Abstract
Centrosomes are dynamic organelles that consist of a pair of cylindrical centrioles, surrounded by pericentriolar material. The pericentriolar material contains factors that are involved in microtubule nucleation and organization, and its recruitment varies during the cell cycle. We report here that proteasome inhibition in HeLa cells induces the accumulation of several proteins at the pericentriolar material, including gamma-tubulin, GCP4, NEDD1, ninein, pericentrin, dynactin, and PCM-1. The effect of proteasome inhibition on centrosome proteins does not require intact microtubules and is reversed after removal of proteasome inhibitors. This accrual of centrosome proteins is paralleled by accumulation of ubiquitin in the same area and increased polyubiquitylation of nonsoluble gamma-tubulin. Cells that have accumulated centrosome proteins in response to proteasome inhibition are impaired in microtubule aster formation. Our data point toward a role of the proteasome in the turnover of centrosome proteins, to maintain proper centrosome function.
INTRODUCTION
The centrosome is a small organelle composed of a pair of centrioles surrounded by pericentriolar material (Nigg, 2002). The pericentriolar material contains a variety of proteins, such as proteins responsible for the nucleation of microtubules and for cell cycle regulation (for reviews, see Bornens, 2002; Doxsey et al., 2005). The protein gamma-tubulin, a member of the tubulin superfamily that is present in all eukaryotes, is one of the best characterized components of the pericentriolar material. Gamma-tubulin is required for the formation and function of the centrosome (Stearns et al., 1991; Zheng et al., 1991; for review, see Raynaud-Messina and Merdes, 2007) and is responsible for microtubule nucleation, bipolar spindle formation, and centriole duplication during cell cycle progression. In the cytosol, gamma-tubulin is assembled in complexes termed the gamma-tubulin ring complexes (γ-TuRCs). These consist of gamma-tubulin and additional subunits, named gamma complex proteins (GCPs; Zheng et al., 1995; for reviews, see Schiebel, 2000; Raynaud-Messina and Merdes, 2007). Recent studies have demonstrated that gamma-tubulin accumulates at the centrosome after proteasome inhibition (Zhao et al., 2003). However, the functional link between proteasome activity and the centrosome is not understood. The ubiquitin-proteasome pathway is responsible for the constitutive degradation of the majority of cellular proteins. It plays an important role in maintaining a constant balance between de novo protein synthesis and proteolysis. The 26S proteasome assembles from ring-shaped regulatory 19S and 20S core particles, which are composed of multiple polypeptide subunits. The composition of the proteasome varies depending on the cellular compartment. Proteasomes are present in the cytoplasm and in the nucleus of all eukaryotic cells, but excluded from the nucleoli (Wojcik and DeMartino, 2003). In the cytoplasm, proteasomes associate with the cytoskeletal network and with the outer surface of the endoplasmic reticulum (Kostova and Wolf, 2003). More recently, elements of the ubiquitin-proteasome pathway have also been localized to the centrosome in interphase cells (Freed et al., 1999; Wigley et al., 1999; Fabunmi et al., 2000; Zhao et al., 2003; Parvin, 2004), and inhibition of proteasome function led to spindle pole fragmentation during mitosis (Ehrhardt and Sluder, 2005).
To understand the role of the proteasome at the centrosome, we examined changes in the pericentriolar material after proteasome inhibition in interphase cells. Here, we report that specific proteasome inhibitors induce accumulation of several centrosome proteins at the pericentriolar material. Blocking proteasome function impaired the ability of the centrosome to form regular microtubule asters.
MATERIALS AND METHODS
Cell Culture
HeLa, U2OS, Cos-7, and DLD-1 cells were cultured in DMEM. Raji cells were cultured in RPMI-1640 medium. Tissue culture medium was supplemented with 100 mM l-glutamine and 10% fetal bovine serum (FBS). Cells were grown in a humidified atmosphere with 5% CO2 at 37°C. HeLa cells stably expressing green fluorescent protein (GFP)-centrin (Piel et al., 2000) were obtained from Dr. Michel Bornens (UMR144, Curie Institute, France) and grown in the same medium supplemented with G-418. Cells were seeded onto 24-well plates containing glass coverslips and treated with proteasome inhibitor or control medium containing DMSO, for 18 h. For release experiments, the drug was washed out, and the cells were allowed to grow in medium supplemented with FBS for the indicated time. For nocodazole and taxol experiments, cells were incubated with 1 μM of the drug for 6–8 h, before proteasome inhibition for an additional 16 h.
Antibodies and Reagents
Primary antibodies used in this study were: anti-gamma-tubulin (mouse GTU-88, Sigma Aldrich, St. Louis, MO; or rabbit serum R75; Julian et al., 1993), anti-α-tubulin (mouse T-5168, Sigma Aldrich), anti-PCM1 rabbit serum (Dammermann and Merdes, 2002), a rabbit polyclonal antibody directed against NEDD1 peptide (279–660; Haren et al., 2006), mouse anti-pericentrin (Dammermann and Merdes, 2002), and rabbit anti-ninein. Mouse antibodies directed to the Rpn7 subunit of the 19S proteasome and the beta2 (MCP168) and the beta7 (MCP205) subunits of the 20S proteasome were purchased from Affiniti Research/BIOMOL International (Exeter, UK). Rabbit polyclonal antibody P1663 against clathrin heavy chain was purchased from Cell Signaling (Beverly, MA). MG-115, lactacystin (Craiu et al., 1997), and epoxomicin were purchased from Calbiochem (La Jolla, CA). Bortezomib a selective dipeptidyl boronic acid proteasome inhibitor (PS-341) was prepared according to previously reported methods (Adams et al., 1998). Nocodazole and taxol were purchased from Sigma Aldrich. Stock solutions of all drugs were prepared at a concentration of 10 mM in DMSO.
Protein Extracts and Immunoblotting
HeLa cells grown on 100-mm dishes were scraped and washed twice in PBS. Total cell lysates were prepared from control and proteasome inhibitor-treated cells as described (Nekrep et al., 2000). To isolate soluble and insoluble fractions, the cell pellet was resuspended in lysis buffer (50 mM HEPES, pH 7.4, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) containing 0.1 mg/ml phosphatase and protease inhibitors (Sigma). Cells were lysed by incubation on ice for 30 min with repeated mixing and cleared by centrifugation, and detergent-soluble fractions were collected. The pellet (detergent-insoluble fraction) was resuspended in 60 mM Tris-HCl, 2 μM SDS, 2.5% 2-mercapthoethanol, and protease inhibitors and sonicated for 20 min. The amount of proteins was quantified with BCA (bicinchoninic acid) assay kit (Bio-Rad Laboratories, Hercules, CA). Equivalent amounts of proteins from both fractions were then resolved by 10% SDS-PAGE and analyzed by Western blotting with specific antibodies, using Odyssey buffers (Odyssey, ScienceTec, Les Ulis, France). After washing under stringent conditions, immune complexes were revealed with Alexa Fluor 680–labeled secondary anti-mouse antibodies (Odyssey, ScienceTec) and visualized by a Li-Cor Odyssey infrared imaging system (700-nm channel; Lincoln, NE).
Two-Dimensional Gel Electrophoresis
Two-dimensional gel electrophoresis (2D gel) was performed according to the protocol provided by the manufacturer of the apparatus (Amersham Pharmacia Biosciences, Piscataway, NJ). Cell extracts were trichloroacetic acid (TCA) precipitated and rehydrated in 200 μl of rehydration buffer for 2 h. Samples were loaded on an 18-cm Immobiline DryStrip, pH 3–10 NL (nonlinear immobilized pH gradient, Amersham Biosciences). Strips were then subjected to passive rehydration for 8 h at 50 μV for each strip on a Multiphor II electrophoresis unit (Amersham Biosciences). After a rehydration step, samples were subjected to separation at 50 V for 6 h, 50–8000 V for 2 h, and 8000 V for an additional 12 h. After equilibration, a total of 70,000 Vh, immobilized pH gradient strips were further processed for second-dimension SDS-PAGE electrophoresis on ExcelGel 2D Homogeneous 12.5%. The resulting gels were immunoblotted to nitrocellulose and processed for Western blot with specific antibody as above.
Immunostaining and Fluorescence Microscopy
Cells grown on coverslips were fixed in methanol at −20°C for 10 min. The cells were then washed three times (5 min each) in PBS and processed for immunostaining using conventional protocols. Alexa-488– or Alexa-568–conjugated secondary antibodies (Molecular Probes, Eugene, OR) were used for detection. Immunofluorescence was analyzed using a Zeiss Axiovert microscope (Thornwood, NY), with 63×/1.4 NA or 100×/1.4 NA objectives, or a Zeiss AxioImager Z1. Z-series were acquired with an Axiocam MRm camera and Axiovision software (Zeiss) for the Axiovert microscope or with a Hamamatsu digital CCD camera (Bridgewater, NJ) and Visilog software (Noesis, Aptos, CA) for the AxioImager microscope. Quantification of fluorescence intensity of centrosomal gamma-tubulin staining was performed using Visilog software. Products of pixel area and pixel intensity of gamma-tubulin at the centrosome are given as relative units after cytoplasmic background substration. Deconvolved immunofluorescence images were obtained with a Deltavision RT workstation (Applied Precision, Issaquah, WA), using the manufacturer's software.
Electron Microscopy
HeLa cells were fixed with 2.5% glutaraldehyde in PHEM buffer (60 mM PIPES, pH 6.9, 25 mM HEPES, 1 mM EGTA, 2 mM MgCl2, pH 7.0). Fixed cells were scraped and pelleted and subsequently were postfixed in 2% osmium tetroxide. After dehydration, cells were embedded in araldite, serially sectioned, and contrasted with uranyl acetate and lead citrate. For immunoelectron microscopy of gamma-tubulin, cells were embedded in LR White, and sections were incubated with monoclonal anti-gamma-tubulin (GTU-88, Sigma-Aldrich) for 3 h at 37°C, followed by gold-conjugated secondary antibody (Molecular Probes) for 90 min. Pictures were taken with a Philips 300 electron microscope (Mahwah, NJ). The density of gold particles was quantified on the sections. Immunoelectron microscopy of microtubules was performed on human osteosarcoma cells (U2OS) after 16-h treatment with 1 μM PS-341 or epoxomicin, followed by microtubule staining with monoclonal anti-α-tubulin, secondary antibody labeled with ultrasmall gold, and silver enhancement, exactly as described in Merdes and De Mey (1990).
Centrosome Isolation
Centrosome enriched fractions were isolated using adaptations of previously described protocols (Mitchison and Kirschner, 1984; Bornens et al., 1987). Briefly, 109 cells were treated with 33 μM nocodazole and 1 μg/ml cytochalasin D for 1 h at 37°C. Cells were lysed in 1 mM Tris-HCl, pH 8.0, 0.1% β-mercaptoethanol, 0.5% NP-40, and 0.5 mM MgCl2 in the presence of protease inhibitor mix. The centrosomes were purified by sucrose gradient. Fractions (50 μl) were collected and were analyzed by Western blotting using antibodies against gamma-tubulin, centrin, and the proteasome.
Proteasome Activity Assay
To monitor the chymotrypsin-like (CTL) and caspase-like enzymatic activities of the proteasome, centrosomes were purified by sucrose gradient as above. Twenty microliters of collected fractions (250 μl) were assayed according to standard protocols (Brown and Monaco, 1993). Briefly, proteins were diluted in a reaction buffer (5 mM HEPES, pH 8, 0.05 mM DTT) and preincubated in 384 well microtiter black plate (Greiner, Germany) with or without the proteasome inhibitor lactacystin (50 μM) for 15 min at 37°C. Reactions were then incubated for an additional 60 min at 37°C with 100 μM of the 7-amido-4-methylcoumarin (AMC)-coupled fluorogenic substrate in 100 mM HEPES, pH 7.6, in the dark. The fluorogenic peptides N-succinyl-Leu-Leu-Val-Tyr-AMC (Suc-LLVY-AMC) and Z-Leu-Leu-Glu-AMC (Z-LLE-AMC) were used to quantify CTL and caspase-like activity, respectively. Hydrolysis of substrates was monitored by fluorescence measurement of the liberated AMC at 380/440-nm excitation/emission (Hoffman et al., 1992; Oberdorf et al., 2001) on a microtiter plate reader (Synergy HT, Biotek Instruments, Burlington, VT). All peptides were purchased from Calbiochem (Nottingham, United Kingdom). The emitted fluorescence plotted against the fraction number shows the proteasome activity in the different fractions.
Nondenaturing PAGE
For in-gel assay, fractions enriched in centrosomes were resolved on a 4% nondenaturing polyacrylamide gel in 1× Tris-borate-EDTA buffer and overlaid by 2% stacking gel in the same buffer. Gels were run at 20 mA for 4 h at 4°C. The running buffer was the same as the gel buffer but without acrylamide. Stacking was removed, and gels were then incubated in 10 ml of reaction buffer (30 mM Tris-HCl, pH 7.8, 5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, and 2 mM ATP) containing 0.2 mM of Suc-LLVY-AMC fluorogenic peptide for 1 h 30 min at 37°C (Hoffman et al., 1992). Purified 20S and 26S proteasomes were included as positive controls. The gels were exposed and photographed using a UV transilluminator (Gel Doc 1000, Bio-Rad, Hercules, CA). The 26S proteasome was purchased from Calbiochem. The 20S proteasome was purified by affinity chromatography from human erythrocytes (Claverol et al., 2002).
Microtubule Nucleation and Depolymerization Assays
Cells grown on glass coverslips were treated with PS-341 (1 μM) 16 h before performance of microtubule nucleation assays. Cell culture plates were incubated on ice for at least 1 h to depolymerize microtubules. To allow microtubule regrowth, coverslips were dipped in warm (37°C) tissue culture medium containing 10% FBS. Regrowth was stopped at indicated time points. Cells were extracted with 0.5% Triton X-100, fixed in 4% paraformaldehyde in PHEM buffer (60 mM PIPES, pH 6.9, 25 mM HEPES, 1 mM EGTA, 2 mM MgCl2, pH 7.0). Cells and immunofluorescence analysis was performed using standard protocols.
RESULTS
Inhibition of the Proteasome Induces Accumulation of Pericentriolar Material
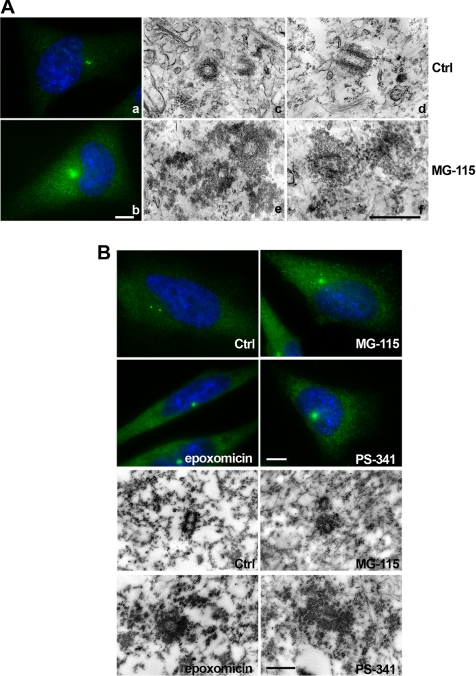
To test the potential role of the proteasome in centrosome assembly, we inhibited the catalytic activity of the proteasome using various proteasome inhibitors. Proteasome inhibition in HeLa cells with MG-115, a reversible peptide aldehyde proteasome inhibitor, results in a significant accumulation of gamma-tubulin staining at the center of the cell after 16 h (Figure 1A, a and b). In addition, a slight increase of the cytoplasmic levels of gamma-tubulin are seen. Shorter treatments of 4 to 8 h were performed with the same inhibitor, but failed to induce any significant accumulation of gamma-tubulin (data not shown). To further characterize the observed effect on gamma-tubulin accumulation, electron microscopy (EM) was carried out. HeLa cells were treated with control medium or with 3 μM MG-115 for 16 h and prepared as indicated in Materials and Methods. Thin sections of control cells showed centriolar cylinders surrounded by a thin layer of pericentriolar material (Figure 1A, c and d, Ctrl). Consistent with the immunofluorescence data, EM of cells treated with proteasome inhibitor (Figure 1A, e and f, MG-115) showed the presence of significantly increased amounts of electron-dense material surrounding the two centrioles, within a radius of ∼300 nm. Because the peptide aldehyde inhibitor MG-115 can also block proteases such as calpain that are not associated with the proteasome (Kim et al., 2004), we compared the effects of MG-115 to other synthetic proteasome inhibitors. Epoxomicin, which specifically inhibits the chymotrypsin-like (96%) and caspase-like (83%) activities, and PS-341 (Bortezomib), a peptide-based inhibitor with a boronic acid pharmacophore (Adams, 2002, 2004) that inhibits mostly chymotrypsin- and trypsin-like activities (Kisselev and Goldberg, 2005) were used. Hela cells were treated with either MG-115 (3 μM), epoxomicin (1 μM), or PS-341 (1 μM) and processed for immunofluorescence staining. Although 16 h treatment with MG-115 resulted in centrosome enlargement in 70% of the cells, epoxomicin or PS-341 induced enlargement in more than 95% of treated HeLa cells (350–400 total cells scored per condition, in at least three independent experiments). Apparently, the cellular response to PS-341 was faster than to MG-115, showing gamma-tubulin accumulation around the centrosome already at earlier time points in 18 or 30% of the treated cells, after 6 or 8 h, respectively. Six to 8 h appears to be the minimum time required for proteasome inhibitors to produce cellular effects; cell lines that stably express the well-characterized proteasome substrate ornithine decarboxylase fused to GFP only show GFP accumulation after several hours of proteasome inhibitor treatment (Ghoda et al., 1989; and unpublished data).
Figure 1.
Proteasome inhibition triggers enlargement and structural modification of the centrosome. HeLa cells were treated with proteasome inhibitors and processed either for immunostaining or EM. (A) Control cells (a) or cells treated with MG-115 (b) were fixed and stained for gamma-tubulin (green) and DNA (blue). Pictures were taken at identical exposures and microscope settings. HeLa cells were processed for EM (c–f). Orthogonal sections (middle) and longitudinal sections (right) of the centrioles and the pericentriolar material are shown for control and MG-115–treated cells. (B) Control cells (Ctrl) and cells treated with proteasome inhibitors MG-115, epoxomicin, or PS-341 were processed and stained with antibody against gamma-tubulin for immunofluorescence (top four panels) or for immunoelectron microscopy of thin sections, using anti gamma-tubulin followed by 40-nm immunogold (bottom four panels). Scale bars in immunofluorescence pictures, 10 μm; scale bars in electron micrographs, 0.5 μm.
Cells treated with proteasome inhibitor exhibited a single large focus of gamma-tubulin protein, which we suspected to be enlarged pericentriolar material. To verify this, immunoelectron microscopy analysis was performed using anti gamma-tubulin, detected by secondary antibody conjugated with colloidal gold. Figure 1B shows an increase of the pericentriolar material in cells that were treated with proteasome inhibitors, and this material stained positively for gamma-tubulin. We quantified gold particles associated with the pericentriolar material and subtracted values for background staining of identical surface areas in the cytoplasm. Our quantifications indicate a threefold increase of gamma-tubulin staining within a radius of 300 nm of the centrioles after MG-115 treatment, a 17-fold increase after epoxomicin, and a 9-fold increase after PS-341. Accumulation of gamma-tubulin upon proteasome inhibition was found in multiple different cell types besides HeLa, as shown in Cos-7 and DLD-1 cells (Supplementary Figure S1A). We further confirmed that the centrosome size increases after treatment with proteasome inhibitors, by sucrose gradient analysis of purified centrosomes from Raji B-cells. In Supplementary Figure S1B, we show that the peak of gamma-tubulin–containing fractions is found at higher sucrose density than in control cells. Because proteasome inhibitors induce a cell cycle arrest at G2/M phase (Wagenknecht et al., 1999; Ling et al., 2002; Supplementary Figure S2A), we wanted to exclude that the increase in pericentriolar material is an indirect effect of proteasome inhibition. We used a variety of therapeutic agents that interfere with the cell cycle. Neither DNA-binding/cleaving agents (adriamycin, bleomycin), inhibitors of topoisomerases I and II (etoposide, camptothecin), antimetabolite (5-fluorouracil), alkylating agent (cisplatin), nor the antimitotic drug (vinorelbine) were able to produce effects similar to those observed with proteasome inhibitors (Supplementary Figure S2B).
Proteasome Inhibition Leads to the Accrual of Many Different Centrosomal Proteins
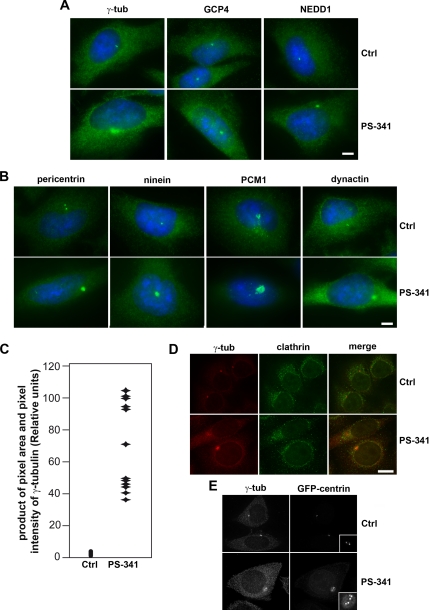
Proteins at the centrosome can be divided into different functional groups. One group of proteins forms complexes with gamma-tubulin and is involved in microtubule nucleation, such as gamma-tubulin itself, proteins of the GCP superfamily, and NEDD1. To test their behavior after proteasome inhibition with the inhibitor PS-341, we followed the localization of GCP4 and NEDD1 in addition to gamma-tubulin. All these proteins show increased accumulation at the pericentriolar material in response to proteasome inhibition (Figure 2A). A different group of proteins has been suggested to form filamentous networks, providing a scaffold of the pericentriolar material, such as pericentrin and ninein. These proteins are equally found enriched at the pericentriolar material after proteasome inhibition (Figure 2B). Other proteins, such as PCM-1 and dynactin have been suggested to play roles in centrosome assembly and protein transport to the pericentriolar material (Dammermann and Merdes, 2002; Kubo and Tsukita, 2003).
Figure 2.
Proteasome inhibition triggers the accumulation of different centrosome proteins. HeLa cells were treated with control medium (Ctrl) or proteasome inhibitor PS-341 and stained for immunofluorescence of various centrosome proteins (green). (A) gamma-Tubulin (γ-tub), GCP4 and NEDD1 (B) pericentrin and ninein, PCM-1 and dynactin, DNA is stained in blue. Bar, 10 μm. Immunofluorescence was analyzed using the Zeiss Axiovert microscope and Axiovision software. (C) The accumulation of gamma-tubulin was quantified in control and PS-341–treated cells using Visilog software. The product of pixel area and pixel intensity are shown for 22 control and 12 PS-341–treated cells. (D) HeLa cells (Ctrl and P-341–treated), stained for clathrin heavy chain and gamma-tubulin. (E) HeLa control (Ctrl) and P-341–treated cells, stably expressing GFP-centrin, were stained for gamma-tubulin (γ-tub). Insets show enlarged views of the centrioles. Scale bars, 10 μm.
Figure 2B shows that these are also found enriched at the pericentriolar material after the inhibition of the proteasome (n > 100). Furthermore, costaining experiments of gamma-tubulin and either GCP4, NEDD1, or PCM-1 shows that the degree of accumulation of these proteins varies (Supplementary Figure S4). To quantify the enrichment of centrosome protein, the product of the pixel area within a 10.57 μm2 circumference surrounding the centrosome and the pixel intensity of gamma-tubulin immunofluorescence in the same area was determined after background subtraction using cell imaging software Visilog (Noesis, Gif sur Yvette, France) and Adobe Photoshop (San Jose, CA). As shown in Figure 2C, an 8- to 10-fold increase was detected in PS-341–treated HeLa cells, consistent with our data obtained by immunoelectron microscopy of gamma-tubulin (Figure 1B).
To test whether proteasome inhibition affects primarily proteins of the centrosome or whether it leads to an unspecific accumulation of all types of proteins, we followed the localization of the membrane skeleton protein clathrin in treated cells. In our experiments, we did not observe any significant accumulation of clathrin to the centrosome in cells after proteasome inhibition (Figure 2D). This indicates that protein accumulation at the centrosome as induced by proteasome inhibitors is selective and does not affect all classes of proteins in the cell. Because proteasome inhibition leads to cell cycle arrest in G2 (Supplementary Figure S2A; Wagenknecht et al., 1999; Ling et al., 2002), we tested whether this arrest correlated with centriole duplication, by examining HeLa cells that stably expressed centrin-GFP as a marker of the centrioles (Paoletti et al., 1996; Piel et al., 2000). In untreated HeLa cells expressing GFP-centrin, two spots that colocalize with gamma-tubulin were detected in most cells (82%, n = 815), indicating the presence of two centrioles in G1 phase. In contrast, after proteasome inhibition four or more centrin spots were detected in 99% of the cells (n = 521), and these spots were often found surrounded by a halo of accumulated centrin (Figure 2E). This indicates that proteasome inhibition leading to G2 arrest does not prevent centriole duplication in the preceding S-phase.
Accumulation of Centrosome Proteins upon Proteasome Inhibition Does Not Require Intact Microtubules
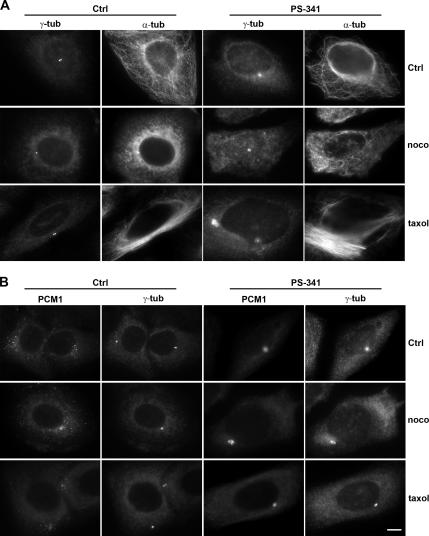
Because several proteins such as pericentrin or ninein assemble at the pericentriolar material after microtubule-dependent transport toward the minus end (Purohit et al., 1999; Dammermann and Merdes, 2002), we wanted to test whether the protein accumulation seen after proteasome inhibition required microtubule-dependent transport and an intact microtubule network. We therefore depolymerized microtubules by treatment with 1 μM nocodazole for 6–8 h, before adding the proteasome inhibitor PS-341 at 1 μM and continuing the incubation in the presence of nocodazole for an additional 16 h. Microtubule depolymerization was confirmed by immunostaining with an anti-α-tubulin antibody (Figure 3A, nocodazole). Despite the absence of microtubules, proteasome inhibition still induced accumulation of gamma-tubulin to the same degree as in cells with an intact microtubule network, as determined by measuring pixel intensity in the pericentriolar area (Figure 3A). We observed a similar accumulation for the protein PCM-1 (Figure 3B). Because we obtained similar data when HeLa cells were treated with the microtubule-stabilizing drug taxol before inhibition of proteasomes, we concluded that the accumulation of gamma-tubulin and PCM1 at the pericentriolar material is independent of microtubules, microtubule dynamics, or microtubule-dependent transport.
Figure 3.
Accumulation of centrosome proteins in response to proteasome inhibition does not require microtubules. (A) Control (Ctrl) and PS-341–treated HeLa cells were immunostained for gamma-tubulin (γ-tub) or α-tubulin (α-tub). The first row shows cells with intact microtubules (α-tub), whereas the two bottom rows show cells treated for 6 h with the microtubule inhibitor nocodazole (noco) or the microtubule-stabilizing agent taxol, before proteasome inhibition with PS-341 for an additional 12 h. (B) HeLa cells treated as in A, immunostained for PCM1 and gamma-tubulin (γ-tub). Scale bar, 10 μm.
Inhibition of Proteasome Activity Leads to Posttranslational Modification of Insoluble Gamma-Tubulin
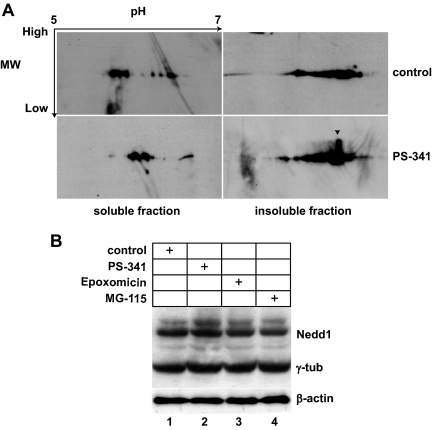
As immunostaining analysis of different centrosome proteins shows an accumulation at the centrosome after proteasome inhibition, we wanted to know if the total amount of centrosome protein was increased. Immunoblot analysis of total HeLa cell lysates with antibodies against gamma-tubulin or NEDD1 revealed equal intensities in controls and treated cells (Figure 4), suggesting that proteasome inhibition does not significantly affect the total amount of gamma-tubulin or NEDD1. However, we detected a slight increase of the isoelectric point of soluble gamma-tubulin, as well as an extra spot of increased molecular weight in the insoluble fraction of gamma-tubulin, suggesting that posttranslational modifications upon proteasome inhibition might occur.
Figure 4.
Inhibition of proteasome activity leads to modification of gamma-tubulin, but does not affect gamma-tubulin or NEDD1 protein levels. (A) Control HeLa cells or HeLa cells treated with proteasome inhibitor PS-341 were fractionated by treatment with a cocktail of detergents followed by centrifugation. Equal amounts of proteins from detergent-soluble (100 μg) and detergent-insoluble material (200 μg) were subjected to 2D gel electrophoresis, followed by immunoblotting for gamma-tubulin. Arrows indicate acidic to basic pH ranges for isoelectric focusing and high-to-low molecular weight separation for SDS-PAGE. The arrowhead in the bottom right image indicates a spot of gamma-tubulin with a shifted molecular weight. (B) Total cell lysates of control HeLa cells and HeLa cells treated with the proteasome inhibitors PS-341, epoxomicin, or MG-115 were analyzed on immunoblots probed with anti-NEDD1 and anti-gamma-tubulin. Immunoblotting for β-actin was performed to test for equal loading (bottom gel).
Active Proteasomes Are Required for Controlling the Size of the Pericentriolar Material
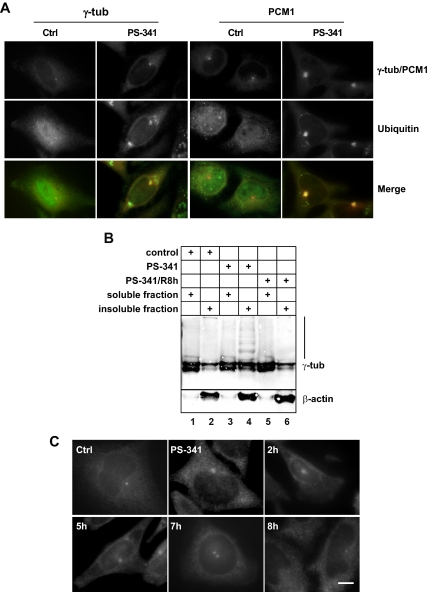
Because centrosome proteins accumulate in the presence of proteasome inhibitors, we wanted to test whether these proteins would be degraded by the proteasome under normal circumstances. Consistent with previously published literature (Wigley et al., 1999; Fabunmi et al., 2000), we found that a fraction of the 26S proteasome cosediments with centrosome proteins and that this fraction has proteolytic activity using fluorogenic peptides as substrates (Supplementary Figure S3). Degradation by the proteasome requires posttranslational modification of the protein substrates by addition of polyubiquitin chains. In control HeLa cells, ubiquitin is localized throughout the cytoplasm and the nucleus with a higher level within the nucleus (Figure 5A). In contrast, after proteasome inhibition with PS-341, ubiquitin is almost excluded from the nucleus and forms aggregates within the cytoplasm. Furthermore, a very distinct colocalization with accumulated centrosome proteins such as gamma-tubulin and PCM1 (Figure 5A) was observed. Because part of gamma-tubulin on 2D Western blots showed a molecular weight shift upon proteasome inhibition (Figure 4A), we tested whether this alteration is due to polyubiquitylation. Figure 5B reveals a ladder of higher molecular weight bands in addition to the 52-kDa band of gamma-tubulin in the insoluble fraction of treated cells, consistent with a polyubiquitylation signature. We also noticed that the relative amount of insoluble gamma-tubulin was increased after proteasome inhibition (Figure 5B, compare lanes 1 and 2 with lanes 3 and 4). Because of the insoluble nature of this modified gamma-tubulin fraction, we were unable to perform immunoprecipitation and verify ubiquitylation by immunoblotting. The ladder of higher molecular weight bands of gamma-tubulin disappeared, and the relative amount of insoluble protein was back to normal once the reversible proteasome inhibitor PS-341 was removed and the cells were allowed to recover for 8 h (Figure 5B, lanes 5 and 6). Consistently, immunofluorescence showed that gamma-tubulin accumulation at the centrosome was reversed in more than 90% of the cells after removal of the proteasome inhibitor PS-341, followed by 7–8 h of recovery (>100 scored cells in three independent experiments; Figure 5C). In contrast, when the nonreversible proteasome inhibitor epoxomicin was used, cells failed to recover and massive cell death was observed (data not shown).
Figure 5.
Ubiquitin colocalizes with accumulated centrosome proteins upon proteasome inhibition. (A) Control HeLa cells (Ctrl) and PS-341 treated cells (PS-341) were stained for gamma-tubulin (γ-tub), PCM1, and ubiquitin (Ub). (B) Immunoblot of 10 μg of detergent-soluble (lanes 1, 3, and 5) and detergent-insoluble fractions (lanes 2, 4, and 6) of HeLa cells were separated on 10% SDS-PAGE and immunoblotted for gamma-tubulin or β-actin. Lanes 1 and 2, control cells; lanes 3 and 4, PS-341–treated cells; lanes 5 and 6, PS-341–treated cells 8 h after washout of the drug. Lane 4 (nonsoluble fraction of PS-341–treated cells) shows a ladder of high-molecular-weight products, staining positively for gamma-tubulin. (C) HeLa cells were treated with control medium (Ctrl), or with PS-341 (PS-341), followed by washout of the drug and recovery for 2, 5, 7, or 8 h. Immunofluorescence of gamma-tubulin is shown. Bar, 10 μm.
Proteasome Activity Is Necessary for Radial Microtubule Organization
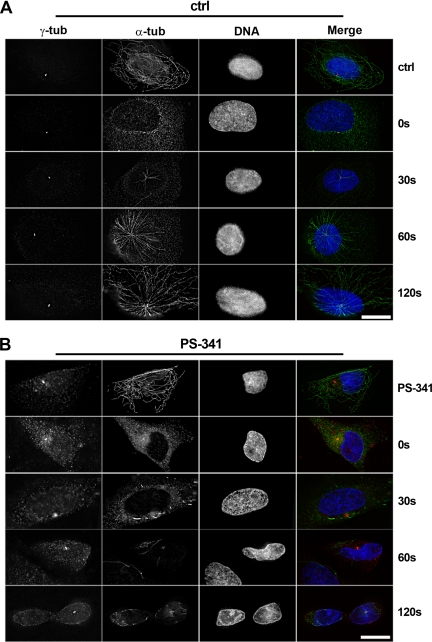
Because treatment of cells with proteasome inhibitors leads to accumulation of large amounts of centrosome proteins at the pericentriolar material, we wanted to test whether the capacity of the centrosome to nucleate microtubules was altered. For this purpose, we performed assays of microtubule regrowth after cold-induced depolymerization in human osteosarcoma cells (U2OS), because U2OS cells possess a radial microtubule network with microtubules growing from well-focused asters. We found that in control cells (n = 3 experiments, >200 scored cells), microtubule asters started to form within 30 s of reheating in 25% of control cells and that a microtubule network radiating from the centrosome was formed within 60 s in 95% of all cells (Figure 6A). In contrast, 92% of cells treated with the proteasome inhibitor PS-341 failed to form microtubule asters within that time frame (n = 3 experiments, >200 scored cells); instead, microtubule formation at several random points in the cytoplasm was seen (Figure 6B). After 120 s of reheating, 10% of PS-341-treated cells displayed small asters, but these contained only very short microtubules (Figure 6B). We conclude that microtubule nucleation from the centrosome is impaired after proteasome inhibition.
Figure 6.
Proteasome inhibition impairs microtubule nucleation in U2OS cells. U2OS cells were treated on ice to depolymerize microtubules, followed by recovery at 37°C to allow microtubule regrowth. Microtubule regrowth was analyzed in control cells (A; Ctrl) or in cells treated with proteasome inhibitor PS-341 (B; PS-341). Deconvolved immunofluorescence images of cells before cold treatment (top rows in A and B), and at 0, 30, 60, and 120 s after recovery from the cold. Cells were immunostained for gamma-tubulin (γ-tub, red) or α-tubulin (α-tub, green). DNA is stained in blue. At least 100 cells were analyzed for each condition, and the lack of centrosome focused aster formation in PS-341–treated cells was consistently observed. Bar, 15 μm.
Analysis of the microtubule network before depolymerization also revealed differences in microtubule organization. Cells treated with proteasome inhibitors epoxomicin (Figure 7) or PS-341 (not shown) lacked a clear microtubule organizing center. Electron microscopic analysis of cells in which the microtubule network was labeled by immunogold/silver staining revealed that these cells had no microtubules anchored to the pericentriolar material, whereas in control cells dense microtubule asters were radiating from the area around the centrosome.
Figure 7.
Proteasome inhibition impairs radial microtubule organization from the centrosome in U2OS cells. (A) Immunofluorescence of α-tubulin in control and epoxomicin-treated cells. (B) EM of control and epoxomicin-treated cells. To visualize microtubules over long distances, thick sections were cut of cells that were stained en bloc with anti-α-tubulin and with secondary antibody coupled to ultrasmall gold, followed by silver enhancement. Insets show enlarged views of the centrioles. Bars, 10 μm in A, 0.5 μm in B.
DISCUSSION
We show that inhibition of the proteasome induces the accumulation of a wide variety of centrosome proteins at the pericentriolar material. Similar accumulation of centrosome components in response to proteasome inhibition has been shown previously for gamma-tubulin and the Nek2 kinase (Zhao et al., 2003; Hames et al., 2005). The accumulated protein is visible as electron-dense material in treated cells. In addition, cells treated with proteasome inhibitors are impaired in centrosomal microtubule nucleation and show an aberrant microtubule network lacking a centrosomal focus. Currently, we cannot exclude that the accumulation of centrosome proteins after prolonged proteasome inhibition is due to indirect effects via other proteins that are regulated by proteasome-dependent degradation.
We considered several possibilities to explain the observed phenotypes: 1) Proteasome inhibition leads to an accumulation of cells in G2 phase, as shown previously (Wagenknecht et al., 1999; Ling et al., 2002). The observed concentration of centrosome proteins might therefore reflect cell cycle–dependent maturation of the centrosome. However, the finding that treated cells are defective in nucleating and anchoring microtubules at the centrosome cannot be explained by simple cell cycle arrest, and different drugs that lead to cell cycle arrest in G2 by inducing DNA damage fail to induce centrosome protein accumulation. 2) Proteasome inhibition interferes with proteins involved in microtubule organization or microtubule-dependent transport. This may cause unspecific aggregation of proteins at the center of the cell. However, although we cannot rule out indirect effects of proteasome inhibition, we think that microtubule-dependent mechanisms are an unlikely explanation for the observed phenotypes because our experiments with the microtubule inhibitor nocodazole indicated that centrosome protein accumulation occurred independently of microtubules. 3) Proteasome inhibition affects the turnover of centrosome proteins and might therefore increase the cytoplasmic levels of these proteins, leading to ectopic nucleation of microtubules in the cytoplasm and competition with centrosomal microtubule nucleation. This interpretation would be consistent with our immunofluorescence data showing that both cytoplasmic and centrosomal amounts of gamma-tubulin increase upon proteasome inhibition. Conflicting with this idea, however, we find that the overall levels of gamma-tubulin do not increase significantly after proteasome inhibition. We believe that the increased cytoplasmic signal of gamma-tubulin is due to a shift from soluble to detergent-resistant forms of gamma-tubulin, as supported by our immunoblot analysis of cell fractions (Figure 5B). This raises the question whether the insoluble gamma-tubulin is fully functional. 4) Our favored interpretation is that centrosome protein accumulation after proteasome inhibition is due to failure of degrading polyubiquitylated protein. This is supported by our immunoblotting data showing a ladder of insoluble forms of gamma-tubulin at an increased molecular weight after proteasome inhibition, consistent with polyubiquitylation of gamma-tubulin. Moreover, ubiquitin localization at the centrosome increases in the presence of proteasome inhibitors (this study, as well as Zhao et al., 2003). Additional support for this idea comes from the detection of ubiquitin ligases such as SCF and parkin at the centrosome (Freed et al., 1999; Zhao et al., 2003). Interestingly, mono-ubiquitylation of gamma-tubulin by BRCA1/BARD1 has been documented, although it is unclear whether this triggers proteolysis of gamma-tubulin (Starita et al., 2004).
Our own data indicate that the accumulation of gamma-tubulin at the centrosome is reversed after the proteasome inhibitors have been removed from the cell, to allow proteasome-dependent degradation to resume. This raises the question about the potential biological role of proteolysis of centrosome proteins. It is possible that proteolysis is necessary upon exit from mitosis, to reduce the amount of centrosome proteins previously accumulated during mitosis, to reestablish a regular microtubule network after spindle disassembly. Furthermore, it is possible that because of the high rate of protein transport and the high dynamics of microtubule assembly and disassembly at the centrosome, a significant number of centrosome proteins may need to be replaced to maintain centrosome function. Replacement may be necessary due to posttranslational modifications to regulate the activity of centrosome protein or due to denaturation of protein. Consistent with this we found that centrosome proteins accumulating upon proteasome inhibition do not lead to increased nucleation or anchoring of microtubules at the pericentriolar material, suggesting that they are not functional, although we cannot exclude that proteasome inhibition impairs microtubule organization without affecting microtubule nucleation. We propose that centrosome proteins that need to be eliminated or replaced are tagged by polyubiquitin, which enables recognition by the proteasome, followed by degradation. This must be in an equilibrium with the translation of new centrosome proteins. Our data provide the first evidence for gamma-tubulin polyubiquitination, suggesting that it is regulated by the proteasome. Because polyubiquitination of proteins may be involved in different cellular function such as localization or protein–protein interactions, further studies will be necessary to understand the role of this posttranslational modification.
For centrosome proteins, ubiquitylation and proteolysis may be a mechanism for regulating their exchange with the cytoplasmic pool, in particular during mitosis when the majority of gamma-tubulin seems to be exchangeable, whereas in interphase, only about half of the centrosome-bound gamma-tubulin exchanges (Khodjakov and Rieder, 1999). In this context, a study by Ehrhardt and Sluder (2005) indicated that inhibition of the proteasome during mitosis can lead to excess accumulation of centrosome proteins and subsequent spindle pole fragmentation. Further work including measurement of protein levels, half life, exchange rates in living cells, and characterization of the ubiquitylation status of these proteins will be necessary to better understand the complex relationship between proteasome activity and centrosome assembly and function. We cannot exclude that proteasome inhibition also provokes the accumulation of other, noncentrosome proteins that also affect centrosome function and microtubule organization, either by hindering physically the nucleation or anchoring of microtubules at the centrosome upon accumulation, or indirectly via regulatory pathways. Deregulation of centrosome assembly or microtubule organization might result in aberrant centrosome numbers, which are frequently correlated with genetic instability and human cancer (Pihan et al., 1998, 2003; Doxsey, 2005; Quintyne et al., 2005). Future studies will reveal whether proteasomes play a role in controlling the homeostasis of centrosome proteins not only during the cell cycle, but also in developmental as well as in oncogenic processes.
Supplementary Material
ACKNOWLEDGMENTS
We thank André Moisand for EM and Drs. Michel Wright, Chantal Etievant, Alain Duflos for helpful comments, as well as Delphine Reberioux for help with Visilog software. We thank Dr. Michel Bornens for providing HeLa cells stably expressing GFP-centrin. Financial support was provided by Centre National de la Recherche Scientifique and Pierre Fabre. C.D. was funded by a Pierre Fabre postdoctoral fellowship.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-12-1140) on December 19, 2007.
REFERENCES
- Adams J. The proteasome as a novel target for the treatment of breast cancer. Breast Dis. 2002;15:61–70. doi: 10.3233/bd-2002-15107. [DOI] [PubMed] [Google Scholar]
- Adams J. The development of proteasome inhibitors as anticancer drugs. Cancer Cell. 2004;5:417–421. doi: 10.1016/s1535-6108(04)00120-5. [DOI] [PubMed] [Google Scholar]
- Adams J., Behnke M., Chen S., Cruickshank A. A., Dick L. R., Grenier L., Klunder J. M., Ma Y. T., Plamondon L., Stein R. L. Potent and selective inhibitors of the proteasome: dipeptidyl boronic acids. Bioorg. Med. Chem. Lett. 1998;8:333–338. doi: 10.1016/s0960-894x(98)00029-8. [DOI] [PubMed] [Google Scholar]
- Bornens M. Centrosome composition and microtubule anchoring mechanisms. Curr. Opin. Cell Biol. 2002;14:25–34. doi: 10.1016/s0955-0674(01)00290-3. [DOI] [PubMed] [Google Scholar]
- Bornens M., Paintrand M., Berges J., Marty M. C., Karsenti E. Structural and chemical characterization of isolated centrosomes. Cell Motil. Cytoskelet. 1987;8:238–249. doi: 10.1002/cm.970080305. [DOI] [PubMed] [Google Scholar]
- Brown M. G., Monaco J. J. Biochemical purification of distinct proteasome subsets. Enzyme Protein. 1993;47:343–353. doi: 10.1159/000468692. [DOI] [PubMed] [Google Scholar]
- Claverol S., Burlet-Schiltz O., Girbal-Neuhauser E., Gairin J. E., Monsarrat B. Mapping and structural dissection of human 20 S proteasome using proteomic approaches. Mol. Cell Proteomics. 2002;1:567–578. doi: 10.1074/mcp.m200030-mcp200. [DOI] [PubMed] [Google Scholar]
- Craiu A., Gaczynska M., Akopian T., Gramm C. F., Fenteany G., Goldberg A. L., Rock K. L. Lactacystin and clasto-lactacystin beta-lactone modify multiple proteasome beta-subunits and inhibit intracellular protein degradation and major histocompatibility complex class I antigen presentation. J. Biol. Chem. 1997;272:13437–13445. doi: 10.1074/jbc.272.20.13437. [DOI] [PubMed] [Google Scholar]
- Dammermann A., Merdes A. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J. Cell Biol. 2002;159:255–266. doi: 10.1083/jcb.200204023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxsey S., Zimmerman W., Mikule K. Centrosome control of the cell cycle. Trends Cell Biol. 2005;15:303–311. doi: 10.1016/j.tcb.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Doxsey S. J. Molecular links between centrosome and midbody. Mol. Cell. 2005;20:170–172. doi: 10.1016/j.molcel.2005.10.010. [DOI] [PubMed] [Google Scholar]
- Ehrhardt A. G., Sluder G. Spindle pole fragmentation due to proteasome inhibition. J. Cell Physiol. 2005;204:808–818. doi: 10.1002/jcp.20335. [DOI] [PubMed] [Google Scholar]
- Fabunmi R. P., Wigley W. C., Thomas P. J., DeMartino G. N. Activity and regulation of the centrosome-associated proteasome. J. Biol. Chem. 2000;275:409–413. doi: 10.1074/jbc.275.1.409. [DOI] [PubMed] [Google Scholar]
- Freed E., Lacey K. R., Huie P., Lyapina S. A., Deshaies R. J., Stearns T., Jackson P. K. Components of an SCF ubiquitin ligase localize to the centrosome and regulate the centrosome duplication cycle. Genes Dev. 1999;13:2242–2257. doi: 10.1101/gad.13.17.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoda L., van Daalen Wetters T., Macrae M., Ascherman D., Coffino P. Prevention of rapid intracellular degradation of ODC by a carboxyl-terminal truncation. Science. 1989;243:1493–1495. doi: 10.1126/science.2928784. [DOI] [PubMed] [Google Scholar]
- Hames R. S., Crookes R. E., Straatman K. R., Merdes A., Hayes M. J., Faragher A. J., Fry A. M. Dynamic recruitment of Nek2 kinase to the centrosome involves microtubules, PCM-1, and localized proteasomal degradation. Mol. Biol. Cell. 2005;16:1711–1724. doi: 10.1091/mbc.E04-08-0688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haren L., Remy M. H., Bazin I., Callebaut I., Wright M., Merdes A. NEDD1-dependent recruitment of the gamma-tubulin ring complex to the centrosome is necessary for centriole duplication and spindle assembly. J. Cell Biol. 2006;172:505–515. doi: 10.1083/jcb.200510028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman L., Pratt G., Rechsteiner M. Multiple forms of the 20 S multicatalytic and the 26 S ubiquitin/ATP-dependent proteases from rabbit reticulocyte lysate. J. Biol. Chem. 1992;267:22362–22368. [PubMed] [Google Scholar]
- Julian M., Tollon Y., Lajoie-Mazenc I., Moisand A., Mazarguil H., Puget A., Wright M. gamma-Tubulin participates in the formation of the midbody during cytokinesis in mammalian cells. J. Cell Sci. 1993;105(Pt 1):145–156. doi: 10.1242/jcs.105.1.145. [DOI] [PubMed] [Google Scholar]
- Khodjakov A., Rieder C. L. The sudden recruitment of gamma-tubulin to the centrosome at the onset of mitosis and its dynamic exchange throughout the cell cycle, do not require microtubules. J. Cell Biol. 1999;146:585–596. doi: 10.1083/jcb.146.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I., Kim C. H., Kim J. H., Lee J., Choi J. J., Chen Z. A., Lee M. G., Chung K. C., Hsu C. Y., Ahn Y. S. Pyrrolidine dithiocarbamate and zinc inhibit proteasome-dependent proteolysis. Exp. Cell Res. 2004;298:229–238. doi: 10.1016/j.yexcr.2004.04.017. [DOI] [PubMed] [Google Scholar]
- Kisselev A. F., Goldberg A. L. Monitoring activity and inhibition of 26S proteasomes with fluorogenic peptide substrates. Methods Enzymol. 2005;398:364–378. doi: 10.1016/S0076-6879(05)98030-0. [DOI] [PubMed] [Google Scholar]
- Kostova Z., Wolf D. H. For whom the bell tolls: protein quality control of the endoplasmic reticulum and the ubiquitin-proteasome connection. EMBO J. 2003;22:2309–2317. doi: 10.1093/emboj/cdg227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A., Tsukita S. Non-membranous granular organelle consisting of PCM-1, subcellular distribution and cell-cycle-dependent assembly/disassembly. J. Cell Sci. 2003;116:919–928. doi: 10.1242/jcs.00282. [DOI] [PubMed] [Google Scholar]
- Ling Y. H., Liebes L., Ng B., Buckley M., Elliott P. J., Adams J., Jiang J. D., Muggia F. M., Perez-Soler R. PS-341, a novel proteasome inhibitor, induces Bcl-2 phosphorylation and cleavage in association with G2-M phase arrest and apoptosis. Mol. Cancer Ther. 2002;1:841–849. [PubMed] [Google Scholar]
- Merdes A., De Mey J. The mechanism of kinetochore-spindle attachment and polewards movement analyzed in PtK2 cells at the prophase-prometaphase transition. Eur. J. Cell Biol. 1990;53:313–325. [PubMed] [Google Scholar]
- Mitchison T., Kirschner M. Microtubule assembly nucleated by isolated centrosomes. Nature. 1984;312:232–237. doi: 10.1038/312232a0. [DOI] [PubMed] [Google Scholar]
- Nekrep N., Jabrane-Ferrat N., Peterlin B. M. Mutations in the bare lymphocyte syndrome define critical steps in the assembly of the regulatory factor X complex. Mol. Cell Biol. 2000;20:4455–4461. doi: 10.1128/mcb.20.12.4455-4461.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg E. A. Centrosome aberrations: cause or consequence of cancer progression? Nat. Rev. Cancer. 2002;2:815–825. doi: 10.1038/nrc924. [DOI] [PubMed] [Google Scholar]
- Oberdorf J., Carlson E. J., Skach W. R. Redundancy of mammalian proteasome beta subunit function during endoplasmic reticulum associated degradation. Biochemistry. 2001;40:13397–13405. doi: 10.1021/bi011322y. [DOI] [PubMed] [Google Scholar]
- Paoletti A., Moudjou M., Paintrand M., Salisbury J. L., Bornens M. Most of centrin in animal cells is not centrosome-associated and centrosomal centrin is confined to the distal lumen of centrioles. J. Cell Sci. 1996;109(Pt 13):3089–3102. doi: 10.1242/jcs.109.13.3089. [DOI] [PubMed] [Google Scholar]
- Parvin J. D. Overview of history and progress in BRCA1 research: the first BRCA1 decade. Cancer Biol. Ther. 2004;3:505–508. doi: 10.4161/cbt.3.6.839. [DOI] [PubMed] [Google Scholar]
- Piel M., Meyer P., Khodjakov A., Rieder C. L., Bornens M. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J. Cell Biol. 2000;149:317–330. doi: 10.1083/jcb.149.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pihan G. A., Purohit A., Wallace J., Knecht H., Woda B., Quesenberry P., Doxsey S. J. Centrosome defects and genetic instability in malignant tumors. Cancer Res. 1998;58:3974–3985. [PubMed] [Google Scholar]
- Pihan G. A., Wallace J., Zhou Y., Doxsey S. J. Centrosome abnormalities and chromosome instability occur together in pre-invasive carcinomas. Cancer Res. 2003;63:1398–1404. [PubMed] [Google Scholar]
- Purohit A., Tynan S. H., Vallee R., Doxsey S. J. Direct interaction of pericentrin with cytoplasmic dynein light intermediate chain contributes to mitotic spindle organization. J. Cell Biol. 1999;147:481–492. doi: 10.1083/jcb.147.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintyne N. J., Reing J. E., Hoffelder D. R., Gollin S. M., Saunders W. S. Spindle multipolarity is prevented by centrosomal clustering. Science. 2005;307:127–129. doi: 10.1126/science.1104905. [DOI] [PubMed] [Google Scholar]
- Raynaud-Messina B., Merdes A. Gamma-tubulin complexes and microtubule organization. Curr. Opin. Cell Biol. 2007;19:24–30. doi: 10.1016/j.ceb.2006.12.008. [DOI] [PubMed] [Google Scholar]
- Schiebel E. gamma-tubulin complexes: binding to the centrosome, regulation and microtubule nucleation. Curr. Opin. Cell Biol. 2000;12:113–118. doi: 10.1016/s0955-0674(99)00064-2. [DOI] [PubMed] [Google Scholar]
- Starita L. M., Machida Y., Sankaran S., Elias J. E., Griffin K., Schlegel B. P., Gygi S. P., Parvin J. D. BRCA1-dependent ubiquitination of gamma-tubulin regulates centrosome number. Mol. Cell. Biol. 2004;24:8457–8466. doi: 10.1128/MCB.24.19.8457-8466.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T., Evans L., Kirschner M. Gamma-tubulin is a highly conserved component of the centrosome. Cell. 1991;65:825–836. doi: 10.1016/0092-8674(91)90390-k. [DOI] [PubMed] [Google Scholar]
- Wagenknecht B., Hermisson M., Eitel K., Weller M. Proteasome inhibitors induce p53/p21-independent apoptosis in human glioma cells. Cell Physiol. Biochem. 1999;9:117–125. doi: 10.1159/000016308. [DOI] [PubMed] [Google Scholar]
- Wigley W. C., Fabunmi R. P., Lee M. G., Marino C. R., Muallem S., DeMartino G. N., Thomas P. J. Dynamic association of proteasomal machinery with the centrosome. J. Cell Biol. 1999;145:481–490. doi: 10.1083/jcb.145.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik C., DeMartino G. N. Intracellular localization of proteasomes. Int. J. Biochem. Cell Biol. 2003;35:579–589. doi: 10.1016/s1357-2725(02)00380-1. [DOI] [PubMed] [Google Scholar]
- Zhao J., Ren Y., Jiang Q., Feng J. Parkin is recruited to the centrosome in response to inhibition of proteasomes. J. Cell Sci. 2003;116:4011–4019. doi: 10.1242/jcs.00700. [DOI] [PubMed] [Google Scholar]
- Zheng Y., Jung M. K., Oakley B. R. Gamma-tubulin is present in Drosophila melanogaster and Homo sapiens and is associated with the centrosome. Cell. 1991;65:817–823. doi: 10.1016/0092-8674(91)90389-g. [DOI] [PubMed] [Google Scholar]
- Zheng Y., Wong M. L., Alberts B., Mitchison T. Nucleation of microtubule assembly by a gamma-tubulin-containing ring complex. Nature. 1995;378:578–583. doi: 10.1038/378578a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.