Abstract
Candida albicans must undergo a switch from white to opaque to mate. Opaque cells then release mating type-specific pheromones that induce mating responses in opaque cells. Uniquely in C. albicans, the same pheromones induce mating-incompetent white cells to become cohesive, form an adhesive basal layer of cells on a surface, and then generate a thicker biofilm that, in vitro, facilitates mating between minority opaque cells. Through mutant analysis, it is demonstrated that the pathways regulating the white and opaque cell responses to the same pheromone share the same upstream components, including receptors, heterotrimeric G protein, and mitogen-activated protein kinase cascade, but they use different downstream transcription factors that regulate the expression of genes specific to the alternative responses. This configuration, although common in higher, multicellular systems, is not common in fungi, and it has not been reported in Saccharomyces cerevisiae. The implications in the evolution of multicellularity in higher eukaryotes are discussed.
INTRODUCTION
Both single cell organisms and the individual cells of multicellular organisms must respond to a variety of environmental signals (Dorsky et al., 2000; Balázsi and Oltvai, 2005). In addition, in higher eukaryotes different cell types frequently respond to the same signal in unique ways (Rincón and Pedraza-Alva, 2003; Bacci et al., 2005; Dailey et al., 2005). In most cases, different signals interact with unique surface receptors that activate different signal transduction pathways, as has been demonstrated in Saccharomyces cerevisiae (Leberer et al., 1997; Gustin et al., 1998; Madhani and Fink, 1998; Elion, 2000). The mitogen-activated protein (MAP) kinase pathways have evolved as highly efficient, multipurpose signal transduction systems. S. cerevisiae uses multiple MAP kinase pathways, each one for a distinct signaling system, including the mating process, the filamentation process, cell wall integrity, ascospore formation, and osmoregulation (Levin and Errede, 1995; Gustin et al., 1998; Saito and Tatebayashi, 2004; Chen and Thorner, 2007). Several of these pathways share a limited number of components, but all are presumed to use different receptors to elicit very different responses. In the mating process of S. cerevisiae, a cells release a-factor that interacts with the a-receptor on α cells, and α cells release α-factor that interacts with the α-receptor on a cells. These alternative signals are then transduced through the same heterotrimeric G protein to activate the same MAP kinase pathway, which in turn activates the same downstream regulators that elicit similar mating responses, including G1 arrest, polarization, and shmooing (Sprague et al., 1983; Bender and Sprague, 1986; Leberer et al., 1997). Other fungi, including Magnaporthe grisea (Dixon et al., 1999; Zhao et al., 2005b) Neurospora crassa (Li et al., 2005), and Cryptococcus neoformans (Davidson et al., 2003; Kraus et al., 2003; Bahn et al., 2005) also use MAP kinase pathways for a variety of responses (Kruppa and Calderone, 2006).
The pathogenic yeast Candida albicans also uses MAP kinase pathways in the mating process (Chen et al., 2002; Magee et al., 2002), filamentation (Liu et al., 1994; Csank et al., 1998; Navarro-Garcia et al., 1998), and osmoregulation (Alonso-Monge et al., 1999; Smith et al., 2004). But C. albicans has one additional and unique response to mating pheromones that so far has not been identified in other yeast (Daniels et al., 2006). To mate, MTL-heterozygous strains of C. albicans must undergo homozygosis to a/a or α/α (Hull et al., 2000; Magee and Magee, 2000), then switch from the mating-incompetent white phenotype to the mating-competent opaque phenotype (Miller and Johnson, 2002; Lockhart et al., 2003a). Although pheromones induce mating responses in opaque a/a and α/α cells, including G1 arrest, polarization, and shmooing, as in S. cerevisiae, they do not induce these responses in white cells (Bennett et al., 2003; Lockhart et al., 2003a,b; Zhao et al., 2005a; Daniels et al., 2006). They do, however, elicit in white cells a dramatic increase in cohesion, adhesion, and biofilm development (Daniels et al., 2006). These changes have been demonstrated, at least in vitro, to result in a thicker white cell biofilm that provides a protective environment for spontaneously arising opaque cells to undergo mating (Daniels et al., 2006; Soll and Daniels, 2007).
Although mutational studies using complementation of auxotrophic traits as an assay for mating indicated that the α-receptor Ste2p; the MAP kinases Cek1p and Cek2p, homologues of S. cerevisiae Kss1p and Fus3p; and a key target transcription factor, Cph1p, the homologue of S. cerevisiae Ste12p, were necessary for mating in C. albicans a/a cells (Chen et al., 2002; Magee et al., 2002; Bennett et al., 2003), the receptors, signal transduction pathways, and downstream transcription factor(s) that mediate the unique white cell response to pheromone remained unknown.
There existed at least three possible scenarios for this pathway. First, the same receptor, G protein complex, MAP kinase pathway, and targeted transcription factor could mediate both the opaque cell mating response and the white response. Second, select components of the signal transduction pathway regulating the opaque pheromone response could be shared with the pathway regulating the white pheromone response. Third, completely different receptors and transduction pathways, with no overlap, could mediate the alternative opaque and white responses. To distinguish between these possible scenarios, we generated deletion derivatives in a natural a/a strain for components mediating the mating response, including the α-pheromone receptor gene, STE2; the gene for the β-subunit of the heterotrimeric G protein, STE4; the genes for the MAP kinases, CEK1 and CEK2; the gene for the downstream trans-acting factor CPH1 (the S. cerevisiae STE12 homologue); and the gene for the downstream cyclin-dependent kinase inhibitor, FAR1. We also generated deletion derivatives in natural α/α strain for the a-pheromone receptor gene, STE3, and for FAR1. Mutant and complemented strains were analyzed for the pheromone response of opaque cells and the pheromone response of white cells.
Our results demonstrate that the pathways regulating the alternative responses in opaque and white cells to the same pheromone share the same receptor, heterotrimeric G protein, and MAP kinase cascade, but they do not share the same downstream transcription factor(s). This represents a configuration in which two different cell types, white and opaque, respond to the same signals, and use the same receptors, heterotrimeric G protein, and the same MAP kinase cascade, but different downstream transcription factors, Cph1p in the opaque cell response and an as-yet-unidentified transcription factor in the white cell response. This configuration, which has no analogous example in S. cerevisiae, is found in a variety of multicellular systems in which the same signal is transduced in different cell types by the same signal transduction pathway, but results in different cellular responses (Rincón and Pedraza-Alva, 2003). We argue that several aspects of the signaling system between opaque and white cells suggest that it may represent an antecedent to multicellularity in higher eukaryotes.
MATERIALS AND METHODS
Strain Maintenance and Growth
Strains used in this study and their origins and genotypes are listed in Supplemental Table S1. Cells of the natural strains P37005 (a/a) (Lockhart et al., 2002), P57072 (α/α) (Lockhart et al., 2002), and WO-1 (α/α) (Slutsky et al., 1987), the derived mutants, and complemented strains were maintained at 25°C on agar containing modified Lee's medium (Bedell and Soll, 1979) or YPD medium (Sherman et al., 1986). For distinguishing between white and opaque phase sectors or colonies, colonies were grown on modified Lee's agar medium supplemented with 5 μg/ml phloxine B, which differentially stained opaque phase cells red (Anderson and Soll, 1987). Before use, white and opaque phase cells were verified microscopically for the unique differences in cell shape and vacuole formation (Anderson and Soll, 1987; Slutsky et al., 1987).
Generation of Null Mutants
In this study, the following null mutants were generated: ste2Δ, ste4Δ, cek1Δ, cek2Δ, cek1Δ cek2Δ, cph1Δ, and far1Δ in the natural a/a strain P37005; and ste3Δ in the natural α/α strain P57072. In addition, a far1Δ mutant was generated in the natural α/α strain WO-1. The recyclable flipper cassette from pSFS2A (Reuss et al., 2004), containing a dominant nourseothricin resistance marker (CaSAT1), was used to create all mutants. The plasmid pSFS2A was a generous gift from Joachim Morschhauser (The University of Würzburg, Germany). The 4.2-kb XhoI–SacII fragment SAT1-2A of the cassette was blunt-ended with T4 polymerase before its use in ligations to create the deletion cassettes.
All of the primers used to create gene deletions are provided in Supplemental Table S2. To obtain a homozygous mutant strain for a particular gene, deletion cassettes I and II were generated in a two-step disruption strategy. Deletion cassette I was constructed as follows: 5′ and 3′ flanking regions of each target gene were amplified by polymerase chain reaction (PCR) by using the primers provided in Supplemental Table S2. The 5′ region and 3′ region were then each digested by SmaI and ligated together using T4 ligase. The 5′-3′ fusion product was amplified by PCR and subcloned into the pGEM-T Easy vector (Promega, Madison, WI). The SAT1-2A fragment was then inserted into the SmaI-digested, dephosphorylated plasmid. This plasmid was digested with SacI plus SphI to generate the deletion cassette, which was then used to transform C. albicans strain P37005, P57072, or WO-1 by electroporation (De Backer et al., 1999). For each gene, two independent transformants were confirmed as heterozygous by both PCR and Southern analysis. The heterozygotes were then subjected to a pop-out strategy in the maltose-containing medium YPM (1% yeast extract, 2% Bacto-peptone, and 2% maltose) to excise the CaSAT1 marker. Deletion cassette II was constructed in a similar manner. The new 5′ and 3′ flanking regions that contained sequences deleted in the first step were amplified by PCR, by using the primers noted for each gene in Supplemental Table S2. The resulting plasmid was digested with SacI and SphI, and it was used to transform the heterozygous mutant derivatives. Two independent null mutants were confirmed by both PCR and Southern analysis for each gene.
Mutant Complementation
Complementation was performed for the mutants ste2Δ, steΔ3, ste4Δ, cek1Δ, cek2Δ, cph1Δ, and far1Δ, generating ste2Δ/STE2, ste3Δ/ste3, ste4Δ/STE4, cek1Δ/CEK1, cek2Δ/CEK2, cph1Δ/CPH1, and far1Δ/FAR1. The CaSAT1 marker was deleted by a pop-out protocol from each null mutant as described for heterozygous mutants. The 5′ and 3′ regions flanking the stop codon were amplified by PCR with the primers noted for each gene in Supplemental Table S2. The 5′-3′ fusion product was amplified by PCR and subcloned into pGEM-T Easy (Promega). For complemented strains, a DNA fragment containing both green fluorescent protein (GFP) and CaSAT1 was amplified by PCR with the primers noted in Supplemental Table S2, using plasmid pK91.6 (Srikantha and Soll, unpublished data) as template. GFP was inserted into the plasmid for future experiments and not used in this study. The GFP–CaSAT1 fragment was digested with BamHI plus BglII, and it was ligated into the Bg1II- or BamHI-digested, dephosphorylated plasmid containing the 5′-3′ fusion product of the gene. This plasmid contained the transformation module for targeting to the gene locus. The in-frame GFP-gene fusion was confirmed by sequencing. This plasmid was digested with XhoI or StuI and used for transformation into the null mutant of each gene. Transformants were verified by both PCR sequencing and Southern analysis.
Opaque Cell Shmooing and Mating
Opaque cells were grown in liquid modified Lee's medium in a rotary water bath shaker (250 rpm) at 25°C to early saturation phase (∼5 × 107 cells/ml) (Lockhart et al., 2003b). Cells were then pelleted, resuspended at 106 cells/ml in fresh medium containing 3 × 10−6 M synthetic 13-mer α-pheromone (Bennett et al., 2003; Panwar et al., 2003) and incubated at 25°C in a shaker (250 rpm). The 13-mer peptide (GFRLTNFGYFEPG), synthesized by Open Biosystems (Huntsville, AL), was dissolved in dimethyl sulfoxide (DMSO). In controls not treated with pheromone, equivalent amount of DMSO was added. Shmooing and conjugation tube growth were monitored microscopically. Cell concentration was also monitored over time. To test for a-pheromone–induced shmoo formation, a transwell assay was performed according to Daniels et al. (2006).
To test for mating (Lockhart et al., 2003a), opaque cells of an a/a or α/α mutant were grown to early saturation phase and mixed with an equal concentration of WO-1 (α/α) or P37005 (a/a) opaque cells, respectively, in liquid culture. The mating mixtures were incubated at 25°C in a rotary shaker (250 rpm), and they were monitored for fusants microscopically over a 48-h period.
White Cell Cohesion and Adhesion Assays
To test for α-pheromone–induced cohesion according to the methods of Daniels et al. (2006), a/a white cells from a saturation phase culture (∼4 × 108 cells/ml) of strain P37005 or mutant derivatives were resuspended in fresh medium at a concentration of 5 × 107/ml. The medium was supplemented with the 13-mer synthetic α-pheromone at a concentration of 3 × 10−6 M. The culture was rotated at 250 rpm at 25°C. Samples were taken from the suspension culture after 6 h, and they were examined microscopically for cell aggregates. To test for a-pheromone–induced cohesion of white cells of strain P57072 and the mutant derivative ste3Δ, a 50:50 mixture of opaque P37005 and WO-1 cells was added to a suspension of either white P57072 or ste3Δ cells so that the former-inducing mixture made up 1% of cells. Opaque P37005 cells (a/a), stimulated by opaque WO-1 (α/α) cells, released a-pheromone.
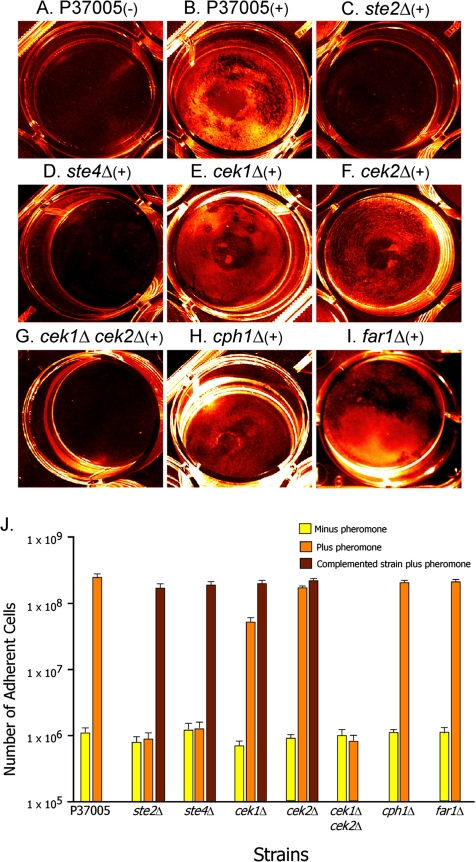
To test for α-pheromone–induced adhesion of white cells of the natural a/a strain P37005 and its mutant derivatives to plastic, the methods of Daniels et al. (2006) were used. Two milliliters of cells (5 × 107/ml) were incubated in a well of a Costar six-cluster well plate (Corning Life Sciences, Lowell, MA) in the presence of 3 × 10−6 M synthetic 13-mer α-pheromone. After 16 h at 25°C, the wells were gently washed with phosphate-buffered solution and photographed. Gray scale images were subsequently pseudocolored for clarity. Three hundred microliters of a 0.05% trypsin-EDTA solution (Invitrogen, Carlsbad, CA) was added to each well. After 15 min, the cells on the dish bottom were released into 300 μl of supplemental Lee's medium containing 10% calf serum, and the number of adhering cells was determined in a hemocytometer. To test for a-pheromone–induced adhesion of α/α strain P57072 and its mutant derivative ste3Δ, 1% opaque cells of a/a strain P37005 and α/α strain WO-1 were added to the well culture. After 16 h at 25°C, adhesion was analyzed as described above.
Biofilm Thickness
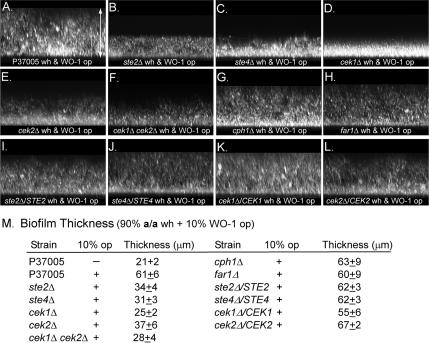
Biofilm enhancement was quantitated in strain P37005 and mutant derivatives according to a protocol described previously (Daniels et al., 2006), with one exception. Although in earlier experiments, a minority mixture (50:50) of opaque a/a and α/α cells was found more stimulatory than opaque α/α cells alone in enhancing a majority (90%) white a/a cell biofilm formation, recent experiments proved that minority opaque α/α cells (WO-1) alone induced near maximum enhancement of majority white biofilms. Therefore, a mixture of 90% white a/a test cells and 10% opaque α/α WO-1 cells (a total of ∼5 × 107 cells in 2.5 ml of RPMI 1640 medium) was distributed on a silicone elastomer square in a well and incubated for 90 min. To test for enhancement of white cell biofilms of strain P57072 and the mutant derivative ste3Δ, opaque a/a cells (P37005) (10%) were added to majority α/α cells in the presence of 3 × 10−6 α-pheromone. The square was then rinsed and incubated in RPMI 1640 medium on a rocker at 29°C for the subsequent 48 h. Biofilms were prepared in triplicate cultures. The biofilm was fixed, stained with calcofluor, and the thickness was measured using Bio-Rad LaserSharp software in a Bio-Rad Radiance 2100 MP laser scanning confocal microscope (LSCM) (Bio-Rad, Hermel, Hamstead, United Kingdom).
Quantitative Fluorescence Analysis of DNA
Two methods were used. In the first method, described in detail in Zhao et al. (2005a), opaque cells were grown to saturation phase, and then they were resuspended in fresh medium at 106 cells/ml. Cells were treated with synthetic α-pheromone (13-mer) in suspension, and then they were fixed after 3 h in 70% ethanol and treated overnight with RNase. Nuclei were stained with 25 μM Sytox Green (Invitrogen). Fluorescent quantitation of the staining of individual nuclei was performed using a confocal method we described in detail previously (Zhao et al., 2005a). Using the projected confocal image, a line profile of pixel intensity was measured across the center of each nucleus. In both control P37005 and far1Δ cell populations, only the nuclei of cells that had formed shmoos were scanned. That represented ∼60–70% of the P37005 cell population and 25% of the far1Δ cell population. In a second method, cell cycle status was determined by fluorescence-activated cell sorting (FACS). Cells were prepared as described above, with modification. RNase treatment was followed by proteinase K digestion. The final cell suspension was then stained overnight with 1 μM Sytox Green. The cells were sonicated briefly to disrupt cell aggregates, and then they were analyzed with a FACScan (BD Biosciences, Mountain View, CA). Cell cycle status was analyzed using ModFitLT version 2.0 software (BD Biosciences).
Northern and Southern Analyses
Northern and Southern analyses were performed as described previously (Lockhart et al., 2003b; Srikantha et al., 2006). For northern analyses, cells from saturation phase cultures were diluted into fresh medium in the absence or presence of 3 × 10−6 M α-pheromone, and they were pelleted after 4 h. Total RNA was extracted using the RNeasy Mini kit (QIAGEN, Valencia, CA). PCR products were used for probing Northern and Southern blots. The primers used to generate the PCR probe for each gene are listed in Supplemental Table S2.
RESULTS
The Pheromone Response Pathway Plays No Role in Switching
The deletion mutants ste2Δ, ste4Δ, cek1Δ, cek2Δ, cek1Δ cek2Δ, cph1Δ, and far1Δ were generated in the natural a/a strain P37005 (Lockhart et al., 2002). The deletion mutant ste3Δ (α/α) was generated in the natural α/α strain P57072 (Lockhart et al., 2002) and a far1Δ deletion mutant was generated in the natural α/α strain WO-1 (Slutsky et al., 1987). Each mutant and complemented derivatives were individually tested for spontaneous white-opaque switching by plating cells from single white colonies at low density on nutrient agar containing phloxine B, which differentially stained opaque cells red (Anderson and Soll, 1987). One thousand derivative colonies were scored for each strain. In every case, red colonies and/or sectors formed after 7 d at low frequencies similar to wild type (∼10−3 opaque colonies). Cells from every tested red colony or sector of each mutant were found to exhibit the unique elongate opaque cell shape (data not shown). When these opaque cells were in turn plated at low density on agar, they formed a majority of opaque colonies and a minority of white colonies, demonstrating reversibility for every mutant. The same was true for the complemented strains ste2Δ/STE2, ste3Δ/STE3, ste4Δ/STE4, cek1Δ/CEK1, cek2Δ/CEK2, cph1Δ/CPH1, and far1Δ/FAR1 (a/a). Zordan et al. (2006) demonstrated previously that switching was unimpaired in deletion mutants of STE2, CEK2, and FAR1 generated in an a cell background in a laboratory strain derived from strain SC5314. Together, these results demonstrate that the genes in the pheromone response pathway are not essential for white-opaque switching. This allowed ready isolation of white and opaque cells for each mutant and complemented strain, which were then tested for the alternative pheromone responses.
Opaque Cell Pheromone Response of ste2Δ, ste3Δ, and ste4Δ
No shmoo formation was observed in opaque cells of parent strain P37005 in the absence of α-pheromone. Seventy-five percent formed shmoos after 4 h of treatment with α-pheromone and 91% after 8 h (Figure 1, A, B, and M). Neither opaque cells of the ste2Δ mutant nor of the ste4Δ mutant formed shmoos in response to α-pheromone (Figure 1, C and D, respectively; and M). The complemented strains ste2Δ/STE2 and ste4Δ/STE4 regained the capacity to form shmoos in response to pheromone, and to the same extent as parental P37005 cells (data not shown). Bennett et al. (2003) also found that STE2 was required for opaque a cells to undergo shmoo formation.
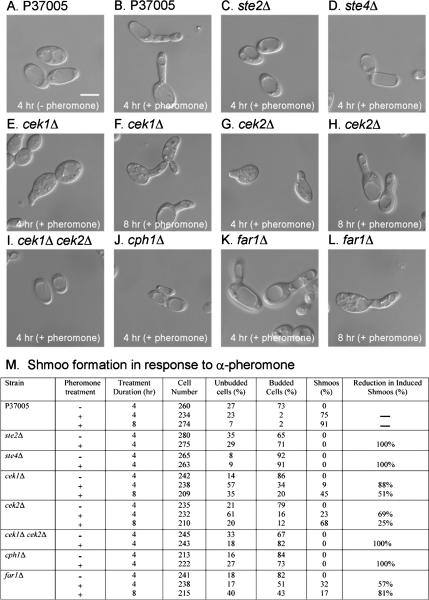
Figure 1.
α-Pheromone does not induce conjugation tube (“shmoo”) formation in opaque cells of the mutants ste2Δ, ste4Δ, the cek1Δ cek2Δ double mutant, or cph1Δ, derived from the natural a/a strain P37005. (A and B) Representative images of P37005 cells in the absence (−) or presence (+) of α-pheromone, respectively. (C, D, I, and J) Representative images of mutants ste2Δ, ste4Δ, cek1Δ cek2Δ, and cph1Δ, which did not form shmoos after 4 h of pheromone treatment. The same was true after 8 h (data not shown). Selected images of shmoo formation after 4 and 8 h for mutants cek1Δ (E and F), cek2Δ (G and H), and far1Δ (K and L). It should be noted that in these cases, the proportion of cells that had shmooed ranged between 9 and 68%; therefore, the images were selected. (M) Quantitation of shmooing for different strains. The “percentage of reduction in induced shmoos” was computed by dividing the difference in percentage of shmooing between P37005 and mutant strain, by percentage of shmooing of P37005, and multiplying by 100%. Bar (A), 5 μm.
Opaque cells of the a-receptor mutant ste3Δ could not be tested with synthetic pheromone, because a-pheromone is not readily synthesized chemically due to extensive posttranslational modification (Chen et al., 1997a,b; Huyer et al., 2006). Therefore, mutant ste3Δ cells were compared with parent P57072 cells for their response to a-pheromone released by opaque a/a cells (P37005) mixed with wild-type opaque α/α cells (P57072) that up-regulated a-pheromone production in the former. This inducing mixture was separated from opaque ste3Δ cells or α/α wild type cells by a micropore filter in a transwell chamber (Figure 2A) (Daniels et al., 2006). Whereas opaque cells of parent strain P57072 cells were induced to form shmoos (Figure 2B), opaque cells of ste3Δ were not (Figure 2C). Opaque cells of the complemented strain ste3Δ/STE3 regained shmoo formation in response to a-pheromone (data not shown).
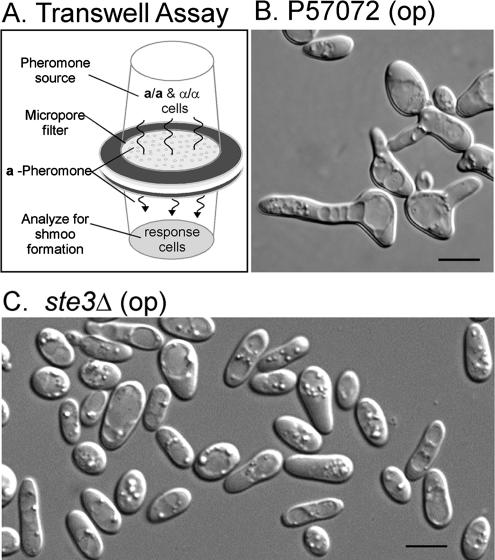
Figure 2.
a-Pheromone does not induce conjugation tube formation in opaque cells of the mutant ste3Δ, derived from the α/α strain P57072. (A) Transwell apparatus for a-pheromone induction of α/α cells. (B) Representative image of opaque P57072 cells after 7 h of incubation in the response well. (C) Representative image of ste3Δ after 7 h in response well. Bar (C), 5 μm.
Opaque cells of the mutants ste2Δ and ste4Δ were also compared with opaque cells of parent strain P37005 for their ability to mate with opaque cells of the α/α strain WO-1, by using microscopically identified fusion as an assay (Lockhart et al., 2003a). Although 27% of opaque cells in a 50:50 mixture of opaque a/a P37005 cells and opaque α/α WO-1 opaque cells fused (Figure 3, A and K), no fusions were observed between opaque ste2Δ or ste4Δ cells, and opaque WO-1 cells (Figure 3, B and C, respectively; and K). Opaque cells of the complemented strains ste2Δ/STE2 and ste4Δ/STE4 regained the capacity to mate with opaque WO-1 cells (data not shown). Using complementation between a and α auxotrophs as a fusion assay, Bennett et al. (2003) had previously demonstrated that STE2 was essential for mating.
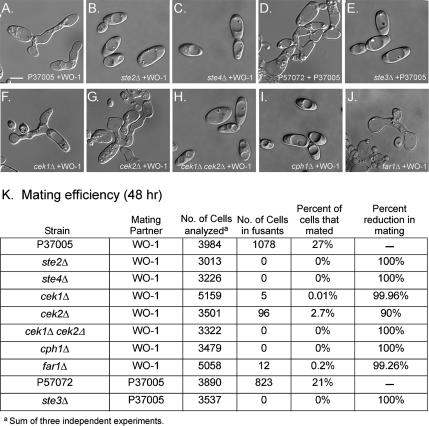
Figure 3.
Opaque cells of the mutants ste2Δ, ste3Δ, ste4Δ, the double mutant cek1Δ cek2Δ, and cph1Δ do not mate (i.e., undergo fusion) with opaque cells of opposite mating type in suspension cultures, whereas mutants cek1Δ, cek2Δ, and far1Δ mate, but at reduced frequency. (A and D) Selected image of mating opaque cells of parent strain P37005 (a/a) with opaque cells of strain WO-1 (α/α), and P57072 (α/α) with P37005 (a/a), respectively. Selected images of mixtures of opaque cells of ste2Δ (B), ste4Δ (C), ste3Δ (E), the double mutant cek1Δ cek2Δ (H), and cph1Δ (I), with mating partners, none of which mated. Selected images of mixtures of opaque cells of cek1Δ (F), cek2Δ (G), and far1Δ (J), with mating partners, which underwent mating. (K) Quantitation of mating efficiency. “Percentage of reduction in mating” was computed as follows. The percentage of opaque cells of the different strains that fused with opaque cells of opposite mating type was subtracted from the percentage of the parent strain that fused with opaque cells of opposite mating type. The difference was then divided by the percentage of parent strain cells that fused, and the fraction multiplied by 100%. The percentage of mating cells of the complemented strains ste2Δ/STE2, ste3Δ/STE3, ste4Δ/STE4, cek1Δ/CEK1, cek2Δ/CEK2, cph1Δ/CPH1, and far1Δ/FAR1 was similar to that of the parent wild type strains from which they were derived (data not shown). Bar (A), 5 μm.
The ste3Δ mutant was also incapable of mating. Whereas the parent α/α strain P57072 mated readily with the natural a/a strain P37005 (Figure 3, D and K), ste3Δ did not (Figure 3, E and K). Opaque cells of the complemented strain ste3Δ/STE3 mated with opaque cells of the a/a strain P37005 (data not shown). Together with the pheromone response data, these results indicated that the α-pheromone receptor, Ste2p and the β-subunit of the heterotrimeric G protein Ste4p were essential for α-pheromone–induced shmooing and fusion of opaque a/a cells, and that the a-pheromone receptor Ste3p was essential for a-pheromone–induced shmooing and fusion of opaque α/α cells.
White Cell Pheromone Response of ste2Δ, ste3Δ, and ste4Δ
White cells of the mutants ste2Δ and ste4Δ were compared with white cells of the parental strain P37005 for the α-pheromone–stimulated white cell response, which included dramatic increases in cohesion, adhesion, and enhanced biofilm development (Daniels et al., 2006). To assess pheromone-induced cohesion, ste2Δ and ste4Δ cells were incubated in suspension either in the absence or in the presence of α-pheromone for 6 h. Cells were then distributed on a slide, and the average number of cells per aggregate was calculated. As we described previously (Daniels et al., 2006), the majority of white P37005 cells remained largely separated or formed small aggregates in the absence of α-pheromone (Figure 4I), but in the presence of α-pheromone, the majority of cells formed large aggregates (Figure 4, A and I). In the absence (data not shown) or presence of α-pheromone, the majority of white ste2Δ and ste4Δ cells remained largely separated or formed small aggregates (Figure 4, B and I, and 4, C and I, respectively). White cells of the complemented strains ste2Δ/STE2 and ste4Δ/STE4 regained the aggregation response to α-pheromone (data not shown).
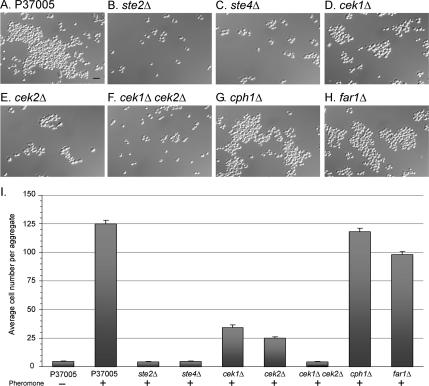
Figure 4.
White cells of the mutants ste2Δ, ste4Δ, and cek1Δ cek2Δ do not form large aggregates in response to pheromone as do wild-type cells, but mutants cph1Δ and far1Δ do. White cells of cek1Δ and cek2Δ form clumps of intermediate size. Cells of each strain from saturation phase cultures were diluted into fresh medium in the absence of pheromone (−) or in the presence of 3 × 10−6 M α-pheromone (+). Samples were incubated 6 h before analysis. (A–H) Representative images of cells from parent and mutant strains in the presence of pheromone. (I) Average number of cells (error bar represents standard deviation) in aggregates. In total, 20 cell aggregates were analyzed for each strain. Bar (A), 5 μm.
To assess pheromone-induced adhesion, ste2Δ and ste4Δ cells were incubated on a plastic surface in the absence or in the presence of 3 × 10−6 M synthetic α-pheromone for 16 h. As described previously (Daniels et al., 2006), in the absence of α-pheromone, white P37005 cells did not form a tight adhesive film on the dish bottom (Figure 5A), but in the presence of pheromone they did (Figure 5B). In contrast, neither white ste2Δ cells nor white ste4Δ cells formed a tight adhesive film on the plastic dish bottom in the absence of pheromone (Figure 5J) or presence of α-pheromone (Figure 5, C and D, respectively; and J). White cells of ste2Δ/STE2 and ste4Δ/STE4 regained the capacity to form an adhesive film in response to α-pheromone (Figure 5J).
Figure 5.
In response to pheromone, white cells of the mutants ste2Δ, ste4Δ, and the double mutant cek1Δ cek2Δ do not form an adhesive film on the bottom of a plastic well. White cells of the individual mutants cek1Δ and cek2Δ form films nearly as dense as wild type, and white cells of the mutants cph1Δ and far1Δ form normal films. Dish bottoms were examined for a cell film after 16 h. Pseudocolor images (in orange) are provided. (A and B) Representative images of the dish bottom of P37005 cultures in the absence (−) and presence (+) of pheromone. (C–I). Representative images of the dish bottom of mutant cultures in the presence (+) of α-pheromone. (J) Quantitation of cells adhering to the dish bottom. The “number of adherent cells” has been computed for the entire bottom of three separate wells. The average number is presented. Bar represents standard error. Data are also presented in J for complemented strains ste2Δ/STE2, ste4Δ/STE4, cek1Δ/CEK1, and cek2Δ/CEK2.
To test for the enhancement of a majority white cell biofilm by minority opaque cells of opposite mating type, a mixture of 10% opaque WO-1 (α/α) cells and 90% ste2Δ (a/a) or ste4Δ white (a/a) cells was incubated on a silicone elastomer surface for 48 h, and biofilm thickness was measured using LSCM (Daniels et al., 2006). In the absence of opaque WO-1 (α/α) cells, white P37005 cells formed a biofilm with an average thickness of 21 ± 2 μm (Figure 6M). In the presence of minority opaque α/α cells, the majority of white a/a P37005 cells formed a biofilm with an average thickness of 61 ± 6 μm, >3 times the thickness of biofilms formed by untreated cells (Figure 6, A and M). In the absence of opaque α/α cells, white ste2Δ or ste4Δ cells formed biofilms of approximately the same thickness as untreated white P37005 cells (data not shown); in the presence of opaque α/α cells, majority white ste2Δ or ste4Δ still formed biofilms approximately half as thick as those formed by stimulated white cells of strain P37005 (Figure 6, B and C, respectively; and M). The majority of white cells of both complemented strains ste2Δ/STE2 and ste4Δ/STE4 regained the capacity to form biofilms comparable with those of stimulated white cells of strain P37005 in the presence of minority opaque α/α cells (Figure 6, I and J, respectively; and M). Together with the cohesion and adhesion data (Figures 4 and 5), these results indicate that the same α-pheromone receptor and heterotrimeric G protein that regulate the α-pheromone–induced opaque cell response also regulate the pheromone-induced white cell response.
Figure 6.
The thickness of biofilms formed by white cells of mutants ste2Δ, ste4Δ, and cek1Δ, cek2Δ, and the double mutant cek1Δ cek2Δ, was not enhanced by minority (10%) opaque cells of opposite mating types, as was the thickness of the biofilms of parent strain P37005, cph1Δ and far1Δ. A mixture of 90% white test cells and 10% opaque α/α cells was inoculated onto a silicone square and incubated for 48 h. The z-series projections were viewed from the side (90° tilt) of stacked multiphoton laser scanning confocal microscope scans. (A–L). Representative z-series projections of parental, mutant and complemented strain biofilms. (M) Average thickness (±standard deviation) of biofilms, computed from 10 measurements that included multiple cultures. Arrow span A represents 75 μm. Enhancement returned in the complemented strains ste2Δ/STE2, ste4Δ/STE4, cek1Δ/CEK1, and cek2Δ/CEK2.
Because of the unavailability of chemically synthesized a-pheromone, we tested whether natural a-pheromone released from opaque cells induced cohesiveness between white cells in a mixture maintained in suspension. In this protocol, majority white α/α P57072 cells (99%) were mixed with minority opaque a/a P37005 (0.5%) and opaque α/α WO-1 (0.5%) cells, the latter opaque cells added to stimulate the former to release a-pheromone. Whereas white α/α P57072 cells were induced by minority opaque a/a cells to form large clumps, majority white ste3Δ cells were not (Supplemental Figure S1). The cohesive response to a-pheromone, similar to that of parent α/α strain P57072, was restored in the complemented strain ste3Δ/STE3 (Supplemental Figure S1).
To test whether a-pheromone induced ste3Δ cells to form a tight adhesive film on a plastic surface, majority white cells (99%) of either strain P57072 or strain ste3Δ, were mixed with a minority (1%) of half opaque P37005 (a/a) cells and opaque WO-1 (α/α) cells. As in the previous strategy, opaque α/α cells stimulate the release of a-pheromone by opaque a/a cells, which then stimulates white-specific responses in white α/α cells (Daniels et al., 2006). Whereas white P57072 cells were induced to form an adhesive film on the plastic well bottom, ste3Δ cells were not (Supplemental Figure S2). Pheromone-induced substrate adhesion was restored in the complemented strain ste3Δ/STE3 (Supplemental Figure S2). Minority opaque a/a cells (P37005) stimulated by α-pheromone also enhanced biofilm formation by majority white P57072 cells, but not majority white ste3Δ cells (Supplemental Figure S3). The former were close to twice as thick as the latter (Supplemental Figure S3). Enhancement of biofilm formation by minority opaque cells similar to that in parent strain P57072 was restored in the complemented strain ste3Δ/STE3 (Supplemental Figure S3). Together, these results demonstrate that the same a-pheromone receptor (Ste3p) regulates the a-pheromone–induced opaque cell response and white cell response, just as the same Ste2p receptor regulates the α-pheromone responses of opaque and white a/a cells.
Opaque Cell Pheromone Response of cek1Δ, cek2Δ, and the Double Mutant
Opaque cells of the individual MAP kinase mutants cek1Δ and cek2Δ, which were generated in the natural a/a strain P37005, formed shmoos in response to α-pheromone, but the response in both cases was delayed, and the proportion of cells that responded after 4 h was reduced (Figure 1, E and G, respectively; and M). The percentage of cells that shmooed in both mutants increased after 8 h of treatment (Figure 1, F and H, respectively; and M). Opaque cells of both mutants also underwent mating with opaque cells of the α/α strain WO-1 (Figure 3, F and G, respectively), but the proportion of fusants was reduced by >99% for cek1Δ and by 90% for cek2Δ (Figure 3K). Pheromone-induced shmooing and mating of opaque cells was restored in the complemented strains cek1Δ/CEK1 and cek2Δ/CEK2 (data not shown).
However, opaque cells of the double mutant cek1Δ cek2Δ neither shmooed in response to α-pheromone (Figure 1, I and M) nor underwent mating with opaque cells of the α/α strain WO-1 (Figure 3, H and K). These results were consistent with those obtained with null mutants of KSS1 and FUS3, the respective orthologues of CEK1 and CEK2, in S. cerevisiae (Elion et al., 1991), and they confirm and extend earlier observations by Chen et al. (2002) on cek1Δ, cek2Δ, and the cek1Δ cek2Δ double mutant of C. albicans strain CAI4, in which complementation was used between auxotrophic a and α strains as an assay for mating.
White Cell Pheromone Responses of cek1Δ, cek2Δ, and the cek1Δ cek2Δ Double Mutant
In the absence of α-pheromone, white cek1Δ and cek2Δ cells formed only small aggregates in suspension cultures (data not shown), as did white P37005 cells (Figure 4I). In response to α-pheromone, both white cek1Δ and white cek2Δ cells formed aggregates that were, on average, larger than in the absence of α-pheromone (Figure 4, D and E, respectively), but still far smaller than those formed by α-pheromone–treated P37005 cells (Figure 4A). The average number of cells per clump for treated white cek1Δ and cek2Δ cells was 35 and 25, respectively, compared with 125 for white P37005 cells (Figure 4I). In response to α-pheromone, white cek1Δ and white cek2Δ cells formed a film on a plastic surface (Figure 5, E and F, respectively). Quantitation revealed that the number of adherent cells in these films was greater than that in unstimulated cultures, but consistently smaller (p < 0.001) than that of pheromone-stimulated P37005 cells (Figure 5J). This adhesive response was regained in the complemented strains cek1Δ/CEK1 and cek2Δ/CEK2 (Figure 5J). A minority of α/α opaque cells of strain WO-1 did not enhance the thickness of biofilms formed by majority cek1Δ or cek2Δ white cells over a 48-h period (Figure 6, D and E, respectively), as it did white P37005 cells (Figure 6M). Enhancement of the thickness of white cell biofilms by minority α/α opaque cells returned in the complemented strains (Figure 6, K and L, respectively; and M).
In the presence of α-pheromone, white cells of the double mutant cek1Δ cek2Δ formed only small clumps (Figure 4, F and I) like untreated cells (data not shown). White cek1Δ cek2Δ cells also did not form a film on a plastic surface in response to α-pheromone (Figure 5, G and J). Finally, minority α/α opaque cells of strain WO-1 did not stimulate an increase in the thickness of a majority a/a white cell biofilm of the cek1Δcek2Δ mutant on a silicone elastomer surface (Figure 6, F and M). These results demonstrate that as was the case for STE2, STE3, and STE4, the partially redundant functions of CEK1 and CEK2 are necessary for both the opaque and white cell responses, suggesting that the response pathways from receptor through the MAP kinase cascade are shared.
Opaque Cell Pheromone Response of cph1Δ
Opaque cells of the trans-acting factor mutant cph1Δ, which was generated in the natural a/a strain P37005, neither formed shmoos in response to α-pheromone (Figure 1, J and M), nor mated with opaque cells of the α/α strain WO-1 (Figure 3, I and K). The complemented strain cph1Δ/CPH1 reacquired these responses (data not shown). These results support and extend earlier observations by Magee et al. (2002) and Chen et al. (2002) in which complementation between auxotrophs was used as an assay to demonstrate that mating depends on CPH1 function.
White Cell Pheromone Response of cph1Δ
Although deletion of CPH1 completely blocked pheromone-induced formation and mating of opaque cells, it did not block the white cell pheromone response. Treatment of white cph1Δ cells with α-pheromone stimulated aggregation in suspension cultures (Figure 4, G and I) to levels comparable with that of treated white P37005 cells (Figure 4, A and I). Treatment with α-pheromone also induced cph1Δ cells to form a tightly adhering film on a plastic surface (Figure 5, H and J) comparable with that formed by treated white P37005 cells (Figure 5, B and J). Finally, minority α/α opaque cells of strain WO-1 stimulated an approximate threefold increase in the thickness of majority white cph1Δ cell biofilms (Figure 6, G and M), an increase comparable with that induced by α/α cells in white P37005 biofilms (Figure 6, A and M). These results clearly demonstrate that the white cell response to pheromone does not require the downstream target Cph1p. Therefore, although the pheromone response pathway from receptor through the MAP kinase pathway is shared, the downstream components of the pathways regulated by the MAP kinases differ.
Opaque Cell Pheromone Response of far1Δ
It was demonstrated previously that white cells do not shmoo (Bennett et al., 2003; Lockhart et al., 2003b) and that they do not arrest in G1 in response to α-pheromone (Zhao et al., 2005a). Therefore, one might not expect FAR1 to play a role in the white cell response, because the role FAR1 plays in the analogous mating process of S. cerevisiae is in the polarization of cells in a gradient of pheromone and G1 arrest (Chang and Herskowitz, 1990; Valtz et al., 1995; Butty et al., 1998). However, in supplemental data to Roberts et al. (2000), it was reported that STE12 was not up-regulated by pheromone in a far1Δ mutant, indicating that Far1p played a role in the up-regulation of pheromone-induced genes. Therefore, we considered the possibility that Far1p may also be involved in the up-regulation of genes by pheromone in the white cell response. The far1Δ mutant of S. cerevisiae shmoos in response to α-pheromone, but the shmoos do not polarize in a gradient of pheromone, and far1Δ cells are not blocked in G1 by pheromone (Chang and Herskowitz, 1990; Dorer et al., 1995; Valtz et al., 1995). The far1Δ mutant of S. cerevisiae is capable of mating, but the frequency of mating is significantly reduced, presumably because far1Δ cells cannot efficiently find partners because they are defective in chemotropism (Valtz et al., 1995).
Opaque cells of the C. albicans far1Δ mutant generated in the a/a strain P37005, exhibited mating-associated abnormalities similar to those in S. cerevisiae. Opaque cells of C. albicans far1Δ shmooed in response to α-pheromone (Figure 1, K and L), but the percentage of shmooing was reduced by 57% after 4 h and 81% after 8 h (Figure 1M). The C. albicans far1Δ mutant also exhibited a strong mating defect (99.26% reduction) compared with the parental strain P37005 (Figure 3, J and K). To test whether bilateral mating between far1Δ (a/a) and a far1Δ (α/α) strain completely blocked mating, we generated a far1Δ mutant in the α/α strain WO-1. More than 4000 cells were scanned in mixtures of opaque far1Δ (a/a) and far1Δ (α/α). No mating was observed (data not shown). These results suggest that FAR1 is not essential for shmooing in response to pheromone, although the frequency is reduced, but FAR1 seems to be essential for fusion. Full shmooing and mating responses were restored in the complemented C. albicans strain far1Δ/FAR1 (data not shown).
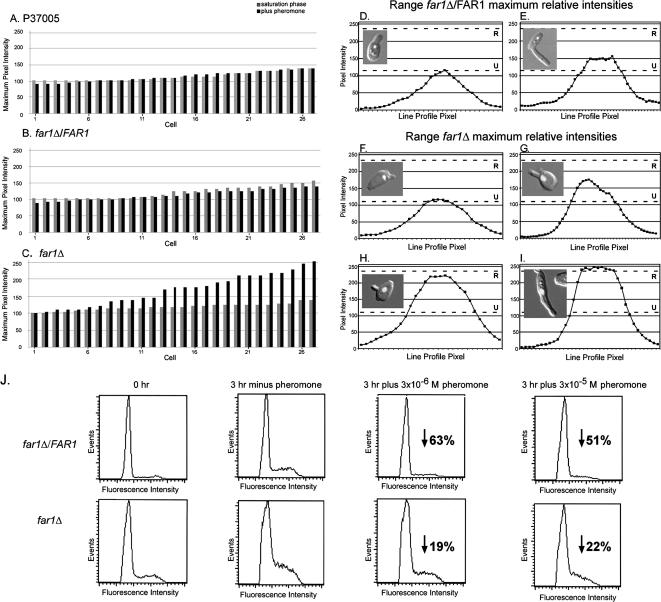
To test whether FAR1 was required for a pheromone-induced block in G1 during shmoo formation in C. albicans, the DNA content of the nuclei of opaque cells of strain P37005, the far1Δ derivative, and the complemented far1Δ/FAR1 strain undergoing shmooing was assessed by measuring the pixel intensity of a line scan through the nucleus of cells stained with Sytox Green according to methods described previously (Zhao et al., 2005a). At saturation phase in liquid culture, the distributions of maximum pixel intensities of the stained nuclei of 27 independently scanned opaque cells of each of strain P37005 (Figure 7A), far1Δ/FAR1 (Figure 7B), and far1Δ (Figure 7C), which in all three cases were primarily unbudded, were similar, ranging between ∼100 and 150 relative units. When opaque cells from saturation phase cultures of P37005, far1Δ/FAR1, or far1Δ were diluted into fresh medium and incubated for 3 h in the absence of α-pheromone, the range of maximum pixel intensities in all cases increased. In the control P37005 or far1Δ/FAR1, the range increased to ∼150–250 relative units, and in far1Δ, to 110–250 relative units (data not shown), indicating a lag in far1Δ cells. If cells from the two control strains P37005 and far1Δ/FAR1 were diluted into fresh medium containing α-pheromone and incubated for 3 h, the increases in DNA content of cells forming shmoos in the population did not occur (Figure 7, A and B). However, if cells from far1Δ were diluted into fresh medium containing α-pheromone, the increase still occurred (Figure 7C). In Figure 7, D and E, examples are presented of representative line scans of the relative DNA content of nuclei of far1Δ/FAR1 cells undergoing shmoo formation in response to pheromone. In Figure 7, F–I, examples are presented of representative line scans of the relative DNA content of nuclei of far1Δ cells undergoing shmoo formation in response to pheromone. Note that the two cells in Figure 7, H and I, are undergoing DNA replication and, hence, are not blocked in G1. Together, these results indicate that just as in S. cerevisiae, FAR1 plays a role in the pheromone-induced G1 block in mating-competent opaque cells.
Figure 7.
Opaque cells of far1Δ, although induced by pheromone to form shmoos, are not arrested in G1. Parental P37005, complemented far1Δ/FAR1, and far1Δ cells were grown to saturation at 25°C in liquid culture (saturation phase), and then they were released into fresh medium in the absence of pheromone (− pheromone) or in the presence of pheromone (+ pheromone) and incubated for 3 h. Cells were then fixed, stained with Sytox Green for DNA, and the intensity of staining in the nucleus of cells forming shmoos quantitated by LSCM (Zhao et al., 2005a). Alternatively, the same cell preparation was analyzed by FACS analysis. (A–C) Distributions of maximum intensity measurements of 27 individual nuclei from saturation phase cells and saturation phase cells diluted into fresh medium containing 3 × 10−6 M α-pheromone and incubated for 3 h, for P37005, far1Δ/FAR1, and far1Δ cells, respectively. (D and E) Examples of line profile scans of the intensity of nuclei of opaque far1Δ/FAR1 cells that formed shmoos, with fluorescent nucleus overlaid on differential interference contrast (DIC) cell images. Similar results were obtained for opaque P37005 cells (data not shown). (F–I) Representative line profile scans of the intensity of nuclei of opaque far1Δ cells that formed shmoos, with fluorescent nucleus overlaid on DIC images. (J) FACS analysis of far1Δ/FAR1 and far1Δ. The percentage of reduction due to the addition of α-pheromone in the proportion of cells undergoing DNA replication is presented at the two tested concentrations of α-pheromone. R, average replicated state estimate; U, average unreplicated state estimate.
We used confocal line scans of individual nuclei because the more common method for assessing cell cycle, FACS, does not allow one to assess the DNA content of a morphologically identified cell. This proved to be an issue because the proportion of cells that formed shmoos differed between saturation phase opaque far1Δ cells and control cells that had been released into fresh medium containing α-pheromone (Figure 1M). Even so, a FACS analysis revealed that when saturation phase far1Δ opaque cells were released into fresh medium containing α-pheromone and incubated for 3 h, a reproducibly higher proportion underwent DNA replication than control far1Δ/FAR1 cells treated similarly. In the absence of pheromone, the proportion of the control population undergoing DNA replication after 3 h was 0.35, and the proportion of far1Δ cells was 0.32 (Figure 7J). In the presence of 3 × 10−6 or 3 × 10−5 M α-pheromone, the proportion in the control population undergoing DNA replication decreased by 63 and 51%, respectively, from that in the absence of pheromone (Figure 7J). For far1Δ cells, the decrease was 19 and 22%, respectively, from that in the absence of pheromone (Figure 7J). Similar results were obtained in repeat experiments. Therefore, the difference in the proportion of opaque cells undergoing DNA replication in the absence and presence of α-pheromone was approximately threefold higher in the far1Δ mutant than in the parental strain (data not shown) or complemented strain (Figure 7J). These results are consistent with the confocal microscopy line scan data of individual cells. However, the small decrease in the proportion undergoing DNA replication that did occur in opaque cells treated with pheromone suggests that regulation other than through FAR1 also has an effect on the cell cycle.
White Cell Pheromone Response of far1Δ
As one would expect given its characteristics, deletion of FAR1 had no effect on the white cell pheromone response. When treated with α-pheromone, white cells of the far1Δ mutant formed large clumps in suspension (Figure 4, H and I), like white parental P37005 cells (Figure 4, A and I), and thick films on plastic surfaces (Figure 5I) with the number of cells adhering comparable with that of films formed by α-pheromone–treated white cells of strain P37005 (Figure 5J). Furthermore, minority opaque α/α cells stimulated approximately a threefold increase in the thickness of majority white cell biofilms of the far1Δ mutant on a silicone elastomer surface (Figure 6, H and M), as they did biofilms of white cells of strain P37005 (Figure 6, A and M). These results demonstrate that Far1p is not involved in the white cell response to pheromone.
Regulation of CPH1 Expression
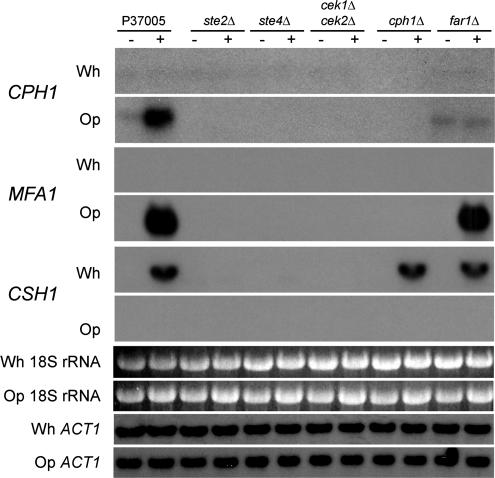
It had been demonstrated previously that CPH1 was up-regulated by α-pheromone in opaque a/a cells (Bennett et al., 2003; Zhao et al., 2005a). In S. cerevisiae, the orthologue to CPH1, STE12, is expressed at a basal level in untreated a cells, and, as in C. albicans, it is up-regulated by α-pheromone (Crosby et al., 2000; Roberts et al., 2000). In the absence of α-pheromone, CPH1 was expressed at a low but reproducible level in opaque cells of strain P37005 (Figure 8). In the presence of α-pheromone, CPH1 was up-regulated (Figure 8). In opaque cells of the mutants ste2Δ, ste4Δ, and cek1Δ cek2Δ, CPH1 expression was undetectable in the absence or presence of pheromone (Figure 8), indicating that basal level expression was dependent on STE2, STE4, and on CEK1 and CEK2. However, CPH1 was expressed at a low basal level in far1Δ cells in the absence of pheromone, indicating that basal level expression of CPH1 was not dependent on FAR1. However, CPH1 expression was not up-regulated in far1Δ cells by pheromone, indicating that up-regulation was dependent on FAR1, as it is in S. cerevisiae (Roberts et al., 2000) (Figure 8). Pheromone induction of CPH1 expression was regained in the complemented strain far1Δ/FAR1 (Supplemental Figure S4).
Figure 8.
Northern analysis of the expression of the downstream opaque cell regulator CPH1, the a-pheromone gene MFA1, and the cell surface hydrophobicity gene CSH1 in the mutant strains ste2Δ, ste4Δ, the double mutant cek1Δ cek2Δ, cph1Δ, and far1Δ. Each gene was probed in white (Wh) and opaque (Op) cells grown to saturation phase in liquid medium at 25°C, and then released and incubated in fresh medium in the absence (−) or presence (+) of α-pheromone for 3 h. White and opaque samples were hybridized in unison on the same blot, and the sequence of images was separated for clarity. It should be noted that longer exposures reveal very low level up-regulation of CSH1 in pheromone-treated ste2Δ and ste4Δ cells, but not cek1Δ cek2Δ, an observation now being explored. To demonstrate equal loading of RNA among lanes, 18S rRNA levels are provided. In addition, hybridization was performed with the white and opaque blots for ACT1, a constitutively expressed actin gene. Furthermore, it should be noted that white and opaque samples were exposed to the probe on the same autoradiograms, scanned, and then digitally separated, so exposure times are equal.
In white cells of strain P37005, CPH1 was expressed at levels that were barely detectable, far below the basal levels observed in opaque cells (Figure 8). In addition, CPH1 was not up-regulated in response to pheromone in white cells (Figure 8). Similar results were obtained for the mutants ste2Δ, ste4Δ, cek1Δ cek2Δ, and far1Δ (Figure 8). These results indicate that the pheromone response pathway functions in the basal level expression of CPH1 in opaque cells, but not white cells.
Molecular Markers for the Alternative Phenotypic Responses to Pheromone
The alternative phenotypic responses to pheromone of white and opaque cells must depend upon the activation of alternative batteries of genes. Activation of such genes must exhibit the same dependencies on the components of the pheromone response pathways as the cell type-specific phenotypic responses. Hence, up-regulation of genes in the opaque battery must depend on an intact pathway from receptor (STE2) through transcription factor (CPH1); alternatively, genes in the white battery must depend on an intact pathway from receptor (STE2) through the MAP kinases (CEK1 and CEK2), but not on CPH1. Genes exhibiting these alternative dependencies were identified.
In S. cerevisiae, there are two genes for the a-pheromone, MFA1 and MFA2 (Gething, 1985; Michaelis and Herskowitz, 1988). The expression of these genes is selectively up-regulated in a cells by α-pheromone (Roberts et al., 2000). C. albicans has only one a-pheromone gene, MFA1, which is also selectively up-regulated in opaque a/a cells by α-pheromone (Dignard et al., 2007). Up-regulation of this gene exhibited the same dependencies on components of the opaque pheromone response pathway as shmooing and mating. MFA1 expression was undetectable by Northern analysis in white cells of strain P37005 and white cells of all tested mutants in the absence or presence of α-pheromone (Figure 8). In opaque P37005 cells, MFA1 was up-regulated by α-pheromone (Figure 8). It was not similarly up-regulated by α-pheromone in the mutants ste2Δ, ste4Δ, cek1Δ cek2Δ, or cph1Δ (Figure 8). However, it was up-regulated by α-pheromone in the mutant far1Δ. MFA1 thus exhibits the dependencies expected of genes up-regulated by pheromone in opaque cells.
Because pheromone induces both cohesiveness and adhesiveness in white cells but not opaque cells, we screened a set of genes encoding cell surface proteins by Northern analysis for an expression pattern consistent with the white cell pheromone response (Sahni, Yi, Srikantha and Soll, unpublished data). The screen revealed that the cell surface hydrophobicity gene CSH1 (Singleton and Hazen, 2004; Singleton et al., 2005) was strongly up-regulated by α-pheromone in white cells, but not opaque cells, of parent strain P37005 (Figure 8). Up-regulation of this gene exhibited the same dependencies on components of the white pheromone response pathway as increased cohesion, adhesion, and biofilm development. CSH1 was expressed at a very low to negligible level in opaque P37005 cells in the absence of α-pheromone, and it was not up-regulated by the addition of pheromone (Figure 8). In white P37005 cells, CSH1 expression was low in the absence of α-pheromone, but it was up-regulated in the presence of α-pheromone (Figure 8). CSH1 was expressed at low levels in white cells of the mutants ste2Δ, ste4Δ, and cek1Δ cek2Δ cells in the absence or presence of α-pheromone, demonstrating that normal pheromone-induced expression depended on the pheromone receptor, heterotrimeric G protein and MAP kinase cascade (Figure 8). However, CSH1, was fully up-regulated by α-pheromone in both the cph1Δ and far1Δ mutants (Figure 8), demonstrating that pheromone-induced expression was independent of CPH1 or FAR1.
Effects of Low and High Concentrations of Pheromone
Suboptimal concentrations of α-pheromone that do not induce shmooing in haploid a cells of S. cerevisiae have been shown to induce invasive growth (Moore, 1983). Therefore, we entertained the possibilities that in C. albicans high concentrations of pheromone may be necessary to induce shmooing in white cells and suboptimal concentrations of α-pheromone may induce the white response or filamentous growth in opaque a/a cells. We discovered early in our studies that the chemically synthesized α-pheromone 13-mer at a concentration of 3 × 10−6 M was sufficient to induce maximum opaque and white cell responses. Furthermore, FACS analysis revealed that concentrations of α-pheromone between 10−5 and 10−6 M caused a maximum G1 block after 3 h (i.e., the DNA of ∼90% of opaque a/a cells of natural strain P37005 remained in the unreplicated state), that a concentration of 10−7 M α-pheromone resulted in <90% of cells in the unreplicated state after 3 h, and that concentrations ranging from 10−8 through 10−10 M, or no added pheromone, resulted in 60–70% of cells in the unreplicated state after 3 h (data not shown). These results supported our use of 3 × 10−6 M α-pheromone as the inducing concentration for both the opaque and white cell responses.
To test whether α-pheromone concentrations higher than 3 × 10−6 M induced shmooing in white cells, we diluted saturation phase white P37005 cells into fresh medium containing a 10-fold higher concentration of α-pheromone, 3 × 10−5 M. FACS analysis revealed no decrease in the proportion of cells undergoing DNA replication (data not shown), and absolutely no shmoo formation (>10,000 white cells were assessed after 3 and 6 h of treatment).
To test whether suboptimal concentrations of pheromone induced filamentation or the white response among opaque cells, saturation phase P37005 opaque cells were diluted into fresh medium containing α-pheromone in the range of 10−7 to 10−10 M. Suboptimal concentrations did not induce cohesion in suspension or adhesion to plastic, and they did not induce either pseudohypha or hypha formation (data not shown). These results demonstrate that high concentrations of pheromone do not block white cells in G1 or induce shmoo formation, and low concentrations do not induce opaque cells to undergo the white cell response or filamentation.
DISCUSSION
We demonstrated previously that the pheromones released by opaque C. albicans cells to induce mating responses in opaque cells of opposite mating type also signal white cells to become both cohesive and adhesive so that they can more readily form incipient biofilms that then develop into mature biofilms twice as thick as those formed by untreated white cells (Daniels et al., 2006). We hypothesized previously, based on in vitro results, that these white cell biofilms might function as protective environments that facilitate mating between minority opaque cells of opposite mating type in nature (Daniels et al., 2006). Using biotinylated α-pheromone, we further demonstrated that white cells bound pheromone to their surfaces and that the binding of pheromone then down-regulated the receptors (Daniels et al., 2006). However, the staining pattern of receptors on the surface differed between white and opaque cells, and reappearance of receptors after down-regulation occurred only in opaque cells (Daniels et al., 2006). Here, we have presented evidence that the same pheromone signal, receptor, heterotrimeric G protein, and MAP kinase cascade, but different downstream regulators, mediate the disparate pheromone responses of opaque and white cells of C. albicans.
In the fungi, including plant pathogens, there are numerous examples of the conservation of components of MAP kinase cascades in different pathways regulating a variety of responses, but in the majority of cases, the signals, receptors, and differing numbers of components along the transduction pathways are distinct (Banuett, 1998; Xu, 2000). In S. cerevisiae, the signals and receptors triggering the mating response of the alternative mating types differ, as also occurs in other yeast (Leberer et al., 1997; Elion, 2000; Davidson et al., 2003; Li et al., 2005). In S. cerevisiae, the signals, receptors, and the majority of components of the signal transduction pathways differ among several environmental responses, including the mating response to pheromones, filamentation, ascospore formation, and osmoregulation (Gustin et al., 1998; Chen and Thorner, 2007). However, we found no definitive example in other fungi of the scenario we have described for white and opaque cells of C. albicans, specifically, that the same signal interacts with the same receptor, activating the same upstream signal transduction pathway, but different downstream regulators in two different cell types, resulting in completely different responses. This is not to say that this scenario does not exist in other fungi, but simply that it has not yet been fully described, if it does. However, in higher eukaryotes, there are several examples of this signaling scenario. For example, both CD4+ and CD8+ T cells respond to the same mitogenic signals, and through the same T cell receptor, MAP kinase kinase (MAPKK) (MKK4/7) and MAP kinase (MAPK) (JNK2), induce interleukin (IL)-2 expression in CD4+ cells, but repress IL-2 expression in CD8+ cells (Rincón and Pedraza-Alva, 2003). In addition, both cell types transduce the same signal through the same receptor, MAPKK (MKK4/7) and MAPK (p38), but the response in CD4+ cells is survival, whereas that in CD8+ cells is apoptosis (Rincón and Pedraza-Alva, 2003). There seem to be a variety of additional examples in higher eukaryotes, especially in developing systems (Bacci et al., 2005; Dailey et al., 2005), of the same signal, receptor, and upstream components in the transduction pathway, but different downstream regulators in different cell types.
Common Upstream Components
Although we have presented evidence that the same upstream components, including receptors, heterotrimeric G protein, and MAP kinase cascade, are shared in the pheromone response pathways of opaque and white cells (Figure 9), we cannot exclude the possibility that additional, parallel pathways involved in the alternative pheromone responses are activated by the same or even different receptors. We can also not exclude the possibility that CEK1 and CEK2 have nonoverlapping and overlapping functions in the opaque and white responses, given that the effects of neither cek1Δ nor cek2Δ were complete for the majority of assayed pheromone responses in both white and opaque cells, in comparison with the double mutant cek1Δ cek2Δ. The cek1Δ mutant exhibited stronger defects than the cek2Δ mutant in a majority of the measured responses. Therefore, the individual roles of CEK1 or CEK2 remain to be elucidated.
Figure 9.
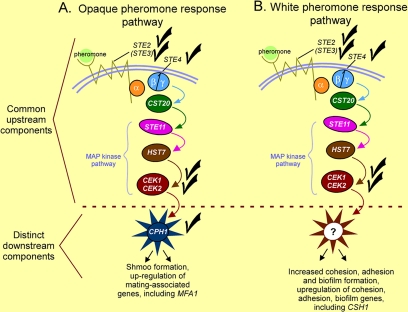
Comparison of the pheromone response pathways of opaque cells (A) and white cells (B). The roles of some of the components (the trimeric G protein α and γ subunits and CST20) in the models were not analyzed here, but they were inferred from the conserved and more thoroughly studied pheromone response pathway of S. cerevisiae. The distinctive points of the comparison are that the components of pathways from receptor through the MAP kinase cascade are shared, whereas the terminal regulatory component, the response-specific transcription factor, differs. Although CPH1 is the downstream regulator in the opaque pheromone response pathway, the downstream regulator in the white pheromone response pathway remains unidentified, hence the question mark. Although FAR1 plays a major role as a downstream regulator in the opaque pheromone response pathway, its functions have no analogies in the white pheromone response pathway, so it has not been represented in the opaque model.
Difference in Downstream Component(s)
Our evidence demonstrates that although the transcription factor Cph1p represents the downstream transcription factor in the opaque pheromone response pathway, it does not seem to play any distinct role in the white pheromone response pathway (Figure 9). Here, we have demonstrated that although MFA1 is up-regulated by the opaque pheromone response pathway through CPH1, CSH1 is up-regulated by the same pheromone response pathway, but through a different downstream transcription regulator, which is yet to be identified. Recent experiments, using both expression arrays, and Northern analyses of both putative cell surface adhesion molecules and transcription factors, have revealed additional pheromone-induced genes in white cells, the up-regulation of which depends on the same pathway that regulates CSH1 (Sahni, Yi, Srikantha, and Soll, unpublished data). Identification of the downstream transcription factor in the white pheromone response pathway represents our immediate challenge.
FAR1, which encodes a cyclin-dependent kinase inhibitor (Chenevert et al., 1994; Valtz et al., 1995), proved to play no apparent role in the white cell response. This was expected because white cells neither become blocked in G1 nor form shmoos in response to pheromone, the former response dependent on FAR1 and the latter influenced by FAR1 both in S. cerevisiae and C. albicans.
Regulation of CPH1
As is the case for the orthologue STE12 in haploid S. cerevisiae (Roberts et al., 2000), CPH1 is up-regulated by pheromone in opaque cells of C. albicans (Zhao et al., 2005a). Here, we have shown that it is not similarly up-regulated by pheromone in white cells. We have found that CPH1 is expressed at a basal level in opaque cells in the absence of pheromone and that basal expression depends on a functional receptor, heterotrimeric G protein, and MAP kinase cascade, as was indicated by the results of Roberts et al. (2000). However, basal expression of CPH1 is not dependent on FAR1. Therefore, the possibility must be considered that basal expression of CPH1 may depend on a complete opaque pheromone response pathway, including CPH1, in which case, expression would depend on autoregulation at the level of transcription.
In contrast, pheromone induction of CPH1 expression in opaque cells is dependent on FAR1, just as is pheromone induction of STE12, its orthologue in S. cerevisiae (see supplement to Roberts et al., 2000). However, this result is paradoxical both for S. cerevisiae and C. albicans. If the target transcription factor of the pheromone response pathway is not up-regulated by pheromone in the far1Δ mutant of both haploid S. cerevisiae and MTL-homozygous C. albicans cells, how does pheromone induce shmoo formation in a significant proportion of far1Δ cells? The answer may lie in posttranslational modification or stability. STE12 activity has been demonstrated to be enhanced posttranslationally by pheromone through FUS3-mediated phosphorylation (Elion et al., 1993) and its stability decreased through the effect of pheromone on ubiquitin-mediated degradation (Esch et al., 2006).
Evolutionary Implications of the Pheromone Response Pathways of C. albicans
We noted previously (Daniels et al., 2006) that signaling of mating-incompetent white cells by mating-competent opaque cells to form a biofilm that facilitates mating, at least in vitro, was very much akin to the types of inductive events in embryogenic development, most notably between germ cells and somatically derived follicle cells (Gilchrist et al., 2004). Our demonstration here that white and opaque cells respond to the same signal through the same receptor, heterotrimeric G protein, and MAP kinase cascade, but different target transcription factors (Figure 9), reveals a configuration more common in higher eukaryotes (Rincón and Pedraza-Alva, 2003; Bacci et al., 2005), adding support to the suggestion (Daniels et al., 2006) that the interactions between opaque and white cells may represent an antecedent to higher eukaryotic multicellularity.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant AI-2392 and the Developmental Studies Hybridoma Bank. We acknowledge The University of Iowa for use of the W. M. Keck Dynamic Image Analysis Facility. We are indebted to Julie Collins for assistance in assembling the manuscript and to Dr. Joachim Morschhaüser for the generous gifts of plasmids.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-07-0688) on January 2, 2008.
REFERENCES
- Alonso-Monge R., Navarro-Garcia F., Molero G., Diez-Orejas R., Gustin M., Pla J., Sanchez M., Nombela C. Role of the mitogen-activated protein kinase Hog1p in morphogenesis and virulence of Candida albicans. J. Bacteriol. 1999;181:3058–3068. doi: 10.1128/jb.181.10.3058-3068.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J. M., Soll D. R. Unique phenotype of opaque cells in the white-opaque transition of Candida albicans. J. Bacteriol. 1987;169:5579–5588. doi: 10.1128/jb.169.12.5579-5588.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacci A., Huguenard J. R., Prince D. A. Modulation of neocortical interneurons: extrinsic influences and exercises in self-control. Trends Neurosci. 2005;28:602–610. doi: 10.1016/j.tins.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Bahn Y. S., Kojima K., Cox G. M., Heitman J. Specialization of the HOG pathway and its impact on differentiation and virulence of Cryptococcus neoformans. Mol. Biol. Cell. 2005;16:2285–2300. doi: 10.1091/mbc.E04-11-0987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balázsi G., Oltvai Z. N. Sensing your surroundings: how transcription-regulatory networks of the cell discern environmental signals. Sci. STKE. 2005;2005:e20. doi: 10.1126/stke.2822005pe20. [DOI] [PubMed] [Google Scholar]
- Banuett F. Signalling in the yeasts: an informational cascade with links to the filamentous fungi. Microbiol. Mol. Biol. Rev. 1998;62:249–274. doi: 10.1128/mmbr.62.2.249-274.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedell G. W., Soll D. R. Effects of low concentrations of zinc on the growth and dimorphism of Candida albicans: evidence for zinc-resistant and -sensitive pathways for mycelium formation. Infect. Immun. 1979;26:348–354. doi: 10.1128/iai.26.1.348-354.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender A., Sprague G. F., Jr Yeast peptide pheromones, a-factor and alpha-factor, activate a common response mechanism in their target cells. Cell. 1986;47:929–937. doi: 10.1016/0092-8674(86)90808-1. [DOI] [PubMed] [Google Scholar]
- Bennett R. J., Uhl M. A., Miller M. G., Johnson A. D. Identification and characterization of a Candida albicans mating pheromone. Mol. Cell. Biol. 2003;23:8189–8201. doi: 10.1128/MCB.23.22.8189-8201.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butty A. C., Pryciak P. M., Huang L. S., Herskowitz I., Peter M. The role of Far1p in linking the heterotrimeric G protein to polarity establishment proteins during yeast mating. Science. 1998;282:1511–1516. doi: 10.1126/science.282.5393.1511. [DOI] [PubMed] [Google Scholar]
- Chang F., Herskowitz I. Identification of a gene necessary for cell cycle arrest by a negative growth factor of yeast: FAR1 is an inhibitor of a G1 cyclin, CLN2. Cell. 1990;63:999–1011. doi: 10.1016/0092-8674(90)90503-7. [DOI] [PubMed] [Google Scholar]
- Chen J., Chen J., Lane S., Liu H. A conserved mitogen-activated protein kinase pathway is required for mating in Candida albicans. Mol. Microbiol. 2002;46:1335–1344. doi: 10.1046/j.1365-2958.2002.03249.x. [DOI] [PubMed] [Google Scholar]
- Chen P., Choi J. D., Wang R., Cotter R. J., Michaelis S. A novel a-factor-related peptide of Saccharomyces cerevisiae that exits the cell by a Ste6p-independent mechanism. Mol. Biol. Cell. 1997a;8:1273–1291. doi: 10.1091/mbc.8.7.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Sapperstein S. K., Choi J. D., Michaelis S. Biogenesis of the Saccharomyces cerevisiae mating pheromone a-factor. J. Cell Biol. 1997b;136:251–269. doi: 10.1083/jcb.136.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R. E., Thorner J. Function and regulation in MAPK signaling pathways: lessons learned from the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2007;1773:1311–1340. doi: 10.1016/j.bbamcr.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenevert J., Valtz N., Herskowitz I. Identification of genes required for normal pheromone-induced cell polarization in Saccharomyces cerevisiae. Genetics. 1994;136:1287–1296. doi: 10.1093/genetics/136.4.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby J. A., Konopka J. B., Fields S. Constitutive activation of the Saccharomyces cerevisiae transcriptional regulator Ste12p by mutations at the amino-terminus. Yeast. 2000;16:1365–1375. doi: 10.1002/1097-0061(200011)16:15<1365::AID-YEA630>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Csank C., Schroppel K., Leberer E., Harcus D., Mohamed O., Meloche S., Thomas D. Y., Whiteway M. Roles of the Candida albicans mitogen-activated protein kinase homolog, Cek1p, in hyphal development and systemic candidiasis. Infect. Immun. 1998;66:2713–2721. doi: 10.1128/iai.66.6.2713-2721.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey L., Ambrosetti D., Mansukhani A., Basilico C. Mechanisms underlying differential responses to FGF signaling. Cytokine Growth Factor Rev. 2005;16:233–247. doi: 10.1016/j.cytogfr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Daniels K. J., Srikantha T., Lockhart S. R., Pujol C., Soll D. R. Opaque cells signal white cells to form biofilms in Candida albicans. EMBO J. 2006;25:2240–2252. doi: 10.1038/sj.emboj.7601099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson R. C., Nichols C. B., Cox G. M., Perfect J. R., Heitman J. A MAP kinase cascade composed of cell type specific and non-specific elements controls mating and differentiation of the fungal pathogen Cryptococcus neoformans. Mol. Microbiol. 2003;49:469–485. doi: 10.1046/j.1365-2958.2003.03563.x. [DOI] [PubMed] [Google Scholar]
- De Backer M. D., Maes D., Vandoninck S., Logghe M., Contreras R., Luyten W. H. Transformation of Candida albicans by electroporation. Yeast. 1999;15:1609–1618. doi: 10.1002/(sici)1097-0061(199911)15:15<1609::aid-yea485>3.3.co;2-p. [DOI] [PubMed] [Google Scholar]
- Dignard D., El Naggar A. L., Logue M. E., Butler G., Whiteway M. Identification and characterization of MFA1, the gene encoding Candida albicans a-factor pheromone. Eukaryot. Cell. 2007;6:487–494. doi: 10.1128/EC.00387-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon K. P., Xu J. R., Smirnoff N., Talbot N. J. Independent signaling pathways regulate cellular turgor during hyperosmotic stress and appressorium-mediated plant infection by Magnaporthe grisea. Plant Cell. 1999;11:2045–2058. doi: 10.1105/tpc.11.10.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorer R., Pryciak P. M., Hartwell L. H. Saccharomyces cerevisiae cells execute a default pathway to select a mate in the absence of pheromone gradients. J. Cell Biol. 1995;131:845–861. doi: 10.1083/jcb.131.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsky R. I., Moon R. T., Raible D. W. Environmental signals and cell fate specification in premigratory neural crest. Bioessays. 2000;22:708–716. doi: 10.1002/1521-1878(200008)22:8<708::AID-BIES4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Elion E. A. Pheromone response, mating and cell biology. Curr. Opin. Microbiol. 2000;3:573–581. doi: 10.1016/s1369-5274(00)00143-0. [DOI] [PubMed] [Google Scholar]
- Elion E. A., Brill J. A., Fink G. R. FUS3 represses CLN1 and CLN2 and in concert with KSS1 promotes signal transduction. Proc. Natl. Acad. Sci. USA. 1991;88:9392–9396. doi: 10.1073/pnas.88.21.9392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion E. A., Satterberg B., Kranz J. E. FUS3 phosphorylates multiple components of the mating signal transduction cascade: evidence for STE12 and FAR1. Mol. Biol. Cell. 1993;4:495–510. doi: 10.1091/mbc.4.5.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esch R. K., Wang Y., Errede B. Pheromone-induced degradation of Ste12 contributes to signal attenuation and the specificity of developmental fate. Eukaryot. Cell. 2006;5:2147–2160. doi: 10.1128/EC.00270-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gething M.-J. Cold Spring Harbor Laboratory. New York: Cold Spring Harbor; 1985. Protein transport and secretion. [Google Scholar]
- Gilchrist R. B., Ritter L. J., Armstrong D. T. Oocyte-somatic cell interactions during follicle development in mammals. Anim. Reprod. Sci. 82–. 2004;83:431–446. doi: 10.1016/j.anireprosci.2004.05.017. [DOI] [PubMed] [Google Scholar]
- Gustin M. C., Albertyn J., Alexander M., Davenport K. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1998;62:1264–1300. doi: 10.1128/mmbr.62.4.1264-1300.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull C. M., Raisner R. M., Johnson A. D. Evidence for mating of the “asexual” yeast Candida albicans in a mammalian host. Science. 2000;289:307–310. doi: 10.1126/science.289.5477.307. [DOI] [PubMed] [Google Scholar]
- Huyer G., Kistler A., Nouvet F. J., George C. M., Boyle M. L., Michaelis S. Saccharomyces cerevisiae a-factor mutants reveal residues critical for processing, activity, and export. Eukaryot. Cell. 2006;5:1560–1570. doi: 10.1128/EC.00161-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus P. R., Fox D. S., Cox G. M., Heitman J. The Cryptococcus neoformans MAP kinase Mpk1 regulates cell integrity in response to antifungal drugs and loss of calcineurin function. Mol. Microbiol. 2003;48:1377–1387. doi: 10.1046/j.1365-2958.2003.03508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruppa M., Calderone R. Two-component signal transduction in human fungal pathogens. FEMS Yeast Res. 2006;6:149–159. doi: 10.1111/j.1567-1364.2006.00024.x. [DOI] [PubMed] [Google Scholar]
- Leberer E., Thomas D. Y., Whiteway M. Pheromone signalling and polarized morphogenesis in yeast. Curr. Opin. Genet. Dev. 1997;7:59–66. doi: 10.1016/s0959-437x(97)80110-4. [DOI] [PubMed] [Google Scholar]
- Levin D. E., Errede B. The proliferation of MAP kinase signaling pathways in yeast. Curr. Opin. Cell Biol. 1995;7:197–202. doi: 10.1016/0955-0674(95)80028-x. [DOI] [PubMed] [Google Scholar]
- Li D., Bobrowicz P., Wilkinson H. H., Ebbole D. J. A mitogen-activated protein kinase pathway essential for mating and contributing to vegetative growth in Neurospora crassa. Genetics. 2005;170:1091–1104. doi: 10.1534/genetics.104.036772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Kohler J., Fink G. R. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266:1723–1726. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- Lockhart S. R., Daniels K. J., Zhao R., Wessels D., Soll D. R. Cell biology of mating in Candida albicans. Eukaryot. Cell. 2003a;2:49–61. doi: 10.1128/EC.2.1.49-61.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart S. R., Pujol C., Daniels K. J., Miller M. G., Johnson A. D., Pfaller M. A., Soll D. R. In Candida albicans, white-opaque switchers are homozygous for mating type. Genetics. 2002;162:737–745. doi: 10.1093/genetics/162.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart S. R., Zhao R., Daniels K. J., Soll D. R. Alpha-pheromone-induced “shmooing” and gene regulation require white-opaque switching during Candida albicans mating. Eukaryot. Cell. 2003b;2:847–855. doi: 10.1128/EC.2.5.847-855.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani H. D., Fink G. R. The riddle of MAP kinase signaling specificity. Trends Genet. 1998;14:151–155. doi: 10.1016/s0168-9525(98)01425-5. [DOI] [PubMed] [Google Scholar]
- Magee B. B., Legrand M., Alarco A. M., Raymond M., Magee P. T. Many of the genes required for mating in Saccharomyces cerevisiae are also required for mating in Candida albicans. Mol. Microbiol. 2002;46:1345–1351. doi: 10.1046/j.1365-2958.2002.03263.x. [DOI] [PubMed] [Google Scholar]
- Magee B. B., Magee P. T. Induction of mating in Candida albicans by construction of MTLa and MTLα strains. Science. 2000;289:310–313. doi: 10.1126/science.289.5477.310. [DOI] [PubMed] [Google Scholar]
- Michaelis S., Herskowitz I. The a-factor pheromone of Saccharomyces cerevisiae is essential for mating. Mol. Cell. Biol. 1988;8:1309–1318. doi: 10.1128/mcb.8.3.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. G., Johnson A. D. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell. 2002;110:293–302. doi: 10.1016/s0092-8674(02)00837-1. [DOI] [PubMed] [Google Scholar]
- Moore S. A. Comparison of dose-response curves for alpha factor-induced cell division arrest, agglutination, and projection formation of yeast cells. Implication for the mechanism of alpha factor action. J. Biol. Chem. 1983;258:13849–13856. [PubMed] [Google Scholar]
- Navarro-Garcia F., Alonso-Monge R., Rico H., Pla J., Sentandreu R., Nombela C. A role for the MAP kinase gene MKC1 in cell wall construction and morphological transitions in Candida albicans. Microbiology. 1998;144:411–424. doi: 10.1099/00221287-144-2-411. [DOI] [PubMed] [Google Scholar]
- Panwar S. L., Legrand M., Dignard D., Whiteway M., Magee P. T. MFα1, the gene encoding the alpha mating pheromone of Candida albicans. Eukaryot. Cell. 2003;2:1350–1360. doi: 10.1128/EC.2.6.1350-1360.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuss O., Vik A., Kolter R., Morschhauser J. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene. 2004;341:119–127. doi: 10.1016/j.gene.2004.06.021. [DOI] [PubMed] [Google Scholar]
- Rincón M., Pedraza-Alva G. JNK and p38 MAP kinases in CD4+ and CD8+ T cells. Immunol. Rev. 2003;192:131–142. doi: 10.1034/j.1600-065x.2003.00019.x. [DOI] [PubMed] [Google Scholar]
- Roberts C. J., Nelson B., Marton M. J., Stoughton R., Meyer M. R., Bennett H. A., He Y. D., Dai H., Walker W. L., Hughes T. R., Tyers M., Boone C., Friend S. H. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science. 2000;287:873–880. doi: 10.1126/science.287.5454.873. [DOI] [PubMed] [Google Scholar]
- Saito H., Tatebayashi K. Regulation of the osmoregulatory HOG MAPK cascade in yeast. J. Biochem. 2004;136:267–272. doi: 10.1093/jb/mvh135. [DOI] [PubMed] [Google Scholar]
- Sherman F., Fink G. R., Hicks J. B. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1986. Laboratory Course Manual for Methods in Yeast Genetics. [Google Scholar]
- Singleton D. R., Fidel P. L., Jr, Wozniak K. L., Hazen K. C. Contribution of cell surface hydrophobicity protein 1 (Csh1p) to virulence of hydrophobic Candida albicans serotype A cells. FEMS Microbiol. Lett. 2005;244:373–377. doi: 10.1016/j.femsle.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Singleton D. R., Hazen K. C. Differential surface localization and temperature-dependent expression of the Candida albicans CSH1 protein. Microbiology. 2004;150:285–292. doi: 10.1099/mic.0.26656-0. [DOI] [PubMed] [Google Scholar]
- Slutsky B., Staebell M., Anderson J., Risen L., Pfaller M., Soll D. R. “White-opaque transition”: a second high-frequency switching system in Candida albicans. J. Bacteriol. 1987;169:189–197. doi: 10.1128/jb.169.1.189-197.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. A., Nicholls S., Morgan B. A., Brown A. J., Quinn J. A conserved stress-activated protein kinase regulates a core stress response in the human pathogen Candida albicans. Mol. Biol. Cell. 2004;15:4179–4190. doi: 10.1091/mbc.E04-03-0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll D. R., Daniels K. J. MAT, mating, switching, and pathogenesis in Candida albicans, Candida dubliniensis and Candida glabrata. In: Heitman J., Kronstad J. W., Taylor J. W., Casselton L. A., editors. Sex in Fungi. Washington, DC: ASM Press; 2007. pp. 215–234. [Google Scholar]
- Sprague G. F., Jr, Blair L. C., Thorner J. Cell interactions and regulation of cell type in the yeast Saccharomyces cerevisiae. Annu. Rev. Microbiol. 1983;37:623–660. doi: 10.1146/annurev.mi.37.100183.003203. [DOI] [PubMed] [Google Scholar]
- Srikantha T., Borneman A. R., Daniels K. J., Pujol C., Wu W., Seringhaus M. R., Gerstein M., Yi S., Snyder M., Soll D. R. TOS9 regulates white-opaque switching in Candida albicans. Eukaryot. Cell. 2006;5:1674–1687. doi: 10.1128/EC.00252-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valtz N., Peter M., Herskowitz I. FAR1 is required for oriented polarization of yeast cells in response to mating pheromones. J. Cell Biol. 1995;131:863–873. doi: 10.1083/jcb.131.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J. R. MAP kinases in fungal pathogens. Fungal Genet. Biol. 2000;31:137–152. doi: 10.1006/fgbi.2000.1237. [DOI] [PubMed] [Google Scholar]
- Zhao R., Daniels K. J., Lockhart S. R., Yeater K. M., Hoyer L. L., Soll D. R. Unique aspects of gene expression during Candida albicans mating and possible G(1) dependency. Eukaryot. Cell. 2005a;4:1175–1190. doi: 10.1128/EC.4.7.1175-1190.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Kim Y., Park G., Xu J. R. A mitogen-activated protein kinase cascade regulating infection-related morphogenesis in Magnaporthe grisea. Plant Cell. 2005b;17:1317–1329. doi: 10.1105/tpc.104.029116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zordan R. E., Galgoczy D. J., Johnson A. D. Epigenetic properties of white-opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop. Proc. Natl. Acad. Sci. USA. 2006;103:12807–12812. doi: 10.1073/pnas.0605138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.