Abstract
We determined composition and relative roles of deubiquitylating proteins associated with the 26S proteasome in mammalian cells. Three deubiquitylating activities were associated with the 26S proteasome: two from constituent subunits, Rpn11/S13 and Uch37, and one from a reversibly associated protein, Usp14. RNA interference (RNAi) of Rpn11/S13 inhibited cell growth, decreased cellular proteasome activity via disrupted 26S proteasome assembly, and inhibited cellular protein degradation. In contrast, RNAi of Uch37 or Usp14 had no detectable effect on cell growth, proteasome structure or proteolytic capacity, but accelerated cellular protein degradation. RNAi of both Uch37 and Usp14 also had no effect on proteasome structure or proteolytic capacity, but inhibited cellular protein degradation. Thus, proper proteasomal processing of ubiquitylated substrates requires Rpn11 plus either Uch37 or Usp14. Although the latter proteins feature redundant deubiquitylation functions, they also appear to exert noncatalyic effects on proteasome activity that are similar to but independent of one another. These results reveal unexpected functional relationships among multiple deubiquitylating proteins and suggest a model for mammalian 26S proteasome function whereby their concerted action governs proteasome function by linking deubiquitylation to substrate hydrolysis.
INTRODUCTION
Regulated proteolysis constitutes a pervasive mechanism for controlling cellular processes by determining the level of proteins that mediate those processes. Much of the regulated proteolysis in eukaryotic cells is catalyzed by the ubiquitin-proteasome system (Glickman and Ciechanover, 2002). Substrates of the ubiquitin-proteasome system (UPS) are marked for destruction by the covalent attachment of a polyubiquitin chain, a processes catalyzed by the concerted action of multiple ubiquitin-conjugating proteins (Pickart, 2001;Hochstrasser, 2006). Polyubiquitin chains composed of four or more ubiquitin moieties target the substrate to the 26S proteasome, a 2400,000-Da protease complex that degrades the modified protein but spares ubiquitin by detaching it from the substrate (Voges et al., 1999;Thrower et al., 2001). Proteolysis and deubiquitylation are catalyzed by distinct multiprotein subcomplexes that compose the 26S proteasome (Gillette and DeMartino, 2007). Proteolysis is accomplished by the 20S proteasome, a 700,000-Da cylinder-shaped particle formed by four axially stacked hepameric rings (Baumeister et al., 1998; Pickart and Cohen, 2004). The subunits of the two inner rings include six proteases whose active sites face an interior lumen where proteolysis occurs (Groll et al., 1997). The outer rings form narrow, reversibly gated pores through which substrates must pass to reach the lumenal degradation chamber (Groll et al., 2001; Köhler et al., 2001). Deubiquitylation of substrates is accomplished by PA700 (aka 19S regulatory particle; Glickman et al., 2004; Gillette and DeMartino, 2007). In addition to deubiquitylation, PA700 plays multiple roles essential for normal 26S proteasome function. Six of PA700's ∼18 different subunits are AAA ATPases arranged in a hexameric ring that directly abuts each the outer rings of the 20S proteasome and together with several additional subunits constitute a PA700 subcomplex termed the “base” (Glickman et al., 1998; Smith and Goldberg, 2006). The ATP-dependent binding of base subunits to the 20S proteasome is required for assembly of the 26S proteasome and relieves the conformational occlusions that otherwise block the narrow pores at the ends of the 20S cylinder (Groll et al., 2001; Smith et al., 2005; Liu et al., 2006). The remaining PA700 subunits constitute a subassembly termed the “lid” (Glickman et al., 1998). Several PA700 subunits, including components of both the base and lid directly or indirectly bind polyubiquitin chains, thereby providing the basis for selective targeting of ubiquitylated proteins to the proteasome (Lam et al., 2002; Verma et al., 2004). PA700 also displays chaperone-like activity that probably destabilizes the tertiary structure of protein substrates as required for their passage through the narrow opened pores at each end of the 20S cylinder (Braun et al., 1999; Strickland et al., 2000). However, passage of even unfolded substrates through the opened pores is impeded sterically by covalently linked polyubiquitin chains. Thus, substrate deubiquitylation is required for efficient proteolysis and is likely mechanistically linked to normal 26S proteasome function (Verma et al., 2002; Yao and Cohen, 2002; Liu et al., 2006).
Despite the importance of substrate deubiquitylation for proteolysis by the 26S proteasome, a comprehensive understanding of the proteins that catalyze this process and the mechanistic details of their action are lacking. Deubiquitylating enzymes (DUBs) constitute a remarkably large group of cellular proteins that play diverse physiological roles in the expanding constellation of cellular processes mediated by ubiquitin (Amerik and Hochstrasser, 2004; Nijman et al., 2005). Thus, most DUBs probably do not participate directly in 26S proteasome function. Nevertheless, several DUBs have been shown to be physically and/or functionally associated with the 26S proteasome. Rpn11/S13 is a constituent stoichiometric subunit of PA700 in all examined species (Voges et al., 1999). It was identified as a DUB only recently because it is a metalloprotease featuring a JAMM/MPN+ motif and thus differs from nearly all previously recognized and characterized DUBs that utilize a conserved cysteine/histidine-based mechanism for catalysis (Verma et al., 2002; Yao and Cohen, 2002). Rpn11/S13 appears to remove polyubiquitin chains from proteins en bloc. Several studies have shown that Rpn11 and its catalytic function is essential for normal growth of budding yeast Saccharomyces cerevisiae (Verma et al., 2002; Yao and Cohen, 2002). Uch37 (Uch L5) was first identified as a constituent stoichiometric subunit of purified PA700 in mammalian cells; orthologues of Uch37 are present in other species such as Schizosaccharomyces pombe and Drosophila melanogaster, but are absent from others such as S. cerevisiae (Hölzl et al., 2000; Li et al., 2000; Stone et al., 2004). Uch37 processively removes ubiquitin from the distant end of polyubiquitin chains and has been suggested to serve an “editing” function that can rescue poorly ubiquitylated proteins from a proteolytic fate (Lam et al., 1997a,b). The yeast protein Ubp6 and its mammalian ortholog, Usp14, also have been identified as components of 26S proteasome, but do not appear to be a constituent subunit. Ubp6/Usp14 contains an N-terminal ubiquitin-like (Ubl) domain that, in yeast, mediates reversible, salt-sensitive binding to the Rpn1/Rpn2 subunits of PA700 (Leggett et al., 2002). The deubiquitylating activity of Ubp6 increases upon binding to PA700, suggesting that the proteasome is a physiological site of Ubp6 function (Leggett et al., 2002; Yao et al., 2006). Moreover, recent evidence shows that Ubp6 inhibits proteasome activity by a mechanism that does not depend on its DUB activity (Hanna et al., 2006). Finally, the yeast gene product, Doa4, is a DUB that associates with the 26S proteasome. Although active site mutants of Doa4 are defective in the degradation of certain substrates of the ubiquitin-proteasome system, Doa4 has been shown to play a role in ubiquitin-mediated, but proteasome-independent, pathways involving vacuolar protein sorting and endocytosis (Papa et al., 1999; Amerik et al., 2000).
A comprehensive understanding of the role of substrate deubiquitylation in the mechanism of 26S proteasome-catalyzed proteolysis will require both qualitative and quantitative inventory of the DUBs associated with the complex and a determination of their relative biochemical and physiological contributions to proteasome function. Despite the highly conserved nature of most elements of 26S proteasome structure and function, deubiquitylation may be an exception. Thus, the features of DUB activity and its relationship to 26S proteasome function, studied most extensively in yeast, may not be directly applicable to analogous features in other cells. In this report we provide a systematic examination of the content of DUBs in mammalian 26S proteasome and an exploration of their relative cellular roles in ubiquitin-dependent proteolysis. Our results indicate that a complex functional interplay among multiple proteasome-associated DUBs imparts unique regulatory features to proteasome function in mammalian cells.
MATERIALS AND METHODS
Antibodies and Reagents
Anti-ubiquitin mAb was from Santa Cruz Biotechnology (Santa Cruz, CA); anti-Usp14 polyclonal antibody was prepared in rabbits against the peptide “N”-CYGPRRVEIMEEESEQ-“C” (Genemed Synthesis, South San Francisco, CA); anti-Uch37 polyclonal antibody was prepared in rabbits against the sequence “N”-CASNQDDWGASPKRYKIENIRRKHN-“C”; anti-Adrm1 was purchased from Biomol (Plymouth Meeting, PA). MG132 was from Calbiochem (La Jolla, CA). Ubiquitin vinylsulphone was from Boston Biochem (Cambridge, MA). Fluorescent peptide substrate Succinyl-Leu-Leu-Val-Tyr-7-amino-4-methylcoumarin (Suc-Leu-Leu-Val-Tyr-Y-AMC) was from Bachem (King of Prussia, PA). All the cell culture media and reagents were from Invitrogen Invitrogen (Carlsbad, CA). All the remaining reagents were from Sigma (St. Louis, MO) unless otherwise specified.
Purification of 26S Proteasome and PA700
26S proteasome, 20S proteasome, and PA700 were purified from bovine red blood cells as described previously (Liu et al., 2007).
SDS-PAGE and Western Blotting
SDS-PAGE and Western blotting were performed with indicated corresponding antibodies by standard methods as described previously and detailed in appropriate figure legends (Liu et al., 2007).
Cell Culture and Preparation of Cell Lysates
HeLa cells were grown at 37°C in Advanced DMEM (Invitrogen) supplemented with 5% fetal bovine serum, Gluta-MAX (Invitrogen), and antibiotic/antimytotic solution. Cells were passaged when they reached ∼90% confluency. For preparation of lysates, cells were washed with cold phosphate-buffered saline and resuspended in 3 volumes of either RIPA buffer (50 mM Tris-HCl, pH 8.1, at 4°C, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) for total cell lysates or buffer A (20 mM Tris-HCl, pH 7.6, at 4°C, 1 mM β-mercaptoethanol, 1 mM ATP, and 5 mM MgCl2) for soluble cell lysates. After 10 min at 4°C, cells were disrupted with 30 passages through a 27-gauge needle. Lysates were centrifuged at 16,000 × g for 10 min; supernatants were supplemented with 10% glycerol and utilized for experiments described in the text.
Glycerol Density Gradient Centrifugation
Density gradient centrifugation was conducted in 10–40% linear glycerol gradients. Gradients contained 50 mM Tris-HCl, pH 7.6, 1 mM dithiothreitol, 1 mM ATP, 5 mM MgCl2, in a volume of 1.95 ml. Samples were centrifuged at 55,000 rpm for 3 h in Beckman Optima TL ultracentrifuge using a TLS55 rotor (Fullerton, CA). Fractions of 100 μl were collected for appropriate analysis as described for individual experiments. Proteins of known size were centrifuged as standards for gradient calibration.
RNA Interference
Small interfering RNAs (siRNAs) were designed using the modified algorithm proposed by Elbashir et al. (2001) and offered on the homepage of Dharmacon Research (Lafayette, CO). The specificities of sequences were confirmed by BLAST search against the NCBI databases (Table 1). The siRNAs were synthesized, purified, and annealed by Dharmacon Research. siRNAs were dissolved in RNase-free water at 20 μM and stored at −20°C. HeLa cells were seeded at 30–50% confluence in either six-well plates or 10-cm dishes 16 h before transfection. For each well of the six-well plate, 5 μl of stock siRNA were mixed with 1.5 μl of Oligofectamine reagent (Invitrogen) and 87.5 μl of Opti-MEM (Invitrogen) medium. After incubation for 20 min at room temperature, cells were washed with Opti-MEM and transfected in volume of 500 μl of Opti-MEM. After 3.5 h, cells were supplemented with 1.5 ml of complete culture medium. For 10-cm dishes, all volume were increased by fivefold. Controls included transfection with siRNA against enhanced green fluorescent protein (EGFP) or against irrelevant double-strand RNA (dsRNA) that had no corresponding sequence in data bases.
Table 1.
siRNA primers
| Gene name | Primer |
|---|---|
| Rpn11 | 5′-CAAGAGCUUUCUUUAAGAA-3′ (positions 230-248 of human mRNA) |
| 5′-GUUGUACCUGCCAGAAUUA-3′ (positions 210-228 of human mRNA) | |
| UCHL5 (Uch37) | 5′-CUUAGAGCAACAUCCUAAU-3′ (positions 1192-1211 of human mRNA) |
| 5′-GAGAAGGACCGAUUGAUUU-3′ (positions 676-694 of human mRNA) | |
| Usp14 | 5′-AGAAAUGCCUUGUAUAUCA-3′ (positions 1135-1153 of human mRNA) |
| 5′-GCAUUGCAGGGAAACUUAU-3′ (positions 3406-3425 of human mRNA) | |
| S4 | 5′-CAUGCACCGUCCAUCGUGU-3′ (positions 877-895 of human mRNA) |
| b5 | 5′-GAAGGUGAUAGAGAUCAAC-3′ (positions 292-310 of human mRNA) |
| EGFP | 5′-GGUGAACUUCAAGAUCCGC-3′ (positions 654-672 of human mRNA) |
RT-PCR. Total RNA was isolated from HeLa cells using Trizol reagent (Invitrogen) according to the manufacturer's instructions. RNA content was determined by measuring A260. RT-PCR was carried out using OneStep RT-PCR kit (Qiagen) using pairs of primers amplifying the genes of interest, as indicated in Table 2. The number of cycles was adjusted to obtain a linear range of reaction products.
RT-PCR
Total RNA was isolated from HeLa cells using Trizol reagent (Invitrogen) according to the manufacturer's instructions. RNA content was determined by measuring A260. RT-PCR was carried out using OneStep RT-PCR kit (Qiagen, Valencia, CA) using pairs of primers amplifying the genes of interest, as indicated in Table 2. The number of cycles was adjusted to obtain a linear range of reaction products.
Table 2.
Primers used for semi-quantitative RT-PCR
| Gene name | Accession number | Forward primer | Reverse primer |
|---|---|---|---|
| Rpn11 | NM_005805 | 5′-AAGGAGATGTTGGAATTAGCC-3′ | 5′-CATAGCTGCTAAACACTGGACA-3′ |
| UCHL5 (Uch37) | NM_015984 | 5′-GAACGCAAAGAAAGCTCAGG-3′ | 5′-AGACAAGACAGGCTGGCACT-3′ |
| Usp14 | NM_005151 | 5′-TCCAACGTTGCAAAGAAATG-3′ | 5′-ATTTGGATCGAAAAGACACC-3′ |
| Actin | NM_001101 | 5′-TTCCTTCCTGGGCATGGAGT-3′ | 5′-ATCCACATCCTGCTGGAAGGT-3′ |
Measurement of Proteasome Activity in Cell Lysates
Hydrolytic activity of the proteasome was measured in soluble cell lysates using peptide substrate Suc-Leu-Leu-Val-Tyr-AMC as described previously. Assays were conducted in triplicate, and mean values were normalized for protein concentration using the method of Bradford (1976) with reagent purchased from Bio-Rad (Richmond, CA).
Measurement of Intracellular Protein Degradation
Intracellular protein degradation was measured by monitoring changes in steady-state levels of GFP from a HeLa cell line that stably expresses Ub-R-GFP, as described previously (Wojcik et al., 2006). This protein is degraded by the proteasome via the N-end Rule pathway.
RESULTS
Mammalian 26S Proteasome Contains Two Constituent and One Reversibly Associated Deubiquitylating Proteins
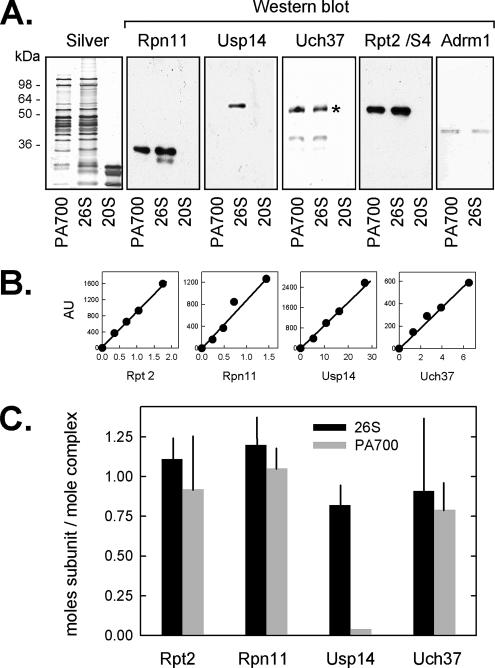
To establish the complement of DUBs present in mammalian 26S proteasome, we subjected 26S proteasome purified from bovine red blood cells to Western blotting with antibodies against three candidate DUBs: Rpn11/S13, Uch37, and Usp14. Each DUB was identified in 26S proteasome at approximately stoichiometric levels (Figure 1). To confirm the specific association of these DUBs with 26S proteasome, we subjected purified 26S proteasome to glycerol density gradient centrifugation and analyzed gradient fractions by Western blotting and proteasome activity assays. Each DUB sedimented coincidently with each other, with other established 26S proteasome subunits, and with proteasome activity (Figure 2, A and C). Similar results were obtained when DUB content of 26S proteasome was analyzed by native PAGE (data not shown). These results indicate that Rpn11/S13, Uch37, and Usp14 are components of the bovine red blood cell 26S proteasome. Several recent studies have identified Adrm1 (Rpn13) as a newly discovered PA700 subunit that binds to Uch37 (Hamazaki et al., 2006; Jorgensoen et al., 2006; Qiu et al., 2006; Yao et al., 2006). Consistent with these results, we also identified Adrm1 as a component of bovine 26S proteasome (Figures 1 and 2).
Figure 1.
Identification of deubiquitylating proteins in 26S proteasome and PA700. (A) Equimolar amounts (5 nmol) of purified PA700, 26S, and 20S were subjected to SDS-PAGE followed by either silver staining or Western blotting using indicated antibodies (asterisk indicates a nonspecific cross-reacting band against the Uch37 antibody). (B) Indicated purified recombinant proteins in quantities indicated on the abscissa were subjected to Western blotting. Resulting blots were quantified and used as standard curves for content of respective proteins in purified 26S proteasome and PA700. Data are expressed as mean values (±SEM) of four independent 26S proteasome or five independent PA700 preparations (C).
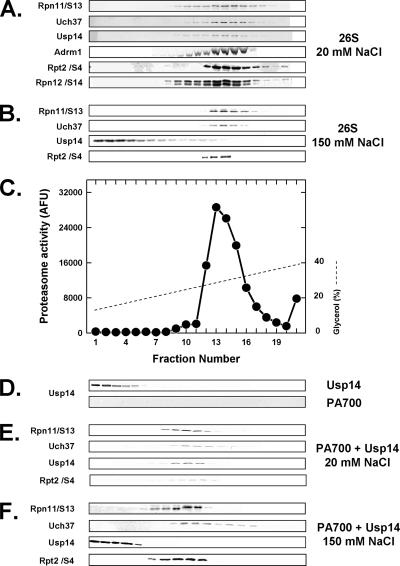
Figure 2.
26S proteasome contains two constitutive and one salt-dissociable deubiquitylating proteins. Purified 26S proteasome (50 μg; A–C), purified PA700 (50 μg; D, bottom, E, and F), and purified recombinant Usp14 (10 μg; D, top) were subjected to glycerol density gradient centrifugation in buffer containing 20 mM Tris-HCl, pH 7.6, 1 mM β-mercaptoethanol, 1 mM ATP, and 5 mM Mg2+ and either 20 mM NaCl (A, C, D, and E) or 150 mM NaCl (B and F). Purified recombinant Usp14 (5 μg) was preincubated with PA700 for 15 min at 25°C in the presence of 20 mM NaCl (E) or 150 mM NaCl (F) before centrifugation. Gradient fractions were Western-blotted for the indicated proteins or assayed for proteasome activity using Suc-Leu-Leu-Val-Tyr-AMC substrate.
Purified PA700, the 19S regulatory complex of 26S proteasome, also contained stoichiometric levels of Rpn11/S13, Uch37, and Adrm1, but had undetectable levels of Usp14 (Figure 1). These results indicate that Usp14 is reversibly associated with 26S proteasome, as shown previously for Ubc6, the yeast homolog of Usp14 (Leggett et al., 2002). To document this feature directly for mammalian Usp14, we performed two types of experiments. First, we treated purified 26S proteasome with 150 mM NaCl or KCl. On subsequent glycerol density gradient centrifugation, Usp14, but not Rpn11 or Uch37, dissociated from the 26S proteasome (Figure 2B and data not shown). This salt concentration had no effect on the global structural stability of 26S proteasome, as evidenced by the normal distribution pattern of other established constituent proteasome subunits (Figure 2 and data not shown). Second, purified recombinant Usp14 was added to purified PA700, which contained no endogenous Usp14 (Figure 2D). In the presence of 20 mM NaCl, Usp14 cosedimented with PA700 upon subsequent glycerol density (Figure 2E); in contrast, in the presence of 150 mM NaCl, Usp14 did not associate with PA700 (Figure 2F). These results confirm the salt-sensitive nature of this association and demonstrate the capacity of isolated PA700 to bind Usp14. Thus, the difference in the content of Usp14 between purified preparations of PA700 and 26S proteasome likely results from the different salt conditions used during their respective purification procedures.
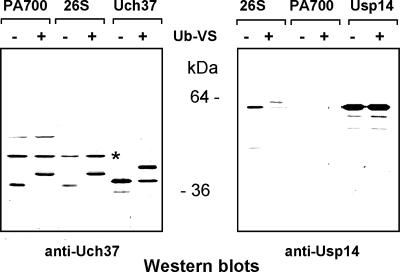
Ubiquitin vinylsulphone (Ub-VS) is an active site covalent labeling reagent for cysteine-type DUBs, such as Usp14 and Uch37, and therefore can be utilized to identify the presence and catalytical competence of DUBs (Borodovsky et al., 2001). Previous work has shown that Usp14/Ubp6 and Uch37 have weak deubiquitylating activity as isolated proteins but are catalytically activated upon incorporation into the 26S proteasome complex (Borodovsky et al., 2001; Leggett et al., 2002; Yao et al., 2006). We treated purified recombinant Uch37 and Usp14, as well as purified PA700 and 26S proteasome complexes with Ub-VS. Ub-VS labeled the purified recombinant proteins weakly, indicating that they were poor catalysts in this state (Figure 3). Ub-VS efficiently labeled Uch37 in both 26S proteasome and PA700, as indicated by the quantitative increase in the Uch37-postitive band by ∼8 kDa. In contrast, Ub-VS labeled Usp14 in 26S proteasome (as indicated by an analogous increase in molecular weight of the Usp14-positive band), but not in PA700 (Figure 3). These results confirm the differential content of Usp14 and Uch37 in 26S proteasome compared with PA700 and support the conclusion that each DUB becomes catalytically competent upon complex formation.
Figure 3.
Uch37 and Usp14 are activated upon association with PA700. Purified PA700, 26S proteasome, recombinant Uch37, or recombinant Usp14 were incubated with (+) or without (−) ubiquitin vinyl sulfone (Ub-VS) as indicated. Samples were subjected to Western blotting with anti-Uch37 (left panel) or anti-Usp14 (right panel). Asterisk (*) indicates a nonUch37 protein that cross-reacts with this antibody.
Distribution of DUBs in HeLa Cell Extracts
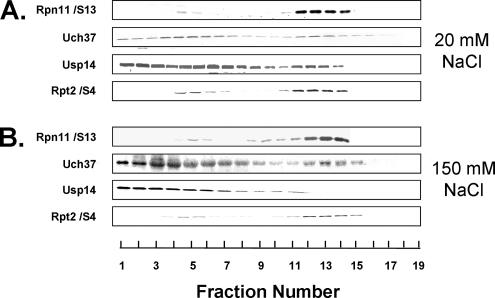
To gain insight into the relative cellular distributions of proteasome-associated DUBs, we prepared extracts of HeLa cells under high- and low-salt conditions and subjected them to glycerol density gradient centrifugation. This method permits assessment of the structural and functional status of the proteasome by assaying gradient fractions for proteasome enzymatic activity and Western blotting gradient fractions for proteasome content (see below). Rpn11, Uch37, and Usp14 migrated coincidently with each other and with 26S proteasome activity under low-salt conditions (Figure 4A). However, in addition to their association with 26S proteasome, substantial portions of total Usp14 and Uch37 were present in lower molecular weight fractions, and in the case of Usp14 this distribution was prominently multimodal (Figure 4A). Quantification of the DUBs in these gradients indicated that each was present in 26S proteasome in approximately stoichiometric levels. These data indicate that most cellular Uch37 and Usp14 are not associated with the proteasome and therefore may have other cellular roles. Exposure of extracts to 150 mM salt had little effect on the distribution patterns of cellular Rpn11/S13 or Uch37 but promoted redistribution of all Usp14 to slower sedimenting fractions (Figure 4B). These results are consistent with and explain findings for these DUBs in purified proteins and indicate that mammalian 26S proteasome contains Rpn11/S13 and Uch37 as constituent subunits and Usp14 as a reversibly associated subunit.
Figure 4.
Distribution of proteasome-associated DUBs in HeLa cell lysates. Soluble lysates of HeLa cells were prepared as described in Materials and Methods and adjusted to final concentrations of with 20 mM NaCl or 150 mM NaCl. Lysates (660 μg) were subjected to glycerol density gradient centrifugation with corresponding NaCl concentrations in the gradients. Gradient fractions were subjected to Western blotting with the indicated antibodies.
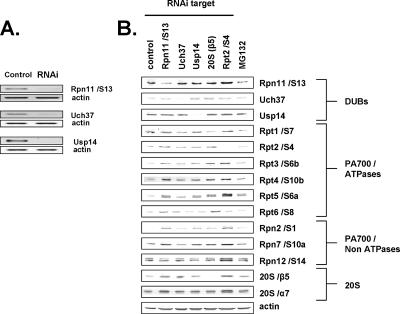
RNAi of Proteasome-associated DUBs
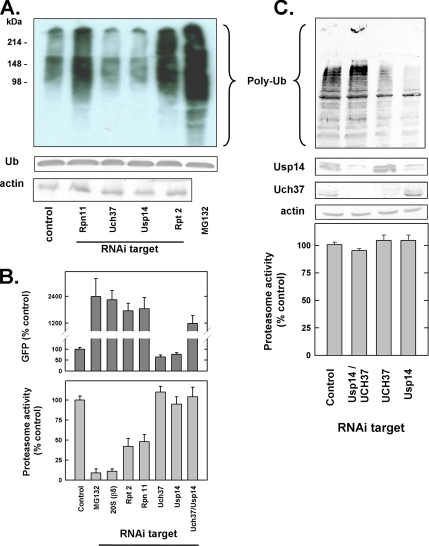
To determine the relative physiological roles of the proteasome-associated DUBs identified above, each was targeted for RNA interference (RNAi) using siRNAs transfected into HeLa cells. RNAi decreased mRNA and protein levels of each DUB (Figure 5), but the reduced content of individual DUBs elicited different effects on cellular features related to proteasome function. For example, RNAi of Rpn11/S13 greatly slowed cell growth, whereas RNAi of Uch37 and Usp14 had no detectable effect on growth (data not shown). RNAi of Rpn11/S13 also caused significant accumulation of polyubiquitylated proteins, a sensitive indicator of inhibited UPS function (Figure 6A). The magnitude of this effect was similar to that caused by RNAi of either Rpt2/S4, an ATPase of PA700 (Figure 6A), or β5, a catalytic subunit of the 20S proteasome (data not shown), but less than that caused by treatment of cells with MG132, a pharmacological inhibitor of the proteasome. In contrast, RNAi of Uch37 or Usp14 reduced the level of polyubiquitylated cellular proteins, suggesting that loss of either of these proteasomal DUBs enhanced rather than inhibited UPS function (see below). To test the effects of RNAi of these DUBs on cellular protein degradation directly, we repeated these experiments in a HeLa cell line engineered to stably express Ub-R-GFP. This protein is degraded rapidly by the UPS via the N-end rule pathway and accumulates upon treatment of cells with the proteasome inhibitor MG132. As expected, RNAi of Rpt2/S4 or Rpn11/S13 also inhibited degradation of this protein (Figure 6B). In contrast, RNAi of Uch37 or Usp14 significantly reduced steady-state levels of GFP. This effect is consistent with the reduced levels of cellular polyubiquitylated proteins described above and suggests that loss of either of these DUBs accelerates cellular proteolysis.
Figure 5.
RNAi of proteasome-associated DUBs. HeLa cells were subjected to RNAi of indicated proteins or EGFP (control) for 24 h as described in Materials and Methods. (A) Total cellular RNA was collected, and 1 ng RNA was used for RT-PCR of indicated proteins as described in Materials and Methods. (B) HeLa cells were subjected to RNAi of the indicated proteins or EGFP (control) for 72 h or treated with MG132 (10 μM) for 16 h. Fifteen micrograms total cell lysate were subjected to Western blotting with the indicated antibodies.
Figure 6.
Effect of RNAi of proteasome-associated DUBs on cellular protein degradation. (A) HeLa cells were subjected to RNAi of the indicated proteins or EGFP (control) for 72 h or treated with MG132 (10 μM) for 16 h. Total cell lysates (40 μg) were subjected to SDS-PAGE and Western blotting with the anti-ubiquitin antibodies. For detection of monoubiquitin (Ub), the membrane was autoclaved in water for 15 min before blocking. (B) Top, HeLa cells stably expressing Ub-R-GFP were subjected to RNAi of the indicated proteins as described above. GFP was quantified in total cell extracts as expressed as a percentage of control cells transfected with siRNA against an irrelevant dsRNA. Results show mean values of four independent plates of cells (±SEM); similar results were obtained in three separate experiments. All values are significantly different from control. Bottom, extracts from HeLa cells, normalized for protein concentration, were subjected to RNAi as above were assayed for proteasome activity using Suc-Leu-Leu-Val-Tyr-AMC as substrate. Activity in control cells was set at 100%, and other activities are expressed as a percentage of that value. Results show mean values of three independent plates of cells (±SEM), and similar results were obtained in three separate experiments. (C) HeLa cells were subjected to RNAi as described above, including simultaneous transfection with siRNAs for Usp14 and Uch37. Extracts normalized for protein concentration were subjected to Western blotting with the indicated antibodies (top) or assayed for proteasome activity as described above (bottom).
To assess the relationship between alterations in cellular proteolysis by RNAi of individual DUBs and the proteolytic capacity of the proteasome, we assayed corresponding cell extracts for hydrolysis of Suc-Leu-Leu-Val-Tyr-AMC, a synthetic peptide hydrolyzed by activated forms of the proteasome in a ubiquitin-independent manner. RNAi of Rpn11/S13 reduced proteasome-catalyzed hydrolysis of Suc-Leu-Leu-Val-Tyr-AMC by ∼50% (Figure 6B). This level of inhibition was approximately similar to that achieved by RNAi of Rpn2/S4 and several other subunits of PA700, but less than that caused by RNAi of the β5 subunit of 20S proteasome, or by MG132 (see below). Thus, RNAi of Rpn11 likely inhibits cellular proteolysis by inhibiting proteasome function. In contrast, RNAi of either Uch37 or Usp14 had no effect on rates of peptide hydrolysis, suggesting that the accelerated proteolysis detected under these conditions was not a consequence of enhanced hydrolytic capacity of the proteasome (Figure 6).
To gain additional insight into the relative roles of these DUBs on cellular protein degradation and proteasome function, we subjected cells to simultaneous RNAi of multiple DUBs. RNAi of Rpn11 and Uch37 or Rpn11 and Usp14 reduced levels of each target protein by greater than 85%, inhibited cellular protein degradation, as determined by accumulation of polyubiquitylated proteins and GFP (Figure 6C), and inhibited hydrolysis of Suc-Leu-Leu-Val-Tyr-AMC by 50–60% (data not shown). These results indicate that loss of Rpn11 dominants the resulting phenotype in these cells. Simultaneous RNAi of Uch37 and Usp14 also caused accumulation of cellular polyubiquitylated proteins and GFP, but had no effect on hydrolysis of the peptide substrate (Figure 6). These surprising results suggest that loss of both Uch37 and Usp14, unlike loss of either alone, inhibits cellular protein degradation, but does so without a concomitant reduction in catalytic capacity of the proteasome.
Relative Effects of Proteasome-associated DUBs on Proteasome Structure and Function
To determine the basis for the effects RNAi of proteasome-associated DUBs on proteasome function described above, we prepared extracts from HeLa cells subjected to RNAi, fractionated them by glycerol density gradient centrifugation, and analyzed fractions as described in Figure 4. The results of this analysis are compiled in Figure 7.
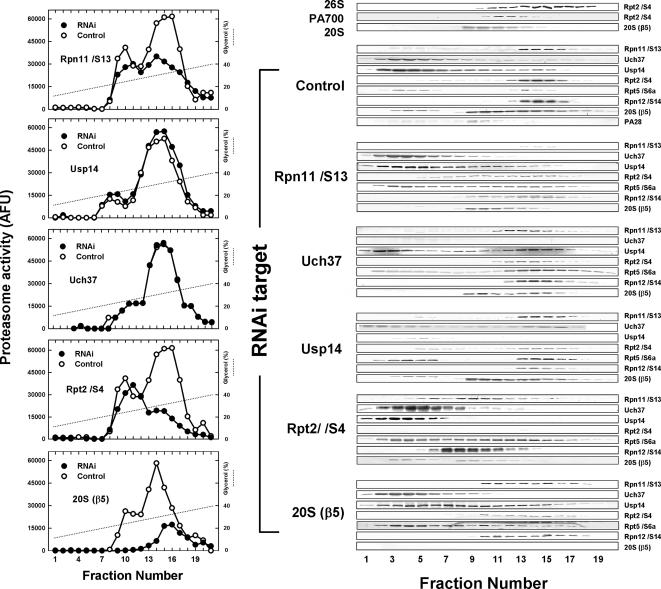
Figure 7.
Relative effects of proteasome-associated DUBs on proteasome structure and function. Soluble extracts were prepared from HeLa cells treated for 3 d with siRNA of the indicated subunits. Extracts, 550–700 μg, were subjected to glycerol density gradient centrifugation, and collected fractions were assayed for Suc-Leu-Leu-Val-Tyr-AMC hydrolyzing activity (left panel) or were separated using SDS-PAGE probed with the indicated antibodies. Shown gradients were obtained from separate centrifugations due to limited capacity of the centrifuge. However, multiple blots for each RNAi target were conducted with samples from the same gradient. Results were qualitatively similar for each profile in at least three independent experiments.
Glycerol density gradient centrifugation of extracts from control HeLa cells resolves two peaks of proteasome activity. The faster sedimenting peak accounts for most hydrolytic activity and corresponds to 26S proteasome, as evidenced by the comigration of subunits of both 20S proteasome and PA700 and a sedimentation position indistinguishable from that of purified 26S proteasome. Native PAGE of these fractions also revealed bands characteristic of 26S proteasome and confirmed its identification (data not shown). The slower sedimenting peak contained lesser amounts of proteasome activity that varied among independent extracts. Nevertheless, this peak usually contained ∼50% of 20S proteasome protein, as shown here for the β5 subunit. Similar distribution patterns were obtained for other 20S subunits and represented both free 20S proteasome, with very low catalytic activity, and 20S proteasome associated with PA28, a proteasome activator that accounts for stimulated proteasome activity in these fractions. Western blotting also revealed some slowly sedimenting β5 subunit with distinctly greater molecular weight on SDS-PAGE than β5 subunits in the 20S and 26S proteasome fractions. This probably represents unprocessed β5 subunits in unassembled 20S proteasome complexes.
RNAi of Rpn11/S/13 caused a marked reduction in 26S proteasome activity but had little effect on 20S proteasome activity. Western blotting revealed that 26S proteasome structure was severely disrupted by loss of this subunit. Thus, the content of PA700 subunits normally present in the portion of the gradient corresponding to 26S proteasome was diminished and redistributed to slower migrating fractions, probably containing subcomplexes of PA700 incompetent for complete PA700 assembly (see below). RNAi of Rpt2/S4 had similar effects on 26S proteasome activity but promoted a qualitatively greater redistribution of PA700 subunits to lower molecular weight complexes. This distinction indicates that RNAi of Rpt2/S4 had a more severe effect on 26S proteasome assembly than did RNAi of Rpn11/S13 and could reflect the different consequences of eliminating a PA700 base subunit (Rpt2/S4) versus a PA700 lid subunit (Rpn11/S13). RNAi of Rpn11/S13 also reduced the amount of 20S proteasome subunits in fractions corresponding to 26S proteasome but shifted them to fractions corresponding to the 20S proteasome. These results indicate that Rpn11/S13 is essential for normal assembly of PA700 and 26S proteasome, but does not affect assembly of 20S proteasome. In contrast to RNAi of Rpn11/S13, RNAi of Uch37 or Usp14 had little detectable effect on the activity or structure of the 26S proteasome. These results are consistent with the reversible nature of Usp14, but were unexpected for a constituent subunit such as Uch37. Thus, lack of Uch37 does not appear to inhibit assembly of PA700 and Uch37-deficient PA700 binds normally to 20S proteasome. Because most cellular Usp14 and Uch37 was extra-proteasomal, it is possible that even substantial reduction of their levels by RNAi could leave residual protein sufficient to populate 26S proteasomes fully. Such an effect could explain the failure of RNAi of these proteins to influence proteasome activity or structure. However, Western blotting revealed undetectable levels of these proteins in gradient fractions corresponding to 26S proteasome and greatly diminished, but detectable levels of the protein in nonproteasome fractions after their respective RNAi. These results argue that the DUB composition of 26S proteasome in these experiments was reduced substantially and suggest that at least some of the extra-proteasomal populations of Uch37 and Usp14 may not be competent or solely destined for proteasome assembly. Additional work will be required to define the precise cellular factors and conditions that govern the assembly of these proteins into the proteasome. Simultaneous RNAi of both Uch37 and Usp14 had no effect on 26S proteasome structure or function, or the relative distribution of the other proteasome subunits, as expected from the results described above.
Finally, we note and emphasize that a precise quantitative comparison of changes in proteasome subunit distribution among the many different RNAi experiments depicted in Figure 7 is complicated by multiple factors. First, the data represent results of independent centrifugations. Although the relative distribution patterns of given proteins were qualitatively reproducible among independent experiments, quantitative comparisons of a given protein among different blots is inappropriate. Second, RNAi of a given proteasome subunit not only reduced expression of the protein target, but often affected the expression of other proteasome subunits. This phenomenon appears to represent a specific compensatory response of cells to the loss of a proteasome subunit, rather than an off-target effect of RNAi and has been documented previously in S. cerevisiae and Drosophila (Wojcik and DeMartino, 2002). Figure 5B illustrates this effect for RNAi of the three DUBs as well as for two other proteasome subunits of the 20S and PA700 subcomplexes (β5 and Rpt2/S4, respectively). The nature of the compensatory response appeared to depend more on the compensating protein than on the identity the RNAi target. In other words, a given proteasome subunit generally responded similarly to RNAi of each target. For example, Rpn11/S13 and Uch37, the two constituent proteasome DUBs, showed increased expression in response to RNAi of most proteasome subunits, including the reversibly associated Usp14. In this regard, they behaved similarly to many other proteasome subunits including the ATPases of the PA700 base, non-ATPases of the base and lid, and at least one examined 20S proteasome subunit. In contrast, cellular levels of Usp14, the reversibly associated DUB, as well as certain other constituent subunits such as Rpn12/S14, were not detectably affected or affected only modestly by RNAi of other proteasome subunits. Curiously, despite the lack of altered Usp14 expression in response to RNAi of other proteasome subunits, RNAi of Usp14 generally promoted increased expression of proteasome subunits that responded to RNAi of constituent subunits. The overall phenomenon of altered proteasome expression in response to changes in levels of individual proteasome components is complex, and an understanding of its significance and mechanism will require additional work. Nevertheless, the current results demonstrate that increased expression of proteasome subunits to the RNAi targets examined here does not cause increased formation of functional proteasomes, presumably because most subunits are required for normal complex assembly. Moreover, this response does not appear to result solely from diminished proteasome activity because MG132 was not a perfect mimic of the general effects.
DISCUSSION
Degradation of cellular proteins by the ubiquitin-proteasome system requires the antithetical processes of substrate ubiquitylation and deubiquitylation. Ubiquitylation of proteolytic substrates serves mainly, it not exclusively, to target the modified proteins to the 26S proteasome (Laney and Hochstrasser, 1999;Thrower et al., 2001). In contrast, deubiquitylation of proteasome substrates contributes several elements intrinsic to the biochemical mechanism of proteolysis and also serves broader physiological roles. For example, removal of polyubiquitin chains from substrates eases transit of polypeptides through the narrow pores of the proteasome en route to the degradation chamber (Yao and Cohen, 2002). DUB activity also may be required to clear substrate-free polyubiquitin chains from binding sites on the proteasome in preparation for binding of new polyubiquitylated substrates. Proteasome-associated DUB activity also may provide an editing function to spare inappropriately or poorly modified substrates from destruction (Lam et al., 1997b). Finally, substrate deubuitylation plays an essential role in cellular ubiquitin homeostasis because the ubiquitin recycled by this process is required to maintain cellular ubiquitin at normal levels (Chernova et al., 2003; Hanna et al., 2007). Therefore, defects in proteasome-localized DUB activity can lead to the paradoxical consequences of impaired rates of substrate proteolysis but enhanced rates of ubiquitin degradation.
Although the recycling of ubiquitin monomers from polyubiquitin chains during proteolysis was recognized in early studies of ubiquitin-dependent protein degradation, the mechanistic details of the process, including the identity and relative roles of specific DUBs that catalyze substrate deubiquitylation, have remain poorly defined. Our data confirm the identification of Rpn11/S13 as a constitutive, stoichiometric subunit of the 26S proteasome. In addition to its function as a DUB, Rpn11 was essential for 26S proteasome structure; thus, the severely impaired function of 26S proteasome in response to RNAi of Rpn11 was at least a consequence of reduced 26S proteasome content. Although several previous studies in yeast, Drosophila, and human cells have shown active-site mutants of Rpn11 to be nonviable, thereby suggesting that Rpn11 DUB activity is essential (Verma et al., 2002; Yao and Cohen, 2002; Lundgren et al., 2004; Gallery et al., 2007), at least one other study has shown an active-site yeast mutant to be viable (Guterman and Glickman, 2004). Our attempts to distinguish between functional and structural effects of RNAi of Rpn11 by complementation with active site mutants of Rpn11 were unsuccessful due to incomplete incorporation of the transiently expressed subunit (Kulich and DeMartino, unpublished observations). Our data also show that Usp14, like its yeast ortholog Ubp6, reversibly associates with 26S proteasome; each protein contains an N-terminal Ubl domain that mediates this association. RNAi of Usp14 had no detectable effect on proteasome structure. In cell-free experiments using either purified proteins or crude cell extracts, physiological salt concentrations were sufficient to dissociate Usp14 from the proteasome. Thus, the factors or conditions that govern association of Usp14 with the proteasome in intact cells are unclear. Although 26S proteasomes from HeLa cells contained approximately stoichiometric levels of Usp14, most cellular Usp14 was not associated with the proteasome. This suggests that Usp14 may play other cellular roles or exist in distinct functional pools. Finally, we identified Uch37 as a stoichiometric component of the 26S proteasome. This designation is consistent with the original identification of Uch37 as a stoichiometric component of purified PA700 (Lam et al., 1997b). Data in several recent reports can be interpreted to indicate that Uch37, like Usp14, reversibly associates with the 26S proteasome (Hamazaki et al., 2006; Jorgensoen et al., 2006; Qiu et al., 2006; Yao et al., 2006). We failed to detect significant dissociation of Uch37 under the same conditions that completely dissociated Usp14, but it is possible that Uch37 can be selectively displaced from 26S proteasome under other conditions. Similarly, RNAi of Uch37, like RNAi of Usp14, had no detectable effect on 26S proteasome structure, suggesting that it is not necessary for stability of the complex. Uch37 shared other features with Usp14, including a substantial nonproteasomal population and an increase in catalytic activity upon binding to the proteasome. These nonproteasomal populations could serve as latent protein reserves or may have other functions related or unrelated to DUB activity. For example, several nonproteasomal proteins have been identified as specific binding partners for Uch37, suggesting that Uch37 may function in multiple cellular contexts (Li et al., 2001; Wicks et al., 2005, 2006).
Uch37 represents one of the few structural distinctions between the otherwise highly conserved yeast and mammalian proteasomes. This distinction suggests that DUB function may also differ between these sources. Our data indicate that Uch37 and Usp14 share several activities that function redundantly. RNAi of either protein resulted in accelerated proteasome-dependent proteolysis and a decreased cellular ubiquitin pool. Previous results have also noted increased protein degradation as either a direct consequence of Uch37 content (Qiu et al., 2006) or an indirect consequence of reduced proteasomal association (Hamazaki et al., 2006; Qiu et al., 2006). Finley and colleagues have shown that Ubp6 inhibited proteasome-dependent degradation of ubiquitylated proteins in yeast (which lack Uch37) by a mechanism that was independent of its DUB activity (Hanna et al., 2006). Our data are consistent with similar functions for both Usp14 and Uch37. In contrast, simultaneous RNAi of both proteins generated a cellular phenotype similar to that of RNAi of Rpn11 including inhibited cell growth, decreased protein degradation, and accumulation of polyubiquitylated proteins. These results indicate that each protein could at least partially compensate for loss of function of the other, but that lack of both proteins compromised normal ubiquitin-dependent proteolysis. Because RNAi of both Uch37 and Usp14 does not influence the proteolytic capacity of the proteasome per se (as judged by hydrolysis of synthetic peptides), the resulting cellular effects on proteolysis likely relate to diminished capacity for substrate deubiquitylation.
We propose that Usp14 and Uch37 complement the function of Rpn11 to satisfy two major requirements of proteasome-associated deubiquitylation. First, the substrate must be freed of conjugated ubiquitin. Failure to remove ubiquitin from substrates both impedes proteolysis and promotes excessive degradation of ubiquitin. Second, polyubiquitin chains, whether attached to substrates or not, must be cleared from their binding sites on the proteasome to create occupancy for new rounds of polyubiquitylated protein binding and proteolysis. We propose that Rpn11 and Usp14/Uch37 divide these necessary functions. Rpn11 cleaves polyubiquitin chains en bloc at or very near their site of substrate conjugation. Although this activity may relieve most of the steric inhibition otherwise detrimental to efficient substrate degradation, it does not clear the proteasome of bound polyubiquitin chains. Uch37 and Usp14, which hydrolyze polyubiquitin chains from their distal ends, accomplish this function and also trim any remaining ubiquitin remnants from the substrates (Lam et al., 1997a,b; Hu et al., 2005). The precise distinctions in catalytic function between Usp14 and Uch37 are not clear, but the activities appear to operate, at least in part, in similar pathways because RNAi of both creates a phenotype not exhibited by RNAi of either alone. Inhibition of proteasome-catalyzed protein degradation in the double knockdown indicates the essential nature of these functions. In addition to their DUB activities, we propose that Usp14 and Uch37 each has noncatalytic proteasome inhibitory functions that are similar to but independent of one another. Such activities, recently demonstrated for Ubp6, may be analogous to a governor on an engine and serve to limit rates of proteolysis to those that most efficiently couple it to corresponding rates of deubiquitylation. Thus, RNAi of either protein could accelerate proteolysis via partial relief of otherwise additive inhibitory effects, but tolerate the loss of one DUB activity via compensatory effects of the other. In contrast, RNAi of both Usp14 and Uch37 inhibits proteolysis because of loss of essential DUB activity that cannot be assumed by Rpn11. Notably, a S. cerevisiae strain with an active site mutation of Rpn11 and a deletion of Ubp6 shows synthetic lethality (Guterman and Glickman, 2004). This feature is highlights both the cooperative nature of Rpn11 with other DUBs and the functional interaction between Usp14 and Uch37, the latter of which is absent in yeast. Future work will determine the temporal relationship between deubiquitylation and other features of substrate processing such as unfolding, translocation, and peptide bond hydrolysis.
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health (DK 46181) and the Welch Foundation (I-500) to G.N.D.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-10-1040) on December 27, 2007.
REFERENCES
- Amerik A., Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim. Biophys. Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Amerik A., Nowak J., Swaminathan S., Hochstrasser M. The Doa4 deubiquitinating enzyme is functionally linked to the vacuolar protein-sorting and endocytic pathways. Mol. Biol. Cell. 2000;11:3365–3380. doi: 10.1091/mbc.11.10.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumeister W., Walz J., Zühl F., Seemüller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- Borodovsky A., Kessler B. M., Casagrande R., Overkleeft H. S., Wilkinson K. D., Ploegh H. L. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J. 2001;20:5187–5196. doi: 10.1093/emboj/20.18.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Braun B. C., Glickman M., Kraft R., Dahlmann B., Kloetzel P.-M., Finley D., Schmidt M. The base of the proteasome regulatory particle exhibits chaperone-like activity. Nat. Cell Biol. 1999;1:221–226. doi: 10.1038/12043. [DOI] [PubMed] [Google Scholar]
- Chernova T. A., Allen K. D., Wesoloski L. M., Shanks J. R., Chernoff Y. O., Wilkinson K. D. Pleiotropic effects of Ubp6 loss on drug sensitivities and yeast prion are due to depletion of the free ubiquitin pool. J. Biol. Chem. 2003;278:52102–52115. doi: 10.1074/jbc.M310283200. [DOI] [PubMed] [Google Scholar]
- Elbashir S. M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Gallery M., Blank J. L., Lin Y., Gutierrez J. A., Pulido J. C., Rappoli D., Badola S., Rolfe M., Macbeth K. J. The JAMM motif of human deubiquitinase Poh1 is essential for cell viability. Mol. Cancer Ther. 2007;6:262–268. doi: 10.1158/1535-7163.MCT-06-0542. [DOI] [PubMed] [Google Scholar]
- Gillette T. G., DeMartino G. N. Proteasomes: machines for all reasons. Cell. 2007;129:659–662. doi: 10.1016/j.cell.2007.05.007. [DOI] [PubMed] [Google Scholar]
- Glickman M. H., Ciechanover A. The ubiquitin-proteasome system: destruction for the sake of construction. Physiol. Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- Glickman M. H., Guterman A. Deubiquitinating enzymes are IN/(trinsic to proteasome function) Curr. Protein Pep. Sci. 2004;5:201–211. doi: 10.2174/1389203043379756. [DOI] [PubMed] [Google Scholar]
- Glickman M. H., Rubin D. M., Coux O., Wefes I., Pfeifer G., Cjeka Z., Baumeister W., Fried V., Finley D. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- Groll M., Bajorek M., Kohler A., Moroder L., Rubin D. M., Huber R., Glickman M. N., Finley D. A gated channel into the proteasome core particle. Nat. Struct. Biol. 2001;11:1062–1067. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- Groll M., Ditzel L., Lowe J., Stock D., Bochtler M., Bartunik H. D., Huber R. Structure of the 20S proteasome from yeast at 2.4A resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- Guterman A., Glickman M. H. Complementary roles for Rpn11 and Ubp6 in deubiquitination and proteolysis by the proteasome. J. Biol. Chem. 2004;279:1729–1738. doi: 10.1074/jbc.M307050200. [DOI] [PubMed] [Google Scholar]
- Hamazaki J., Iemura S., Natsume T., Yashiroda H., Tanaka K., Muratua S. A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. EMBO J. 2006;25:4524–4536. doi: 10.1038/sj.emboj.7601338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J., Hathaway N. A., Tone Y., Crosas B., Elsasser S., Kirkpatrick D. S., Leggett D. S., Gygi S. P., King R. W., Finley D. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell. 2006;127:99–111. doi: 10.1016/j.cell.2006.07.038. [DOI] [PubMed] [Google Scholar]
- Hanna J., Meides A., Zhang D. P., Finley D. A ubiquitin stress response is distinct from the proteasome stress response and alters proteasome composition. Cell. 2007;129:747–759. doi: 10.1016/j.cell.2007.03.042. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Lingering mysteries of ubiquitin-chain assembly. Cell. 2006;124:27–34. doi: 10.1016/j.cell.2005.12.025. [DOI] [PubMed] [Google Scholar]
- Hölzl H., et al. The regulatory complex of Drosophila melanogaster 26S proteasomes: subunit composition and localization of a deubiquitylating enzyme. J. Cell Biol. 2000;150:119–129. doi: 10.1083/jcb.150.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M., Li P., Song L., Jeffrey P. D., Chenova T. A., Wilkinson K. D., Cohen R. E., Shi Y. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. EMBO J. 2005;24:3747–3756. doi: 10.1038/sj.emboj.7600832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensoen J. P., Lauridsen A.-M., Kristensen P., Dissing K., Johnsen A. H., Hendil K. B., Hartmann-Petersen R. Adrm1, a putative cell adhesion regulation protein, is a novel proteasome-associated factor. J. Mol. Biol. 2006;360:1043–1052. doi: 10.1016/j.jmb.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Köhler A., Cascio P., Leggett D. S., Woo K. M., Goldberg A. L., Finley D. The axial channel of the proteasome core particle is gated by the Rpt2 ATPase and controls both substrate entry and product release. Mol. Cell. 2001;7:1143–1152. doi: 10.1016/s1097-2765(01)00274-x. [DOI] [PubMed] [Google Scholar]
- Lam Y. A., DeMartino G. N., Pickart C. M., Cohen R. E. Specificity of the ubiquitin isopeptidase in the PA700 regulatory complex of 26S proteasomes. J. Biol. Chem. 1997a;272:28438–28446. doi: 10.1074/jbc.272.45.28438. [DOI] [PubMed] [Google Scholar]
- Lam Y. A., Lawson T. G., Velayultham M., Zweierm J. L., Pickart C. M. A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature. 2002;416:763–767. doi: 10.1038/416763a. [DOI] [PubMed] [Google Scholar]
- Lam Y. A., Xu W., DeMartino G. N., Cohen R. E. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature. 1997b;385:737–740. doi: 10.1038/385737a0. [DOI] [PubMed] [Google Scholar]
- Laney J. D., Hochstrasser M. Substrate targeting in the ubiquitin system. Cell. 1999;97:427–430. doi: 10.1016/s0092-8674(00)80752-7. [DOI] [PubMed] [Google Scholar]
- Leggett D. S., Hanna J., Borodovsky A., Crosas B., Schmidt M., Baker R. T., Waltz T., Ploeugh H., Finley D. Multiple associated proteins regulate proteasome structure and function. Mol. Cell. 2002;10:498–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- Li T., Duan W., Yang H., Lee M. K., Bte M. F., Lee B. H., Teo T. S. Identification of two proteins, S14 and UIP1, that interact with UCH37. FEBS Lett. 2001;488:201–205. doi: 10.1016/s0014-5793(00)02436-4. [DOI] [PubMed] [Google Scholar]
- Li T., Naqvi N. I., Yanf H., Teo T. S. Identification of a 26S proteasome-associated UCH in fission yeast. Biochem. Biophys. Res. Commun. 2000;272:270–275. doi: 10.1006/bbrc.2000.2767. [DOI] [PubMed] [Google Scholar]
- Liu C. W., Li X., Thompson D., Wooding K., Chang T., Tang Z., Yu H., Thomas P. J., DeMartino G. N. ATP binding and ATP hydrolysis play distinct roles in the function of 26S proteasome. Mol. Cell. 2006;24:39–50. doi: 10.1016/j.molcel.2006.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. W., Li X., Thompson D., Wooding K., Chang T., Tang Z., Yu H., Thomas P. J., DeMartino G. N. ATP binding and ATP hydrolysis play distinct roles in the function of 26S proteasome. Mol. Cell. 2007;24:39–50. doi: 10.1016/j.molcel.2006.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundgren J., Masson P., Realini C., Young P. Use of RNA interference and complementation to study the function of the Drosophila and human 26S proteasome subunits. Mol. Cell. Biol. 2004;23:5320–5330. doi: 10.1128/MCB.23.15.5320-5330.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijman S.M.B., Luna-Vargas M.P.A., Velds A., Brummelkamp T. R., Dirac A.M.G., Sixma T. K., Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Papa F. R., Amerik A., Hochstrasser M. Interaction of the Doa4 deubiquitinating enzyme with the yeast 26S proteasome. Mol. Biol. Cell. 1999;10:741–756. doi: 10.1091/mbc.10.3.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart C. M. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001;70:530–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Pickart C. M., Cohen R. E. Proteasomes and their kin: proteases in the machine age. Nat. Rev. Mol. Cell Biol. 2004;5:177–187. doi: 10.1038/nrm1336. [DOI] [PubMed] [Google Scholar]
- Qiu X.-B., Ouyang S.-Y., Li C.-J., Miao S., Wang L., Goldberg A. L. hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. EMBO J. 2006;25:5742–5753. doi: 10.1038/sj.emboj.7601450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. M., Goldberg A. L. Proteasomes and their associated ATPases: a destructive combination. J. Struct. Biol. 2006;156:72–83. doi: 10.1016/j.jsb.2006.04.012. [DOI] [PubMed] [Google Scholar]
- Smith D. M., Kafri G., Cheng Y., Ng D., Wala T., Goldberg A. L. ATP binding to PAN or the 26S ATPases causes association with the 20S proteasome, gate opening, and translocation of unfolded proteins. Mol. Cell. 2005;20:687–698. doi: 10.1016/j.molcel.2005.10.019. [DOI] [PubMed] [Google Scholar]
- Stone M., Hartmann-Petersen R., Seeger M., Bach-Otschir D., Wallace M., Gordon C. Uch2/Uch37 is the major deubiquitinating enzyme associated with the 26S proteasome in fission yeast. J. Mol. Biol. 2004;344:697–706. doi: 10.1016/j.jmb.2004.09.057. [DOI] [PubMed] [Google Scholar]
- Strickland E., Hakala K., Thomas P. J., DeMartino G. N. Recognition of misfolding proteins by PA700, the regulatory subcomplex of the 26S proteasome. J. Biol. Chem. 2000;275:5565–5572. doi: 10.1074/jbc.275.8.5565. [DOI] [PubMed] [Google Scholar]
- Thrower J. S., Hoffman L., Rechsteiner M., Pickart C. M. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2001;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R., Aravind L., Oania R., McDonald W. H., Yates J. R., Koonin E. V., Deshaies R. J. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- Verma R., Ogura N., Graumann J., Deshaies R. J. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell. 2004;118:99–110. doi: 10.1016/j.cell.2004.06.014. [DOI] [PubMed] [Google Scholar]
- Voges D., Zwickl P., Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- Wicks S. J., Grocott T., Haros K., Maillard M., ten Dijke P., Chantry A. Reversible ubiquitination regulates the Smad/TGF-beta signalling pathway. Biochem. Soc. Trans. 2006;34:761–763. doi: 10.1042/BST0340761. [DOI] [PubMed] [Google Scholar]
- Wicks S. J., Haros K., Maillard M., Song L., Cohen R. E., Dijke P. T., Chantry A. The deubiquitinating enzyme UCH37 interacts with Smads and regulates TGF-beta signalling. Oncogene. 2005;24:8080–8084. doi: 10.1038/sj.onc.1208944. [DOI] [PubMed] [Google Scholar]
- Wojcik C., DeMartino G. N. Analysis of Drosophila 26S proteasome using RNA interference. J. Biol. Chem. 2002;277:6188–6197. doi: 10.1074/jbc.M109996200. [DOI] [PubMed] [Google Scholar]
- Wojcik C., Rowicka M., Kudlicki A., Nowis D.M.E., Kujawa M., DeMartino G. N. Valosin-containing protein (p97) is a regulator of endoplasmic reticulum stress and of the degradation of N-end rule and ubiquitin-fusion degradation pathway substrates in mammalian cells. Mol. Biol. Cell. 2006;14:4606–4618. doi: 10.1091/mbc.E06-05-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T., Cohen R. E. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- Yao T., Song L., DeMartino G. N., Florens L., Swanson S. K., Washburn M. P., Conaway R. C., Conaway J. W., Cohen R. E. Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat. Cell Biol. 2006;8:994–1002. doi: 10.1038/ncb1460. [DOI] [PubMed] [Google Scholar]