Abstract
Recent results suggest that cytoplasmic mRNAs can form translationally repressed messenger ribonucleoprotein particles (mRNPs) capable of decapping and degradation, or accumulation into cytoplasmic processing bodies (P-bodies), which can function as sites of mRNA storage. The proteins that function in transitions between the translationally repressed mRNPs that accumulate in P-bodies and mRNPs engaged in translation are largely unknown. Herein, we demonstrate that the yeast translation initiation factor Ded1p can localize to P-bodies. Moreover, depletion of Ded1p leads to defects in P-body formation. Overexpression of Ded1p results in increased size and number of P-bodies and inhibition of growth in a manner partially suppressed by loss of Pat1p, Dhh1p, or Lsm1p. Mutations that inactivate the ATPase activity of Ded1p increase the overexpression growth inhibition of Ded1p and prevent Ded1p from localizing in P-bodies. Combined with earlier work showing Ded1p can have a positive effect on translation, these results suggest that Ded1p is a bifunctional protein that can affect both translation initiation and P-body formation.
INTRODUCTION
Proper control of translation and mRNA degradation is important in the regulation of gene expression. Moreover, translation and mRNA degradation often show an inverse relationship (Coller and Parker, 2004). Insight into the relationships between translation and mRNA decay has come from an understanding of the pathways of mRNA degradation (reviewed in Garneau et al., 2007; Meyer et al., 2004; Parker and Song, 2004; Coller and Parker, 2004). Both major mRNA decay pathways initiate with deadenylation of the poly(A) tail with the predominant deadenylase being the Ccr4p/Pop2p/Not complex. Deadenylation can lead to 3′ to 5′ degradation by the exosome complex, but is often followed by decapping by the Dcp1p/Dcp2p decapping enzyme and 5′ to 3′ degradation by the exonuclease Xrn1p.
The process of translation repression and mRNA decapping involve the formation of translationally repressed messenger ribonucleoprotein particles (mRNPs), which contain factors involved in translation repression and mRNA degradation (reviewed in Anderson and Kedersha, 2006; Eulalio et al., 2007; Parker and Sheth, 2007). These translationally repressed mRNPs can also accumulate in conserved cytoplasmic RNA granules termed processing bodies (P-bodies), which are in dynamic exchange with translating mRNAs. P-bodies and the mRNPs assembled within them are of significant interest because they have been implicated in translation repression (Holmes et al., 2004; Coller and Parker, 2005), normal mRNA decay (Sheth and Parker, 2003; Cougot et al., 2004), nonsense-mediated decay (Unterholzner and Izaurralde, 2004; Sheth and Parker, 2006), micro-RNA (miRNA)-mediated repression (Liu et al., 2005; Pillai et al., 2005), mRNA storage (Brengues et al., 2005; Bhattacharyya et al., 2006), and more recently, RNA viral life cycles (Beliakova-Bethell et al., 2006; Beckham et al., 2007). In addition, P-bodies are related to some germinal and neuronal RNA granules (Barbee et al., 2006; Seydoux and Braun, 2006), which have critical roles in proper embryonic development and neuronal function. Despite the significance of conserved P-body-like complexes and their component mRNPs in diverse biological contexts, the machinery that promotes mRNA movement between polysomes and translationally repressed mRNPs that can accumulate in P-bodies is not understood.
Several observations in the literature suggest that Ded1p might have a role in modulating the distribution of mRNAs between P-bodies and polysomes. Ded1p is a member of the DEAD/DExH family of ATPases (Iost et al., 1999), which generally function to remodel RNA or RNP complexes (reviewed in Linder, 2006). Ded1p has been suggested to promote translation initiation in yeast because it is a high copy suppressor of conditional alleles in the cap binding protein, eIF4E (de la Cruz et al., 1997), mutant alleles of Ded1p inhibit translation in extracts (Chuang et al., 1997), and translation in vitro can be stimulated by the addition of recombinant Ded1p to extracts (Coller and Parker, 2005).
Several observations also raise the possibility that Ded1p and its orthologues have roles in translation repression, P-body formation, and/or mRNA degradation. For example, specific alleles of Ded1p selectively reduce the translation of the Brome Mosaic Virus RNA2 similar to defects in the Lsm1-7p complex, Pat1p, or Dhh1p (Noueiry et al., 2000, 2003). Because the Lsm1-7p complex, Pat1p, and Dhh1p all function in decapping and affect the distribution of mRNAs between P-bodies and polysomes (Coller and Parker, 2005; Teixeira and Parker, 2007), this suggests that Ded1p might function in decapping and/or translation repression. Similarly, the Drosophila melanogaster Ded1p ortholog, Belle, is required for optimal silencing by RNA interference (RNAi; Ulvila et al., 2006), and the Schizosaccharomyces pombe ortholog enhances RNAi when overexpressed (Raponi and Arndt, 2002). Because RNAi can involve the formation of P-bodies (reviewed in Valencia-Sanchez et al., 2006), these latter observations raise the possibility that Ded1p might also be involved in translation repression and/or mRNA degradation. Finally, orthologues of Ded1p are found in P-body–related maternal granules in D. melanogaster (Johnstone et al., 2005) and neuronal granules in mammals (Kanai et al., 2004), which are biochemically and functionally related to P-bodies (Barbee et al., 2006). Taken together, these results suggest that Ded1p might be a conserved component of P-bodies and related RNA granules, thereby affecting the distribution of mRNAs between P-bodies and polysomes.
In this work, we examine the contributions of Ded1p to translation repression, P-body formation and mRNA turnover in yeast. We demonstrate that Ded1p and its ortholog in D. melanogaster localize to P-bodies and related neuronal granules, respectively. Moreover, Ded1p appears to contribute to formation of P-bodies because overexpression of Ded1p results in increased size and number of P-bodies and inhibition of growth in a manner dependent on Pat1p, Dhh1p, and Lsm1p, whereas depletion of Ded1p shows defects in P-body formation. In contrast, overexpression or depletion of Ded1p has little effect on mRNA degradation, suggesting its primary function is in regulation of translation. Combined with earlier work showing Ded1p can have a positive effect on translation, these results suggest that Ded1p is a bifunctional protein that mediates both translation initiation and repression.
MATERIALS AND METHODS
Yeast Strains and Growth Conditions
The genotypes of all strains used in this study are listed in Table 1. The fusion proteins include the full-length protein and are at least partially functional (Sheth and Parker 2003; data not shown). Yeast crosses were carried out using standard laboratory procedures.
Table 1.
Yeast strains used in study
| Strain | Genotype | Reference |
|---|---|---|
| yRP840 | MATaleu2-3,112, trp1, ura3-52, his4-539, cup1::LEU2/PGK1pG/MFA2pG | Hatfield and Parker (1996) |
| yRP1069 | MATaleu2, trp1-Δ1, lys2, cup1::LEU2/PGK1pG/MFA2pG DCP1::ura3 | Hatfield and Parker (1996) |
| yRP1346 | MATaura3, trp1, lys2, leu2, his4, DCP2::TRP1 | Dunckley and Parker (1999) |
| yRP1725 | MATaleu2-3,112, trp1, ura3-52, his4, cup1::LEU2/PGK1pG/MFA2pG, DHH1-GFP-Neo | Sheth and Parker (2003) |
| yRP1727 | MATaleu2, trp1, ura3, his4, cup1::LEU2/PGK1pG/MFA2pG, DCP2-GFP | Sheth and Parker (2003) |
| yRP1728 | MATaleu2-3,112, trp1, ura3-52, his4, cup1::LEU2/PGK1pG/MFA2pG, PAT1-GFP-NEO | Sheth and Parker (2003) |
| yRP2054 | MATaura3Δ0, leu2Δ0, his3Δ1, met15Δ0, xrn1::KanMX | Invitrogen |
| yRP2061 | MATα ura3, lys2, leu2, trp1, cup1::LEU2/PGK1pG/MFA2pG, dcp2::TRP1, HIS4 | She et al. (2006) |
| yRP2065 | BY4741: MATaura3Δ0, leu2Δ0, his3Δ1, met15Δ0 | Invitrogen |
| yRP2066 | MATaura3Δ0, leu2Δ0, his3Δ1, met15Δ0, dhh1::KanMX | Invitrogen |
| yRP2067 | MATaura3Δ0, leu2Δ0, his3Δ1, met15Δ0, pat1::KanMX | Invitrogen |
| yRP2068 | MATaura3Δ0, leu2Δ0, his3Δ1, met15Δ0, lsm1::KanMX | Invitrogen |
| yRP2080 | MATaura3Δ0, leu2Δ0, his3Δ1, met15Δ0, DCP2-GFP-NEO | Invitrogen |
| yRP2210 | MATaura3Δ0, leu2Δ0, his3Δ1, met15Δ0, XRN1-GFP-NEO | Invitrogen |
| yRP2433 | MATahis3–1, met15-0, leu2–0, URA3::CMV-tTA pDED1::kanR-tet07-TATA | Open Biosystems |
| yRP2434 | MATaura3Δ0, leu2Δ0, his3Δ1, met15Δ0, ccr4::KanMX | Invitrogen |
| yRP2435 | MATaura3Δ0, leu2Δ0, his3Δ1, met15Δ0, pop2::KanMX | Invitrogen |
| yRP2436 | YTC127: MATaura3-52, lys2–801, ade2-101, trp1-Δ1, his3Δ200, leu2-Δ1, ded1::TRP1pDED1009(DED1/CEN/LEU2/pRS315) | Chuang et al. (1997) |
Strains were grown on standard yeast extract/peptone medium or the appropriate selective media with either 2% dextrose (Dex), 2% sucrose, or 2% galactose (Gal) as a carbon source. Strains were grown at 30°C.
Tet-off DED1 strain was grown according to Gari et al. (1997) with modifications. Briefly, cells were grown in YEPD or selective media overnight to saturation. Cultures were then split in half and received either doxycycline to a final concentration of 10 μg/ml or the equivalent volume of the 70% EtOH vehicle. After overnight growth at 30°C, the cultures were back-diluted to an OD600 of 0.1 and supplemented with fresh doxycycline to a final concentration of 10 μg/ml or fresh vehicle. Cells were harvested for analysis at an OD600 of 0.3–0.4.
Galactose induction experiments were performed as previously described (Coller and Parker, 2005). Briefly, cells were grown in selective media with 2% sucrose until early midlog an OD600 of 0.25–0.3. Cells were transferred to selective media with 2% galactose and incubated at 30°C for 2 h.
Preparation of Cells for Microscopy
Cells were grown to an OD600 of 0.3–0.35 in appropriate media. For glucose depletion, cells were washed in selective media without dextrose and resuspended in the same media for 10 min. Cells were then washed with selective media without dextrose, resuspended in the same medium, and examined. For cells stressed in water, the cells were treated as described above, except that water was used instead of selective media without glucose. In all other cases, cells were washed once and resuspended in selective media with 2% dextrose and immediately observed.
To analyze the effects of DED1 overexpression on P-bodies, green fluorescent protein (GFP)-tagged strains (Table 1) were transformed with a Gal-DED1 plasmid (Open Biosystems, Huntsville, AL) or 2-μm control vector (pRS426). Cells were grown in selective media with 2% sucrose to an OD600 of 0.25–0.3. Cultures were split in half: one-half was resuspended in selective media with 2% sucrose and the other, in selective media with 2% galactose. Cells were incubated for 2 h before being washed twice and resuspended in the same media and observed.
For observation at high OD600, cells were grown in selective media with 2% dextrose to stationary phase. Cells were then washed and resuspended in selective media with 2% dextrose and immediately observed.
Observations were made using Nikon PCM 2000 confocal microscope (Melville, NY) using a 100× objective with a 3× zoom using Compix software (Sewickley, PA). The average percentage of P-body–containing cells was calculated for all conditions. Each percentage, out of 200 cells, represents the average from three to five experiments.
Plasmids
Plasmids used are summarized in Table 2. Plasmid pRP1557 DED1-GFP was constructed in multiple stages. GFP was amplified from plasmid yEGFP1 (Cormack et al., 1997) using primers X1: 5′CAAACAACTCTTCTTGGTTGTCTAAAGGTGAAGAATTATTC3′; and X2: 5′CCTCACCCTAGTTTGTCTGAAATCATTTGTACAATTCATCCATACC3′ to generate fragment 1. The 3′UTR segment of DED1 was amplified from pDED1/TRP (Noueiry et al., 2000) using primers X3: 5′GGTATGGATGAATTGTACAAATGATTTCAGACAAACTAGGGTGAGG3′; and X4: 5′CCTCTTCGCTATTACGCCA3′ to generate fragment 2. The C terminal portion of the DED1 ORF was amplified from pDED1/TRP using primers X5: 5′GAACAATTCTTCACCTTTAGACCACCAAGAAGAGTTGTTTG3′; and X6: 5′CAGAGATTACCGTAAGGCCGG3′ to generate fragment 3. Fragments 1 and 2 were gel-purified and extended by mutually primed synthesis using primers X1 and X4. The resulting PCR product was gel-purified, mixed with gel-purified fragment 3, and extended by mutually primed synthesis using primers X4 and X6. The final PCR product was cut with NheI and SpeI and cloned into the NheI and SpeI sites of pDED1/TRP. The final construct was verified by sequencing. Note that the construct is under control of the DED1 promoter and contains the 3′ untranslated region of DED1. Plasmid pRP1558 was made by moving the 2530-base pair KpnI-SacI fragment from pRP1557 into pRS416 (Sikorski and Hieter, 1989).
Table 2.
Plasmids used in study
| Plasmid number | Description | Reference |
|---|---|---|
| pRP247 | pRS426 (URA3/2 μ) | Sikorski and Hieter (1989) |
| pRP250 | pRS416 (URA3/CEN) | Sikorski and Hieter (1989) |
| pRP590 | MFA2pG(LEU2/URA3/CEN) | Muhlrad et al. (1994) |
| pRP603 | PGK1pG (LEU2/URA3/CEN) | Muhlrad et al. (1995) |
| pRP1155 | DCP2-RFP (LEU2/CEN) | Sheth and Parker (2003) |
| pRP1175 | DCP2-GFP (LEU2/CEN) | Coller and Parker (2005) |
| pRP1275 | DCP2-GFP (URA3/CEN) | Coller and Parker (2005) |
| pRP1557 | DED1-GFP (TRP1/CEN) | This work |
| pRP1558 | DED1-GFP (URA3/CEN) | This work |
| pRP1559 | Gal-DED1 (URA3/2 μ) | Open Biosystems |
| pRP1560 | DED1 (URA3/CEN) | This work |
| pRP1561 | DED1 (URA3/2 μ) | This work |
| pRP1562 | ded1-DAAD-GFP (URA3/CEN) | This work |
| pRP1563 | DED1 (LEU2/CEN) | Chuang et al. (1997) |
| pRP1564 | Gal-ded1-DAAD (URA3/2 μ) | This work |
Plasmids pRP1560 and pRP1561 were constructed by removing the KpnI-SacI fragment, which contains the DED1 gene, from plasmid pDED1009/pRP1563 (Chuang et al., 1997) into pRS416/pRP250 or pRS426/pRP247 (Sikorski and Hieter, 1989).
The mutant DAAD plasmids were constructed by quick-change mutagenesis as described by Wang and Malcolm (1999). Briefly, the glutamic acid residue of the DEAD motif was changed to alanine by using primers oCJBDed06F: 5′ G GTT CTA GAT GCT GCT GAT AGA ATG TTG GAT ATG GGT TTC GAA CC 3′; and oCJBDed07R: 5′ GG TTC GAA ACC CAT ATC CAA CAT TCT ATC AGC AGC ATC TAG AAC C 3′. The DAAD mutation was introduced into plasmids pRP1558 and pRP1559 to generate pRP1562 and pRP1564, respectively. All mutations were confirmed by DNA sequencing.
Immunofluorescence in Drosophila Neurons
Localization of Belle was determined by indirect immunofluorescence using anti-sera against Belle in Drosophila neurons expressing a dFMR-YFP transgene as a marker of neuronal granules. Neuronal culturing, staining, and microscopy were performed as described earlier (Barbee et al., 2006).
Growth Assays
To analyze the effects of overexpression of DED1 on growth of wild-type or mutant strains, cells were transformed with either a 2-μm control (pRS426/pRP247), the Gal-DED1 (Open Biosystems; pRP1559), or Gal-ded1-DAAD (pRP1564) plasmid. Cells were grown overnight in selective media containing 2% glucose. Cells were back diluted to an OD600 of 0.1 and allowed to double twice. All cultures were spotted at the same concentration onto selective media containing either 2% glucose or galactose. The plates were incubated at 30°C for 6 d.
35S Incorporation
Cultures were grown in the appropriate media to an OD600 of 0.3–0.4, and cells were resuspended in 5 ml of selective media minus methionine. Cyclohexamide samples included cyclohexamide to a final concentration of 10 μg/ml and were incubated for 10 min at 30°C with shaking. For dextrose depletion experiments, the cells were grown in selective media with 2% dextrose to an OD600 of 0.3–0.4. Cells were resuspended in selective media without methionine or dextrose and incubated for 10 min at 30°C with shaking.
At the zero time point, 50 mM methionine was added to each culture along with 1 μCi/ml 35S-labeled promix (Amersham Pharmacia Biotech, Piscataway, NJ). A 1-ml sample was immediately removed and added to 1 ml of 20% trichloroacetic acid (TCA). Remaining cells were placed in 30°C water bath with shaking. After 10 min, a 1-ml aliquot was removed and mixed with 1 ml of 20% TCA.
The TCA-containing samples were boiled for 20 min at 100°C, cooled for 15 min, and filtered through GF/C filters (Whatman, Clifton, NJ). The filters were then washed three times with 5 ml of 10% TCA and one time with 10 ml of 95% EtOH. The filters were dried and placed in 5 ml of scintillation cocktail (Bio-safe II from Research Products International, Mt. Prospect, IL). Labeled methionine/cysteine incorporation into protein was quantitated and normalized to wild-type values. Experiments were repeated between three and five times.
RNA Analysis
Effects of DED1 depletion on RNA stability were achieved using a Tet-off DED1 strain (Open Biosystems) that was transformed with either Gal-MFA2pG LEU2/URA3 (pRP590) or Gal-PGK1 LEU2/URA3 (pRP603) plasmid. Cells were grown as discussed above (Gari et al., 1997). Time points after transcriptional repression by the addition of glucose (Decker and Parker, 1993) were collected.
To analyze the effects of overexpression of DED1 on RNA stability, strain yRP840 was transformed with either Gal-DED1 plasmid (pRP1559; Open Biosystems) or a 2μ control plasmid (pRS426/pRP247). Strain yRP840 contains genomic integration of MFA2pG and PGK1pG inducible with galactose. Cells were grown to an OD600 of 0.25–0.3 in selective media with 2% sucrose. Cultures were harvested and resuspended in selective media with 2% galactose to induce MFA2pG and PGK1pG transcription and the overexpression of DED1. After 2-h induction, the cells were resuspended in selective media with 4% dextrose to shut-off transcription from the Gal promoter; time points were collected after the addition of dextrose.
RNA was prepared by glass bead phenol:chloroform:let preparation (Caponigro et al., 1993). Formaldehyde agarose Northern blots were run, and the RNA was transferred to nylon membranes, which were probed with 32P-labeled oligonucleotides complementary to MFA2pG, PGK1pG, and the loading control SCR1 (oRP140, oRP141, and oRP100, respectively; Decker and Parker, 1993). Blots were scanned using Typhoon 9410 (Amersham Pharmacia Biotech) and quantitated using ImageQuant software (Molecular Dynamics, Sunnyvale, CA).
Polysome Analysis
Polysome analysis was performed with modifications as previously described (Brengeus et al., 2005). Briefly, cellular lysates were prepared from 200 ml of a culture grown under the conditions described above. Cells were pelleted by centrifugation at 7000 rpm for 5 min at 4°C and washed in 20 ml of lysis buffer (20 mM Tris-HCl, pH 8.0, 140 mM KCl, 5 mM MgCl2, 0.5 mM dithiothreitol, 1% Triton-X, 0.1 mg/ml cyclohexamide, 1 mg/ml heparin). Cells were resuspended in 400 μl volume of lysis buffer and 400 μl of glass beads. Cells were vortexed at full speed for 2 min and incubated on ice for 2 min; this step was repeated three times. Clarification of the lysate was performed by centrifugation at 2000 rpm for 2 min at 4°C, and the supernatant was transferred to a new tube. Approximately 20 A254 units were loaded on top of a 15–50% sucrose gradient that contained lysis buffer lacking detergent or cushion. Samples were sedimented in a Beckman SW41 rotor (Fullerton, CA) at 4°C for 2.5 h at 39,000 rpm, and fractions were collected. The A254 value was monitored using a continuous flow cell UV detector.
SDS-PAGE and Western Analysis
SDS-PAGE and Western blot analysis were performed using standard methods. Anti-Ded1p antibody (USO 58) was used at 1:1000 (generous gift from T. Chang, The Ohio State University, Columbus, OH). Stabilized goat anti-rabbit horseradish peroxidase conjugate was used at 1:5000 (Pierce, Rockford, IL). Blots were developed using ECL Western Blotting Detection Kit (GE Healthcare, Waukesha, WI).
RESULTS
Ded1p and Its Orthologues Localize to P-bodies
To examine the possible functions of Ded1p, we first wanted to determine if Ded1p could accumulate in yeast P-bodies. For this experiment, we constructed a Ded1p-GFP fusion protein expressed from a centromeric plasmid under the DED1 promoter. The fusion protein is functional by the criteria that it can complement the lethality of a ded1Δ strain (data not shown).
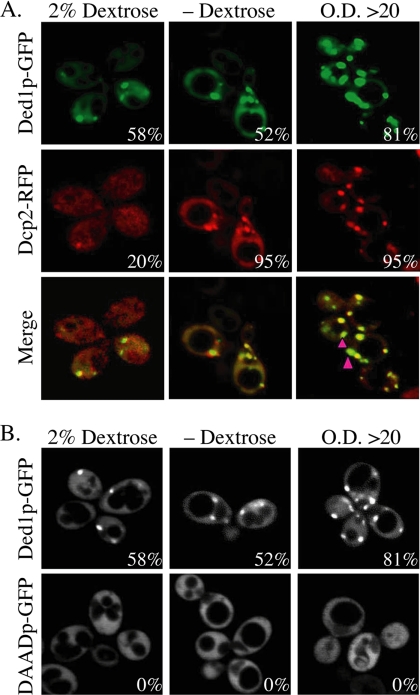
We found that Ded1p-GFP and the P-body marker Dcp2p-RFP colocalize in cytoplasmic foci (Figure 1). Furthermore, similar to Dcp2p and other P-body markers (Teixeira et al., 2005), the size and/or number of Ded1p-containing P-bodies was increased under conditions of general translational repression induced by glucose deprivation or higher cell densities (Figure 1). We also observed that the stoichiometry between Dcp2p and Ded1p could vary between different foci. For example, at high cell densities some foci show high concentrations of Ded1p-GFP with lower levels of Dcp2p-RFP (Figure 1, pink arrows). This difference in relative abundance of Dcp2p and Ded1p argues that these proteins are not stoichiometric components of a shared complex and may represent different types of mRNPs that can accumulate in P-bodies. These results indicate that Ded1p can be a component of yeast P-bodies and thereby might influence the process of translational control and mRNA degradation.
Figure 1.
Ded1p-GFP accumulation in P-bodies is dependent on ATP-hydrolysis. (A) P-body formation of wild-type strain yRP2065 containing Ded1p-GFP (pRP1558) and Dcp2p-RFP (pRP1155) with or without dextrose or at high OD600. (B) yRP2065 containing Ded1p-GFP (pRP1558) or Ded1p-DAAD-GFP (pRP1562). Percentage of foci-containing cells is indicated.
To determine if the localization of Ded1p in P-bodies was conserved in metazoans, we examined the subcellular distribution of the Ded1p ortholog, Belle (Johnstone et al., 2005), in D. melanogaster neurons, which contain large RNA granules related to P-bodies in neurites (Barbee et al., 2006). In early embryogenesis, Belle is distributed throughout the cytoplasm; in later stages, Belle localizes to the posterior pole of oocytes (Johnstone et al., 2005). To determine whether Belle can localize to RNA granules related to P-bodies, we used antisera against Belle to observe the localization of Belle in neurons expressing dFMR-yellow fluorescent protein (YFP), a marker of neuronal RNA granules (Barbee et al., 2006). We observed that Belle colocalizes with dFMR in foci in the neurites (Figure 2A). These dFMR-containing granules contain mRNA and canonical P-body components and are sites of translation repression (Barbee et al., 2006), suggesting they are structurally and functionally related to P-bodies. Belle and dFMR each form large aggregates in the neuronal cell body, which partially colocalize (Figure 2B). Unlike granules in the neurites, Staufen-containing granules in the cell body do not contain RNA (Köhrmann et al., 1999); thus Belle- and dFMR-containing aggregates within the cell body may not depend on RNA for their formation, unlike P-bodies. The colocalization of Belle and dFMR in foci in neurites and previous work showing the localization of the mammalian Ded1 ortholog, DDX3, in RNA granules in neurons (Kanai et al., 2004), argue that Ded1p and its orthologues are conserved components of P-bodies and related RNP granules.
Figure 2.
Belle accumulates in RNA granules in Drosophila neurites. Localization of Belle in neurites (A) and cell bodies (B) as determined by indirect immunofluorescence in Drosophila neurons expressing a dFMR-YFP transgene as a marker of neuronal granules. Neuronal culturing, staining, and microscopy were performed as described earlier (Barbee et al., 2006).
Ded1p ATPase Activity Is Required for Accumulation in P-bodies
As a member of the DEAD-box family of RNA helicases, Ded1p is known to hydrolyze ATP in a manner stimulated by RNA (Iost et al., 1999). Moreover, the ATPase activity is known to be required for the essential Ded1p function in vivo, because a mutation altering the canonical DEAD-box element to DAAD, which inactivates ATP hydrolysis by Ded1p, is unable to complement a ded1Δ strain for viability (de la Cruz et al., 1997; Iost et al., 1999). Given this, we created a Ded1-GFP fusion protein carrying the DAAD mutation and thereby asked if ATPase activity of Ded1p was required for Ded1p to accumulate in P-bodies. We observed that Ded1p-(DAAD)-GFP no longer accumulated in P-bodies (Figure 1), although the Ded1p-(DAAD)-GFP fusion protein was expressed at levels comparable to the wild-type Ded1p-GFP fusion protein (data not shown). This observation indicates that the ATPase activity of Ded1p is required either for its entry into or stable persistence in P-bodies.
Depletion of Ded1p Affects Translation and P-body Formation
Given the connection between Ded1p and P-bodies described above we examined how depletion of Ded1p from yeast cells affected the processes of translation, translation repression, P-body formation, and mRNA degradation. To deplete Ded1p we utilized a strain with the DED1 coding region under the control of a promoter repressed by doxycycline (Open Biosystems). This “Tet-off” DED1 strain was treated with doxycycline, which led to the loss of Ded1p, as revealed by Western analysis (data not shown). Comparison of the Ded1p-depleted culture to the Ded1p-containing culture led to several key observations.
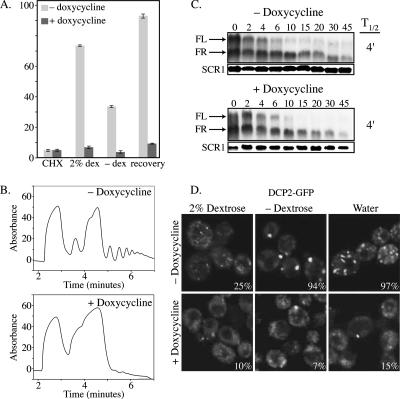
First, we observed that strains depleted of Ded1p showed a substantial decrease in translation rate as assessed by the incorporation of [35S]methionine and by polysome analysis (Figure 3, A and B). On repression of DED1 expression by doxycycline, overall translation is decreased comparable to cyclohexamide treatment (Figure 3A). Polysome analysis shows that depletion of Ded1p results in a loss of polysomes and an increase in the 80S peak (Figure 3B). This is consistent with earlier work suggesting Ded1p has a role in promoting translation initiation and that defects in Ded1p function can lead to the accumulation of unproductive 80S couples (de la Cruz et al., 1997; Chuang et al., 1997).
Figure 3.
Depletion of Ded1p inhibits translation and P-body formation, but not mRNA decay. (A) 35S incorporation in Tet-off DED1 strain (yRP2433) in the absence or presence 10 μg/ml doxycycline. Cells were assayed with 10 μg/ml cyclohexamide in the presence of 2% dextrose (CHX), the presence of 2% dextrose only (dex), or the absence of dextrose (−dex). Recovery refers to a 10-min incubation after the reintroduction of 2% dextrose to cells starved of dextrose. (B) Polysome analysis of Tet-off DED1 strain (yRP2433) in the presence or absence of 10 μg/ml doxycycline. (C) RNA stability is unaffected by depletion of Ded1p. Tet-off strain (yRP2433) transformed with a plasmid carrying MFA2pG (pRP590). (D) Tet-off DED1 strain (yRP2433) carrying Dcp2p-GFP (pRP1175) in the presence or absence of 10 μg/ml doxycycline. Cells were analyzed in the presence of dextrose, in the absence of dextrose, or in water. Percentage of cells with foci is indicated.
Second, we observed that strains depleted of Ded1p showed mRNA decay rates similar to wild type for the MFA2pG or PGK1pG reporter mRNAs (Figure 3C; data not shown). This indicates that Ded1p is not generally required for mRNA decay, although we cannot determine if Ded1p might affect the degradation of a subclass of mRNAs. The absence of an effect on mRNA decay rates after depletion of Ded1p is notably different from defects in general translation initiation factors such as the cap binding protein eIF4E or subunits of eIF3, which show a clear increase in mRNA decay rates when translation is compromised by these defects (Schwartz and Parker, 1999). This suggests that Ded1p has functions distinct from more canonical translation initiation factors.
Finally, we observed that strains depleted of Ded1p showed a decrease in P-bodies, as judged by the accumulation of Dcp2p-GFP in foci, most notable during dextrose deprivation or osmotic stress (Figure 3D). This suggests that Ded1p might have a role in P-body formation. This observation also raises the possibility that Ded1p might have a role in translation repression under stress. However, the large decrease in translation seen in the Ded1p depleted cells even when growing in dextrose limit our ability to determine if Ded1p is required for translation repression during stress.
Taken together, these observations indicate that strains depleted of Ded1p show relatively normal mRNA decay rates, but have reduced translation rates, and a defect in the ability to form P-bodies. These results suggest that in addition to the previously described positive role in translation Ded1p might also have a function in P-body formation.
Overexpression of Ded1p Inhibits Growth and Promotes P-body Formation
The results above suggested that Ded1p is a component of P-bodies and could have a role in P-body formation and possibly the translational control of some mRNAs, similar to other components of P-bodies such as Dhh1p and Pat1p (Coller and Parker, 2005). Previous results have shown that overexpression of DHH1 or PAT1 can inhibit translation and drive P-body formation (Coller and Parker, 2005). Given this, we examined the effects of overexpressing DED1 from the Gal promoter on growth, translation, P-body formation, and mRNA stability.
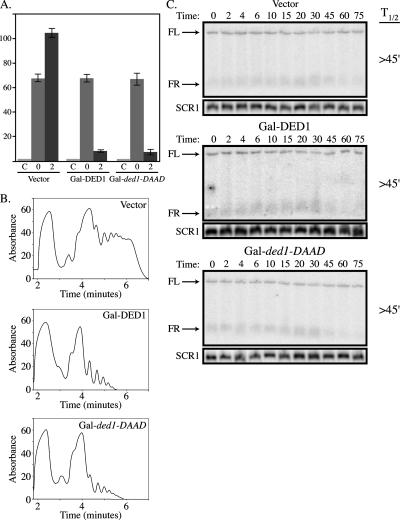
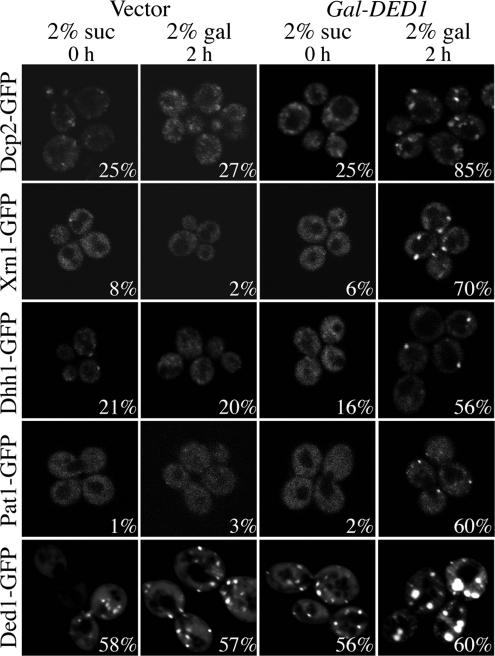
Strikingly, we observed that overexpression of DED1 from Gal UAS on a high copy plasmid led to an inhibition of cell growth (see Figure 6). Moreover, we observed that overexpression of Ded1p or ded1p-DAAD decreased translation rates as judged by the incorporation of 35S methionine and by polysome analysis (Figure 4, A and B). Additionally, overexpression of Ded1p increased P-bodies as judged by the subcellular distribution of GFP-tagged Dcp2p, Dhh1p, Xrn1p, Pat1p, or Ded1p itself (Figure 5). In contrast, we observed that overexpression of DED1 had no significant effect on the degradation rates of the MFA2pG and PGK1pG reporters (Figure 4C; data not shown). Because Ded1p overexpression decreases translation and drives mRNAs into P-bodies in a manner that does not accelerate mRNA degradation, these data are consistent with Ded1p promoting translation repression.
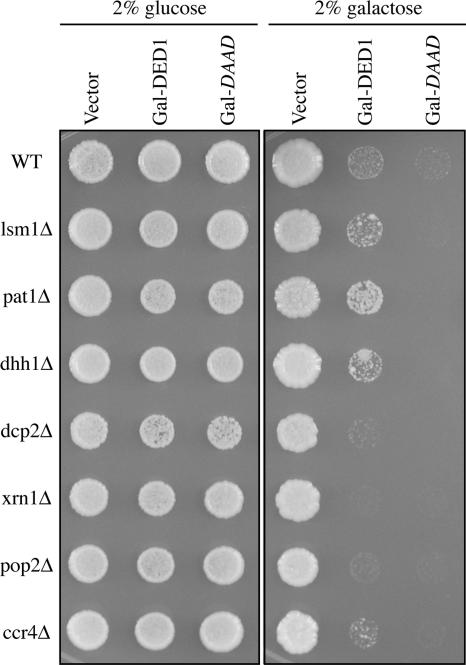
Figure 6.
Overexpression of DED1 confers a growth defect that is partially suppressed by deletion of PAT1, DHH1, or LSM1. Wild-type (yRP2065) or deletion strains (see Table 1) were transformed with either 2μ control vector (pRP247), Gal-DED1 (pRP1559), or Gal-ded1-DAAD (pRP1564) and grown on selective media containing either 2% dextrose or 2% galactose.
Figure 4.
Overexpression of DED1 or the ATP-hydrolysis mutant inhibits translation, but does not affect mRNA stability. (A) Wild-type strain yRP840 was transformed with either a 2μ vector (pRP247), Gal-DED1 (pRP1559), or mutant Gal-ded1-DAAD (pRP1564). 35S incorporation was measured in the presence of cyclohexamide (C), before galactose induction of overexpression (0), and after 2 h of induction (2). (B) Polysome analysis of the strains as described in A after a 2-h induction in 2% galactose. (C) Decay analysis of PGK1pG mRNA in the strains described in A after a 2-h induction in 2% galactose.
Figure 5.
Overexpression of DED1 can promote the formation of P-bodies. For the Ded1-GFP strain (bottom row), yRP840 was transformed with DED1-GFP (pRP1557) and either 2μ control vector (pRP247) or Gal-DED1 (pRP1559). For all other strains, a strain containing a GFP-tagged RNA metabolism factor (see Table 1) was transformed with either 2μ control vector (pRP247) or Gal-DED1 (pRP1559). All strains were monitored in 2% sucrose (suc) before induction (0 h) and after 2 h (2 h) of induction in selective media containing 2% galactose (gal).
In addition to its effect on translation and P-body formation, we observed that overexpression of DED1 causes growth inhibition (Figure 6). To examine if the inhibition of growth was related to P-bodies and/or mRNA decay, we examined how overexpression of DED1 affected the growth of cells lacking components of P-bodies and/or degradative enzymes. Overexpression of DED1 still inhibited the growth of dcp2Δ, xrn1Δ, pop2Δ, and ccr4Δ strains (Figure 6), arguing that DED1 overexpression does not inhibit growth by triggering premature mRNA degradation, which is consistent with overexpression of DED1 not altering the decay rates of the mRNAs we examined. In fact, deletion of DCP2, XRN1, or POP2 exacerbated the growth defect conferred by overexpression of DED1, suggesting that these degradative enzymes normally antagonize Ded1p function. More importantly, we observed that deletion of PAT1, LSM1, or DHH1 allowed partial suppression of the growth inhibition due to overexpression of DED1, with the pat1Δ being a more effective suppressor than lsm1Δ or dhh1Δ (Figure 6). Because Pat1p, Dhh1p, and Lsm1p function in translation repression, P-body formation, and mRNA decapping, this provides evidence that the growth inhibition induced by Ded1p overexpression is related to translation repression and/or P-body formation. Consistent with this interpretation, we observed that the pat1Δ strain show reduced formation of P-bodies compared with the wild-type strain, in response to Ded1p overexpression (data not shown).
Overexpression Inhibits Growth by Acting Upstream of P-Bodies
The results above indicate that overexpression of Ded1p inhibits cell growth in a manner that leads to the accumulation of mRNPs in P-bodies. In principle, the repression of translation could affect mRNA upstream of P-bodies, or trap mRNA in an mRNP that accumulates in the P-body complex. Because the DAAD allele of Ded1p blocks ATP-hydrolysis and prevents Ded1p accumulation in P-bodies, we used this allele to determine if the inhibition of growth by Ded1p requires formation of P-bodies.
We observed that overexpression of the DAAD allele of Ded1p inhibited cell growth to a greater extent than overexpression of the wild-type DED1 (Figure 6) and also inhibited translation as assessed by the incorporation of labeled amino acids or polysome analysis (Figure 4, A and B). Moreover, overexpression of Ded1p-DAAD still inhibited growth in the deletion strains, including those deletions that were previously identified as suppressors of the defect conferred by overexpression of Ded1p (Figure 6), suggesting that cooperation between Ded1p and translation repression factors requires the ATP hydrolysis activity of Ded1p or its presence in P-bodies. Because the Ded1p-DAAD protein does not accumulate in P-bodies (Figure 1), the simplest interpretation of these observations is that the inhibition of growth by Ded1p overexpression occurs upstream of the formation of mRNAs into P-bodies. However, we cannot rule out the formal possibility that when the Ded1p-DAAD allele is overexpressed to high levels it becomes concentrated in P-bodies.
Ded1p Fractionates Primarily with RNPs
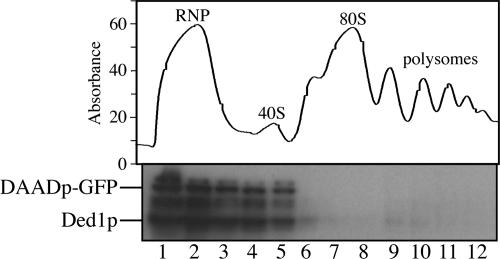
The observations above suggest that Ded1p overexpression inhibits cell growth at a stage upstream of P-body formation. Moreover, given that Ded1p can function in translation initiation, these results raise the possibility that Ded1p has a cyclical function where it first associates with initiation complexes or translating mRNPs in polysomes to enhance translation, but can also promote the movement of mRNAs and itself into P-bodies, in a manner requiring ATP-hydrolysis by Ded1p. We examined the association of Ded1p and the DAAD allele with initiation complexes and polysomes by sucrose gradients. We observed that both wild-type Ded1p and the DAAD allele of ded1 were present in top fractions of a sucrose gradient (Figure 7), which contain a mixture of RNPs including P-body components, 40S subunits, and mRNPs. Longer exposures of the Western blotting show that small amounts of Ded1p fractionate with polysomes (data not shown), suggesting that Ded1p may transiently interact with polysomes to affect translation initiation or translation repression. Like other P-body components, such as Dcp2p (data not shown), Ded1p does fractionate with the mRNP fraction, suggesting that Ded1 behaves like a P-body component. Although only wild-type Ded1p colocalizes to P-bodies, both Ded1p and ded1-DAADp associate with the RNP fraction. This likely reflects the complexity of the RNP fraction. These data indicate that Ded1p fractionates primarily in the fractions containing RNPs and 40S subunits.
Figure 7.
Ded1p fractionates with RNPs and the 40S subunit. A polysome analysis of wild type strain yRP840 transformed with ded1-DAAD-GFP (pRP1562) and a Western blot of collected fractions probed with anti-Ded1p antibody.
DISCUSSION
Several observations argue that Ded1p and its orthologues in other species can be components of P-bodies and P-body-like RNP granules. First, we observe Ded1p is a component of P-bodies in Saccharomyces cerevisiae (Figure 1). Second, we observe that the ortholog in D. melanogaster, Belle, is a component of neuronal RNA granules (Figure 2), which are functionally related to P-bodies (Barbee et al., 2006). This is consistent with earlier work that has observed Ded1p orthologues in germinal granules in D. melanogaster (Johnstone et al., 2005), which are also related to P-bodies (reviewed in Parker and Sheth, 2007; Seydoux and Braun, 2006), and previous work showing the localization of the mammalian Ded1 ortholog, DDX3, in RNA granules in neurons (Kanai et al., 2004). Taken together, these observations indicate that Ded1p and its orthologues are conserved components of translationally repressed mRNPs and thereby may play a role in modulating translation.
Several observations suggest that one function of Ded1p is to enhance the rate of translation initiation. This was first suggested by biochemical evidence showing that depletion of Ded1p from extracts reduced their ability to translate exogenously provided mRNA (Chuang et al., 1997). Moreover, Ded1p is a high copy suppressor of conditional alleles in eIF4E (de la Cruz et al., 1997) and shows genetic interactions with other translation factors (Tseng-Rogenski et al., 2003). Finally, addition of recombinant Ded1p to cell-free extracts enhances their ability to translate exogenous mRNA (Coller and Parker, 2005). Moreover, we observe that depletion of Ded1p led to decreases in translation as assessed by 35S methionine incorporation and polysome analysis (Figure 3). Thus, one function of Ded1p appears to be to enhance translation rate, by a yet-to-be-defined mechanism.
We now present several lines of evidence suggesting that Ded1p may have an additional role in the formation of P-bodies. This was first implied by the presence of Ded1p and its orthologues in P-bodies and related granules (Figures 1 and 2; Kanai et al., 2004). Moreover, depletion of Ded1p compromises the ability of yeast cells to form P-bodies (Figure 3D), suggesting that Ded1p affects the formation of P-bodies. Because P-bodies are generally devoid of initiation factors, this dual role of Ded1p in translation initiation and P-body formation is unique. In addition, overexpression of DED1 leads to growth inhibition, inhibition of translation, and an increased number of P-bodies (Figures 4–6). Importantly, the lethality conferred by overexpression of DED1 is suppressed by pat1Δ and to a lesser extent by lsm1Δ and dhh1Δ (Figure 6). Because Pat1p, Dhh1p, and Lsm1p function in aspects of translation repression and mRNA decapping (Coller and Parker, 2004, 2005), this observation suggests that growth inhibition by Ded1p overexpression is due to hyperactivation of translation repression, arguing that Ded1p has a role in translation repression. Moreover, because RNAi can involve the formation of P-bodies (for discussion see Valencia-Sanchez et al., 2006), a role for Ded1p in translational repression and P-body formation is also consistent with the D. melanogaster ortholog Belle being required for optimal RNAi in S2 cells (Ulvila et al., 2006) and overexpression of the S. pombe ortholog increasing RNAi (Raponi and Arndt, 2002). Thus, Ded1p appears to be a bifunctional protein with roles in both translation initiation and P-body formation and thereby may play a role in controlling the distribution of mRNAs between translation and the untranslating mRNPs, which accumulate in P-bodies.
Ded1p appears to function in translation repression upstream of P-body formation. Overexpression of Ded1p increases P-bodies and confers a translation and growth defect that is dependent on the presence of translation repression factors (Figure 6), suggesting that overexpression of Ded1p results in hyperactivation of translation repression. Overexpression of a ded1 mutant defective for ATP hydrolysis, ded1-DAAD, similarly causes growth and translation inhibition (Figure 6), yet ded1p-DAAD cannot accumulate in P-bodies (Figure 1), suggesting that P-body formation is not necessarily required for the defects conferred by hyperactive Ded1p. Thus, ded1p-DAAD is likely stalled in a complex upstream of P-bodies and hydrolysis of ATP is required to join P-bodies. One possibility is that Ded1p functions to help commit an mRNA either to translation or to a repression state that leads to accumulation in P-bodies.
Although the observations that both depletion and overexpression of Ded1p lead to a decrease in translation seem contradictory, their effects on P-body formation are different, suggesting different mechanisms of reducing translation. We hypothesize that these differences reflect a dual function for Ded1p in promoting translation and repressing translation. Depletion of Ded1p reduces translation (Figure 3), consistent with previous work suggesting that Ded1p promotes translation initiation (Chuang et al., 1997; de la Cruz et al., 1997; Tseng-Rogenski et al., 2003; Coller and Parker, 2005), but prevents P-body formation (Figure 3D), suggesting Ded1p promotes P-body formation. In contrast, overexpression of Ded1p also decreases translation (Figure 4), but increases P-body formation (Figure 5) and inhibits growth in a manner dependent on the presence of translation repression factors, such as Pat1p (Figure 6), suggesting that Ded1p cooperates with repression factors. We hypothesize that overexpression of Ded1p causes an increase in Ded1p-driven translation repression activity to the detriment of the cell. By deleting other factors that promote translation repression, the hyperactivity of translation repression by Ded1p is balanced. A second possible explanation for the growth inhibition conferred by Ded1p overexpression is that the increased levels of Ded1p titrates away a limiting translation initiation factor(s). Although we cannot currently rule out this possibility, it is unlikely that deletion of translation repression factors would increase the availability of limiting translation initiation factors. Thus we favor the hypothesis that both depletion and overexpression of Ded1p causes a decrease in translation because Ded1p functions in both translation initiation and translation repression and/or P-body formation. Other proteins that localize to P-bodies can have both positive and negative roles in translation. For example, Pat1p is typically a translation repressor (Coller and Parker, 2005), but is required for the translation of some specific mRNAs (Noueiry et al., 2003). Thus, the possibility that Ded1p has both positive and negative roles in translation is not without precedent.
Ded1p is an attractive protein for modulating the distribution of mRNAs between polysome and P-bodies for several reasons. First, as discussed above Ded1p appears to have both positive and negative roles in translation. Second, the S. pombe ortholog of Ded1p is known to be modified and/or degraded during various cellular stresses that affect translation, suggesting it might be involved in an important step of regulation (Liu et al., 2002). In addition, the S. pombe ortholog interacts with the Chk1 kinase and may be modified by this kinase under some conditions (Liu et al., 2002). Future work identifying the nature of Ded1p modifications and their functional significance may shed light on the control of translation and translation repression.
A possible cyclical model for Ded1p function can now be proposed with the following salient points. First, Ded1p interacts with the translation machinery to enhance some aspect of translation initiation. This is also consistent with physical interactions of Ded1p seen with translation initiation factors in various genomic screens (Krogan et al., 2004; Collins et al., 2007; Gavin et al., 2006) and the role of Ded1p in enhancing translation per se (Chuang et al., 1997; de la Cruz et al., 1997). In a subsequent functional step, Ded1p may play a role in repressing the translation of some mRNAs and promoting their movement from translation to P-bodies. As Ded1p is a DEAD-box ATPase, it may interact with mRNA within P-bodies to promote translation repression or to maintain localization of the mRNA in P-bodies. However, we have not ruled out that Ded1p localizes to P-bodies independently of its ability to bind RNA.
The observation that Ded1p and its orthologues are in P-bodies provides possible explanations for the connection of the mammalian ortholog DDX3 to viral life cycles. For example, the core protein for hepatitis C virus (HCV) particles physically interacts with Ddx3p and colocalizes in cytoplasmic foci, which our data would now suggest are P-bodies (Mamiya and Worman, 1999; You et al., 1999). This is consistent with recent results that P-bodies might play important roles in the replication of plus-strand RNA viruses (Beckham et al., 2007). Strikingly, the replication of HCV is increased by interaction of the viral RNA with the liver-specific miRNA, miR122, which binds to the 5′ noncoding region of the HCV genome (Jopling et al., 2005). Because miRNAs and RISC can target RNAs to P-bodies (Liu et al., 2005; Pillai et al., 2005), this raises the speculative model that entry of HCV RNAs into P-bodies might be important for efficient replication and be assisted by DDX3 in some manner. Consistent with that possibility, recent results demonstrate that DDX3 is required for HCV replication in cultured cells (Ariumi et al., 2007).
A second intriguing connection between DDX3 and mammalian viruses has come from the observation that DDX3 is required for the export and translation of unspliced HIV-1 from the nucleus (Yedavalli et al., 2004). This is striking because Ded1p/DDX3 and other components of P-bodies may be nuclear-shuttling proteins and have been proposed to play a role in the export of mRNAs directly into the P-body state (Parker and Sheth, 2007). Interestingly, analysis of the Ty3 retrotransposon suggests that its viral particles associate with and may assemble at P-bodies (Beliakova-Bethell et al., 2006). Taken together, these data raise the possibility that Ded1p first engages the unspliced HIV-1 RNA in the nucleus, leading to export in a manner that targets the HIV-1 RNA to a P-body after export, which might be important for subsequent packaging. Given this, a more detailed understanding of Ded1p function should not only inform as to the mechanisms of mRNA translational control but is likely to provide insight into the life cycle of important pathogenic mammalian viruses.
ACKNOWLEDGMENTS
We thank the members of the Parker lab and Anne Webb for helpful discussions and for assistance with the manuscript. We also thank T. Chang for plasmids, strains, and antisera. This work was supported by the Howard Hughes Medical Institute and the National Institutes of Health Grants GM45443, DA15495, and DA17749 and the Science Foundation of Ireland.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-09-0954) on December 26, 2007.
REFERENCES
- Anderson P., Kedersha N. RNA granules. J. Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariumi Y., Kuroki M., Abe K. I., Dansako H., Ikeda M., Wakita T, Kato N. DDX3 DEAD box RNA helicase is required for hepatitis C virus (HCV) RNA replication. J. Virol. 2007 doi: 10.1128/JVI.01517-07. Sept 12 Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbee S. A., et al. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron. 2006;52:997–1009. doi: 10.1016/j.neuron.2006.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beliakova-Bethell N., Beckham C., Giddings T. H., Winey M., Parker R., Sandmeyer S. Virus-like particles of the Ty3 retrotransposon assemble in association with P-body components. RNA. 2006;12:94–101. doi: 10.1261/rna.2264806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckham C. J., Light H. R., Amar Nissan T., Ahlquist P., Parker R., Noueiry A. Interactions between Brome Mosaic Virus RNAs and cytoplasmic processing bodies. J. Virol. 2007;81:9759–9768. doi: 10.1128/JVI.00844-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S. N., Habermacher R., Martine U., Closs E. I., Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Brengues M., Teixeira D., Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caponigro G., Muhlrad D., Parker R. A small segment of the MAT alpha 1 transcript promotes mRNA decay in Saccharomyces cerevisiae: a stimulatory role for rare codons. Mol. Cell. Biol. 1993;13:5141–5148. doi: 10.1128/mcb.13.9.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang R. Y., Weaver P. L., Liu Z., Chang T. H. Requirement of the DEAD-Box protein ded1p for messenger RNA translation. Science. 1997;275:1468–1471. doi: 10.1126/science.275.5305.1468. [DOI] [PubMed] [Google Scholar]
- Coller J., Parker R. Eukaryotic mRNA decapping. Annu. Rev. Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- Coller J., Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins S. R., Kemmeren P., Zhao X. C., Greenblatt J. F., Spencer F., Holstege F. C., Weissman J. S., Krogan N. J. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol. Cell Proteomics. 2007;6:439–450. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- Cormack B. P., Bertram G., Egerton M., Gow N.A.R., Falkow S., Brown A.J.P. Yeast-enhanced green fluorescent protein (yEGFP) a reporter of gene expression in Candida albicans. Microbiology. 1997;143(Pt. 2):303–311. doi: 10.1099/00221287-143-2-303. [DOI] [PubMed] [Google Scholar]
- Cougot N., Babajko S., Seraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J., Iost I., Kressler D., Linder P. The p20 and Ded1 proteins have antagonistic roles in eIF4E-dependent translation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1997;94:5201–5206. doi: 10.1073/pnas.94.10.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker C. J., Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- Eulalio A., Behm-Ansmant I., Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nat. Rev. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- Gari E., Piedrafita L., Aldea M., Herrero E. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast. 1997;13:837–848. doi: 10.1002/(SICI)1097-0061(199707)13:9<837::AID-YEA145>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Garneau N. L., Wilusz J., Wilusz C. J. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- Gavin A. C., et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Holmes L. E., Campbell S. G., De Long S. K., Sachs A. B., Ashe M. P. Loss of translational control in yeast compromised for the major mRNA decay pathway. Mol. Cell. Biol. 2004;24:2998–3010. doi: 10.1128/MCB.24.7.2998-3010.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iost I., Dreyfus M., Linder P. Ded1p, a DEAD-box protein required for translation initiation in Saccharomyces cerevisiae, is an RNA helicase. J. Biol. Chem. 1999;274:17677–17683. doi: 10.1074/jbc.274.25.17677. [DOI] [PubMed] [Google Scholar]
- Johnstone O., Deuring R., Bock R., Linder P., Fuller M. T., Lasko P. Belle is a Drosophila DEAD-box protein required for viability and in the germ line. Dev. Biol. 2005;277:92–101. doi: 10.1016/j.ydbio.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Jopling C. L., Yi M., Lancaster A. M., Lemon S. M., Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- Kanai Y., Dohmae N., Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Köhrmann M., Luo M., Kaether C., DesGroseillers L., Dotti C. G., Kiebler M. A. Microtubule-dependent recruitment of Staufen-green fluorescent protein into large RNA-containing granules and subsequent dendritic transport in living hippocampal neurons. Mol. Biol. Cell. 1999;10:2945–2953. doi: 10.1091/mbc.10.9.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan N. J., et al. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell. 2004;13:225–239. doi: 10.1016/s1097-2765(04)00003-6. [DOI] [PubMed] [Google Scholar]
- Linder P. Dead-box proteins: a family affair—active and passive players in RNP-remodeling. Nucleic Acids Res. 2006;34:4168–4180. doi: 10.1093/nar/gkl468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. Y., Nefsky B. S., Walworth N. C. The Ded1 DEAD box helicase interacts with Chk1 and Cdc2. J. Biol. Chem. 2002;277:2637–2643. doi: 10.1074/jbc.M109016200. [DOI] [PubMed] [Google Scholar]
- Liu J., Valencia-Sanchez M. A., Hannon G. J., Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamiya N., Worman H. J. Hepatitis C virus core protein binds to a DEAD box RNA helicase. J. Biol. Chem. 1999;274:15751–15756. doi: 10.1074/jbc.274.22.15751. [DOI] [PubMed] [Google Scholar]
- Meyer S., Temme C., Wahle E. Messenger RNA turnover in eukaryotes: pathways and enzymes. Crit. Rev. Biochem. Mol. Biol. 2004;39:197–216. doi: 10.1080/10409230490513991. [DOI] [PubMed] [Google Scholar]
- Muhlrad D., Decker C. J., Parker R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′→3′ digestion of the transcript. Genes Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- Muhlrad D., Decker C. J., Parker R. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol. Cell. Biol. 1995;15:2145–2156. doi: 10.1128/mcb.15.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noueiry A. O., Chen J., Ahlquist P. A mutant allele of essential, general translation initiation factor DED1 selectively inhibits translation of a viral mRNA. Proc. Natl. Acad. Sci. USA. 2000;97:12985–12990. doi: 10.1073/pnas.240460897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noueiry A. O., Diez J., Falk S. P., Chen J., Ahlquist P. Yeast Lsm1p-7p/Pat1p deadenylation-dependent mRNA-decapping factors are required for brome mosaic virus genomic RNA translation. Mol. Cell. Biol. 2003;23:4094–4106. doi: 10.1128/MCB.23.12.4094-4106.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R., Sheth U. P bodies and the control of mRNA translation and degradation. Mol. Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Parker R., Song H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- Pillai R. S., Bhattacharyya S. N., Artus C. G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- Raponi M., Arndt G. M. Dominant genetic screen for cofactors that enhance antisense RNA-mediated gene silencing in fission yeast. Nucleic Acids Res. 2002;30:2546–2554. doi: 10.1093/nar/30.11.2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D. C., Parker R. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:5247–5256. doi: 10.1128/mcb.19.8.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux G., Braun R. E. Pathway to totipotency: lessons from germ cells. Cell. 2006;127:891–904. doi: 10.1016/j.cell.2006.11.016. [DOI] [PubMed] [Google Scholar]
- She M., Decker C. J., Chen N., Tumati S., Parker R., Song H. Crystal structure of and functional analysis of Dcp2p from Schizosaccharomyces pombe. Nat. Struct. Mol. Biol. 2006;13:63–70. doi: 10.1038/nsmb1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U., Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U., Parker R. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell. 2006;125:1095–1109. doi: 10.1016/j.cell.2006.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D., Parker R. Analysis of P-body assembly in Saccharomyces cerevisiae. Mol. Biol. Cell. 2007;18:2274–2287. doi: 10.1091/mbc.E07-03-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D., Sheth U., Valencia-Sanchez M. A., Brengues M., Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng-Rogenski S. S., Chong J. L., Thomas C. B., Enomoto S., Berman J., Chang T. H. Functional conservation of Dhh1p, a cytoplasmic DExD/H-box protein present in large complexes. Nucleic Acid Res. 2003;31:4995–5002. doi: 10.1093/nar/gkg712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulvila J., Parikka M., Kleino A., Sormunen R., Ezekowitz R. A., Kocks C., Ramet M. Double-stranded RNA is internalized by scavenger receptor-mediated endocytosis in Drosophila S2 cells. J. Biol. Chem. 2006;281:14370–14375. doi: 10.1074/jbc.M513868200. [DOI] [PubMed] [Google Scholar]
- Unterholzner L., Izaurralde E. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol. Cell. 2004;16:587–596. doi: 10.1016/j.molcel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- Valencia-Sanchez M. A., Liu J., Hannon G. J., Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- Wang W., Malcolm B. A. Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using QuikChange site-directed mutagenesis. BioTechniques. 1999;26:680–682. doi: 10.2144/99264st03. [DOI] [PubMed] [Google Scholar]
- Yedavalli V. S., Neuveut C., Chi Y. H., Kleiman L., Jeang K. T. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell. 2004;119:381–392. doi: 10.1016/j.cell.2004.09.029. [DOI] [PubMed] [Google Scholar]
- You L. R., Chen C. M., Yeh T. S., Tsai T. Y., Mai R. T., Lin C. H., Lee Y. H. Hepatitis C virus core protein interacts with cellular putative RNA helicase. J. Virol. 1999;73:2841–2853. doi: 10.1128/jvi.73.4.2841-2853.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]