Abstract
Phosphatidylinositol (PI) is a component of membrane phospholipids, and it functions both as a signaling molecule and as a compartment-specific localization signal in the form of polyphosphoinositides. Arachidonic acid (AA) is the predominant fatty acid in the sn-2 position of PI in mammals. LysoPI acyltransferase (LPIAT) is thought to catalyze formation of AA-containing PI; however, the gene that encodes this enzyme has not yet been identified. In this study, we established a screening system to identify genes required for use of exogenous polyunsaturated fatty acids (PUFAs) in Caenorhabditis elegans. In C. elegans, eicosapentaenoic acid (EPA) instead of AA is the predominant fatty acid in PI. We showed that an uncharacterized gene, which we named mboa-7, is required for incorporation of PUFAs into PI. Incorporation of exogenous PUFA into PI of the living worms and LPIAT activity in the microsomes were greatly reduced in mboa-7 mutants. Furthermore, the membrane fractions of transgenic worms expressing recombinant MBOA-7 and its human homologue exhibited remarkably increased LPIAT activity. mboa-7 encodes a member of the membrane-bound O-acyltransferase family, suggesting that mboa-7 is LPIAT. Finally, mboa-7 mutants had significantly lower EPA levels in PI, and they exhibited larval arrest and egg-laying defects.
INTRODUCTION
Various kinds of fatty acids are distributed in membrane phospholipids in mammalian cells and tissues (Lands and Crawford, 1976; Holub and Kuksis, 1978; MacDonald and Sprecher, 1991). The fatty acyl residues of individual phospholipids seem to be under strict metabolic regulation. In general, saturated fatty acids are esterified at the sn-1 position, whereas polyunsaturated fatty acids (PUFAs), such as arachidonic acid (AA), are commonly found at the sn-2 position. Three-fourths or more of the phosphatidylinositol (PI) fraction in rat liver and brain constitutes the 1-stearoyl-2-arachidonoyl species (Holub and Kuksis, 1971; Baker and Thompson, 1972). In contrast, the total pool of precursor phosphatidic acid (PA) in rat liver and brain has a fatty acid composition that does not resemble that of PI, showing a low AA content (Possmayer et al., 1969; Akesson et al., 1970; Baker and Thompson, 1972). Selectivity could be expressed during de novo synthesis at the level of formation of cytidine 5′-diphosphate (CDP)-diacylglycerol from PA and cytidine 5′-triphosphate or in the use of CDP-diacylglycerol in the final reaction. In fact, CDP-diacylglycerol synthase, which prefers 1-stearoyl-2-arachidonoyl PA as a substrate in vitro, has been cloned, although expression is restricted to testis, retina, and brain (Saito et al., 1997).
An alternative mechanism for species selection has been proposed on the basis of turnover studies in rat brain in vivo (Baker and Thompson, 1972). [3H]AA and [14C]glycerol injected intracerebrally were incorporated almost exclusively into brain phospholipids. Comparison of PI and PA radioactivity suggest that the initial flux of AA into PI was independent of de novo synthesis from PA. The rapid turnover of the sn-2 acyl chains of glycerophospholipids was originally described by Lands (Lands, 1958; Lands, 1960; Lands and Merkl, 1963; Merkl and Lands, 1963), who proposed that acyl moieties of membrane phospholipids are rapidly metabolized by the action of phospholipase A2s and subsequently by lysophospholipid acyltransferases (Lands' cycle). To date, at least two lysophosphatidylcholine acyltransferases (LPCAT) were cloned from mammals: LPCAT1) catalyzes the incorporation of saturated fatty acids into lysophosphatidylcholine (lysoPC) (Chen et al., 2006; Nakanishi et al., 2006), and LPCAT2 catalyzes the incorporation of acetic acid and AA into lysoPC (Shindou et al., 2007). These enzymes have conserved motifs in common with the enzymes involved in the de novo phospholipid synthesis pathway (Kennedy pathway) such as acylglycerolphosphate acyltransferase and lysophosphatidic acid acyltransferase (LPAAT). Very recently, a novel lysophospholipid acyltransferase was identified from yeast that catalyzes the transfer of unsaturated fatty acids into most major lysophospholipids (Benghezal et al., 2007; Jain et al., 2007; Riekhof et al., 2007; Tamaki et al., 2007). In contrast to LPCAT1 and LPCAT2, this enzyme harbors a membrane-bound O-acyltransferase (MBOAT) motif present in a variety of acyltransferases of bacteria and mammals (Hofmann, 2000). Recent work on the lipid remodeling of glycosylphosphatidylinositol (GPI) anchors in yeast showed that Gup1p is required for the addition of a C26 fatty acid in the sn-2 position of GPI anchors (Bosson et al., 2006). Gup1p is also a member of MBOAT family.
High levels of lysophosphatidylinositol acyltransferase (LPIAT) and LPCAT have been detected in the microsomal fraction of various mammalian tissues and cells (Keenan and Hokin, 1962; Keenan and Hokin, 1964; Inoue et al., 1984). These experiments led to speculation that deacylation–reacylation reactions are important in establishing fatty acid pairing, and particularly in regulating the major molecular species of PI (Baker and Thompson, 1973; Holub, 1976). LPCAT and LPIAT show different substrate specificities for acyl donors, and they can be separated chromatographically (Sanjanwala et al., 1989; Yamashita et al., 2003), indicating that LPIAT and LPCAT are distinct enzymes. However, the gene(s) involved in LPIAT reaction have not been reported.
C. elegans, a free-living nematode, is a good model for studying the physiological functions of unsaturated fatty acids. Unlike mammals, C. elegans possesses all the required fatty acid biosynthetic enzymes, and it has no dietary requirements for essential fatty acids (Napier and Michaelson, 2001; Wallis et al., 2002). Moreover, a series of mutants have been isolated that are unable to synthesize specific PUFAs due to mutations in genes encoding fatty acid desaturases (fat genes, Watts and Browse, 2002). C. elegans membrane fatty acids can be manipulated by dietary supplementation with fatty acids (Watts et al., 2003; Watts and Browse, 2006). In contrast to mammals in which AA is a major PUFA in membrane phospholipids and eicosapentaenoic acid (EPA) is a minor component, C. elegans possesses abundant EPA but not AA (Satouchi et al., 1993; Wallis et al., 2002; Watts and Browse, 2002). The major PUFA attached to PI in wild-type worms is EPA and not AA. However, AA becomes an abundant PUFA in PI in fat-1 mutants in which no n-3 PUFAs, including EPA, are produced (our unpublished data).
We used C. elegans fat mutants to establish a screening system to identify genes required for the incorporation of exogenous PUFAs into endogenous phospholipids. The fat-4 fat-1 double mutants lack both n-3 and Δ5 fatty acid desaturation activities; therefore, they fail to produce AA (20:4n-6) and n-3 PUFAs such as EPA (20:5n-3) (Kahn-Kirby et al., 2004). The fat-4 fat-1 mutants exhibit slow growth at low temperatures, and the growth defects are rescued by dietary supplementation with AA or EPA. Because it has generally been assumed that PUFAs such as AA and EPA are incorporated into phospholipids via a deacylation–reacylation remodeling pathway rather than a de novo synthesis pathway, it was reasonable to hypothesize that RNA interference (RNAi)-mediated knockdown of genes involved in the incorporation of exogenous PUFAs into phospholipids via the remodeling pathway would cause growth retardation even in the presence of PUFAs in the medium. On the basis of this assumption, we performed comprehensive RNAi analysis with fat-4 fat-1 mutants. In the present work, we showed that a previously uncharacterized gene encoding a 453-amino acid protein belonging to the MBOAT family is required for the incorporation of PUFA into PI.
MATERIALS AND METHODS
Materials
[1-14C]Stearic acid, [1-14C]oleic acid, [1-14C]linoleic acid, [1-14C]linolenic acid, [1-14C]eicosatrienoic acid, [1-14C]eicosatetraenoic acid, [1-14C]arachidonic acid, [1-14C]eicosapentaenoic acid, and [1-14C]arachidonoyl-coenzyme A (CoA) were purchased from American Radiolabeled Chemicals (St. Louis, MO). PI and lysoPI from porcine liver were purchased from Serdary Research Laboratories (London, ON, Canada). Lyso-phosphatidylserine (PS) from porcine brain, lysoPE from porcine liver, lysoPC from egg yolk, and sn-1-oleoyl-lysoPA were purchased from Avanti Polar Lipids (Alabaster, AL). AA and EPA were purchased from Cayman Chemical (Ann Arbor, MI). 1,2-Dipalmitoyl PI was purchased from Cayman Chemical. Rhizopus delemer lipase was purchased from Seikagaku (Tokyo, Japan). Phospholipase A2 from honey bee venom was purchased from Sigma-Aldrich (St. Louis, MO).
General Methods and Strains
Maintenance and genetic manipulation of C. elegans were carried out as described previously (Brenner, 1974). The C. elegans wild-type strain was Bristol N2, and Escherichia coli OP50, a bacterium that does not synthesize or require PUFAs (Tanaka et al., 1996), was used as the sole food source. The following mutations were used: fat-1(wa9), fat-3(wa22), fat-4(wa14) (Watts and Browse, 2002), eri-1(mg366) (Kennedy et al., 2004), and mboa-7(gk399). mboa-7(gk399) mutation was backcrossed at five times before further analysis.
Large-Scale RNAi Screening for Genes Involved in Incorporation of Exogenous AA into Phospholipids
Feeding RNAi was performed as described previously (Kamath et al., 2001). In total, 5720 bacterial RNAi feeding strains from the Ahringer library (Ashrafi et al., 2003) were tested with wild-type, fat-4 fat-1, and fat-3 fat-1 mutants under 25 μM AA-supplemented conditions at 15°C. We selected RNAi clones that caused growth defects only in fat-4 fat-1 and/or fat-3 fat-1 mutants even under AA-supplemented conditions. We used AA instead of EPA as an exogenous PUFA source due to its lower cost. Because fat-4 fat-1 mutants lacking endogenous AA and EPA do not exhibit embryonic or larval lethality, phospholipids having these fatty acyl chains seemed not to be essential for survival of C. elegans. Therefore, we excluded genes required for survival in wild-type animals. For dietary supplementation of plates with AA or EPA, an 80 mM stock solution of fatty acids in ethanol was added to nematode growth medium (NGM) at a final concentration of 25 μM. Plates were poured, covered, and allowed to dry in the dark at room temperature overnight. All screenings were carried out at 15°C in the RNAi-hypersensitive eri-1(mg366) background (Kennedy et al., 2004).
Cloning of C. elegans mboa-7 and a Human Orthologue
Full-length mboa-7 cDNA was amplified by polymerase chain reaction (PCR) from a C. elegans cDNA library with primers mboa-7-F, 5′-CAG GTC TGC AGA TGG AAA ATA TCC TTG GCT T-3′ and mboa-7-R, 5′-CTT CCG ATT TTT GAG CTT TTT TCG GC-3′. The PCR-amplified mboa-7 cDNA was cloned into pPD95.67 at the PstI and SmaI sites. Human mboa-7 (h-mboa-7) and mouse LPIAT (m-mboa-7) cDNAs were amplified from HeLa cell and mouse liver cDNA libraries, respectively, and cloned into pCAGGS-MCS vector (Niwa et al., 1991) at the EcoRI and XhoI sites. The following primers were used for cloning of h-mboa-7 and m-mboa-7: h-mboa-7-F1, 5′-CAG CTG AAT TCA TGT CGC CTG AAG AAT GGA C-3′; h-mboa-7-R1, 5′-GTG ACC TCG AGC TCC TCC CGG AGC TTC TCC G-3′; m-mboa-7-F, 5′-ACG GTT GAA TTC ATG ACA CCC GAA GAA TGG AC-3′; and m-mboa-7-R, 5′-CAG CAG CTC GAG CTC TTC CCG GAG CTT TTC-3′. mboa-7, h-mboa-7, and m-mboa-7 sequences have been submitted to the GenBank databases under accession numbers EU016382, EU016381, and EU016380, respectively.
Preparation of Transgenic Worms
DNA injection into the C. elegans germ line was carried out as described previously (Mello et al., 1991). The array xhEx1[mboa-7::GFP] contained the plasmids pKE1 (Pmboa-7::mboa-7::GFP) and pRF4 [rol-6(su1006)]. The arrays xhEx2[mboa-7H350A::GFP] and xhEx3[mboa-7H350A::GFP] contained the plasmids pKE2 (Pmboa-7::mboa-7H350A::GFP) and pRF4. The array xhEx4 [h-mboa-7::GFP] contained the plasmids pKE3 (Pmboa-7::h-mboa-7::GFP) and pFXneges-1EGFP (Pges-1::EGFP). pKE1, pKE2, and pKE3 were prepared as follows.
pKE1.
A 6594-base pair genomic fragment containing 4000 base pairs up-stream of the initiation codon and part of the coding sequence truncated at the 3′ end was amplified by PCR from wild-type worms with primers mboa-7 genome-F, 5′-ACT GAC TGC AGT AAA ACC TTC TTA AAC C-3′, and mboa-7 genome-R, 5′-CTT CCG ATT TTT GAG CTT TTT TCG GC-3′. The fragment was cloned in frame with the green fluorescent protein (GFP) gene into the pPD95.67 vector (a kind gift from Dr. Andrew Fire, Stanford University School of Medicine) at the PstI and SmaI sites.
pKE2.
The two fragments containing a point mutation to change His 350 to Ala were amplified by PCR from the plasmid pKE1 with primers mboa-7H350A-F1, 5′-TAA AAT CAT GGG ATC CCA CAC TGG AAG TGA AGT CT-3′ and mboa-7H350A-R1, 5′-CAT ATG TTC CGG CCC AGA CAG CA-3′, and mboa-7H350A-F2, 5′-TGC TGT CTG GGC CGG AAC ATA TG-3′ and mboa-7H350A-R2, 5′-ATC TGG GTA TCT CGA GAA GCA TTG AAC ACC ATA AC-3′ (underlined nucleotides represent the His-to-Ala mutation). The two overlapping fragments were fused by PCR using primers mboa-7H350A-F1 and mboa-7H350A-R2 and integrated into the BamHI and XhoI sites of the pKE1 vector using an In-Fusion Dry-Down PCR cloning kit (Clontech, Mountain View, CA).
pKE3.
Full-length h-mboa-7 cDNA was amplified by PCR from h-mboa-7 pCAGGS-MCS vector (see above) with primers h-mboa-7-F1, 5′-CCT GGT CTA GAT GTC GCC AGA AGA ATG GAC CTA CCT CG-3′ and h-mboa-7-R1, 5′-GCT CGG GAT CCT CCT CCC GGA GCT TCT CCG GGG C-3′. The fragment was cloned into the pPD95.67 vector at the XbaI and BamHI sites, yielding a vector pKE4. Next, 4000 base pairs upstream of the initiation codon of mboa-7 was amplified from the pKE1 vector with primers Pmboa-7-F, 5′-ACT GAC TGC AGT AAA ACC TTC TTA AAC C-3′ and Pmboa-7-R, 5′-TTC CAT CTA GAT ACC TGA AAC AAA AAA CGA ATA C-3′. The fragment was cloned into the pKE4 vector at the PstI and XbaI sites.
Phenotypic Analysis of mboa-7 Mutants
Adult wild-type and mboa-7(gk399) worms were allowed to lay eggs for 2–3 h at 20°C, and the progeny were scored for embryonic lethality and larval arrest. Unhatched eggs were examined 24 h after being laid, and hatched but arrested larvae were examined 48 h after being laid. For growth rate analysis, first larval stage worms were synchronized after bleach treatment of gravid hermaphrodites, and they were incubated on AA- or EPA-supplemented NGM plates at 15°C. Growth rate was scored by counting adult worms under a stereomicroscope. The average number of unlaid fertilized eggs that accumulated inside of adult animals was quantified as described previously (Koelle and Horvitz, 1996).
Lipid Analysis
Lipids of synchronized young adult worms were extracted by the method of Bligh and Dyer (1959). For fatty acid analysis, phospholipids were separated by one-dimensional thin-layer chromatography (TLC) on silica gel 60 plates (Merck Biosciences, Darmstadt, Germany) in chloroform:ethanol:water:triethylamine (30:35:7:35, vol/vol). Phospholipids were identified by comigration with known standards. The area of silica gel corresponding to each phospholipid was scraped off the plates and treated with dehydrated methanol:acetyl chloride (10:1) to extract the fatty acid methyl esters. The fatty acid methyl ester derivatives were analyzed with a GC 353B gas chromatograph equipped with a flame ionization detector (GL Sciences, Tokyo, Japan) and a TC-FFAP capillary column (60 m × 0.25 mm internal diameter, 0.25 μm; GL Sciences). The oven temperature was programmed to increase from 170°C to 230°C at 5°C/min followed by a hold of 15 min. The injector and detector temperatures were both set at 250°C. Helium was used as carrier gas at 1 ml/min. Individual fatty acids were identified by comparing the retention times with those of known fatty acid standards. Electrospray ionization tandem mass spectrometry (ESI-MS/MS) analysis was performed on a Quattro microtandem quadrupole mass spectrometer (Waters-Micromass, Manchester, United Kingdom) equipped with an ESI interface. PI species were detected by negative ionization, and the mobile phase consisted of acetonitrile:methanol:water (6:7:2, vol/vol/vol) containing 0.1% ammonium formate, pH 6.4. Dry argon was used as the collision gas (2.4 × 10−3 mbar; 1 bar ≈ 105 Pa). Collision gas-induced fragmentation of PI species generated a common dehydrated inositol phosphate fragment, with m/z −241, and parent ion scans of this m/z −241 fragment provided diagnostic determination of PI. The data were acquired and processed with MassLynx NT 4.0 software (Waters-Micromass). PI molecular species were determined by analysis of fatty acyl fragments generated by collision gas-induced fragmentation under negative ionization. Quantitation of individual PI species was obtained based on the ion current response of the PI species relative to that of the internal standard 1,2-dipalmitoyl PI (16:0/16:0 PI). It is noted that due to different ionization efficiency, ion counts obtained for different PI species do not allow us to estimate the relative abundance of the different PI species with regard to each other.
Preparation of sn-2-acyl LysoPI
sn-2-acyl lysoPI was prepared as follows. PI (3–5 μmol) from porcine liver dissolved in 1 ml of diethyl ether was incubated with 100 μl of 100 mM CaCl2, 600 μl of 50 mM Tris-malate, pH 5.7, and 400 μl of enzyme solution containing 16 mg of Rhizopus delemer lipase for 1 h at 40°C while stirring vigorously. After the incubation, the reaction was terminated by adding 1 ml of methanol. Remaining PI and fatty acid were removed by three extractions with 4 ml of diethyl ether:petroleum ether (1:1, vol/vol). 2-Acyl lysoPI remaining in the lower layer was extracted by the method of Bligh and Dyer (1959).
Acyltransferase Assay
Synchronized young adult worms (1 g, wet weight) were suspended in 4 ml of 50 mM potassium phosphate buffer, pH 7.0, containing 0.15 M KCl, 1 mM EDTA, 1 mM dithiothreitol, 1 mM phenylmethanesulfonyl fluoride (PMSF), and 0.25 M sucrose (homogenizing buffer), and they were sonicated three times on ice for 30 s. The C. elegans homogenate was centrifuged at 11,000 × g for 30 min at 4°C. The resulting supernatant was further centrifuged at 105,000 × g for 60 min. The pellet was suspended in homogenizing buffer (without EDTA, dithiothreitol, and PMSF) and immediately used for the enzyme assay described below. HeLa cells were harvested, washed with ice-cold phosphate-buffered saline, and sonicated three times on ice for 3 s in homogenizing buffer. The membrane fraction of HeLa cells was prepared as described above.
In vitro fatty acid incorporation assay was performed as described previously (Tanaka et al., 1999), with slight modifications. Each assay contained 32 nmol of lysophospholipid, 320 μg of protein of the membrane fraction of C. elegans, and the desired 20 nmol of fatty acid in a total volume of 0.8 ml of assay buffer (0.15 M KCl, 5 mM MgCl2, 0.25 M sucrose, 7.5 mM ATP, 0.4 mM CoA, and 50 mM potassium phosphate buffer, pH 6.8). Fatty acids were added as a fatty acid–albumin complex (Chen and Nilsson, 1993), and the specific activity was 55 mCi/mmol. The reaction was incubated at 20°C for 5 min, and then it was stopped by mixing with 2 ml of methanol. The lipids were extracted and separated by TLC as described above. The area of silica gel for each phospholipid was scraped off the plate, and radioactivity was measured.
[14C]Arachidonoyl-CoA:lysophospholipid acyltransferase activity was measured in a similar manner except that 10 nmol of [14C]arachidonoyl-CoA instead of 14C-free fatty acid and increasing concentrations of various lysophospholipids were used. In this assay, MgCl2, ATP, and CoA were omitted from the assay buffer. Acyltransferase activity of the HeLa cell microsomes was measured similarly, except that the incubation temperature was 37°C, and 80 μg of microsomal protein was used.
Cell Culture and Transfection
HeLa cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum and 100 U/ml penicillin, 100 mg/ml streptomycin, and 2 mM l-glutamine. The small interfering RNAs (siRNAs) for human mboa-7 were transfected into HeLa cells with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. The h-mboa-7–specific siRNA sequence spans nucleotide positions 1082–1106 (5′-AUG GUC AGG AAG CUC AGG UAG UAG C-3′).
Quantitative Real-Time Reverse Transcription (RT)-PCR
Total RNA from HeLa cells was extracted with ISOGEN (Nippongene, Toyama, Japan) and reverse transcribed with the SuperScript First-strand Synthesis System for RT-PCR (Invitrogen). Oligonucleotide primers for PCR were designed with Primer Express software (Applied Biosystems, Foster City, CA). The sequences of the oligonucleotides used in PCR reaction were h-mboa-7-F2, 5′-GCC CTC CCT GAT GGA GAC A-3′ and h-mboa-7-R2, 5′-GTA GGT GCG GTA GCG GAA GA; and glyceraldehyde-3-phosphate dehydrogenase (GAPDH)-F-3′ and 5′-GCC AAG GTC ATC CAT GAC AAC T; GAPDH-R-3′ and 5′-GAG GGG CCA TCC ACA GTC TT-3′. GAPDH was amplified as an internal control, and expression of experimental transcript each sample was normalized to that of GAPDH.
In Vivo Incorporation of Exogenous Fatty Acids into C. elegans and HeLa Cells
For incorporation of exogenous fatty acids into C. elegans, synchronized first-stage larvae (800–1200 animals) were cultured with 1 μCi of each 14C-fatty acids for 54 h on NGM plates at 20°C until they reached the young adult stage. Lipids of young adult worms were extracted by the method of Bligh and Dyer (1959), and they were separated by one-dimensional TLC on silica gel 60 plates (Merck Biosciences) in chloroform:ethanol:water:triethylamine (30:35:7:35, vol/vol). Incorporation of 14C-fatty acid into individual phospholipids was expressed as the percentage of radioactivity incorporated into total lipids.
HeLa cells (1.5 × 105) were plated in six-well dishes and incubated for 24 h. siRNAs for h-mboa-7 were then transfected into HeLa cells with Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's protocol. The final concentration of siRNA was 40 nM. After incubation for 48 h, the cells were washed and incubated in DMEM containing 0.1% bovine serum albumin for 24 h. Then, 100 nmol per dish of [14C]AA (0.1 μCi) or [14C]EPA (0.1 μCi) was added to the medium as an albumin complex (Chen and Nilsson, 1993). After 1-h incubation, the cell lipids were extracted and separated by TLC as described above. Incorporation of 14C-fatty acid into individual phospholipids was expressed as the percentage of radioactivity incorporated into total lipids.
RESULTS
Identification of Genes Involved in the Incorporation of Exogenous AA into PI
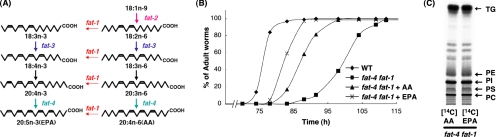
fat-4 fat-1 mutants, which do not produce AA(20:4n-6) and n-3 PUFAs such as EPA(20:5n-3), were viable and fertile (Figure 1A; Watts and Browse, 2002; Kahn-Kirby et al., 2004), but they exhibited slow growth at 15°C (Figure 1B). Dietary supplementation with 25 μM AA or EPA led to incorporation of these PUFAs into the phospholipid fractions of fat-4 fat-1 mutants (Figure 1C), and it rescued the growth defects of this strain (Figure 1B).
Figure 1.
(A) C. elegans PUFA synthetic pathway from bacterial fatty acids. Lipid structures are abbreviated as in 20:4n-6, which has 20 carbons and four double bonds, the first occurring at the n-6 position. fat-1, n-3 desaturase; fat-2, Δ12 desaturase; fat-3, Δ6 desaturase; fat-4, Δ5 desaturase. (B) Rescue of the growth defects of fat-4 fat-1 mutants by dietary supplementation with AA or EPA. Synchronized populations of first-stage larvae were propagated on E. coli OP50 supplemented with AA or EPA, and they were allowed to grow at 15°C. At least 150 worms were scored for each assay and scored as adult if they form vulva. Representative plots of multiple experiments are shown. WT, wild-type. (C) One-dimensional thin-layer chromatographic separation of total lipids from fat-4 fat-1 mutants radiolabeled with [14C]AA (left) and [14C]EPA (right).
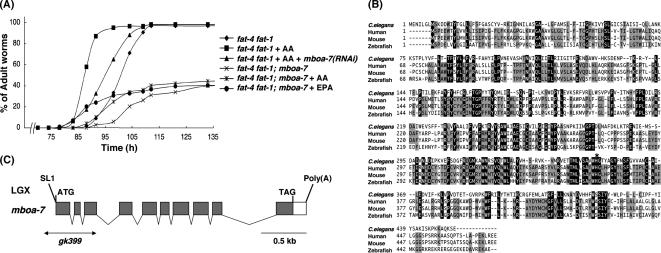
To identify genes that are involved in the incorporation of exogenous PUFAs into phospholipids in C. elegans, we performed a large-scale RNAi screen by using fat-4 fat-1 mutants under AA-supplemented conditions (see Materials and Methods). In this screen, we found that knockdown of a previously uncharacterized gene, F14F3.3, caused weak growth defects despite supplementation with AA (Figure 2A). F14F3.3 encodes a protein with structural similarity to those of MBOATs, which transfer acyl groups to proteins, sterols, or diacylglycerol (Figure 2B; Hofmann, 2000). Our genome database search revealed that C. elegans genome possesses 10 MBOAT family members (Table 1; WormBase [http://www.wormbase.org/]), three of which are homologous to the enzymes transferring acyl groups to specific proteins such as Wnt and Hedgehog (Hofmann, 2000). We named the other seven members membrane-bound O-acyltransferase-1-7 (mboa-1-7); mboa-1 and mboa-2 are homologous to acyl-CoA:cholesterol acyltransferase and diacylglycerol O-acyltransferase 1, respectively; and mboa-3, mboa-4, and mboa-5 show significant homology to human MBOAT1, MBOAT2. mboa-6 corresponds to human MBOAT5. C. elegans F14F3.3 identified in this study encodes homologous sequence to human BB1 (Fukunaga-Johnson et al., 1996), which also belongs to MBOAT family, and it was named mboa-7. We considered mboa-7 a candidate gene for phospholipid fatty acid remodeling, and we examined the growth rate of mboa-7 deletion mutants [mboa-7(gk399); Figure 2C]. In the fat-4 fat-1 mutant background, mboa-7 mutants exhibited severe growth defects even under conditions of AA or EPA supplementation (Figure 2A), suggesting that mboa-7 is required for use of exogenous PUFAs in C. elegans.
Figure 2.
(A) Knockdown and knockout of mboa-7 inhibit growth rescue by dietary supplementation with AA or EPA. Growth of each strain was scored as described in Figure 1B, except that E. coli HT115 was used as a food source. All experiments were performed at 15°C. (B) Multiple sequence alignment of C. elegans mboa-7 and homologous sequences in human (BB1/LENG4; Fukunaga-Johnson et al., 1996; Wende et al., 2000), mouse, and zebrafish. Residues identical in all four sequences are shaded in black, and residues identical in three proteins are shaded in gray. The numbers on the left indicate amino acid positions. The histidine residue indicated by asterisk is the predicted active site of the MBOAT motif in mboa-7 (Hofmann, 2000; Bosson et al., 2006). Accession numbers for the sequences used were as follows: C. elegans, EU016382; human, EU016381; mouse, EU016380; and zebrafish, NP_609029. (C) Genomic structure of mboa-7 (F14F3.3). The mboa-7 gene is located on chromosome X. Gray boxes indicate coding exons, and white boxes indicate 5′ and 3′ untranslated sequences. The positions of the ATG initiation codon, stop codon (TAG), the trans-spliced leader SL1, and the poly(A) tail are shown. The extent of the deletion in mboa-7(gk399) is indicated by a horizontal line.
Table 1.
C. elegans MBOAT familya
| Gene | Sequence name | Human homologue |
|---|---|---|
| mboa-1 | B0395.2 | ACAT-1, ACAT-2 |
| mboa-2 | H19N07.4 | DGAT1 |
| mboa-3 | C54G7.2 | MBOAT1, MBOAT2 |
| mboa-4 | C08F8.4 | MBOAT1, MBOAT2 |
| mboa-5 | ZK550.1 | MBOAT1, MBOAT2 |
| mboa-6 | R155.1 | MBOAT5 |
| mboa-7 | F14F3.3 | BB1/LENG4 |
| mom-1 | T07H6.2 | Porcupine |
| hhat-1 | ZC101.3 | HHAT |
| hhat-2 | Y57G11C.17 | HHAT |
a BB1, genes overexpressed by human bladder and breast cancer cell lines (Fukunaga-Johnson et al., 1996); LENG4, leukocyte receptor cluster member 4 (Wende et al., 2000); and HHAT, Hedgehog acyltransferase.
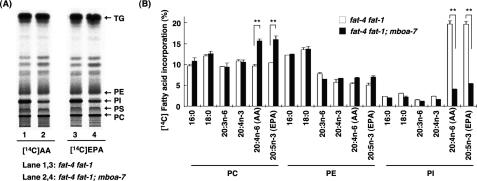
Next, we examined the incorporation of exogenous [14C]AA and [14C]EPA into the lipid fraction of mboa-7 mutants in the fat-4 fat-1 mutant background in which endogenous AA and EPA are absent. In mboa-7(+/+) animals, both [14C]AA and [14C]EPA were incorporated into all the major membrane phospholipids, especially PI and phosphatidylcholine (PC) (Figure 3, A and B). In contrast, in mboa-7 mutants, incorporation of [14C]AA and [14C]EPA into PI was reduced, and uptake of radioactive AA and EPA into the PC fraction was increased. Incorporation of other fatty acids into PI was not affected significantly (Figure 3B, 16:0, 18:0, 20:3n-6, and 20:4n-3). These data indicated that mboa-7 is involved in the selective incorporation of AA and EPA into PI.
Figure 3.
Incorporation of exogenous [14C]AA and [14C]EPA into phospholipids of mboa-7 mutants. (A) One-dimensional thin layer chromatographic separation of total lipids from fat-4 fat-1 and fat-4 fat-1; mboa-7 mutants radiolabeled with exogenous [14C]AA (left) and [14C] EPA (right). (B) Incorporation of various radiolabeled fatty acids into phospholipids of fat-4 fat-1 and fat-4 fat-1; mboa-7 mutants. The amount of incorporation was expressed as the percentage of radioactivity incorporated into total lipids. All experiments were carried out at 20°C. Note that the radiolabeled fatty acids are not metabolized to AA or EPA in vivo due to the lack of Δ5 and n-3 fatty acid desaturation activities. Each bar represents the mean ± SEM of at least three independent experiments. **p < 0.01.
mboa-7 Is Required in LPIAT Activity
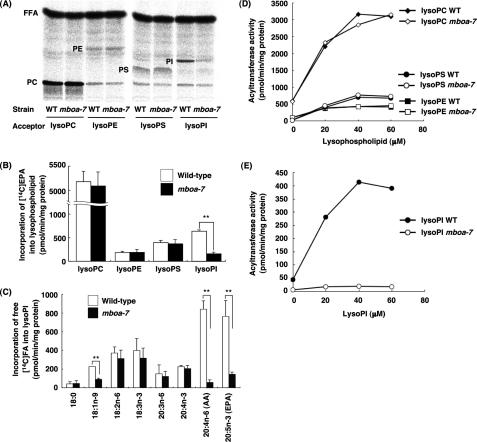
It is generally thought that incorporation of PUFAs into cellular phospholipids requires acyl-CoA:lysophospholipid acyltransferase activity (MacDonald and Sprecher, 1991). To examine the possibility that the mboa-7 gene product is associated with lysophospholipid acyltransferase activity, we measured lysophospholipid acyltransferase activity by using membrane fractions of wild-type and mboa-7 mutants. First, we used free 14C-fatty acids as acyl donors (see Materials and Methods). In this system, free 14C-fatty acids are converted to [14C]acyl-CoAs by the endogenous acyl-CoA synthase, and then they are incorporated into lysophospholipids by acyltransferases during the incubation. As shown in Figure 4, A and B, significant incorporation of [14C]EPA into lysoPC, lysoPI, lysoPS, and lysoPE were detected in the membrane fraction of wild-type worms, whereas incorporation of [14C]EPA into lysoPI was greatly reduced in the membrane fraction of mboa-7 mutants. We then analyzed the acyl donor selectivity using lysoPI as an acyl acceptor. The wild-type membrane fraction showed a preference for [14C]AA and [14C]EPA as acyl donors for lysoPI similar to the situation in mammalian cells (Figure 4C; Baker and Thompson, 1973; Holub, 1976). However, incorporation of [14C]AA and [14C]EPA into lysoPI was significantly reduced in the membrane fraction of mboa-7 mutants. Incorporation of [14C]oleic acid (18:1n-9) into lysoPI was also reduced, but the extent was not so significant compared with [14C]AA and [14C]EPA. Incorporation of [14C]EPA into lysoPC, lysoPE, and lysoPS (Figure 4, A and B), and incorporation of other 14C-free fatty acids such as 18:2n-6, 18:3n-3, and 20:4n-3 into lysoPI (Figure 4C) were not affected in the membrane fraction of mboa-7 mutants, suggesting that acyl-CoA synthesis from free fatty acid and CoA seemed to be normal in the membranes of mboa-7 mutants.
Figure 4.
The membrane fraction of mboa-7 mutants lacks LPIAT activity. (A and B) In vitro fatty acid incorporation assay using the membrane fractions of wild-type and mboa-7 mutants and different lysophospholipids as acyl acceptors. [14C] EPA was used as an acyl donor. (A) One-dimensional thin layer chromatographic separation of total lipids of the reaction mixture after incubation. FFA, free fatty acid. (B) Each bar represents the mean ± SEM of at least three independent experiments. **p < 0.01. (C) In vitro fatty acid incorporation into lysoPI with different radiolabeled fatty acids as acyl donors. Each bar represents the mean ± SEM of at least three independent experiments. **p < 0.01. (D) Acyl-CoA:lysophospholipid acyltransferase activity in the membrane fractions of wild-type and mboa-7 mutants. [14C]Arachidonoyl-CoA (12.5 μM) and the indicated concentration of lysoPC, lysoPE, or lysoPS were used. (E) [14C]Arachidonoyl-CoA:lysoPI acyltransferase activity in the membrane fractions of wild-type and mboa-7 mutants. [14C]Arachidonoyl-CoA (12.5 μM) and the indicated concentration of lysoPI were used.
Then, we examined incorporation of [14C]arachidonoyl-CoA into various lysophospholipids (Figure 4, D and E). Incorporation of [14C]arachidonoyl-CoA was elevated with increasing concentrations of lysophospholipids, and it reached a plateau in the membrane of wild-type worms. Incorporations of [14C]arachidonoyl-CoA into lysoPC, lysoPE, and lysoPS were not affected in the membranes of mboa-7 mutants; however, incorporation of [14C]arachidonoyl-CoA into lysoPI was specifically reduced at any lysoPI concentration. These results indicate that the mboa-7 is required for LPIAT activity with a preference for AA and EPA as the acyl donor.
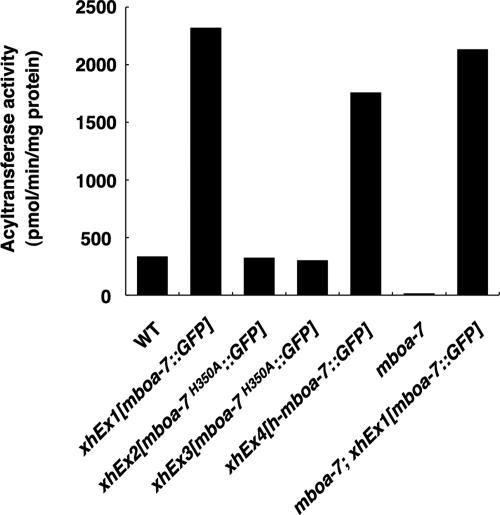
To reveal that the mboa-7 gene product (MBOA-7) shows LPIAT activity, we generated transgenic worms expressing GFP-tagged MBOA-7 (xhEx1[mboa-7::GFP]; see Materials and Methods), and we examined the enzyme activity of MBOA-7::GFP. Expression of MBOA-7::GFP fully rescued the phenotypes of mboa-7(gk399) mutants in the mboa-7 mutant background (see below), indicating that MBOA-7::GFP is functional in vivo. Consistent with the results described above, the membrane fraction of MBOA-7::GFP–expressing worms showed remarkably increased LPIAT activity with [14C]arachidonoyl-CoA as an acyl donor (Figure 5, xhEx1[mboa-7::GFP]). Next, we mutated histidine 350 of MBOA-7 (histidine to alanine; H350A), the predicted active residue of the membrane-bound O-acyltransferase family (Figure 2B; Hofmann, 2000; Bosson et al., 2006), and generated the transgenic worms expressing the MBOA-7H350A:: GFP (xhEx2[mboa-7H350A::GFP], and xhEx3[mboa-7H350A::GFP]). As shown in Figure 5, the membrane fractions of these two MBOA-7H350A::GFP transgenic lines did not show increased LPIAT activity compared with those of wild-type worms. These data strongly suggest that mboa-7 gene encodes LPIAT.
Figure 5.
Recombinant MBOA-7 and h-MBOA-7 (a human homologue of MBOA-7) expressed in C. elegans show remarkably increased LPIAT activity. [14C]Arachidonoyl-CoA:lysoPI acyltransferase activity in the membrane fractions of the indicated strains were measured. 40 μM lysoPI and 12.5 μM [14C]arachidonoyl-CoA were used as an acyl donor and an acyl acceptor, respectively. The membrane fraction of MBOA-7::GFP-expressing worms showed remarkably increased LPIAT activity both in wild-type and mboa-7 mutants (xhEx1[mboa-7::GFP] and mboa-7; xhEx1[mboa-7::GFP], respectively). Human MBOA-7::GFP also catalyzed LPIAT activity (xhEx4[h-mboa-7::GFP]). MBOA-7H350A::GFP, which is mutated at the predicted active site of the membrane-bound O-acyltransferase motif of MBOA-7, had no LPIAT activity (xhEx2[mboa-7H350A::GFP] and xhEx3[mboa-7H350A::GFP]).
All the acyltransferase assays described above were performed using sn-1-acyl-2-lysoPI (sn-2-lysoPI) as an acyl acceptor. Next, we examined whether MBOA-7 incorporates PUFA into the sn-1 position of lysoPI. For this study, the substrate sn-2-acyl-1-lysoPI (sn-1-lysoPI) was prepared by phospholipase A1 digestion of PI (see Materials and Methods), and it was immediately used for acyltransferase assay. The wild-type membrane fraction showed significant LPIAT activity by using sn-1-lysoPI as a substrate (black plus gray bars in the Supplemental Figure S1A), and the enzyme activity was reduced to ∼65% in the membranes of mboa-7 mutants. Because sn-1-lysoPI is known to easily isomerize to form sn-2-lysoPI (see Supplemental Figure S1B; Rouser et al., 1966), it was possible that 14C-acyl donor was incorporated into sn-2-lysoPI and sn-1-lysoPI. To check the positional specificity, [14C]PI produced after the incubation was treated again with bee venom phospholipase A2 (for details, see the legend to Supplemental Figure S1). By measuring the radioactivity of 14C-free fatty acid and [14C]lysoPI produced after the phospholipase A2 treatment, we can estimate the incorporation of 14C-acyl donor into sn-1-lysoPI (sn-1-LPIAT activity, black bar) and sn-2-lysoPI (sn-2-LPIAT activity, gray bar) (Supplemental Figure S1B). The sn-2-LPIAT activity was observed in the microsomes of wild-type worms, whereas the activity was almost undetectable in those of mboa-7 mutants, indicating that sn-2-LPIAT activity in wild-type worms was catalyzed by MBOA-7 and that significant isomerization of sn-1-lysoPI occurred probably during the incubation. In contrast to sn-2 LPIAT activity, similar levels of sn-1 LPIAT activity (black bar) were detected in wild-type and mboa-7 mutants. These results indicate that MBOA-7 prefers sn-2-lysoPI as an acyl acceptor rather than sn-1-lysoPI and that sn-1-LPIAT other than MBOA-7 exists in C. elegans.
PI Molecular Species in mboa-7 Mutants
To address possible involvement of MBOA-7 in regulating the distribution of PI molecular species, we next analyzed phospholipid fatty acid composition of wild-type and mboa-7 mutants by gas chromatography (GC) (Table 2). First, we measured PC, PE, and PI content of wild-type and mboa-7 mutants, and we found that phospholipid contents were not affected in the mboa-7 mutants (PC, 33.0% in wild-type vs. 34.6% in mboa-7 mutants; PE, 39.3% in wild-type vs. 43.7% in mboa-7 mutants; and PI, 1.9% in wild-type vs. 2.0% in mboa-7 mutants). GC analysis of PC, PE, and PI revealed that the mboa-7 mutation significantly reduced the EPA content of PI but not PC or PE. In mboa-7 mutants, the amount of EPA in PI was reduced to 8.9% of total PI fatty acids compared with 26.2% in wild-type animals. Concomitantly, palmitic acid (16:0) and oleic acid (18:1n-9) in PI increased in mboa-7 mutants (17.5% in wild-type vs. 26.4% in mboa-7 mutants and 7.5% in wild-type vs. 15.2% in mboa-7 mutants, respectively). Reduction of EPA-containing PI molecular species was also confirmed by ESI-MS/MS analysis (Supplemental Figure S2, A and B). Consistent with the GC analysis, the amounts of EPA-containing PI molecular species such as 18:0/20:5, 18:1/20:5, 18:0(alkyl)/20:5, and 16:0/20:5 were reduced in mboa-7 mutants. Instead, other PI molecular species such as 18:1/18:1, 18:0(alkyl)/18:1, 16:0/18:1, and 16:1/18:1 increased in mboa-7 mutants. Collectively, these results indicate that mboa-7 is a major determinant of PI molecular species in C. elegans.
Table 2.
Fatty acid composition of wild-type and mboa-7 mutants
| Wild-type |
mboa-7 |
|||||
|---|---|---|---|---|---|---|
| PC | PE | PI | PC | PE | PI | |
| 16:0 | 1.3 ± 0.1 | 3.4 ± 0.3 | 17.5 ± 0.3 | 1.3 ± 0.1 | 3.4 ± 0.1 | 26.4 ± 2.6 |
| 18:0 | 2.5 ± 0.2 | 8.9 ± 0.2 | 21.1 ± 3.0 | 2.6 ± 0.2 | 7.0 ± 0.2 | 18.7 ± 3.2 |
| 18:1n-9 | 1.9 ± 0.1 | 1.6 ± 0.1 | 7.5 ± 1.7 | 1.3 ± 0.0 | 1.4 ± 0.0 | 15.2 ± 2.0 |
| 18:1n-7 | 26.9 ± 1.7 | 31.8 ± 2.1 | 12.0 ± 3.5 | 32.6 ± 0.8 | 37.7 ± 0.2 | 12.7 ± 2.6 |
| 18:2n-6 | 7.7 ± 0.2 | 7.4 ± 0.0 | 2.8 ± 0.9 | 4.2 ± 0.1 | 5.6 ± 0.0 | 5.1 ± 0.5 |
| 18:3n-6 | 2.5 ± 0.2 | 1.5 ± 0.2 | N.D. | 2.1 ± 0.0 | 1.6 ± 0.1 | N.D. |
| 18:3n-3 | 0.8 ± 0.1 | 0.3 ± 0.0 | N.D. | 0.3 ± 0.0 | 0.2 ± 0.0 | N.D. |
| 20:3n-6 | 4.4 ± 0.1 | 2.5 ± 0.0 | N.D. | 4.6 ± 0.1 | 3.1 ± 0.1 | N.D. |
| 20:4n-6 | 3.2 ± 0.1 | 1.5 ± 0.1 | N.D. | 3.9 ± 0.2 | 1.8 ± 0.1 | N.D. |
| 20:4n-3 | 6.4 ±0.0 | 2.7 ± 0.0 | N.D. | 5.7 ± 0.0 | 2.8 ± 0.0 | N.D. |
| 20:5n-3 | 32.9 ± 0.5 | 15.3 ± 1.3 | 26.2 ± 2.3 | 33.0 ± 0.6 | 13.0 ± 0.7 | 8.9 ± 0.6 |
| Others | 9.5 ± 1.1 | 23.0 ± 0.9 | 12.9 ± 1.1 | 8.4 ± 0.3 | 22.4 ± 0.5 | 12.9 ± 7.5 |
Data are mol% and represent the average ± SEM of three independent experiments. Others are the total mol% of 16:1n-7, 17:0, 17:iso, 17:Δ, 18:0DMA, and 19:Δ.
a N.D., not detectable.
Human Homologue of C. elegans mboa-7
C. elegans mboa-7 is homologous to human BB1, which also belongs to the MBOAT family (Figure 2B; Fukunaga-Johnson et al., 1996; Hofmann, 2000). BB1 was reported to be up-regulated in metastatic breast and bladder carcinomas (Fukunaga-Johnson et al., 1996), but the function of this gene has not yet been elucidated. To confirm that human BB1/h-mboa-7 is involved in incorporation of AA into PI, we performed knockdown experiments by using gene-specific siRNAs in HeLa cells. In cells transfected with h-mboa-7 siRNA duplex, h-mboa-7 mRNA levels were reduced to ∼30% of those in control siRNA-transfected cells at 72 h after the transfection (Supplemental Figure S3A). [14C]AA added to the culture was incorporated into PC, PE, and PI of control HeLa cells time-dependently for at least 3 h (data not shown). Incorporation of [14C]AA into cellular PI was specifically reduced from 13.1% of the total radioactivity in control cells to 6.7% in the siRNA-transfected cells (Supplemental Figure S3B). In contrast, incorporation of [14C]AA into PC was increased in siRNA-transfected cells. Incorporation of [14C]EPA into PI was also reduced similar to that of [14C]AA in siRNA-transfected cells, although uptake of [14C]EPA into PI was substantially lower than that of [14C]AA in control cells (4.1 vs. 13.1%) (data not shown).
Acyltransferase activity was also measured in microsomal fractions from siRNA-transfected HeLa cells by using [14C]arachidonoyl-CoA. As shown in Supplemental Figure S3C, incorporation of [14C]arachidonoyl moiety into lysoPI was specifically reduced in the membrane fraction of the siRNA-transfected cells, whereas incorporation of [14C]arachidonoyl moiety into lysoPC and lysoPS was unaffected. Incorporation of [14C]arachidonoyl moiety into lysoPE was very low in the membrane of control HeLa cells under the present assay conditions. These results indicate that human BB1/h-mboa-7 is also involved in incorporation of AA into PI.
To investigate whether human BB1/h-mboa-7 possesses LPIAT activity, we measured LPIAT activity by using membrane fractions of transgenic worms expressing h-mboa-7:: GFP (xhEx4[h-mboa-7::GFP]; see Materials and Methods). The membrane fractions of the human MBOA-7::GFP–expressing worms showed increased LPIAT activity (Figure 5), suggesting that human BB1, a homologue of C. elegans mboa-7, is human LPIAT.
Phenotypic Analysis of mboa-7 Mutants
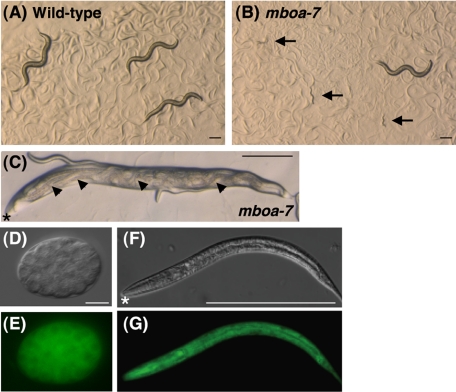
Finally, to elucidate the physiological functions of LPIAT, we analyzed the phenotypes of mboa-7 mutants and examined the expression pattern of mboa-7. Mutation of mboa-7 caused larval arrest at an early developmental stage with incomplete penetrance (Figure 6B; Table 3). Under normal conditions (20°C), ∼22% of mboa-7 mutants were arrested at an early larval stage (Table 3), and the penetrance of the defects increased under high temperature conditions (48% at 25°C). We also found that 14% of mboa-7 mutants accumulated unlaid eggs that hatched internally and formed “bags of worms” (Figure 6C), suggesting that adult mboa-7 mutants display egg-laying defects (Trent et al., 1983). Wild-type young adult animals had an average of 14 fertilized eggs in the uterus. In contrast, in mboa-7 mutants, the average number of eggs in the uterus was 21.9, and two thirds of mboa-7 mutants had >20 eggs in the uterus, whereas wild-type animals rarely possessed >20 eggs (Table 3). Vulval structure of mboa-7 mutants seemed normal as judged by differential interference contrast microscopy. These data suggest that LPIAT activity is required for early larval development and egg-laying behavior in C. elegans.
Figure 6.
Phenotypes of mboa-7 mutants. (A, B) For comparison of growth, wild-type and mboa-7 embryos were placed onto culture plates, incubated at 20°C, and photographed after 72 h of growth. Arrows indicate mboa-7 mutants that have arrested development at an early larval stage. Bar, 200 μm. (C) Some of the mboa-7 mutants show bags of worms phenotype where embryos hatch within the mother, leaving a cuticle sack containing multiple wriggling larvae (arrowheads). Asterisks indicate the anterior of the mother worm. Bar, 200 μm. (C–G) Expression of MBOA-7::GFP in an early embryo (D and E) and a second-stage larva (F and G). Nomarski micrographs (D and F) and corresponding MBOA-7::GFP expression (E and G). MBOA-7::GFP was ubiquitously expressed throughout development. Asterisks indicate the anterior of the larva (F and G). Bar, 10 μm (D and E) and 200 μm (F and G).
Table 3.
Summary of mboa-7 mutant defects
| Strain | Larval arrest (%) | No. of eggs in uterus | % of worms with >20 eggs in uterus |
|---|---|---|---|
| Wild-type | 0.2 ± 0.2 | 14.0 ± 0.6 | 2.5 (1/60) |
| mboa-7 | 21.7 ± 1.7 | 21.9 ± 0.7 | 70.0 (21/30) |
| mboa-7;xhEx1[mboa-7::GFP] | 2.1 ± 1.6 | 10.8 ± 0.5 | 0 (0/30) |
Phenotypes of mboa-7 were analyzed as described in Materials and Methods. Data represents the mean ± SEM of at least three independent experiments. Larval arrest refers to the percentage of larvae that have arrested development at an early larval stage (n > 500 for each strain). MBOA-7::GFP is expressed under the control of its own promoter. All strains were raised at 20°C.
We next examined expression of MBOA-7::GFP driven by the mboa-7 promoter. MBOA-7::GFP fully rescued the larval arrest and egg-laying defects of mboa-7 mutants described above (Table 3). The MBOA-7::GFP fusion protein was expressed ubiquitously throughout development from early embryo to larval and adult stages (Figure 6, D–G). Strong expression was observed in pharyngeal muscles, body wall muscles, vulval cells, distal tip cells, intestinal cells, and spermatheca at adult stage (data not shown).
DISCUSSION
In the present study, we used a comprehensive RNAi screen of C. elegans to clone the previously uncharacterized gene named mboa-7 involved in the PI remodeling pathway originally reported in 1962 (Keenan and Hokin, 1962). Incorporation of exogenous EPA into PI of living worms and LPIAT activity in microsomes were greatly reduced in mboa-7 mutants. mboa-7 encodes a member of the MBOAT family, including sterol acyltransferase and the lysophospholipid acyltransferase recently identified from yeast, suggesting that mboa-7 encodes LPIAT. Moreover, mboa-7 mutants showed significant reductions in the EPA content of PI but not other phospholipids and they exhibited defects in early larval development and egg-laying behavior. These data show that mboa-7 is a major determinant of PI molecular species and that specific PI molecular species are critical for certain biological functions.
Previous studies revealed that LPIAT purified from rat liver microsomes (Yamashita et al., 2003) or bovine heart microsomes (Sanjanwala et al., 1989) shows absolute specificity for lysoPI as the acyl acceptor and no activity with lysoPC, lysoPE, and lysoPS. Consistent with these observations, the incorporation of exogenous EPA or AA into PI in mboa-7 mutants and the in vitro incorporation of AA or EPA into LysoPI in the microsomal fraction of mboa-7 mutants were specifically reduced compared with those in wild-type worms. Acyl donor specificity for LPIAT with lysoPI as a substrate has been reported in the microsomal fraction of various tissues and cells (Baker and Thompson, 1973; Holub, 1976; Kameyama et al., 1983; Inoue et al., 1984; Sanjanwala et al., 1989), and it differs minimally depending on the source and the state of the enzyme. In general, acyl-CoAs with more than three double bonds are particularly good substrates. EPA (20:5n-3) and AA (20:4n-6) were incorporated into lysoPI in the microsomes of wild-type C. elegans and the incorporation of both fatty acids was greatly reduced in the microsomes of mboa-7 mutant, indicating that the incorporation of these PUFAs into the PI fraction is mediated by mboa-7. These biochemical features may explain the enrichment of EPA in PI of wild-type worms and that of AA in PI of fat-1 mutants in which AA is a dominant PUFA (see Introduction).
Furthermore, we found that incorporation of fatty acids such as 18:2n-6, 18:3n-3, and 20:4n-3 into lysoPI was unaffected in the mboa-7 mutants, suggesting the existence of other lysophospholipid acyltransferases that have different acyl donor specificities. It is not clear at present whether another lysoPI-specific acyltransferase with different acyl donor specificity exists or if lysophospholipid acyltransferase with no head group specificity catalyzes the transfer of these fatty acids into PI exists. Very recently, a novel lysophospholipid acyltransferase was identified from yeast that catalyzes the transfer of unsaturated fatty acids into major lysophospholipids such as lysoPE, lysoPC, lysoPS, lysoPI, and lysoPA (Benghezal et al., 2007; Jain et al., 2007; Riekhof et al., 2007; Tamaki et al., 2007). Because other MBOAT family genes were found in C. elegans, one or more of these gene products may express the remaining LPIAT activity in mboa-7 mutants.
The physiological significance of phospholipid fatty acid remodeling remains to be elucidated because the genes responsible for the reactions have not been identified and appropriate animal models deficient of such genes have not been analyzed. In the present study, we showed that C. elegans mboa-7 mutants had significantly lower EPA levels in PI, demonstrating that mboa-7 regulates PI molecular species, supporting the hypothesis proposed by Lands in the 1960s (Lands and Crawford, 1976; Lands, 2000). We also found that mboa-7 mutants showed defects in early larval development and egg-laying behavior, indicative of the importance of specific PI molecular species in certain physiological functions. It has been assumed that specific PI molecular species are critical for normal phosphoinositide function (Hodgkin et al., 1998; Arnhold et al., 1999; Shirai et al., 1999), which plays important roles in signal transduction, such as the phospholipase C-signaling pathway (Rebecchi and Pentyala, 2000; Rhee, 2001). Interestingly, mutants of egl-8, a phospholipase C isoform in C. elegans, exhibit egg-laying defects similar to that observed in our mboa-7 mutants (Trent et al., 1983, Lackner et al., 1999). Both egl-8 and mboa-7 mutants have apparently normal vulvae, and they exhibit an induction of egg laying in response to exogenous serotonin (Bastiani et al., 2003; our unpublished data). The mboa-7 mutants will provide a useful tool for studying the physiological links between PI molecular species and PI signaling.
Although the EPA content of PI is significantly reduced in the mboa-7 mutants, an appreciable amount of EPA is still present in PI. It is possible that other LPIAT may exist in C. elegans or that EPA can be incorporated into PI via different pathways. LPAAT may catalyze the incorporation of PUFA into the sn-2 position of lysoPA during de novo phospholipid synthesis. Both mammalian LPAATα and LPAATβ show relatively broad substrate specificity for acyl-CoAs and incorporate AA into lysoPA (Stamps et al., 1997; Eberhardt et al., 1997). Thus, it is possible that PI-containing AA can be synthesized once AA is incorporated into PA via the de novo pathway. Alternatively, other lysophospholipid acyltransferases, such as lysoPC acyltransferase (Sanjanwala et al., 1988) and lysoPS acyltransferase (Holub, 1980), are also known to prefer PUFA for acylation at the sn-2 position. Once phospholipids other than PI acquire PUFA via a remodeling system, these lipids can be converted by the action of phospholipase C or D into diacylglycerol or PA, respectively, both of which serve as a precursor of PI synthesis.
The mboa-7 gene product belongs to the large and diverse MBOAT family (Hofmann, 2000). All member proteins have several membrane-spanning regions, typically between eight and 10, and they share a region of detectable sequence similarity. Biochemically characterized members of the family encode enzymes that transfer organic acids, typically fatty acids, onto hydroxyl groups of membrane-embedded targets. As mentioned above, several groups very recently cloned a yeast gene named ALE1 (Riekhof et al., 2007), SLC4 (Benghezal et al., 2007), or LPT1 (Jain et al., 2007; Tamaki et al., 2007) that encodes an acyltransferase of the MBOAT family. Deletion of this gene in yeast results in strong reduction of the activities of various lysophospholipid acyltransferases such as LPEAT, LPCAT, LPSAT, LPIAT, and LPAAT, suggesting that this newly identified acyltransferase is involved in the metabolism of a variety of lysophospholipids in the cells. In this context, mboa-7 seems to be involved only in the remodeling of PI according to the data presented in this work. In contrast, it is well known that in GPI-anchored proteins, fatty acyl chains at the sn-2 position of the diacylglycerol moiety receive remodeling. In yeast, the gene called GUP1 is proposed to catalyze the incorporation of a C26:0 fatty acid at the sn-2 position of the diacylglycerol moiety in GPI-anchored proteins (Bosson et al., 2006). GUP1 is also a member of MBOAT family, but the human and C. elegans orthologues have not been cloned so far. According to our preliminary observations, mboa-7 is not involved in the remodeling of GPI-anchored proteins. However, it will be interesting to compare these two molecules to elucidate the domain involved in the recognition of acyl-CoA molecules and the head group of lysophospholipids.
In conclusion, we found that MBOA-7, another member of the MBOAT family, catalyzes the incorporation of AA and EPA into lysoPI. Substrates of several members of the MBOAT family are still unclear, and we are now testing whether they possess lysophospholipid acyltransferase activity.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Andrew Fire (Stanford University School of Medicine) for vector pPD95.67. We also gratefully acknowledge technical assistance from Hideko Fukuda and Yuko Funakoshi. Some of the strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Center for Research Resources of the National Institutes of Health.
Abbreviations used:
- AA
arachidonic acid
- CoA
coenzyme A
- EPA
eicosapentaenoic acid
- GC
gas chromatography
- GFP
green fluorescent protein
- NGM
nematode growth medium
- PA
phosphatidic acid
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PI
phosphatidylinositol
- PS
phosphatidylserine
- PUFA
polyunsaturated fatty acid
- siRNA
small interfering RNA
- WT
wild-type
- X:Yn-Z
fatty acid chain of X carbon atoms and Y methylene-interrupted cis bonds (Z indicates the position of the terminal double bond relative to the methyl end of the molecules).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-09-0893) on December 19, 2007.
REFERENCES
- Akesson B., Elovson J., Arvidson G. Initial incorporation into rat liver glycerolipids of intraportally injected (3H)glycerol. Biochim. Biophys. Acta. 1970;210:15–27. doi: 10.1016/0005-2760(70)90057-3. [DOI] [PubMed] [Google Scholar]
- Arnhold J., Benard S., Kilian U., Reichl S., Schiller J., Arnold K. Modulation of luminol chemiluminescence of fMet-Leu-Phe-stimulated neutrophils by affecting dephosphorylation and the metabolism of phosphatidic acid. Luminescence. 1999;14:129–137. doi: 10.1002/(SICI)1522-7243(199905/06)14:3<129::AID-BIO526>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Ashrafi K., Chang F. Y., Watts J. L., Fraser A. G., Kamath R. S., Ahringer J., Ruvkun G. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature. 2003;421:268–272. doi: 10.1038/nature01279. [DOI] [PubMed] [Google Scholar]
- Baker R. R., Thompson W. Positional distribution and turnover of fatty acids in phosphatidic acid, phosphinositides, phosphatidylcholine and phosphatidylethanolamine in rat brain in vivo. Biochim. Biophys. Acta. 1972;270:489–503. doi: 10.1016/0005-2760(72)90114-2. [DOI] [PubMed] [Google Scholar]
- Baker R. R., Thompson W. Selective acylation of 1-acylglycerophosphorylinositol by rat brain microsomes. Comparison with 1-acylglycerophosphorylcholine. J. Biol. Chem. 1973;248:7060–7065. [PubMed] [Google Scholar]
- Bastiani C. A., Gharib S., Simon M. I., Sternberg P. W. Caenorhabditis elegans Galphaq regulates egg-laying behavior via a PLCbeta-independent and serotonin-dependent signaling pathway and likely functions both in the nervous system and in muscle. Genetics. 2003;165:1805–1822. doi: 10.1093/genetics/165.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benghezal M., Roubaty C., Veepuri V., Knudsen J., Conzelmann A. SLC1 and SLC4 encode partially redundant acyl-Coenzyme A 1-acylglycerol-3-phosphate O-acyltransferases of budding yeast. J. Biol. Chem. 2007;282:30845–30855. doi: 10.1074/jbc.M702719200. [DOI] [PubMed] [Google Scholar]
- Bligh E. G., Dyer W. J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Bosson R., Jaquenoud M., Conzelmann A. GUP1 of Saccharomyces cerevisiae encodes an O-acyltransferase involved in remodeling of the GPI anchor. Mol. Biol. Cell. 2006;17:2636–2645. doi: 10.1091/mbc.E06-02-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Nilsson A. Desaturation and chain elongation of n-3 and n-6 polyunsaturated fatty acids in the human CaCo-2 cell line. Biochim. Biophys. Acta. 1993;1166:193–201. doi: 10.1016/0005-2760(93)90097-s. [DOI] [PubMed] [Google Scholar]
- Chen X., Hyatt B. A., Mucenski M. L., Mason R. J., Shannon J. M. Identification and characterization of a lysophosphatidylcholine acyltransferase in alveolar type II cells. Proc. Natl. Acad. Sci. USA. 2006;103:11724–11729. doi: 10.1073/pnas.0604946103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhardt C., Gray P. W., Tjoelker L. W. Human lysophosphatidic acid acyltransferase. cDNA cloning, expression, and localization to chromosome 9q34.3. J. Biol. Chem. 1997;272:20299–20305. doi: 10.1074/jbc.272.32.20299. [DOI] [PubMed] [Google Scholar]
- Fukunaga-Johnson N., Lee S. W., Liebert M., Grossman H. B. Molecular analysis of a gene, BB1, overexpressed in bladder and breast carcinoma. Anticancer Res. 1996;16:1085–1090. [PubMed] [Google Scholar]
- Hodgkin M. N., Pettitt T. R., Martin A., Michell R. H., Pemberton A. J., Wakelam M. J. Diacylglycerols and phosphatidates: which molecular species are intracellular messengers? Trends Biochem. Sci. 1998;23:200–204. doi: 10.1016/s0968-0004(98)01200-6. [DOI] [PubMed] [Google Scholar]
- Hofmann K. A superfamily of membrane-bound O-acyltransferases with implications for wnt signaling. Trends Biochem. Sci. 2000;25:111–112. doi: 10.1016/s0968-0004(99)01539-x. [DOI] [PubMed] [Google Scholar]
- Holub B. J. Specific formation of arachidonoyl phosphatidylinositol from 1-acyl-sn-glycero-3-phosphorylinositol in rat liver. Lipids. 1976;11:1–5. doi: 10.1007/BF02532576. [DOI] [PubMed] [Google Scholar]
- Holub B. J. The biosynthesis of phosphatidylserines by acylation of 1-acyl-sn-glycero-3-phosphoserine in rat liver. Biochim. Biophys. Acta. 1980;618:255–262. doi: 10.1016/0005-2760(80)90031-4. [DOI] [PubMed] [Google Scholar]
- Holub B. J., Kuksis A. Structural and metabolic interrelationships among glycerophosphatides of rat liver in vivo. Can. J. Biochem. 1971;49:1347–1356. doi: 10.1139/o71-195. [DOI] [PubMed] [Google Scholar]
- Holub B. J., Kuksis A. Metabolism of molecular species of diacylglycerophospholipids. Adv. Lipid Res. 1978;16:1–125. doi: 10.1016/b978-0-12-024916-9.50007-x. [DOI] [PubMed] [Google Scholar]
- Inoue M., Murase S., Okuyama H. Acyl coenzyme A:phospholipid acyltransferases in porcine platelets discriminate between omega-3 and omega-6 unsaturated fatty acids. Arch. Biochem. Biophys. 1984;231:29–37. doi: 10.1016/0003-9861(84)90359-x. [DOI] [PubMed] [Google Scholar]
- Jain S., Stanford N., Bhagwat N., Seiler B., Costanzo M., Boone C., Oelkers P. Identification of a novel lysophospholipid acyltransferase in Saccharomyces cerevisiae. J. Biol. Chem. 2007;282:30562–30569. doi: 10.1074/jbc.M706326200. [DOI] [PubMed] [Google Scholar]
- Kahn-Kirby A. H., Dantzker J. L., Apicella A. J., Schafer W. R., Browse J., Bargmann C. I., Watts J. L. Specific polyunsaturated fatty acids drive TRPV-dependent sensory signaling in vivo. Cell. 2004;119:889–900. doi: 10.1016/j.cell.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Kamath R. S., Martinez-Campos M., Zipperlen P., Fraser A. G., Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2:1–10. doi: 10.1186/gb-2000-2-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameyama Y., Yoshioka S., Imai A., Nozawa Y. Possible involvement of 1-acyl-glycerophosphorylinositol acyltransferase in arachidonate enrichment of phosphatidylinositol in human platelets. Biochim. Biophys. Acta. 1983;752:244–250. [PubMed] [Google Scholar]
- Keenan R. W., Hokin L. E. The identification of lysophosphatidylinositol and its enzymic conversion to phosphatidylinositol. Biochim. Biophys. Acta. 1962;60:428–430. doi: 10.1016/0006-3002(62)90425-0. [DOI] [PubMed] [Google Scholar]
- Keenan R. W., Hokin L. E. The enzymatic acylation of lysophosphatidylinositol. J. Biol. Chem. 1964;239:2123–2129. [PubMed] [Google Scholar]
- Kennedy S., Wang D., Ruvkun G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature. 2004;427:645–649. doi: 10.1038/nature02302. [DOI] [PubMed] [Google Scholar]
- Koelle M. R., Horvitz H. R. EGL-10 regulates G protein signaling in the C. elegans nervous system and shares a conserved domain with many mammalian proteins. Cell. 1996;84:115–125. doi: 10.1016/s0092-8674(00)80998-8. [DOI] [PubMed] [Google Scholar]
- Lackner M. R., Nurrish S. J., Kaplan J. M. Facilitation of synaptic transmission by EGL-30 Gqalpha and EGL-8 PLCbeta: DAG binding to UNC-13 is required to stimulate acetylcholine release. Neuron. 1999;24:335–346. doi: 10.1016/s0896-6273(00)80848-x. [DOI] [PubMed] [Google Scholar]
- Lands W. E. Metabolism of glycerolipides; a comparison of lecithin and triglyceride synthesis. J. Biol. Chem. 1958;231:883–888. [PubMed] [Google Scholar]
- Lands W. E. Metabolism of glycerolipids. 2. The enzymatic acylation of lysolecithin. J. Biol. Chem. 1960;235:2233–2237. [PubMed] [Google Scholar]
- Lands W. E. Stories about acyl chains. Biochim. Biophys. Acta. 2000;1483:1–14. doi: 10.1016/s1388-1981(99)00177-8. [DOI] [PubMed] [Google Scholar]
- Lands W. E., Crawford C. G. Enzymes of membrane phospholipid metabolism in animals. In: Martonosi A., editor. The Enzymes of Biological Membranes. vol. 2. New York: Plenum Press; 1976. pp. 3–85. [Google Scholar]
- Lands W. E., Merkl I. Metabolism of glycerolipids. III. Reactivity of various acyl esters of coenzyme A with alpha'-acylglycerophosphorylcholine, and positional specificities in lecithin synthesis. J. Biol. Chem. 1963;238:898–904. [PubMed] [Google Scholar]
- MacDonald J. I., Sprecher H. Phospholipid fatty acid remodeling in mammalian cells. Biochim. Biophys. Acta. 1991;1084:105–121. doi: 10.1016/0005-2760(91)90209-z. [DOI] [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D., Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkl I., Lands W. E. Metabolism of glycerolipids. IV. Synthesis of phosphatidylethanolamine. J. Biol. Chem. 1963;238:905–906. [PubMed] [Google Scholar]
- Nakanishi H., Shindou H., Hishikawa D., Harayama T., Ogasawara R., Suwabe A., Taguchi R., Shimizu T. Cloning and characterization of mouse lung-type acyl-CoA:lysophosphatidylcholine acyltransferase 1 (LPCAT1). Expression in alveolar type II cells and possible involvement in surfactant production. J. Biol. Chem. 2006;281:20140–20147. doi: 10.1074/jbc.M600225200. [DOI] [PubMed] [Google Scholar]
- Napier J. A., Michaelson L. V. Genomic and functional characterization of polyunsaturated fatty acid biosynthesis in Caenorhabditis elegans. Lipids. 2001;36:761–766. doi: 10.1007/s11745-001-0782-9. [DOI] [PubMed] [Google Scholar]
- Niwa H., Yamamura K., Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Possmayer F., Scherphof G. L., Dubbelman T. M., van Golde L. M., van Deenen L. L. Positional specificity of saturated and unsaturated fatty acids in phosphatidic acid from rat liver. Biochim. Biophys. Acta. 1969;176:95–110. doi: 10.1016/0005-2760(69)90078-2. [DOI] [PubMed] [Google Scholar]
- Rebecchi M. J., Pentyala S. N. Structure, function, and control of phosphoinositide-specific phospholipase C. Physiol. Rev. 2000;80:1291–1335. doi: 10.1152/physrev.2000.80.4.1291. [DOI] [PubMed] [Google Scholar]
- Rhee S. G. Regulation of phosphoinositide-specific phospholipase C. Annu. Rev. Biochem. 2001;70:281–312. doi: 10.1146/annurev.biochem.70.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riekhof W. R., Wu J., Jones J. L., Voelker D. R. Identification and characterization of the major lyso-phosphatidylethanolamine acyltransferase in Saccharomyces cerevisiae. J. Biol. Chem. 2007;282:28344–28352. doi: 10.1074/jbc.M705256200. [DOI] [PubMed] [Google Scholar]
- Rouser G., Siakotos A. N., Fleischer S. Quantitative analysis of phospholipids by thin-layer chromatography and phosphorus analysis of spots. Lipids. 1966;1:85–86. doi: 10.1007/BF02668129. [DOI] [PubMed] [Google Scholar]
- Saito S., Goto K., Tonosaki A., Kondo H. Gene cloning and characterization of CDP-diacylglycerol synthase from rat brain. J. Biol. Chem. 1997;272:9503–9509. doi: 10.1074/jbc.272.14.9503. [DOI] [PubMed] [Google Scholar]
- Sanjanwala M., Sun G. Y., Cutrera M. A., MacQuarrie R. A. Acylation of lysophosphatidylcholine in bovine heart muscle microsomes: purification and kinetic properties of acyl-CoA:1-acyl-sn-glycero-3-phosphocholine O-acyltransferase. Arch. Biochem. Biophys. 1988;265:476–483. doi: 10.1016/0003-9861(88)90152-x. [DOI] [PubMed] [Google Scholar]
- Sanjanwala M., Sun G. Y., MacQuarrie R. A. Purification and kinetic properties of lysophosphatidylinositol acyltransferase from bovine heart muscle microsomes and comparison with lysophosphatidylcholine acyltransferase. Arch. Biochem. Biophys. 1989;271:407–413. doi: 10.1016/0003-9861(89)90290-7. [DOI] [PubMed] [Google Scholar]
- Satouchi K., Hirano K., Sakaguchi M., Takehara H., Matsuura F. Phospholipids from the free-living nematode Caenorhabditis elegans. Lipids. 1993;28:837–840. doi: 10.1007/BF02536239. [DOI] [PubMed] [Google Scholar]
- Shindou H., Hishikawa D., Nakanishi H., Harayama T., Ishii S., Taguchi R., Shimizu T. A single enzyme catalyzes both platelet-activating factor production and membrane biogenesis of inflammatory cells. Cloning and characterization of acetyl-CoA:LYSO-PAF acetyltransferase. J. Biol. Chem. 2007;282:6532–6539. doi: 10.1074/jbc.M609641200. [DOI] [PubMed] [Google Scholar]
- Shirai R., Morita K., NishiKawa A., Nakatsu N., Fukui Y., Morisaki N., Hashimoto Y. The structural requirement of phosphatidylinositols as substrate of phosphatidylinositol 3-kinase. Tetrahedron Lett. 1999;40:1693–1696. [Google Scholar]
- Stamps A. C., Elmore M. A., Hill M. E., Kelly K., Makda A. A., Finnen M. J. A human cDNA sequence with homology to non-mammalian lysophosphatidic acid acyltransferases. Biochem. J. 1997;326:455–461. doi: 10.1042/bj3260455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki H., Shimada A., Ito Y., Ohya M., Takase J., Miyashita M., Miyagawa H., Nozaki H., Nakayama R., Kumagai H. LPT1 encodes a membrane-bound O-acyltransferase involved in the acylation of lysophospholipids in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 2007;282:34288–34299. doi: 10.1074/jbc.M704509200. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Ikita K., Ashida T., Motoyama Y., Yamaguchi Y., Satouchi K. Effects of growth temperature on the fatty acid composition of the free-living nematode Caenorhabditis elegans. Lipids. 1996;31:1173–1178. doi: 10.1007/BF02524292. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Izuwa S., Tanaka K., Yamamoto D., Takimoto T., Matsuura F., Satouchi K. Biosynthesis of 1,2-dieicosapentaenoyl-sn-glycero-3-phosphocholine in Caenorhabditis elegans. Eur. J. Biochem. 1999;263:189–195. doi: 10.1046/j.1432-1327.1999.00480.x. [DOI] [PubMed] [Google Scholar]
- Trent C., Tsuing N., Horvitz H. R. Egg-laying defective mutants of the nematode Caenorhabditis elegans. Genetics. 1983;104:619–647. doi: 10.1093/genetics/104.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis J. G., Watts J. L., Browse J. Polyunsaturated fatty acid synthesis: what will they think of next? Trends Biochem. Sci. 2002;27:467. doi: 10.1016/s0968-0004(02)02168-0. [DOI] [PubMed] [Google Scholar]
- Watts J. L., Browse J. Genetic dissection of polyunsaturated fatty acid synthesis in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2002;99:5854–5859. doi: 10.1073/pnas.092064799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts J. L., Browse J. Dietary manipulation implicates lipid signaling in the regulation of germ cell maintenance in C. elegans. Dev. Biol. 2006;292:381–392. doi: 10.1016/j.ydbio.2006.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts J. L., Phillips E., Griffing K. R., Browse J. Deficiencies in C20 polyunsaturated fatty acids cause behavioral and developmental defects in Caenorhabditis elegans fat-3 mutants. Genetics. 2003;163:581–589. doi: 10.1093/genetics/163.2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wende H., Volz A., Ziegler A. Extensive gene duplications and a large inversion characterize the human leukocyte receptor cluster. Immunogenetics. 2000;51:703–713. doi: 10.1007/s002510000187. [DOI] [PubMed] [Google Scholar]
- Yamashita A., et al. Reverse reaction of lysophosphatidylinositol acyltransferase. Functional reconstitution of coenzyme A-dependent transacylation system. J. Biol. Chem. 2003;278:30382–30393. doi: 10.1074/jbc.M303391200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.