Abstract
We previously identified a multisubunit complex (CENP-H/I complex) in kinetochores from human and chicken cells. We showed that the CENP-H/I complex is divided into three functional classes. In the present study, we investigated CENP-O class proteins, which include CENP-O, -P, -Q, -R, and -50 (U). We created chicken DT40 cell knockouts of each of these proteins, and we found that all knockout lines were viable, but that they showed slow proliferation and mitotic defects. Kinetochore localization of CENP-O, -P, -Q, and -50 was interdependent, but kinetochore localization of these proteins was observed in CENP-R–deficient cells. A coexpression assay in bacteria showed that CENP-O, -P, -Q, and -50 proteins form a stable complex that can associate with CENP-R. Phenotype analysis of knockout cells showed that all proteins except for CENP-R were required for recovery from spindle damage, and phosphorylation of CENP-50 was essential for recovery from spindle damage. We also found that treatment with the proteasome inhibitor MG132 partially rescued the severe mitotic phenotype observed in response to release from nocodazole block in CENP-50–deficient cells. This suggests that CENP-O class proteins are involved in the prevention of premature sister chromatid separation during recovery from spindle damage.
INTRODUCTION
Accurate chromosome segregation during mitosis is essential for the correct transmission of genetic material. A kinetochore is assembled at the centromere of each chromatid of a replicated chromosome, and it forms a dynamic interface with microtubules of the mitotic spindle (Cleveland et al., 2003; Fukagawa, 2004). Although chromosome segregation errors cause genetic diseases, including some cancers (Yuen et al., 2005), the mechanism by which kinetochores interact with microtubules of the spindle apparatus during cell division is not fully understood.
New insight regarding the mechanism of chromosome segregation has come from the discovery and characterization of kinetochore proteins (Chan et al., 2005). In recent years, many kinetochore proteins have been identified in vertebrate cells by various approaches (Cheeseman et al., 2004; Obuse et al., 2004a,b; Minoshima et al., 2005; Foltz et al., 2006; Izuta et al., 2006; McAinsh et al., 2006; Meraldi et al., 2006; Okada et al., 2006). Characterization of the cellular functions of each of these proteins and the protein–protein network within cells will lead to an understanding of how kinetochores assemble and function.
We previously isolated a large protein complex (CENP-H/I complex), which contains at least 11 components, including CENP-H and CENP-I, and we found that this complex can be divided into three functional classes, based on phenotype analyses (Okada et al., 2006). Initial studies of the CENP-H/I complex in chicken and human cells suggested that it is required for proper chromosome alignment and segregation (Okada et al., 2006). To further understand the function of the CENP-H/I complex, it is necessary to characterize each class in detail. Here, we present results of functional analyses of CENP-O class proteins, which include CENP-O, -P, -Q, -R, and -50 (U). We created chicken DT40 cell knockouts (KOs) of each of these proteins, and we expressed the proteins in Escherichia coli. The results suggest that these proteins form a stable complex that is required for proper chromosome segregation. We previously showed that CENP-50 is essential for recovery from spindle damage (Minoshima et al., 2005). In the present study, we found that all CENP-O class proteins except for CENP-R were required for recovery from spindle damage. Furthermore, we found that treatment with the proteasome inhibitor MG132 partially rescued the severe mitotic phenotype that occurs in response to release from nocodazole block in CENP-50–deficient cells. These results suggest that CENP-O class proteins are involved in the prevention of premature sister chromatid separation during the recovery from spindle damage.
MATERIALS AND METHODS
Molecular Biology, Cell Culture, and Transfection
All plasmids were constructed by standard methods. Disruption constructs for each gene were generated such that genomic fragments encoding several exons were replaced with a histidinol-, neomycine-, or puromycin-resistance cassette under control of the β-actin promoter. Target constructs were transfected with a Gene Pulser II electroporator (Bio-Rad, Hercules, CA). All molecular biological experiments, including Southern blot analysis and Western blot analysis, were followed by standard methods.
DT40 cells were cultured and transfected as described previously (Fukagawa et al., 2004). All DT40 cells were cultured at 38°C in Dulbecco's modified medium supplemented with 10% fetal calf serum, 1% chicken serum, 2-mercaptoethanol, penicillin, and streptomycin.
Immunofluorescence and Image Acquisition
Immunofluorescent staining of whole cells was performed as described previously (Okada et al., 2006). Cells were collected onto slides with a cytocentrifuge, and then they were fixed in 3% paraformaldehyde in phosphate-buffered saline (PBS) for 15 min at room temperature and permeabilized in 0.5% NP-40 in PBS for 10 min at room temperature or 100% methanol for 15 min at −20°C, rinsed three times in 0.5% bovine serum albumin (BSA) in PBS, and incubated for 1 h at 37°C with primary antibody. Binding of primary antibody was then detected with fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit immunoglobulin (Ig)G (Jackson ImmunoResearch Laboratories, West Grove, PA) diluted to an appropriate concentration in 0.5% BSA in PBS. Affinity-purified rabbit polyclonal antibodies were generated against recombinant chicken CENP-Q and chicken CENP-R as described previously (Fukagawa et al., 1999). Other antibodies used in this study were described previously (Nishihashi et al., 2002; Hori et al., 2003; Minoshima et al., 2005; Okada et al., 2006). Chromosomes and nuclei were counterstained with 4,6-diamidino-2-phenylindole (DAPI) at 0.2 μg/ml in Vectashield Antifade (Vector Laboratories, Burlingame, CA). All immunofluorescence images were collected with a cooled charge-coupled device camera (CoolSNAP HQ, Photometrics Image Point; Photometrics, Tucson, AZ) mounted on an IX71 inverted microscope (Olympus, Tokyo, Japan) with a 60× objective lens (PlanApo 60×, numerical aperture [NA] 1.40) together with a filter wheel. All subsequent analysis and processing of images were performed using IPLab software (Signal Analytics, Vienna, VA).
Live-Cell Observations
For live-cell imaging, a histone H2B-red fluorescent protein (RFP) plasmid is transfected into CENP-Q–deficient cells to visualize chromosomes. Fluorescently stained living cells were observed with an IX71 inverted microscope with an oil immersion objective lens (PlanApo 60×, NA 1.40) in temperature-controlled box to keep temperature at 38°C. Time-lapse images were recorded at 3-min intervals with an exposure time of 0.2–0.3 s.
Spindle Damage Recovery Assay
Cells were treated with 1.7 μM nocodazole for 10–12 h, and cells were washed with fresh medium several times. Cells were put into fresh medium in the presence or absence of 10 μM MG132, and they were cultured at 38°C. At the indicated time points, cells were collected and examined by Hoechst33342 staining. Morphology of chromosomes and nuclei of the cells were observed and scored. More than 1000 cells were scored for each time point.
Protein Expression and Analysis
Coexpression of CENP-O class proteins was performed by cloning the cDNAs for each protein into pST39 (a gift from S. Tan, The Pennsylvania State University, University Park, PA; Tan, 2001). The cDNA for CENP-R was cloned into pCOLADuet-1 (Novagen, Madison, WI). Protein expression in BL21-codon Plus (DE3)-RIL E. coli was induced with 0.5 mM isopropyl β-d-thiogalactoside for 2 h at 37°C, and the proteins were purified using nickel-nitrilotriacetic acid (Ni-NTA) beads (QIAGEN, Valencia, CA). The eluted protein from the Ni-NTA beads with elution buffer (50 mM Na phosphate, pH 8.0, 0.3 M NaCl, 0.3 M imidazole, 0.1% NP-40, and 1 mM β-mercaptoethanol) was fractionated on a Superose 6 gel filtration column in elution buffer. DT40 cells (1 × 109 cells) that express CENP-50-FLAG or CENP-P-FLAG were lysed in 5 ml of lysis buffer (50 mM Na phosphate, pH 8.0, 0.3 M NaCl, 0.1% NP-40, 5 mM β-mercaptoethanol, complete protease inhibitor [Roche Diagnostics, Mannheim, Germany], and 20 U/ml DNase I [Takara, Kyoto, Japan]), centrifuged at 20,000 × g for 10 min at 4°C, and then a supernatant fraction was collected. Anti-FLAG M2-beads (Sigma, Tokyo, Japan) was incubated with the supernatant fraction for 2 h at 4°C and washed with lysis buffer and eluted with lysis buffer in the presence of 3× FLAG peptide (Sigma). The eluted protein complex was fractionated on a Superose 6 gel filtration column in 50 mM Na phosphate, pH 8.0, 0.3 M NaCl, 0.3 M imidazole, 0.1% NP-40, and 1 mM β-mercaptoethanol at 4°C.
RESULTS
KO Cell Lines of CENP-O Class Proteins Are Viable but Show Slow Proliferation Rates
We previously proposed the CENP-O class of proteins containing CENP-O, -P, -Q, -R, and -50, and we reported the creation of DT40 cell line KOs for CENP-O, CENP-P, and CENP-50 (Minoshima et al., 2005; Okada et al., 2006). To further elucidate the function of CENP-O class proteins, we generated DT40 cell line KOs of the other two proteins in the class (CENP-Q and CENP-R), and we compared the phenotypes of all of these KO cell lines. CENP-Q- or CENP-R-KO constructs were generated such that genomic fragments encoding several exons were replaced with drug-resistance cassettes (Supplemental Figure S1, A and B). We then transfected each construct into DT40 cells, and we isolated homozygous CENP-Q−/− or CENP-R−/−/− clones. We confirmed that CENP-Q or CENP-R protein was not expressed by Western blot analysis with anti-CENP-Q or anti-CENP-R antibody (Supplemental Figure S1, Ac and Bc). All KO cell lines of CENP-O class were viable. We then compared the proliferation rates of these KO cell lines with that of wild-type (WT) DT40 cells (Figure 1A). The proliferation rate of CENP-O–, -P–, -Q–, or -50–deficient cell lines was slower than that of WT cells. The doubling times of the KO cell lines were ∼1–4 h longer than that of WT cells (WT, 9 h; CENP-50-KO, 10.2 h; CENP-O-KO, 10.4 h; CENP-P-KO, 10.4 h; CENP-Q-KO, 13.0 h; and CENP-R KO, 9.5 h). As discussed below, we think that these slow proliferation rates were due to increases of G2/M and apoptotic populations. The proliferation rate of CENP-R–deficient cells was similar to that of WT cells, suggesting that CENP-R may play a different role from that of other CENP-O class proteins.
Figure 1.
KO cells of CENP-O class proteins are viable, but they show mitotic defects. (A) Representative growth curves for WT cells; single-KO cells of CENP-O, -P, -Q, -R, or -50; and double-KO cells of CENP-P and CENP-50. Numbers of cells not stained with trypan blue were counted. +tet indicates addition of tetracycline to the culture medium. (B) Cell cycle distribution in WT cells; single-KO cells of CENP-O, -P, -Q, -R, or -50; and double-KO cells of CENP-P and CENP-50. Cells were stained with FITC-anti-BrdU (y-axis, log scale) to detect BrdU incorporation (DNA replication) and with propidium iodide to detect total DNA (x-axis, linear scale). The bottom left box represents G1 phase cells, the top box represents S phase cells, and the bottom right box represents G2/M phase cells. The numbers given in the boxes indicate the percentage of gated events. +tet indicates addition of tetracycline to the culture medium. (C) Numbers of mitotic and apoptotic cells in WT and KO cells of CENP-O class proteins. More than 1000 cells were scored in each KO cell line. (D) Abnormal mitotic morphology of CENP-Q–deficient cells. Cells were stained with FITC-conjugated anti-α-tubulin (green). DNA was counterstained with DAPI (blue). Several chromosomes are not aligned at the metaphase plate (arrows). Multiple spindles are occasionally present (arrowheads). (E) Chromosome dynamics in control (WT) and CENP-Q–deficient cells observed by time-lapse microscopy of living cells. Selected images of chromosomes from prophase to telophase are shown. CENP-Q–deficient cells take longer to complete mitosis than control cells. Misalignment of chromosomes was observed frequently in CENP-Q-deficient cells. (F) Quantitation of the time for progression from prophase to anaphase in WT (n = 38) and CENP-Q–deficient (n = 47) cells as determined by time-lapse microscopy of living cells.
To determine relations between CENP-O class proteins, we created a double-KO cell line (4-46), in which both CENP-P and CENP-50 proteins were not expressed by Western blot analysis (Supplemental Figure S1C). Cells from the 4-46 line were viable, and the proliferation rate (11.8 h) was slower than that of WT cells (Figure 1A). We did not observe strong additive effects by creation of a double mutant, suggesting that at least four proteins in the CENP-O class (CENP-O, -P, -Q, and -50) function in a single pathway.
To determine the mechanism for the slow proliferation of CENP-O–, -P–, -Q–, or -50–deficient cells and cells deficient in both CENP-P and CENP-50, we performed cell-cycle analysis of CENP-O class KO cells and WT cells. We measured cellular DNA content and DNA synthesis by fluorescence-activated cell sorting (FACS) after pulse labeling with bromodeoxyuridine (BrdU) (Figure 1B). We did not observe significant differences in the distribution of G1 phase fractions between CENP-O class KO cells and WT cells. However, the number of S phase fractions in each CENP-O class KO cell line was decreased, and the number of G2/M phase fractions was increased compared with that of WT cells. Twelve percent of WT cells were in G2/M phase, whereas ∼20% of CENP-O–, -P–, -Q–, or -50–deficient cells were in G2/M phase, suggesting that it takes a longer time for CENP-O–, -P–, -Q–, or -50–deficient cells to progress through G2/M phase. There was also no additive accumulation of G2/M phase in cells deficient in CENP-P and CENP-50, supporting the idea that CENP-O, -P, -Q, and -50 proteins function in a single pathway. The cell cycle distribution of CENP-R–deficient cells was similar to that of WT cells, consistent with the result that the growth curve for CENP-R–deficient cells was similar to that for WT cells (Figure 1, A and B).
We previously showed that a slight accumulation of the G2/M phase fraction in CENP-50–deficient cells is due to a defect in normal mitotic progression (Minoshima et al., 2005). The doubling time of DT40 cells is ∼8–12 h. Therefore, differences in population of G2/M phase between CENP-O class KO cells and WT cells of several percentages points correspond to approximately a 1-h difference on the growth curve. We then measured numbers of mitotic and apoptotic cells in cells with KOs of CENP-O class proteins (Figure 1C), and we observed increases of several percentage points for mitotic and apoptotic cells. These increases are consistent with the mitotic delay observed by live-cell observation described below (Figure 1, E and F). We also observed that some CENP-Q–deficient cells (∼8.5% observed by live-cell imaging) showed abnormal mitotic behavior, with chromosomes that failed to congress normally to the metaphase plate (Figure 1D), although many of the cells went through the cell cycle. These results also suggest that the G2/M increase observed in CENP-Q–deficient cells is due mainly to mitotic defects. However, we cannot exclude the possibility that cells with KOs of CENP-O class proteins have a defect in mitotic entry.
To analyze mitotic defects in individual cells, we transfected the histone H2B-RFP plasmid into CENP-Q–deficient cells to visualize nuclei and chromosomes and observed the behavior of individual cells by microscopy at 38°C. Microscopic images were typically obtained at 3-min intervals, and chromosomes within individual cells were observed through an entire mitotic cycle. Time-lapse observations of WT and CENP-Q–deficient cells are shown in Figure 1E and the Supplemental Movies. Selected frames of the chromosome dynamics from prophase to telophase are shown. We observed well-ordered metaphase plates (12 min) and normal cell division in WT cells. It took WT cells an average of 22.4 ± 5.6 min (±SD, n = 38) to progress from prophase to telophase. Some CENP-Q–deficient cells did not congress chromosomes to the metaphase plate (63 min in Figure 1E), and these cells took an average of 39.1 ± 19.3 min (±SD, n = 47) to complete mitosis from prophase (Figure 1F), twice that of WT cells and similar to that of CENP-50–deficient cells (Minoshima et al., 2005). Thus, cells with KOs of CENP-O class proteins went through the cell cycle, and kinetochore-microtubule interaction and chromosome movements occurred. However, it took longer for KO cells to complete mitosis, and we observed increased numbers of apoptotic cells in KO cells cultures. Considering these data, we concluded that CENP-O class proteins are required for proper kinetochore function, although cells with KOs of CENP-O class proteins are viable.
Role of CENP-O Class Proteins in Kinetochore Assembly
CENP-O class proteins localize constitutively to the kinetochore throughout the cell cycle, and we are interested in how each CENP-O class protein is involved in kinetochore assembly. We generated antibodies against each CENP-O class protein, and we assessed the localization of these proteins in each KO cell line. We observed typical kinetochore staining with each CENP-O class antibody in WT cells, whereas kinetochore staining was abolished in CENP-O–, -P–, -Q–, or -50–deficient cells (Figure 2A). However, we still observed clear kinetochore staining of CENP-O, -P, -Q, and -50 in CENP-R–deficient cells (Figure 2A). These results indicate that kinetochore localization of CENP-O, -P, -Q, and -50 is interdependent and that localization of CENP-R occurs downstream of that of CENP-O, -P, -Q, and -50. These localization data strongly support our idea that CENP-R has a different role from the other CENP-O class proteins.
Figure 2.
Role of CENP-O class proteins in kinetochore assembly. (A) Immunofluorescence analysis of WT and CENP-50–, -O–, -P–, -Q–, or -R–deficient cells. Cells were stained with anti-CENP-50, anti-CENP-O, anti-CENP-P, anti- CENP-Q, or anti-CENP-R antibody. Antibody signals were detected with FITC-conjugated secondary antibody (green). DNA was counterstained with DAPI (blue). Typical interphase (I) and mitotic (M) figures are shown. (B) Immunofluorescence analysis of CENP-H–conditional knockout (KO) cells (5-5). Cells were stained with anti-CENP-Q or anti-CENP-R antibody (green). +tet indicates addition of tetracycline to the culture medium. DNA was counterstained with DAPI (blue). (C) Immunofluorescence analysis of WT, CENP-Q-KO, or CENP-R-KO cells. Cells were stained with anti-CENP-H or anti-CENP-I antibody (green). DNA was counterstained with DAPI (blue). (D) Western blot analysis of CENP-50–, -O–, -P–, -Q–, or -R–deficient cells with anti-CENP-50, anti-CENP-O, anti-CENP-P, anti-CENP-Q, anti-CENP-R, or anti-CENP-H antibody. The amount of CENP-50 is decreased in CENP-Q–deficient cells, and the amount of CENP-P is decreased in CENP-O–deficient cells. The amount of CENP-R is decreased in all mutants, and CENP-H is stable in all mutants. A Coomassie Brilliant Blue (CBB)-stained gel is also shown. (E) Model of the association of CENP-O class proteins. CENP-O, -P, -Q, and -50 form a stable complex. In particular, CENP-50 is close to CENP-Q, and CENP-O is close to CENP-P. This stable complex functions downstream of CENP-H class proteins and upstream of CENP-R.
CENP-O class proteins were originally identified as members of the CENP-H/I complex, and we previously showed that some members of CENP-O class do not localize to kinetochores in CENP-H- or CENP-I–deficient cells (Minoshima et al., 2005; Okada et al., 2006), suggesting that localization of CENP-O class proteins occurs downstream of that of CENP-H and CENP-I. To test this idea, we assessed the localization of CENP-Q and CENP-R in CENP-H–deficient cells (Figure 2B). We also assessed the localization of CENP-H and CENP-I in CENP-Q– or CENP-R–deficient cells (Figure 2C). We observed that kinetochore staining of CENP-Q and CENP-R were abolished in CENP-H–deficient cells (+tet, Figure 2B), but CENP-H and CENP-I kinetochore staining was clear in CENP-Q– or CENP-R–deficient cells, indicating that localization of CENP-O class proteins occurs downstream of that of CENP-H and CENP-I. Thus, CENP-O class proteins are required for their own kinetochore localization. However, these proteins do not strongly affect the localization of essential kinetochore proteins CENP-A, CENP-C, CENP-H, CENP-I, and Ndc80 (Minoshima et al., 2005; Okada et al., 2006; unpublished results). These results support the idea that CENP-O class proteins are required for proper kinetochore function.
We also found that the amounts of both CENP-P and CENP-O were decreased in cells deficient in either CENP-O or CENP-P and that the amounts of both CENP-50 and CENP-Q were decreased in cells deficient in either CENP-50 or CENP-Q (Figure 2D). The amount of CENP-R was decreased in all cells with KOs of CENP-O class proteins (Figure 2D). These results suggest that CENP-P is closely associated with CENP-O and that CENP-50 is closely associated with CENP-Q, although kinetochore localization of these proteins is interdependent (Figure 2E). With respect to the stabilities of the CENP-O class proteins, we suggest that CENP-O, -P, -Q, and -50 form a discrete complex that functions downstream of CENP-H class proteins and upstream of CENP-R (Figure 2E).
CENP-O Class Proteins Form a Discrete Complex
KO cells of CENP-O class proteins, with the exception of CENP-R, show similar phenotypes. Our present results in KO cells also suggest that CENP-O class proteins form a discrete complex (Figure 2E). Therefore, we were interested to determine whether these proteins physically form a stable complex. To examine this possibility, we coexpressed CENP-O, CENP-P, and CENP-50 proteins and 6x histidine-tagged (6xHis) CENP-Q protein in bacteria with a polycistronic system (Tan, 2001; Kline et al., 2006) (Figure 3A). Samples purified with nickel affinity beads were subjected to gel filtration chromatography, and each fraction was analyzed by Western blot analysis with each CENP-O class antibody. The four proteins cofractionated as a single peak (Figure 3B). These results indicated that the four proteins form a stable complex. To test whether the stable complex associates with CENP-R, we coexpressed the above-mentioned four proteins with CENP-R simultaneously. The four proteins and CENP-R protein copurified with nickel affinity beads (Figure 3C), suggesting that CENP-R can associate with the stable complex.
Figure 3.
CENP-O class proteins form a discrete complex. (A) Coexpression of CENP-50, CENP-P, 6xHis- CENP-Q, and CENP-O with a polycistronic system in E. coli cells. The CENP-R construct that was cotransformed with the polycistronic constructs was used in C. (B) Migration of the recombinant complex on a Superose 6 gel filtration column. Western blot analysis of the recombinant complex was also performed. Standards are thyroglobulin (669 kDa), ferritin (440 kDa), aldolase (158 kDa), and BSA (67 kDa). (C) Coexpression of CENP-50, CENP-P, 6xHis-CENP-Q, CENP-O, and CENP- R. CENP-R shows the same behavior as CENP-50, CENP-P, CENP-Q-6xHis, and CENP-O. (D) Purified complex from DT40 cells expressing CENP-50-FLAG was loaded onto a Superose 6 gel filtration column, and Western blot analysis was performed with anti-CENP-50, anti-CENP-O, anti-CENP-P, anti-CENP-Q, or anti-CENP-R antibody. The low-mobility bands observed with anti-CENP-R (arrowhead) indicate that CENP-R is phosphorylated (data not shown). Migration profiles for each protein are similar to that of the purified complex from E. coli cells (B and C). (E) Purified complex from DT40 cells expressing CENP-P-FLAG was loaded onto a Superose 6 gel filtration column, and Western blot analysis was performed with anti-CENP-50 and anti-CENP-P antibodies.
To confirm that the stable complex forms in DT40 cells, we created a cell line (3xFLAG-CENP-50) in which expression of CENP-50 was replaced with that of CENP-50-FLAG. Whole-cell extract was prepared from 3xFLAG-CENP-50 cells, and immunoprecipitation was performed with anti-FLAG antibody. Immunoprecipitates were subjected to gel filtration chromatography, and each fraction was analyzed by Western blot analysis with several antibodies against CENP-O class proteins. CENP-O class proteins cofractionated with one peak nearly coincident with that observed for the recombinant protein complex purified from bacterial cells (Figure 3, B–D). CENP-50 was detected in both the peak fraction (Figure 3D, 14) and smaller fractions (Figure 3D, 15 and 16). Excess CENP-50-FLAG by overexpression in DT40 cells may not form a complex. Therefore, we created another cell line (3xFLAG-CENP-P) in which expression of CENP-P was replaced with that of CENP-P-FLAG, and we performed similar experiments. The strongest CENP-50 band was detected in the peak fraction (Figure 3E, 14). Thus, biochemical experiments in both E. coli and DT40 cells showed that CENP-O class proteins form a stable complex.
CENP-O Class Proteins Are Required for Recovery from Spindle Damage
Although we propose that CENP-O class proteins are required for proper kinetochore function, mutant cells lacking expression of CENP-O class proteins are viable. However, we found that some KO cells of CENP-O class proteins died after chromosome missegregation (Figure 1C). We previously showed that CENP-50 deficiency adversely affects mitotic exit after prolonged mitotic delay. If CENP-O class proteins, including CENP-50, function cooperatively, other CENP-O class KO cells should show a similar phenotype. We then treated CENP-O–, -P–, -Q–, -R–, or -50–deficient cells and WT cells with nocodazole for 12 h, washed the cells, and placed them into fresh media. Then, we examined the entry of cells into the next cell cycle (Figure 4A). As shown previously (Minoshima et al., 2005), WT cells progressed to G1 phase immediately, and <10% were mitotic at 6 h after release from nocodazole block (Figure 4A). In contrast, ∼50% of CENP-O–, -P–, -Q–, or -50–deficient cells showed mitotic figures at 4 h after release, and 30–50% of these cells were still mitotic at 6 h after release (Figure 4A). We also observed that most cells that did not exit mitosis after several hours eventually died (data not shown). The percentage of mitotic CENP-R–deficient cells at 4 h after release was ∼30%, slightly higher than that of WT cells but lower than that of CENP-O–, -P–, -Q–, or -50–deficient cells (Figure 4A).
Figure 4.
CENP-O class proteins are required for recovery from spindle damage. (A) WT cells and KO cells of CENP-O class proteins were treated with nocodazole for 12 h. After removal of nocodazole by washing, cells were cultured, and the percentages of mitotic, interphase, and apoptotic cells at the indicated times were quantified. More than 60% of CENP-50–deficient cells remained in the mitotic state at 4 h after release from nocodazole block, whereas ∼70% of control (WT) cells were at interphase at the same time point. (B) Chromosome morphology of WT and CENP-50-KO cells, just after nocodazole block (top), 4 h after release from nocodazole block (middle), or treated with MG132 for 4 h just after release from nocodazole block (bottom). Red shows α-tubulin, green shows CENP-H-GFP, and blue shows chromosomes. In CENP-50-KO cells, many chromosomes are not aligned at the metaphase plate 4 h after release from nocodazole block. However, the chromosomes start to align in response to MG132 treatment. (C) Index for chromosome alignment is shown. (a) Explanation how to measure the index. Aligned chromosomes are in Area A inside the poles. Area in which unaligned chromosomes are scattered is defined as area B. Regions of area A and area B were quantified and index for chromosome alignment is shown as area B/area A. If all chromosomes aligned between poles, index should be 1. (b) Chromosome alignment index is shown in various experimental conditions. (D) Staining of spindle checkpoint proteins Mad2 and BubR1 in CENP-50-KO cells 4 h after release from nocodazole block. Cy3-staining shows Mad2 or BubR1, green shows CENP-H-GFP, and blue shows chromosomes.
The observed phenotype indicates that CENP-O, -P, -Q, and -50 are required for recovery from spindle damage. We propose that CENP-50 functions cooperatively with CENP-O, -P, and -Q (and in part with CENP-R). An important question is how CENP-50 and other CENP-O class proteins are involved in the recovery from spindle damage. We previously reported that the distance between sister kinetochores in CENP-50–deficient cells is increased after long nocodazole treatment (Minoshima et al., 2005). To gain further information regarding the function of CENP-O class proteins, we added the proteasome inhibitor MG132 in the culture of cells released from nocodazole block. In the absence of MG132, we observed severe mitotic defects 4 h after release from nocodazole block in CENP-50-KO cells (Figure 4B, right middle). We observed many chromosomes (∼10 in each mitosis) that did not align at the metaphase plate and they were scattered (Figure 4B, right middle). In the presence of MG132, most chromosomes aligned at the metaphase plate 4 h after release from nocodazole block in CENP-50-KO cells (Figure 4, B, right, bottom, and C). We also quantified aligned chromosomes in the presence or absence of MG132 (Figure 4C). These results indicate that MG132 treatment rescued the chromosome misalignment phenotype that occurred during release from nocodazole block in CENP-50–deficient cells. MG132 is a proteasome inhibitor that acts to protect proteins involved in sister chromatid adhesion from degradation. Our present data suggest that CENP-50 is involved in the prevention of premature sister chromatid separation during the release from nocodazole block. We observed that mitotic cells showed positive mitotic checkpoint signals during the release from nocodazole block (Figure 4D). Therefore, CENP-50-KO cells do not show silencing of the spindle checkpoint pathway during the release from nocodazole block. We would like to note that although MG132 treatment rescued the chromosome misalignment phenotype that occurred during release from nocodazole block in CENP-50–deficient cells, cells were still mitotic even after removing of MG132 (data not shown). This suggests that that some irreversible mitotic defects also occurred during the nocodazole block.
Phosphorylation of CENP-50 Is Essential for Recovery from Spindle Damage
Our data suggested that CENP-50 is involved in the prevention of premature sister chromatid separation and for recovery from spindle damage. We found that CENP-50 is highly phosphorylated in cells treated with nocodazole (Figure 5Aa). Therefore, the recovery from spindle damage may be related to phosphorylation of CENP-50. There are many potential phosphorylation sites in the amino acid CENP-50 sequence (Supplemental Figure S2). Recently, Kang et al. (2006) reported that human CENP-50 (referred as PBIP1) is a target of polo-like kinase 1 (PLK1) in human HeLa cells. We also found that CENP-O class proteins copurified with PLK1 by a proteomics approach (Hori, Cheeseman, and Fukagawa, unpublished observations). Kang et al. (2006) also reported that S77 and T78 in human CENP-50 are critical sites for association or phosphorylation by PLK1. We found that S77 and T78 in human CENP-50 are highly conserved sites, corresponding to S62 and T63 in chicken CENP-50 (Figure 5, A–C). We created a mutant cDNA of chicken CENP-50 in which two potential serine/threonine phosphorylation sites are substituted with alanine (62.63A cDNA) and transfected it into tetracycline-repressible conditional-KO cells of CENP-50 (17-15). We isolated a CENP-50 KO cell line expressing 62.63A cDNA (62.63A/17-15 cells), and we confirmed that phosphorylation of CENP-50 was decreased in these cells (Figure 5, A and B). We also transfected WT cDNA into 17-15 cells and isolated a cell line expressing WT cDNA (WT/17-15 cells). We then analyzed the recovery of these cells from spindle damage. More than 70% of 62.63A/17-15 cells showed mitotic figures at 4 h after release from nocodazole block (Figure 5B, right), similar to 17-15 cells (+tet; Figure 5B, left), but WT/17-15 cells rescued by WT cDNA expression recovered completely from spindle damage (Figure 5B, middle), suggesting that phosphorylation of S62 and T63 in chicken CENP-50 is essential for complete recovery.
Figure 5.
Phosphorylation of CENP-50 by PLK1 is essential for recovery from spindle damage. (Aa) Low-mobility bands were detected by Western blot analysis with anti-CENP-50 antibody. The amounts of the low-mobility bands (P) were increased in cells enriched in M phase by nocodazole treatment for 12 h (WT, −λ-protein phosphatase [PPase], +nocodazole [Noc]) compared with those in asynchronous cells (WT, −λ-PPase, −Noc). These low-mobility bands were abolished by treatment of the cell lysates with λ-PPase (+λ-PPase), indicating that CENP-50 is phosphorylated. The arrow indicates the unphosphorylated form of CENP-50. (b) Western blot analysis of WT cells (WT) and 62.63A/17-15 cells (62.63A) in the presence of tetracycline, in which mutant CENP-50 cDNA (with substitution of two potential serine/threonine phosphorylation sites with alanine) was transfected into 17-15 cells (CENP-50–conditional KO cells). The amounts of the low-mobility CENP-50 bands were reduced in 62.63A/17-15 cells. Arrowhead and arrow indicate low- and high-mobility bands, respectively. Arrowhead with asterisk indicates hyperphosphorylation caused by M phase arrest with nocodazole treatment. Anti-tubulin antibody was used as a loading control. (c) CENP-50 amino acid sequences of human, mouse, and chicken. S62 and T63 of chicken CENP-50 corresponds S77 and T78 of human sequence. (B) Spindle damage recovery assay as described in Figure 4 (A) for 17-15 cells (CENP-50–conditional KO cells),WT/17-15 cells, in which WT CENP-50 cDNA was transfected into 17-15 cells, and 62.63A/17-15 cells. +tet indicates addition of tetracycline to the culture medium. (C) Western blot analysis of WT cells (WT) and 62.63A/17-15 cells (62.63A) in the presence of tetracycline. Anti-CENP-50, anti-CENP-O, anti-CENP-P, anti CENP-Q, and anti-CENP-R antibodies were used. Anti-tubulin antibody was used as a loading control. Arrows indicate each protein. Arrowheads indicate phosphorylated bands. Phosphorylated bands denoted by arrowhead with asterisk were reduced in 62.63A/17-15 cells. (D) Immunofluorescence analysis of 62.63A/17-15 cells with anti-CENP-50, anti-CENP-O, anti-CENP-P, anti-CENP-Q, and anti-CENP-R antibodies. Arrowheads indicate mitotic cells. (E) (a) Western blot analysis of cells expressing CENP-50 cDNA mutant 16×A or cDNA 16×A + 62.63A with anti-CENP-50 antibodies. Western blot data shows that mobility of CENP-50 with 16×A or 16×A + 62.63A is faster than that of phosphorylated CENP-50. (b) Results of spindle damage recovery assay for cells expressing CENP-50 mutant with 16×A or 16×A + 62.63A are also shown.
CENP-50 functions in association with other CENP-O class proteins. Therefore, it is possible that 62.63A mutation of CENP-50 affects kinetochore localization or protein amounts of CENP-O class proteins. Degradation or mislocalization of CENP-O class proteins may affect the ability to recover from spindle damage. Therefore, we performed Western blot analysis to examine the stability of CENP-O class proteins in 62.63A/17-15 cells (Figure 5C). No significant change in the amounts of CENP-O class proteins was observed in 62.63A/17-15 cells compared with WT cells. Interestingly, we detected a slow-mobility band for CENP-Q, which may correspond to phosphorylated protein. Because we CENP-Q is associated with CENP-50, PLK1 may phosphorylate CENP-Q. The amount of phosphorylated CENP-Q band was reduced in 62.63A/17-15 cells, suggesting that PLK1 localization may be altered in these cells. We also examined the localization of CENP-O class proteins in 62.63A/17-15 cells. Mutant CENP-50 (62.63A) proteins localized to the kinetochore throughout the cell cycle, and kinetochore localization of other CENP-O class proteins was not changed in 62.63A/17-15 cells (Figure 5D), indicating that kinetochore localization of CENP-O class proteins was not affected by 62.63A mutation of CENP-50.
As mentioned, there are many potential phosphorylation sites in the CENP-50 sequence (Supplemental Figure S2). Because we found that CENP-50 phosphorylation was reduced by treatment of a cyclin-dependent kinase (CDK) inhibitor, we focused on potential CDK phosphorylation sites (T99, S112, S145, S148, and S151) and predicted additional phosphorylation sites around the CDK sites by using the NetPhos 2.0 software (http://www.cbs.dtu.dk/services/NetPhos/). In total, 16 potential serine/threonine phosphorylation sites in the chicken CENP-50 sequence were predicted (Supplemental Figure S2). We created a mutant cDNA of chicken CENP-50 in which all 16 of these sites were substituted with alanine (16×A cDNA), and we transfected it into tetracycline-repressible 17-15 cells. We isolated a cell line expressing 16×A cDNA(16×A/17-15 cells), and we confirmed that the amount of the phosphorylated band of CENP-50 was reduced (Figure 5E). However, 16×A/17-15 cells showed recovery from spindle damage, similar to WT cells (Figure 5E). We added the 62.63A mutations to the 16×A cDNA (16×A + 62.63A cDNA) and isolated a cell line expressing 16×A + 62.63A cDNA (16×A + 62.63A/17-15 cells), in which we detected defects in the recovery from spindle damage. These data suggest that S62 and T63 in CENP-50, which are putative phosphorylation sites, are essential for the recovery from spindle damage.
Kinetochore Localization of PLK1 Is Dependent on CENP-O Class Proteins, Especially in the G2 Phase
We showed that the amount of phosphorylated CENP-Q was reduced in 62.63A/17-15 cells expressing CENP-50 cDNA with mutated phosphorylation sites (Figure 5C). Therefore, PLK1 localization may be altered in cells lacking phosphorylation of CENP-50. We created a GFP-fused PLK1 construct under the control of the cytomegalovirus promoter and transfected it into WT cells, CENP-50-KO cells, and 62.63A/17-15 cells (Figure 6). PLK1 localizes to both the centrosomes and kinetochores. However, we focused on the kinetochore localization of PLK1 in this article, because CENP-O class proteins localize to kinetochores not to the centrosomes. PLK1 initially localized to the kinetochores in G2 phase and continued to localize to the kinetochores in prometaphase, but kinetochore-associated signals were very faint when the chromosomes aligned at the metaphase plate (WT in Figure 6). In CENP-50–deficient cells, kinetochore localization of PLK1 in G2 nuclei was not visible. In CENP-50–deficient prometaphase cells, kinetochore localization of PLK1 was weaker that in WT prometaphase cells, but it was visible (Figure 6). In 62.63A/17-15 cells, kinetochore localization of PLK1 was not visible in G2 nuclei and faint in prometaphase (Figure 6). These data suggest that phosphorylation of S62 and T63 in CENP-50 is required for the kinetochore localization of PLK1, especially in the G2 phase. Our data also suggest that there are several localization pathways of PLK1 to kinetochore in prometaphase and that one pathway is dependent on phosphorylation of S62 and T63 in CENP-50.
Figure 6.
Kinetochore localization of PLK1 is dependent on CENP-O class proteins in G2 phase. PLK1-GFP was expressed in WT, CENP-50-KO, and 62.63A/17-15 cells. Kinetochore localization is denoted by arrowheads, and centrosome localization is denoted by arrows. Kinetochore localization observed in WT G2 cells was abolished in CENP-50-KO G2 cells and 62.63A/17-15 G2 cells. In prometaphase, kinetochore localization of PLK1 was weak, but visible in CENP-50-KO and 62.63A/17-15 cells. +tet indicates addition of tetracycline to the culture medium.
DISCUSSION
We previously isolated a CENP-H/I complex containing at least 11 components, and we found that this complex can be divided into three functional classes based on phenotype analysis (Okada et al., 2006). We defined CENP-O, -P, -Q, -R, and -50 as CENP-O class proteins. In the present study, we created single KO cell lines for CENP-Q and CENP-R and a double-KO cell line for CENP-P and CENP-50. We analyzed all KO cell lines for CENP-O class proteins. With a combination of genetic and biochemical approaches, we showed that CENP-O class proteins form a stable complex. Protein stability experiments with KO cells indicated that CENP-P is closely associated with CENP-O, CENP-50 is closely associated with CENP-Q, and CENP-R functions downstream of all other CENP-O class proteins (Figure 2E). The elucidation of molecular structure of CENP-O class proteins will likely aid in the understanding of kinetochore assembly.
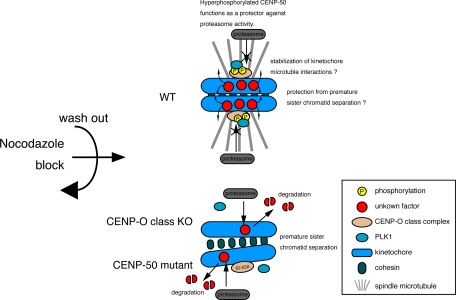
CENP-O class proteins are not required for cell proliferation under normal conditions. However, they seem to be required for recovery from spindle damage. We previously reported that CENP-50 is involved in sister chromatid adhesion during spindle checkpoint activation (Minoshima et al., 2005). In the present study, we showed that all CENP-O class proteins except for CENP-R are essential for recovery from spindle damage, and that treatment with the proteasome inhibitor MG132 partially rescued the severe mitotic phenotype observed in response to release from nocodazole block in CENP-50–deficient cells. We also showed that phosphorylation of S62 and T63 in CENP-50 is essential for the recovery from spindle damage. We therefore propose a model for the function of CENP-O class proteins in the recovery from spindle damage (Figure 7). In this model, CENP-O class proteins are phosphorylated by PLK1 in response to spindles damage. When cells are cultured in fresh media, phosphorylated CENP-O class proteins by PLK1 protect several factors involved in sister chromatid adhesion from degradation by proteasome activity. We think that CENP-O class proteins function to protect against premature sister chromatid separation until the microtubules attach properly to the kinetochores. In CENP-O class KO cells or cells expressing mutant CENP-50, proteasomes degrade several factors before the microtubules attach properly to the kinetochores (Figure 7). This model may explain why we observed an increased distance between sister kinetochores in CENP-50–deficient cells after long nocodazole treatment (Minoshima et al., 2005). There are reports in which the pathway of cohesin removal, which requires PLK1, is proteolysis independent (Tang et al., 2004; Kitajima et al., 2005; Resnick et al., 2006). Our findings related to CENP-O class proteins are distinct from the proteolysis-independent pathway of cohesin removal.
Figure 7.
Model of the functional role of CENP-O class proteins during release from spindle damage. CENP-O class proteins are phosphorylated by PLK1 in response to spindle damage. When cells are cultured in fresh media, phosphorylated CENP-O class by PLK1 protect several factors involved in sister chromatid adhesion from degradation by proteasome activity. CENP-O class proteins may function to protect against premature sister chromatid separation until microtubules properly attach to the kinetochores. In KO cells of CENP-O class protein or cells expressing mutant CENP-50, in which PLK1 is not targeted to the kinetochores, it is easy to access several targets for proteosomes before the microtubules properly attach to the kinetochores, and sister chromatid adhesion is reduced.
We sometimes observed mitotic defects in nonnocodazole treated CENP-Q–deficient cells (Figure 1). We think that these defects were similar defects observed in nocodazole-treated/washout cells with KO of CENP-O class proteins. Although spindle damage does not happen frequently in fresh media, cells occasionally could have some spindle damage. At that time, CENP-O class proteins function in recovery from spindle damage. CENP-O class proteins seem to fine-tune proper chromosome segregation rather than to act as essential structural components for kinetochore assembly. Therefore, CENP-O class KO cells are viable when cultured in fresh media. However, RNA interference analysis for CENP-50 (U) shows defects in chromosome alignment and colony formation in human HeLa cells (Foltz et al., 2006). We have created CENP-50-KO mice, and we observed that these mice die during early embryogenesis (our unpublished data). Cell division early during embryogenesis must be performed under strict conditions for completion of several developmental steps such as differentiation and proliferation. The function of CENP-O class proteins may be essential under these conditions. It will be interesting to determine how the mechanism of chromosome segregation differs between differentiated and undifferentiated cells. We think that CENP-O class proteins are key players in relation to this question. Determination of the molecular and biochemical properties of the CENP-O class complex will be necessary to understand the biological function of this complex.
Supplementary Material
ACKNOWLEDGMENTS
We thank K. Suzuki, M. Takahashi, and K. Kita for technical assistance. We also thank S. Tan for pST39 plasmid and I. M. Cheeseman for unpublished information about PLK1. This work was supported by grants-in-aid for scientific research from the Ministry of Education, Science, Sports and Culture of Japan.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/10.1091/mbc.E07-06-0556) on December 19, 2007.
REFERENCES
- Chan G. K., Liu S. T., Yen T. J. Kinetochore structure and function. Trends Cell Biol. 2005;15:589–598. doi: 10.1016/j.tcb.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Cheeseman I. M., Niessen S., Anderson S., Hyndman F., Yates J.R., III, Oegema K., Desai A. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 2004;18:2255–2268. doi: 10.1101/gad.1234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland D. W., Mao Y., Sullivan K. F. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- Foltz D. R., Jansen L. E., Black B. E., Bailey A. O., Yates J. R., III, Cleveland D. W. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- Fukagawa T. Assembly of kinetochore in vertebrate cells. Exp. Cell Res. 2004;296:21–27. doi: 10.1016/j.yexcr.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Fukagawa T., Nogami M., Yoshikawa M., Ikeno M., Okazaki T., Takami Y., Nakayama T., Oshimura M. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat. Cell Biol. 2004;6:784–791. doi: 10.1038/ncb1155. [DOI] [PubMed] [Google Scholar]
- Fukagawa T., Pendon C., Morris J., Brown W. CENP-C is necessary but not sufficient to induce formation of functional centromere. EMBO J. 1999;18:4196–4209. doi: 10.1093/emboj/18.15.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T., Haraguchi T., Hiraoka Y., Kimura H., Fukagawa T. Dynamic behavior of Nuf2-Hec1 complex that localizes to the centrosome and centromere and is essential for mitotic progression in vertebrate cells. J. Cell Sci. 2003;116:3347–3362. doi: 10.1242/jcs.00645. [DOI] [PubMed] [Google Scholar]
- Izuta H., Ikeno M., Suzuki N., Tomonaga T., Nozaki N., Obuse C., Kisu Y., Goshima N., Nomura F., Nomura N., Yoda K. Comprehensive analysis of the ICEN (interphase centromere complex) components enriched in the CENP-A chromatin of human cells. Genes Cells. 2006;11:673–684. doi: 10.1111/j.1365-2443.2006.00969.x. [DOI] [PubMed] [Google Scholar]
- Kang Y. H., et al. Self-regulated Plk1 recruitment to kinetochores by the Plk1-PBIP1 interaction is critical for proper chromosome segregation. Mol. Cell. 2006;24:409–422. doi: 10.1016/j.molcel.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Kitajima T. S., Hauf S., Ohsugi M., Yamamoto T., Watanabe Y. Human Bub1 defines the persistent cohesion site along the mitotic chromosome by affecting Shugoshin localization. Curr. Biol. 2005;15:353–359. doi: 10.1016/j.cub.2004.12.044. [DOI] [PubMed] [Google Scholar]
- Kline S. L., Cheeseman I. M., Hori T., Fukagawa T., Desai A. The human Mis12 complex is required for kinetochore assembly and proper chromosome segregation. J. Cell Biol. 2006;173:9–17. doi: 10.1083/jcb.200509158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAinsh A. D., Meraldi P., Draviam V. M., Tosa A., Sorger P. K. The human kinetochore proteins Nnf1R and Mcm21R are required for accurate chromosome segregation. EMBO J. 2006;25:4033–4049. doi: 10.1038/sj.emboj.7601293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P., McAinsh A. D., Rheinbay E., Sorger P. K. Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol. 2006;7:R23. doi: 10.1186/gb-2006-7-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoshima Y., Hori T., Okada M., Kimura H., Haraguchi T., Hiraoka Y., Bao Y. C., Kawashima T., Kitamura T., Fukagawa T. The constitutive centromere component CENP-50 is required for recovery from spindle damage. Mol. Cell. Biol. 2005;25:10315–10328. doi: 10.1128/MCB.25.23.10315-10328.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihashi A., Haraguchi T., Hiraoka Y., Ikemura T., Regnier V., Dodson H., Earnshaw W. C., Fukagawa T. CENP-I is essential for centromere function in vertebrate cells. Dev. Cell. 2002;2:463–476. doi: 10.1016/s1534-5807(02)00144-2. [DOI] [PubMed] [Google Scholar]
- Obuse C., Yang H., Nozaki N., Goto S., Okazaki T., Yoda K. Proteomics analysis of the centromere complex from HeLa interphase cells: UV-damaged DNA binding protein 1 (DDB-1) is a component of the CEN-complex, while BMI-1 is transiently co-localized with the centromeric region in interphase. Genes Cells. 2004a;9:105–120. doi: 10.1111/j.1365-2443.2004.00705.x. [DOI] [PubMed] [Google Scholar]
- Obuse C., Iwasaki O., Kiyomitsu T., Goshima G., Toyoda Y., Yanagida M. A conserved Mis12 centromere complex is linked to heterochromatic HP1 and outer kinetochore protein Zwint-1. Nat. Cell Biol. 2004b;6:1135–1141. doi: 10.1038/ncb1187. [DOI] [PubMed] [Google Scholar]
- Okada M., Cheeseman I. M., Hori T., Okawa K., McLeod I. X., Yates J.R., III, Desai A., Fukagawa T. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- Resnick T. D., Satinover D. L., MacIsaac F., Stukenberg P. T., Earnshaw W. C., Orr-Weaver T. L., Carmena M. INCENP and Aurora B promote meiotic sister chromatid cohesion through localization of the Shugoshin MEI-S332 in Drosophila. Dev. Cell. 2006;11:57–68. doi: 10.1016/j.devcel.2006.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S. A modular polycistronic expression system for overexpressing protein complexes in Escherichia coli. Protein Expr. Purif. 2001;21:224–234. doi: 10.1006/prep.2000.1363. [DOI] [PubMed] [Google Scholar]
- Tang Z., Sun Y., Harley S. E., Zou H., Yu H. Human Bub1 protects centromeric sister-chromatid cohesion through Shugoshin during mitosis. Proc. Natl. Acad. Sci. USA. 2004;101:18012–18017. doi: 10.1073/pnas.0408600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen K. W., Montpetit B., Hieter P. The kinetochore and cancer: what's the connection? Curr. Opin. Cell Biol. 2005;17:576–582. doi: 10.1016/j.ceb.2005.09.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.