldehyde-fixed, propidium iodide-stained cells are shown as histograms, and the positions of the G1 phase peak, S phase, and G2/M phase peak are marked by horizontal lines. For comparison, untreated HeLa cells were used (left). (B) Immunoblots of cellular extracts of HeLa, A549 and MDA-MB-231 cells in attachment (interphase, I) and round-up (mitotic stage, M) cells were probed using anti-Sp1 antibodies. Cyclin B1 detected by using anti-cyclin B1 antibodies was used as M phase marker, and actin was used as an equal loading control. (C) Mitotic cell extracts were treated with alkaline phosphatase (CIP) to dephosphorylate Sp1, and incubated at 37°C. For comparison, alkaline phosphatase at 4°C has no enzyme activity to dephosphorylate Sp1. The samples were then analyzed using immunoblotting with anti-Sp1 antibodies. (D) In the normal cultured HeLa cells, the round-up and attached cells were separated, and the cell cycle stage was then checked by DAPI. The arrows 1–3 represent that cells stayed in mitosis stage, and arrow 4 represents that cells stayed in interphase. (E). Equal cell number from mixture (control total cells, lane 1), mitosis (round-up cells, lane 2) and interphase (attachment cells, lane 3) stages was used to detect the Sp1 phosphorylated level by using immunoblot of anti-Sp1 antibodies. (F) After three independent experiments, the level of Sp1 phosphorylation was quantified. Only statistically significant p values are shown (**p < 0.01 and ***p < 0.001). (G) Cells in interphase and mitotic stages synchronized by nocodazole (lanes 2 and 3) or control HeLa cells (lane 1) were pooled, and the cellular extracts were then probed using anti-Sp3 antibodies. Sp1 detected by using anti-Sp1 antibodies was used as a positive control, cyclin B1 as an M phase marker and actin as an equal loading control.
JNK1 Activity Attributes to the High Phosphorylation of Sp1 in Mitotic Stage
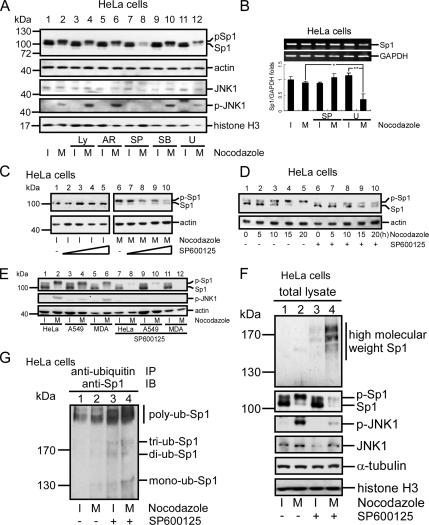
To examine which kinase was activated during mitosis to phosphorylate the Sp1, the kinase inhibitors Ly294002, AR-A014418, SP600125, SB203580, and U0126 were used to inhibit PI3-kinase, glycogen synthase kinase (GSK), JNK, p38, and ERK, respectively, and we examined their effects on the phosphorylated level of Sp1 (Figure 3A). The band shift indicating the phosphorylation still occurred in the mitotic stage after cells were treated with Ly294002, AR-A014418, and SB203580 (Figure 3A, lanes 4, 6, and 10). However, U0126 partially suppressed and SP600125 strongly inhibited the shift (Figure 3A, lanes 8 and 12). It is interesting that, with the SP600125 treatment, not only the phosphorylated level of Sp1 was reduced but also the Sp1 protein level was significantly reduced. We examined next whether this effect of SP600125 on the Sp1 level was due to an inhibition at the transcription level. RNA was thus prepared from interphase or mitotic cells with or without the SP600125 or U0126 treatment, and then it was used to determine the Sp1 mRNA level by RT-PCR (Figure 3B). Results indicated that there was no significant alternation in the Sp1 mRNA level with or without the SP600125 treatment in mitosis, but that the Sp1 mRNA was reduced after the U0126 treatment in mitosis (Figure 3B). These data indicated that the reduction of Sp1 resulted not from a repression of the Sp1 transcription caused by SP600125, but by that from the U0126 treatment. To further address this finding by examining the protein levels, cells were treated with SP600125 and divided into mitosis and interphase groups by nocodazole treatment. No obvious differences in Sp1 level were found with or without the SP600125 treatment in the interphase cells (Figure 3C, lanes 1–5). However, both the Sp1 protein level and its phosphorylated form were dose-dependently decreased in the mitotic cells (Figure 3C, lanes 6–10). In contrast, in cells that were treated with nocodazole or cotreated with SP600125 and collected at different times, the phosphorylated level of Sp1 was time-dependently increased as more and more cells reached mitotic phase (Figure 3D, lanes 1–5), but with the SP600125 treatment, the total Sp1 protein level was also significantly reduced (Figure 3D, lanes 6–10). In addition to HeLa cells, lung cancer cells (A549) and a breast cancer cell line (MDA) were examined for the Sp1 levels in response to the SP600125 treatment (Figure 3E). Data revealed that the level of Sp1 was almost completely abolished under the SP600125 treatment during mitosis in these cells (Figure 3E, lanes 8, 10, and 12). Thus, we concluded that the total level and the phosphorylated level of Sp1 in the mitotic stage are affected by JNK1. Because Sp1 was degraded via the ubiquitination pathway, the cells were treated with nocodazole for collection of interphase and mitotic stage cells and the effect of SP600125 on the ubiquitination of Sp1 was then analyzed (Figure 3F). In mitotic cells, Sp1 ubiquitination was present in the SP600125-treated cells (Figure 3F, lane 4). These results indicated that Sp1 might be shielded from ubiquitination-dependent degradation through a high phosphorylation by JNK1 in the mitotic stage. To further address the JNK activity in regulating the Sp1 stability, cells with nocodazole treatment or SP600125 cotreatment were harvested for immunoprecipitation by using anti-ubiquitin antibodies, followed by immunoblotting with anti-Sp1 antibodies to examine the level of ubiquitinated Sp1 (Figure 3G). Data revealed that the ubiquitinated Sp1 level, especially the poly-ubiquitin-Sp1, was increased in the presence of SP600125 during mitotic phase (Figure 3G, lane 4).
Figure 3.
JNK1 activation in mitosis was important for Sp1 stabilization. HeLa cells were treated with 45 ng/ml nocodazole or cotreated with various kinase inhibitors: Ly294002 (Ly) (inhibits PI3K), AR-A014418 (AR) (inhibits GSK), SP600125 (SP) (inhibits JNK), SB203580 (SB) (inhibits P38), and U0126 (U) (inhibits ERK). (A) Cells were pretreated with the indicated inhibitors for 30 min and then treated with 45 ng/ml nocodazole for 16 h. Attachment cells (I) and round-up cells (M) were pooled separately. These cellular extracts were analyzed using immunoblotting with anti-Sp1, anti-JNK, and anti-p-JNK antibodies. Actin and histone H3 were used as an equal loading control. (B) Total RNA was extracted from interphase or mitotic HeLa cells with or without SP600125 or U0126 treatment, and the Sp1 mRNA level was then determined by RT-PCR. Level of GAPDH mRNA was the internal control. From three independent experiments, the level of Sp1 mRNA was quantified. The p values reaching statistical significance are marked on the graph (*p < 0.05 and **p < 0.01). (C) HeLa cells were treated with different doses (0, 1.25, 2.5, 5, or 10 μM) of SP600125 and 45 ng/ml nocodazole. Interphase and mitotic cell lysates were collected separately, and the expression of Sp1 and actin was analyzed using the same methods as described in A. (D) HeLa cells were pretreated with or without SP600125 for 30 min, and then they were treated with 45 ng/ml nocodazole. After treatment, cells were collected at different time intervals (0, 5, 10, 15, or 20 h). These samples were immunoblotted using anti-Sp1 and anti-actin antibodies. (E) Different cell lines, HeLa, A549, and MDA-MB-231, were treated with 45 ng/ml nocodazole and 10 mM SP600125 (lanes 7–12). The attachment cells and round-up cells were then collected separately, and the level of Sp1 was determined by immunoblot of anti-Sp1 antibodies. Expression of activated JNK determined by anti-p-JNK antibodies was used as SP600125 treatment control and the actin level was used as an internal control. Lanes 1–6 show that HeLa, A549 and MAD-MB-231 cells were only treated with nocodazole as no SP600125 treatment control. (F) Cells were treated with nocodazole or cotreated with SP600125. Western blot of cell lysates using anti-JNK and anti-p-JNK antibodies confirmed JNK protein level and its activity. The same cell lysates were used for immunoblotting with anti-Sp1 antibodies to detect the level of ubiquitinated Sp1, α-tubulin, and histone H3 were used as an equal loading control. (G) The same lysates were used to do the immunoprecipitation with anti-ubiquitin antibodies, and the immunoblotting was performed with anti-Sp1 antibodies. Polyubiquitinated Sp1 is marked with a black line.
JNK Phosphorylates Sp1
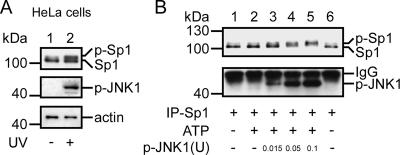
Another method was used to prove that JNK phosphorylates Sp1 and thereby confers the stability of Sp1. Cells were first exposed to UV light to activate JNK1, and then they were examined for the total protein level and the phosphorylated level of Sp1 (Figure 4A). UV exposure indeed activated JNK1 and then increased the level of phosphorylated Sp1 (Figure 4A, lane 2). UV exposure also increased the total level of Sp1. To study the JNK1-induced phosphorylation of Sp1, we used an in vitro kinase assay (Figure 4B). Sp1 was immunoprecipitated from cells, and then it was incubated with various doses of activated JNK1. The results indicated that activated JNK1 dose-dependently induced the degree of band-shifts (Figure 4B, lanes 3–5). Furthermore, to rule out the possibility that JNK1 activation in the mitotic stage was caused by nocodazole treatment rather than being a natural process, we treated cells with nocodazole (Figure 5A) or thymidine (Figure 5B), and then we analyzed varieties of proteins by using immunoblotting. Judging from the contents of cyclin E, a G1/S marker, and cyclin B1, a G2/M marker, we found that cells stayed mostly in the G1/S phase 12–16 h after removal of nocodazole. Also, during this period, the cyclin E level was high, but the following were absent: Sp1 band-shift or JNK1 activation (Figure 5A, lanes 1 and 2). Twenty-two to 24 h after the treatment, cells stayed mostly in the G2/M phase. During this period, the cyclin B1 level was high, JNK was activated, and the phosphorylation of Sp1 increased (Figure 5A, lanes 4 and 5). To further confirm that Sp1 was phosphorylated by activated JNK1 in mitosis, cells were treated with thymidine to collect cells of different stages in cell cycle (Figure 5B). Results indicated that, 0–8 h after thymidine treatment, most of Sp1 still stayed at the bottom band (lanes 1–4). Ten, 11, and 12 h after the treatment, part of Sp1 shifted to the top band (lanes 5–7). This result was consistent with that shown in Figure 5A. Because of these results, the possibility that nocodazole-activated JNK1 was ruled out. In summary, these results were consistent with the idea that JNK1 was activated during the mitotic stage and the active form of JNK1 induced the Sp1 phosphorylation to shield the Sp1 from ubiquitin-dependent degradation.
Figure 4.
JNK1 phosphorylated Sp1 during mitosis. (A) HeLa cells were directly exposed or not exposed to UV radiation for 10 s and recultured in an incubator for 1 h to activate JNK. Cell lysates were immunoblotted using anti-Sp1 and anti-p-JNK antibodies. Actin was used as an equal loading control. (B) Sp1 was purified using immunoprecipitation with anti-Sp1 antibodies from HeLa cells, and was incubated with different doses of activated JNK (0, 0.015, 0.05, or 0.1 U) for a JNK kinase assay (see Materials and Methods). Products were immunoblotted using anti-Sp1 and anti-p-JNK antibodies. The IgG signal is from immunoprecipitated Sp1 antibodies.
Figure 5.
Elevated JNK activation increased the phosphorylation of Sp1. (A) HeLa cells were obtained using synchronization for 16 h with 45 ng/ml nocodazole. Mitotic cells were shaken-off, washed with PBS, and replated in fresh medium. The released cells were collected at different time intervals (12, 16, 20, 22, or 24 h). These samples were immunoblotted using anti-Sp1 and anti-p-JNK antibodies. Anti-JNK antibodies were used to indicate that the total JNK in each of these samples was equal. Cyclin B1 was used as a mitotic phase marker, and cyclin E was used as a G1/S phase marker. Actin was used as an equal loading control. (B) HeLa cells were treated with double thymidine to synchronize cells in G1/S phase and then washed with PBS and recultured in fresh medium to release the cell cycle. Cells were collected at different time intervals (0, 4, 6, 8, 10, 11, or 20 h). Cell lysates were prepared and immunoblotted using antibodies against Sp1, JNK, p-JNK, cyclin B1, and actin.
JNK1 Phosphorylates Sp1 at Thr278 and Thr739
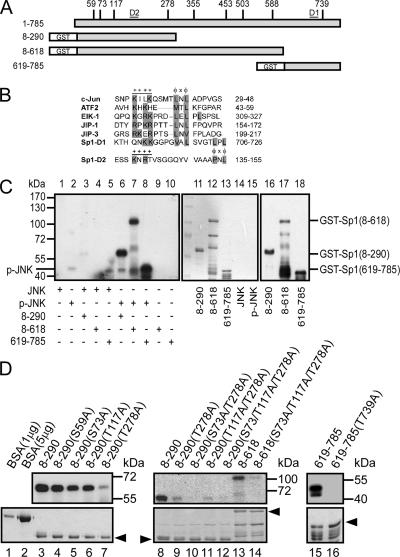
We next studied which residues on Sp1 had been phosphorylated by JNK1. We first searched the amino acid sequence of the Sp1 to screen for the candidates that JNK might phosphorylate (Figure 6A), and we found nine residues as such: the 59th (Ser), 73rd (Ser), 117th (Thr), 278th (Thr), 355th (Thr), 453rd (Thr), 503rd (Thr), 588th (Ser), and 739th (Thr) amino acids. Therefore, three truncated fragments, GST-Sp1 (8-618), GST-Sp1 (8-290), and GST-Sp1 (618-785), were used to perform the JNK in vitro kinase assay (Figure 6C). Activated JNK phosphorylated three of the proteins (Figure 6C, lanes 6–8), but inactivated JNK phosphorylated none (Figure 6C, lanes 3–5). Next, we did a point mutation within the three truncated fragments and performed another JNK in vitro kinase assay (Figure 6D). There was just little decrease in the phosphorylated level when the GST-Sp1 (8-290) fragment was mutated at the 73rd and 117th residues (Figure 6D, lanes 5 and 6). However, the phosphorylation signal was nearly abolished when the 278th amino acid was mutated (Figure 6D, lane 7). Furthermore, the mutant studies indicated that the 278th might be the only residue within the 1st to 618th of Sp1 that could be phosphorylated by activated JNK (Figure 6D, lanes 8–14). We also found that JNK phosphorylated residue 739 in the C terminus of Sp1. The phosphorylation signal was abolished when the 739th amino acid was mutated (Figure 6D, lanes 15 and 16). Because all other JNK substrates, such as the transcription factors c-Jun, ATF2, and Elk-1, and JNK scaffold proteins, such as JIP-1 and JIP-3, had been shown to contain mitogen-activated protein kinase (MAPK) docking sites (Ho et al., 2006), we inspected the amino acid sequence of Sp1 for potential d-sites. Two putative D-sites (hereafter D1 and D2) were found near the Thr739 and Thr278 amino acid N termini (Figure 6B). These results indicated that JNK phosphorylated Sp1 at the 278th and 739th amino acids.
Figure 6.
JNK phosphorylated the Thr278/739 sites of Sp1. (A) The potential JNK-phosphorylated Thr/Ser sites of Sp1 are indicated (S59, S73, T117, T278, T355, T453, T503, S588, and T739), and different GST-Sp1 deletion constructs are also indicated. (B) Known D-sites in JNK-binding proteins aligned with the putative D-sites in Sp1. D1 and D2 are located in residues 706-726 and residues 135-155, and they are marked in A. Consensus basic (+) and hydrophobic (φ) residues are shown in a gray background. (C) The different GST-Sp1 fragments (amino acids [aa] 8-290, aa 8-618, and aa 619-785) were combined with [γ-32P]ATP and activated JNK for an in vitro kinase assay (lanes 6–8). The expressions of these fragments were detected using Coomassie Blue (lanes 11–15) or immunoblotted using anti-Sp1 antibodies (lanes 16–18). (D) The potential JNK-phosphorylated serine/threonine sites were mutated to alanine as indicated. These mutated GST-Sp1 proteins were expressed in E. coli and purified. The yields of these mutants were detected using Coomassie Blue staining (bottom). Products were analyzed as described in C. Bovine serum albumin (BSA) was used to compare the concentration of proteins in Coomassie Blue staining.
JNK-induced Phosphorylation of Sp1 Confers the Sp1 Stability Necessary for the Transcriptional Activity of 12(S)-Lipoxygenase and Cell Growth
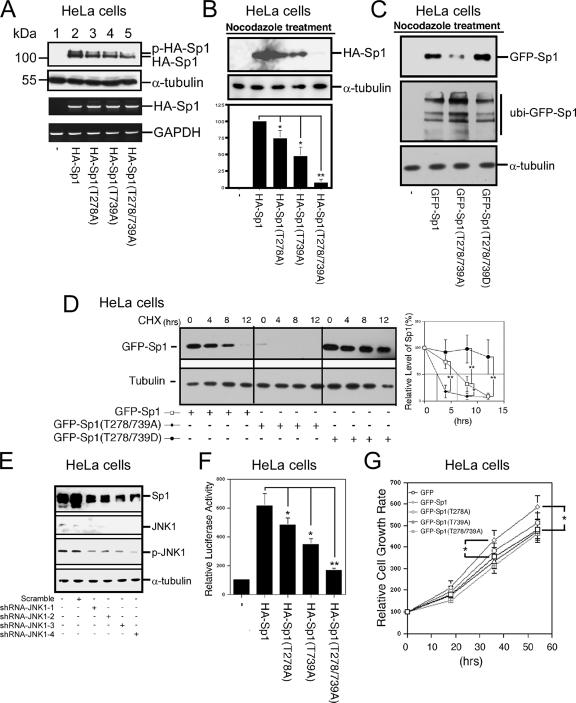
To directly study the relationship between JNK-induced phosphorylation of Sp1 and the Sp1 stability, different mutants, including HA-Sp1, HA-Sp1 (T278A), HA-Sp1 (T739A), and HA-Sp1 (T278/739A), were constructed and transfected into HeLa cells for examinations of protein levels (Figure 7A). We found that the level of Sp1 in HA-Sp1- (T278A) and HA-Sp1 (T739A)–overexpressing cells was reduced by ∼40% compared with that in wild-type HA-Sp1–expressing cells (Figure 7A, lanes 2–4). However, the level of Sp1 was reduced by ∼70% with the HA-Sp1 (T278A/T739A) overexpression (Figure 7A, lane 5). In addition, to rule out the possibility that this reduction was due to different transcriptional activities, we studied the respective RNA levels by using RT-PCR (Figure 7A), and we found that all of the plasmids used possessed the same transcriptional activity. To further address this possibility that phosphorylation of Sp1 by JNK in mitosis might be involved in Sp1 stability, wild-type Sp1, Sp1(T278A), Sp1(T739A), and Sp1(T278/739A) were overexpressed in HeLa cells. Cells were then confined in mitosis by nocodazole and assayed for Sp1 levels (Figure 7B). Data revealed that the levels of HA-Sp1(T278A) and HA-Sp1(T739A) were partially reduced, but the HA-Sp1(T278/739A) level was almost completely abolished during mitosis. In addition, to directly study the relationship between Sp1 stability and Sp1 phosphorylation, GFP-Sp1(T278/739D) was constructed to mimic the phosphorylated form of Sp1, and it was used to study its stability in mitosis (Figure 7C). Results show that GFP-Sp1(T278/739D) level was increase, and the ubiquitin-GFP-Sp1 level was decreased compared with the wild-type Sp1. This result provides the direct evidence that the Sp1 stability can be maintained by JNK via phosphorylation during mitosis. To further strengthen that Sp1 phosphorylated by JNK1 in mitosis was involved in the stability of Sp1, expression vectors including GFP-Sp1, GFP-Sp1(T278/739A), and GFP-Sp1(T278/739D) were overexpressed in HeLa cells, and cells were then synchronized to the mitotic stage. The Sp1 stability was then determined in the presence of cycloheximide (Figure 7D). Data indicated that the half lives of GFP-Sp1, GFP-Sp1(T278/739A), and GFP-Sp1(T278/739D) were ∼6, 2.5, and >12 h, respectively. In addition, JNK1 was directly inhibited by short hairpin RNA (shRNA)-JNK1 to characterize the Sp1 level (Figure 7E). Data also revealed that, based on the level of JNK1 and phospho-JNK1 as an internal control, Sp1 level was really decreased obviously. These results directly prove that the two JNK1-phosphorylated residues are important for the Sp1 stability. The biological functions of Sp1 and its mutants were also assayed for the transcriptional activity of the target gene 12(S)-lipoxygenase (Figure 7F). Gene expression increased by ∼6-fold with the expression of wild-type Sp1, but the gene expression with the HA-Sp1(T278A), HA-Sp1(T739A), or HA-Sp1(T278/739A) overexpression increased by 4.7-, 4.1-, and 1.6-fold, respectively. To study the effect of Sp1 on cell proliferation, wild-type Sp1 and various Sp1 mutants were transfected and examined for cell number counts after 18, 36, and 54 h of incubation (Figure 7G). Results indicated that overexpression of GFP-Sp1 increased the cell proliferation, but GFP-Sp1 (T278A), GFP-Sp1 (T739A), and GFP-Sp1 (T278/739A) did not significantly increase cell proliferation.
Figure 7.
The Sp1 mutated at 278/739 was more unstable than wild-type Sp1 in cells. (A) HeLa cells were transfected with HA-Sp1, HA-Sp1(T278A), HA-Sp1(T739A), or HA-Sp1(T278/739A). The cell extracts were prepared from these HeLa cells and immunoblotted using anti-HA and anti-α-tubulin antibodies. The cells were transfected with control vector (lane 1). Total RNA was extracted from these HeLa cells and underwent RT-PCR using specific primers for HA-Sp1 (Sp1-forward and Sp6 reverse) and GAPDH. HA-Sp1 mRNA was used as an equal control for transfection efficiency. (B) pHA, pHA-Sp1, pHA-Sp1 (T278A), pHA-Sp1 (T739A), and pHA-Sp1 (T278/739A) were transfected into HeLa cells for 12 h, and cells were then arrested at mitosis stage by nocodazole treatment (45 ng/ml) for 16 h. Equal mitotic cell number was used to determine the Sp1 level by immunoblot with anti-HA antibodies, and the tubulin was used as the internal control. From three independent experiments, the level of HA-Sp1, HA-Sp1(T278A), HA-Sp1(T739A), and HA-Sp1(T278/739A) was quantified and normalized with that of tubulin. Only statistically significant p values are shown (*p < 0.05 and **p < 0.01). (C) pGFP, pGFP-Sp1, pGFP-Sp1(T278/739A), and pHA-Sp1 (T278/739D) were transfected into HeLa cells for 12 h, and cells were then arrested at mitosis stage by nocodazole treatment (45 ng/ml) for 16 h. Equal mitotic cell number was used to determine the Sp1 and ubiquitinated Sp1 level by immunoblot with anti-GFP antibodies, and the tubulin was used as the internal control. (D) pGFP, pGFP-Sp1, pGFP-Sp1(T278/739A), and pHA-Sp1 (T278/739D) were transfected into HeLa cells for 12 h, and cells were then arrested at mitosis stage by nocodazole treatment (45 ng/ml) for 16 h. Half-life of GFP-Sp1 and its mutants was determined in the presence of cycloheximide. After three independent experiments, the level of Sp1 protein was quantified. Statistically significant differences are indicated by the corresponding p values of Student's t test (*p < 0.05 and **p < 0.01). (E) Four shRNA-JNK1s (shRNA-JNK1-1, shRNA-JNK1-2, shRNA-JNK1-3, and shRNA-JNK1-4) were transfected into HeLa cells individually for 48 h. Cells were harvested and analyzed with immunoblot of anti-Sp1, anti-JNK1, anti-phospho-JNK1, and anti-tubulin antibodies. (F) HA-Sp1, HA-Sp1(T278A), HA-Sp1(T739A), or HA-Sp1(T278/739A) were cotransfected with pXP 7-1, the promoter of 12(S)-lipoxygenase, into HeLa cells for 24 h, and the luciferase activity was then analyzed. All of the experiments were done three times independently. The p values reaching statistical significance are marked on the graph (*p < 0.05 and **p < 0.01). (G) pGFP, pGFP-Sp1, pGFP-Sp1 (T278A), pGFP-Sp1 (T739A), and pGFP-Sp1 (T278/739A) were transfected into HeLa cells, and the cell number was then counted after 18-, 36-, and 54-h incubation. All of the experiments were independently performed three times, and the statistical analysis was performed by Student's t test.
Sp1 Accumulation and JNK Activation Occurred in MNU-induced Mammalian Cancer Tumor
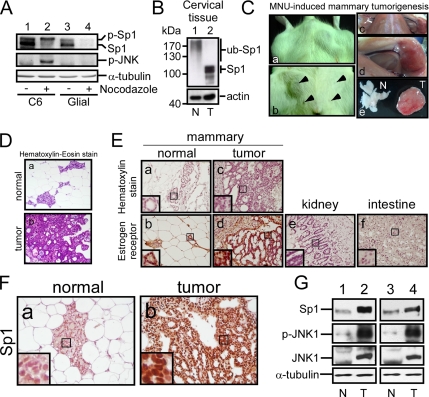
Our data indicated so far that the JNK activation affects the Sp1 accumulation (Figures 3 and 7). Sp1 is known to be increased in many tumor cells (Tellez and Bar-Eli, 2003; Wang et al., 2003; Hosoi et al., 2004; Safe and Abdelrahim, 2005). To study whether the Sp1 accumulation is related to JNK activation during mitosis in tumorigenesis, the rat glioma C6 cells and rat primary glial cells were divided into mitosis and interphase groups by nocodazole treatment (Figure 8A). In glioma C6 cells, Sp1 was consistent with the previous result that had most of Sp1 to be phosphorylated form during mitosis and the level of Sp1 could be maintained. Here, we also found that JNK was activated in mitotic stage. However, in primary glial cells, the Sp1 protein level was significantly reduced, and JNK was inactivated during mitosis. In addition, the amount of Sp1 was accumulated in glioma C6 cells and cervical cancer tissue was compared with primary glial cells and normal cervical tissue (Figure 8, A and B). In an animal study, MNU injection is associated with the formation of mammary tumors, at an incidence of >90%, which are predominantly adenocarcinomas of a ductal origin with the presence of only very few benign tumors (Russo et al., 1990). In our study, tumors were raised in 12 of 17 rats within 8 wk after the MNU injection (Figure 8C). Previous study indicated that MNU predominantly induces adenocarcinoma in mammals (Russo et al., 1990). Thus, the mammary cancer marker, estrogen receptors, was examined by immunohistochemistry (Figure 8E). Lack of estrogen receptor staining in intestine and kidney tissues served as the negative control (Figure 8E, e and f). The signal of estrogen receptors was found only in normal mammary tissues and in MNU-induced tumor (Figure 8E, b and d). Furthermore, we found that amount of Sp1 was accumulated inside the MNU-induced tumors compared with that observed in mammary normal cells (Figure 8, F and G). In addition, the phosphorylation level of JNK1 was increased in the MNU-induced tumor (Figure 8G). Thus, a correlation may exist between the Sp1 accumulation and the JNK1 activity in mammary tumors.
Figure 8.
Sp1 accumulation and JNK activation in glioma cells and MNU-induced tumor. (A) Rat glioma C6 cell line (lanes 1 and 2) and primary glial cells from postnatal day 0 to day 1 rat pups (lanes 3 and 4) were treated with nocodazole for 24 h, and then they were divided into interphase and mitotic cells. These cellular extracts were probed using anti-Sp1 and p-JNK antibodies. The α-tubulin was used as an equal loading control. (B) The normal cervical tissue (N) and cervical cancer tissue (T) were subjected to immunoblot analysis by using antibodies against Sp1. Actin was used as an equal loading control (C) Rats intraperitoneal injected with 50 mg/kg MNU were killed after 8 wk, and the normal and tumor tissues were then harvested. The arrow indicated the nipple position. (D) H&E staining was performed to determine the tissue and tumor type. (E) Estrogen receptor recognized by its antibodies inside the tissue was considered as a mammalian cancer marker. The intestine and kidney tissues were used as the negative control. (F) Sp1 level was determined in the normal and tumor mammalian tissues by immunohistochemistry by using anti-Sp1 antibodies. (G) The level of Sp1, JNK1, and p-JNK1 in normal and tumor tissues was studied by immunoblot with anti-Sp1, anti-JNK1, and anti-phospho-JNK antibodies, respectively.
DISCUSSION
In the present study, the mechanism whereby cells transfer Sp1 to the daughter cells was characterized. We found that JNK1, which is activated during mitosis, induced a high degree of Sp1 phosphorylation during mitosis. Although previous study has indicated that Sp1 displayed the phosphorylated form in PC3 cells with the constitutively active JNK pathway, it was still not clear whether Sp1 is phosphorylated by JNK (Benasciutti et al., 2004). In the present study, we also found that JNK1 phosphorylated Sp1 at Thr278 and Thr739, and the high level of phosphorylation shielded Sp1 from ubiquitin-dependent degradation. Sp1 accumulation in many tumors might thus be resulted from the Sp1 phosphorylation by activated JNK1.
Sp1 was one of the first transcription factors purified and cloned from mammalian cells (Dynan and Tjian, 1983; Kadonaga et al., 1987), and it is generally considered as a factor that determines the core activity of promoter by a direct interaction with factors at the basal transcription machinery, including cooperation with several transcriptional activators, such as CRSP, p300/CBP, steroidogenic factor-1, and TAFII130 (Xiao et al., 2000; Taatjes et al., 2002; Huang et al., 2004; Sugawara et al., 2004). Previous studies have shown that diverse kinase pathways can phosphorylate different serine or threonine residues on Sp1. For example, DNA-dependent protein kinase phosphorylated Ser131 of Sp1 and induced Sp1 transcriptional activity (Chun et al., 1998). Casein kinase II phosphorylated Thr579 of Sp1 and repressed its DNA binding ability (Armstrong et al., 1997). Protein kinase A (PKA) phosphorylated the N terminus of Sp1, increasing both the DNA binding activity and the transcriptional activity of Sp1 (Rohlff et al., 1997). In this study, we established that Sp1 was phosphorylated at Thr278 and Thr739 by JNK (Figure 6). Although the phosphorylation signal was reduced very little when the 73rd and 117th residues were mutated (Figure 6D, lanes 5 and 6), these residues located near the MAPK docking site, Sp1-D2, might provide some nonspecific phosphorylation signal in JNK in vitro kinase assay. All previous studies of the phosphorylation of Sp1 focused on the regulation of DNA binding affinity that mediates the transcriptional activity of target genes. In the present study, we found that the Sp1 phosphorylated by JNK1 in mitosis was related to the stability of the Sp1. There is evidence to support this finding. First, the Sp1 band-shift occurred during the mitotic period (Figure 2). The CIP assay proved that this band-shift was caused by the high level of Sp1 phosphorylation. When cells were treated with SP600125, the band-shift disappeared and the level of Sp1 reduced significantly through the ubiquitin-dependent pathway (Figure 3). In Figure 3G, there was still a little Sp1 that could be ubiquitinated, and degraded in the absence of SP600125 treatment during interphase. Previous studies have revealed that glycosylation of Sp1 might be related to the Sp1 stability (Han and Kudlow, 1997), and Rpt6 phosphorylated by PKA could affect the Sp1 degradation (Zhang et al., 2007). Therefore, not only JNK1 activation but also other mechanisms (e.g., PKA signaling) might be involved in the regulation of the Sp1 stability in different cell cycle phases. Although both JNKs and ERK1/2 were involved in regulating the Sp1 level, our results (Figure 3B) indicated that ERK1/2 contributes to the synthesis of Sp1 protein, whereas JNKs increases the Sp1 stability. Furthermore, only JNK1, but not ERK1/2, was activated during mitosis. In addition, in terms of why the two bands were reduced under SP600125 treatment, it might be because unphosphorylated Sp1 cannot be phosphorylated, but phosphorylated Sp1 can be dephosphorylated by phosphatase continually under SP600125-treated cells. Second, with the overexpression of Sp1, Sp1 (T278A), Sp1 (T739A), and Sp1 (T278/739A), the protein level of wild-type Sp1 was higher than that of Sp1 mutants in normal cells (Figure 7A), but during mitosis, the Sp1 level was nearly abolished when the JNK1-phosphorylated residues T278/739 were mutated (Figure 7B). Third, the protein level of mutant Sp1(T278/739D), mimicking the phosphorylation form of Sp1, was also significantly increased compared with the native Sp1 (Figure 7, C and D). Fourth, Sp1 level was really decreased under JNK1 knockdown by shRNA-JNK1 (Figure 7E). Fifth, in primary glial cells, JNK was inactivated during mitosis and the Sp1 protein level was significantly reduced (Figure 8A). Based on these results, we conclude that the JNK-induced Sp1 phosphorylation is involved in stabilizing Sp1, facilitating the distribution of Sp1 to daughter cells. Several reports have mentioned an association between the phosphorylation and the stability of proteins. For example, inhibitor of nuclear factor-κB can be degraded in a JNK-dependent ubiquitination in the regulation of cyclooxygenase-2 expression (Ki et al., 2006). JNK inflammatory signaling can mediate ASK1 degradation in the presence of tumor necrosis factor (He et al., 2006). In addition, in Drosophila, JNK activation induces the DTRAF1 degradation (Kuranaga et al., 2002). These studies suggest that the activation of JNK induces protein degradation. Conversely, the inactive JNK fragment regulates the p53 stability, and a peptide corresponding to the JNK binding site on p53 efficiently blocks the ubiquitination of p53 (Fuchs et al., 1998). Although these studies reported that JNK is related to the ubiquitin-dependent degradation, our findings suggested that JNK activation increases Sp1 stability. Therefore, activation of JNK might have another role in protecting the degradation of proteins, including Sp1. We did not check whether JNK2, in addition to JNK1, is able to phosphorylate Sp1, but the high phosphorylation of Sp1 may be contributed by JNK1, because our results as shown in Figures 3A and 5A revealed that only JNK1, not JNK2, could be activated in mitosis.
Other studies have shown that abnormal Sp1 activation might augment the growth and metastatic potential of tumor cells through the overexpression of many downstream genes of Sp1, including VEGF (Abdelrahim et al., 2004; Hosoi et al., 2004; Safe and Abdelrahim, 2005). The role of Sp1 as an essential transcription factor for many genes that regulate cell growth, angiogenesis, and survival has been proved in pancreatic, gastric, and colorectal cancers (Han and Kudlow, 1997; Wang et al., 2003; Abdelrahim et al., 2004; Hosoi et al., 2004). In this study, we found that an overexpression of Sp1 caused the activation of 12(S)-lipoxygenase that correlates with tumorigenesis and cell proliferation (Figure 7G). These effects were nevertheless reversed by mutations of the JNK1-phosphorylated residues (T278/738A) (Figure 7). In addition, a related study has indicated that overexpression of the dominant-negative form of JNK reduces the basal transcriptional activation of the Sp1 target gene, human urokinase-minimal promoter (Benasciutti et al., 2004). We also used MNU to induce the formation of mammary adenocarcinoma, and we found Sp1 accumulation and JNK1 phosphorylation, which might result from the increase in the total JNK1 level (Figure 8, F and G), indicating that the level of Sp1 may thus be important for tumor formation. In addition, we also found that the protein level of Sp1 was significantly increased in glioma C6 cells and human cervical tumors compared with primary glial cells and normal cervical tissue (Figure 8, A and B). Other studies have indicated that glycosylation of Sp1 is related to its degradation by proteasome (Han and Kudlow, 1997). In addition, the N-terminal region of Sp1 is important for proteasome-dependent degradation in vitro (Su et al., 1999). The proteasome subunit Rpt6/Sug1/p45 interacts with Sp1 and might be involved in its degradation (Su et al., 2000). Our results showed that during interphase, most of the Rpt6 localized in the cytoplasm, and it had no contact with Sp1 that localized in the nucleus. However, when cells entered mitosis and the nuclear envelope disappeared, Rpt6 colocalized with Sp1 (Hung, unpublished data). Therefore, in tumor cells, Sp1 might need a mechanism to be shielded from ubiquitination-dependent degradation when cells enter the mitotic phase of the cell cycle. According to the results of our study, we proposed a novel model that the JNK1 activation during tumorigenesis phosphorylates Sp1 at Thr278 and Thr739, thus causing Sp1 accumulation.
The present study focused primarily on the distribution and stability of Sp1 in the mitotic stage of cells. Using indirect immunofluorescence analysis, we studied asynchronous HeLa cells during mitosis. Sp1 departed the condensed chromatin at prophase until early telophase, and its protein level could be seen in different phases (Figure 1). Similar Sp1 distribution results were recently reported in the MCF-7 human breast cancer cell line (He and Davie, 2006). In the present study, we presented evidence to support posttranslational modification might be involved in regulating Sp1 stability in the mitotic period. After treating HeLa, A549, MDA-MB-231, and C6 cells with nocodazole to keep a large number of cells at the G2/M stage, a change in the electrophoretic position of Sp1 on the SDS-polyacrylamide gel were observed (Figure 2B). Although previous studies indicated that the phosphorylation of Sp1 regulates the DNA binding ability of Sp1 (Chu and Ferro, 2005; Stark and Assaraf, 2006), our results showed that, during mitosis Sp1 in phosphorylated form dissociated from DNA (Figure 1). Together, these findings indicated that the phosphorylation modification might influence the DNA binding ability of Sp1 and then cause Sp1 to move rapidly off chromatin in mitosis. However, which phosphorylated residue(s) is important for Sp1 localization during mitosis needs further study. Previous studies indicated that Sp1 is significantly increased in many tumor cells and may thus regulate many genes related to tumorigenesis (Tellez and Bar-Eli, 2003; Wang et al., 2003; Hosoi et al., 2004; Safe and Abdelrahim, 2005). Herein, we demonstrated that JNK1 could be activated during mitosis, thus phosphorylating Sp1, increasing Sp1's stability, and leading to Sp1 accumulation during tumorigenesis. Sp1 stability in mitosis and in tumorigenesis might be an important therapeutic target for the tumor therapy.
ACKNOWLEDGMENTS
We thank Dr. Tsung-Ping Su for critical reviewing of the manuscript and Dr. Wen-Chun Hung for providing the pGEX-6P-Sp1. This work was supported by the National Cheng Kung University program for Promoting Academic Excellence and Developing World class Research Centers, and grant NSC 95-2320-B-006-079-MY2 from the National Science Council of the Republic of China.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-09-0881) on January 16, 2008.
REFERENCES
- Abdelrahim M., Safe S. Cyclooxygenase-2 inhibitors decrease vascular endothelial growth factor expression in colon cancer cells by enhanced degradation of Sp1 and Sp4 proteins. Mol. Pharmacol. 2005;68:317–329. doi: 10.1124/mol.105.011825. [DOI] [PubMed] [Google Scholar]
- Abdelrahim M., Smith R., 3rd, Burghardt R., Safe S. Role of Sp proteins in regulation of vascular endothelial growth factor expression and proliferation of pancreatic cancer cells. Cancer Res. 2004;64:6740–6749. doi: 10.1158/0008-5472.CAN-04-0713. [DOI] [PubMed] [Google Scholar]
- Armstrong S. A., Barry D. A., Leggett R. W., Mueller C. R. Casein kinase II-mediated phosphorylation of the C terminus of Sp1 decreases its DNA binding activity. J. Biol. Chem. 1997;272:13489–13495. doi: 10.1074/jbc.272.21.13489. [DOI] [PubMed] [Google Scholar]
- Benasciutti E., Pages G., Kenzior O., Folk W., Blasi F., Crippa M. P. MAPK and JNK transduction pathways can phosphorylate Sp1 to activate the uPA minimal promoter element and endogenous gene transcription. Blood. 2004;104:256–262. doi: 10.1182/blood-2003-08-2661. [DOI] [PubMed] [Google Scholar]
- Bonello M. R., Khachigian L. M. Fibroblast growth factor-2 represses platelet-derived growth factor receptor-α (PDGFR-α) transcription via ERK1/2-dependent Sp1 phosphorylation and an atypical cis-acting element in the proximal PDGFR-α promoter. J. Biol. Chem. 2004;279:2377–2382. doi: 10.1074/jbc.M308254200. [DOI] [PubMed] [Google Scholar]
- Bouwman P., Philipsen S. Regulation of the activity of Sp1-related transcription factors. Mol. Cell. Endocrinol. 2002;195:27–38. doi: 10.1016/s0303-7207(02)00221-6. [DOI] [PubMed] [Google Scholar]
- Chu S., Ferro T. J. Sp 1, regulation of gene expression by phosphorylation. Gene. 2005;348:1–11. doi: 10.1016/j.gene.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Chun R. F., Semmes O. J., Neuveut C., Jeang K. T. Modulation of Sp1 phosphorylation by human immunodeficiency virus type 1 Tat. J. Virol. 1998;72:2615–2629. doi: 10.1128/jvi.72.4.2615-2629.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. J. Signal transduction by the JNK group of MAP kinases. Cell. 2000;103:239–252. doi: 10.1016/s0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- Deniaud E., Baguet J., Mathieu A. L., Pages G., Marvel J., Leverrier Y. Overexpression of Sp1 transcription factor induces apoptosis. Oncogene. 2006;25:7096–7105. doi: 10.1038/sj.onc.1209696. [DOI] [PubMed] [Google Scholar]
- Druckrey H., Preussmann R., Ivankovic S., Schmahl D. Organotropic carcinogenic effects of 65 various N-nitroso-compounds on BD rats. Z. Krebsforsch. 1967;69:103–201. [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell. 1983;35:79–87. doi: 10.1016/0092-8674(83)90210-6. [DOI] [PubMed] [Google Scholar]
- Firestone G. L., Bjeldanes L. F. Indole-3-carbinol and 3–3′-diindolylmethane antiproliferative signaling pathways control cell-cycle gene transcription in human breast cancer cells by regulating promoter-Sp1 transcription factor interactions. J. Nutr. 2003;133:2448S–2455S. doi: 10.1093/jn/133.7.2448S. [DOI] [PubMed] [Google Scholar]
- Fojas de Borja P., Collins N. K., Du P., Azizkhan-Clifford J., Mudryj M. Cyclin A-CDK phosphorylates Sp1 and enhances Sp1-mediated transcription. EMBO J. 2001;20:5737–5747. doi: 10.1093/emboj/20.20.5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs S. Y., Adler V., Buschmann T., Yin Z., Wu X., Jones S. N., Ronai Z. JNK targets p53 ubiquitination and degradation in nonstressed cells. Genes Dev. 1998;12:2658–2663. doi: 10.1101/gad.12.17.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs S. Y., Dolan L., Davis R. J., Ronai Z. Phosphorylation-dependent targeting of c-Jun ubiquitination by Jun N-kinase. Oncogene. 1996;13:1531–1535. [PubMed] [Google Scholar]
- Fuchs S. Y., Xie B., Adler V., Fried V. A., Davis R. J., Ronai Z. c-Jun NH2-terminal kinases target the ubiquitination of their associated transcription factors. J. Biol. Chem. 1997;272:32163–32168. doi: 10.1074/jbc.272.51.32163. [DOI] [PubMed] [Google Scholar]
- Han I., Kudlow J. E. Reduced O-glycosylation of Sp1 is associated with increased proteasome susceptibility. Mol. Cell. Biol. 1997;17:2550–2558. doi: 10.1128/mcb.17.5.2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S., Davie J. R. Sp1 and Sp3 foci distribution throughout mitosis. J. Cell Sci. 2006;119:1063–1070. doi: 10.1242/jcs.02829. [DOI] [PubMed] [Google Scholar]
- He Y., Zhang W., Zhang R., Zhang H., Min W. SOCS1 inhibits tumor necrosis factor-induced activation of ASK1-JNK inflammatory signaling by mediating ASK1 degradation. J. Biol. Chem. 2006;281:5559–5566. doi: 10.1074/jbc.M512338200. [DOI] [PubMed] [Google Scholar]
- Higuchi H., Grambihler A., Canbay A., Bronk S. F., Gores G. J. Bile acids up-regulate death receptor 5/TRAIL-receptor 2 expression via a c-Jun N-terminal kinase-dependent pathway involving Sp1. J. Biol. Chem. 2004;279:51–60. doi: 10.1074/jbc.M309476200. [DOI] [PubMed] [Google Scholar]
- Ho D. T., Bardwell A. J., Grewal S., Iverson C., Bardwell L. Interacting JNK-docking sites in MKK7 promote binding and activation of JNK mitogen-activated protein kinases. J. Biol. Chem. 2006;281:13169–13179. doi: 10.1074/jbc.M601010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoi Y., Watanabe T., Nakagawa K., Matsumoto Y., Enomoto A., Morita A., Nagawa H., Suzuki N. Up-regulation of DNA-dependent protein kinase activity and Sp1 in colorectal cancer. Int. J. Oncol. 2004;25:461–468. [PubMed] [Google Scholar]
- Huang Y. C., Chen J. Y., Hung W. C. Vitamin D3 receptor/Sp1 complex is required for the induction of p27Kip1 expression by vitamin D3. Oncogene. 2004;23:4856–4861. doi: 10.1038/sj.onc.1207621. [DOI] [PubMed] [Google Scholar]
- Hung J. J., Wang Y. T., Chang W. C. Sp1 deacetylation induced by phorbol ester recruits p300 to activate 12(S)-lipoxygenase gene transcription. Mol. Cell. Biol. 2006;26:1770–1785. doi: 10.1128/MCB.26.5.1770-1785.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadonaga J. T., Carner K. R., Masiarz F. R., Tjian R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell. 1987;51:1079–1090. doi: 10.1016/0092-8674(87)90594-0. [DOI] [PubMed] [Google Scholar]
- Ki S. H., Choi M. J., Lee C. H., Kim S. G. Galpha 12 specifically regulates COX-2 induction by sphingosine 1-phosphate: role for JNK-dependent ubiquitination and degradation of IκBα. J. Biol. Chem. 2006;282:1938–1947. doi: 10.1074/jbc.M606080200. [DOI] [PubMed] [Google Scholar]
- Kotsopoulos J., Medline A., Renlund R., Sohn K. J., Martin R., Hwang S. W., Lu S., Archer M. C., Kim Y. I. Effects of dietary folate on the development and progression of mammary tumors in rats. Carcinogenesis. 2005;26:1603–1612. doi: 10.1093/carcin/bgi117. [DOI] [PubMed] [Google Scholar]
- Kuranaga E., Kanuka H., Igaki T., Sawamoto K., Ichijo H., Okano H., Miura M. Reaper-mediated inhibition of DIAP1-induced DTRAF1 degradation results in activation of JNK in Drosophila. Nat. Cell Biol. 2002;4:705–710. doi: 10.1038/ncb842. [DOI] [PubMed] [Google Scholar]
- Liaw Y. W., Liu Y. W., Chen B. K., Chang W. C. Induction of 12-lipoxygenase expression by phorbol 12-myristate 13-acetate in human epidermoid carcinoma A431 cells. Biochim. Biophys. Acta. 1998;1389:23–33. doi: 10.1016/s0005-2760(97)00090-8. [DOI] [PubMed] [Google Scholar]
- Milanini-Mongiat J., Pouyssegur J., Pages G. Identification of two Sp1 phosphorylation sites for p42/p44 mitogen-activated protein kinases: their implication in vascular endothelial growth factor gene transcription. J. Biol. Chem. 2002;277:20631–20639. doi: 10.1074/jbc.M201753200. [DOI] [PubMed] [Google Scholar]
- Mortensen E. R., Marks P. A., Shiotani A., Merchant J. L. Epidermal growth factor and okadaic acid stimulate Sp1 proteolysis. J. Biol. Chem. 1997;272:16540–16547. doi: 10.1074/jbc.272.26.16540. [DOI] [PubMed] [Google Scholar]
- Nagata Y., Nishida E., Todokoro K. Activation of JNK signaling pathway by erythropoietin, thrombopoietin, and interleukin-3. Blood. 1997;89:2664–2669. [PubMed] [Google Scholar]
- Reisinger K., Kaufmann R., Gille J. Increased Sp1 phosphorylation as a mechanism of hepatocyte growth factor (HGF/SF)-induced vascular endothelial growth factor (VEGF/VPF) transcription. J. Cell Sci. 2003;116:225–238. doi: 10.1242/jcs.00237. [DOI] [PubMed] [Google Scholar]
- Rohlff C., Ahmad S., Borellini F., Lei J., Glazer R. I. Modulation of transcription factor Sp1 by cAMP-dependent protein kinase. J. Biol. Chem. 1997;272:21137–21141. doi: 10.1074/jbc.272.34.21137. [DOI] [PubMed] [Google Scholar]
- Russo J., Gusterson B. A., Rogers A. E., Russo I. H., Wellings S. R., van Zwieten M. J. Comparative study of human and rat mammary tumorigenesis. Lab. Invest. 1990;62:244–278. [PubMed] [Google Scholar]
- Ryu H., Lee J., Olofsson B. A., Mwidau A., Dedeoglu A., Escudero M., Flemington E., Azizkhan-Clifford J., Ferrante R. J., Ratan R. R. Histone deacetylase inhibitors prevent oxidative neuronal death independent of expanded polyglutamine repeats via an Sp1-dependent pathway. Proc. Natl. Acad. Sci. USA. 2003;100:4281–4286. doi: 10.1073/pnas.0737363100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabapathy K., Hochedlinger K., Nam S. Y., Bauer A., Karin M., Wagner E. F. Distinct roles for JNK1 and JNK2 in regulating JNK activity and c-Jun-dependent cell proliferation. Mol. Cell. 2004;15:713–725. doi: 10.1016/j.molcel.2004.08.028. [DOI] [PubMed] [Google Scholar]
- Safe S., Abdelrahim M. Sp transcription factor family and its role in cancer. Eur. J. Cancer. 2005;41:2438–2448. doi: 10.1016/j.ejca.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Spengler M. L., Brattain M. G. Sumoylation inhibits cleavage of Sp1 N-terminal negative regulatory domain and inhibits Sp1-dependent transcription. J. Biol. Chem. 2006;281:5567–5574. doi: 10.1074/jbc.M600035200. [DOI] [PubMed] [Google Scholar]
- Stark M., Assaraf Y. G. Loss of Sp1 function via inhibitory phosphorylation in antifolate-resistant human leukemia cells with down-regulation of the reduced folate carrier. Blood. 2006;107:708–715. doi: 10.1182/blood-2005-07-2743. [DOI] [PubMed] [Google Scholar]
- Su K., Roos M. D., Yang X., Han I., Paterson A. J., Kudlow J. E. An N-terminal region of Sp1 targets its proteasome-dependent degradation in vitro. J. Biol. Chem. 1999;274:15194–15202. doi: 10.1074/jbc.274.21.15194. [DOI] [PubMed] [Google Scholar]
- Su K., Yang X., Roos M. D., Paterson A. J., Kudlow J. E. Human Sug1/p45 is involved in the proteasome-dependent degradation of Sp1. Biochem. J. 2000;348:281–289. [PMC free article] [PubMed] [Google Scholar]
- Sugawara T., Nomura E., Nakajima A., Sakuragi N. Characterization of binding between SF-1 and Sp 1, predominant interaction of SF-1 with the N-terminal region of Sp1. J. Endocrinol. Invest. 2004;27:133–141. doi: 10.1007/BF03346258. [DOI] [PubMed] [Google Scholar]
- Suske G. The Sp-family of transcription factors. Gene. 1999;238:291–300. doi: 10.1016/s0378-1119(99)00357-1. [DOI] [PubMed] [Google Scholar]
- Taatjes D. J., Naar A. M., Andel F., 3rd, Nogales E., Tjian R. Structure, function, and activator-induced conformations of the CRSP coactivator. Science. 2002;295:1058–1062. doi: 10.1126/science.1065249. [DOI] [PubMed] [Google Scholar]
- Tellez C., Bar-Eli M. Role and regulation of the thrombin receptor (PAR-1) in human melanoma. Oncogene. 2003;22:3130–3137. doi: 10.1038/sj.onc.1206453. [DOI] [PubMed] [Google Scholar]
- Tighe A., Johnson V. L., Albertella M., Taylor S. S. Aneuploid colon cancer cells have a robust spindle checkpoint. EMBO Rep. 2001;2:609–614. doi: 10.1093/embo-reports/kve127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Tsukada J., Higashi T., Mizobe T., Matsuura A., Mouri F., Sawamukai N., Ra C., Tanaka Y. Growth suppression of human mast cells expressing constitutively active c-kit receptors by JNK inhibitor SP600125. Genes Cells. 2006;11:983–992. doi: 10.1111/j.1365-2443.2006.01005.x. [DOI] [PubMed] [Google Scholar]
- Wang L., Wei D., Huang S., Peng Z., Le X., Wu T. T., Yao J., Ajani J., Xie K. Transcription factor Sp1 expression is a significant predictor of survival in human gastric cancer. Clin. Cancer Res. 2003;9:6371–6380. [PubMed] [Google Scholar]
- Wells L., Vosseller K., Hart G. W. Glycosylation of nucleocytoplasmic proteins: signal transduction and O-GlcNAc. Science. 2001;291:2376–2378. doi: 10.1126/science.1058714. [DOI] [PubMed] [Google Scholar]
- Wong C. F., Barnes L. M., Dahler A. L., Smith L., Popa C., Serewko-Auret M. M., Saunders N. A. E2F suppression and Sp1 overexpression are sufficient to induce the differentiation-specific marker, transglutaminase type 1, in a squamous cell carcinoma cell line. Oncogene. 2005;24:3525–3534. doi: 10.1038/sj.onc.1208372. [DOI] [PubMed] [Google Scholar]
- Xiao H., Hasegawa T., Isobe K. p300 collaborates with Sp1 and Sp3 in p21(waf1/cip1) promoter activation induced by histone deacetylase inhibitor. J. Biol. Chem. 2000;275:1371–1376. doi: 10.1074/jbc.275.2.1371. [DOI] [PubMed] [Google Scholar]
- Zhang F., Hu Y., Huang P., Toleman C. A., Paterson A. J., Kudlow J. E. Proteasome function is regulated by cyclic AMP-dependent protein kinase through phosphorylation of Rpt6. J. Biol. Chem. 2007;282:22460–22471. doi: 10.1074/jbc.M702439200. [DOI] [PubMed] [Google Scholar]