Abstract
Yarrowia lipolytica Pex23p and Saccharomyces cerevisiae Pex30p, Pex31p, and Pex32p comprise a family of dysferlin domain–containing peroxins. We show that the deletion of their Pichia pastoris homologues, PEX30 and PEX31, does not affect the function or division of methanol-induced peroxisomes but results in fewer and enlarged, functional, oleate-induced peroxisomes. Synthesis of Pex30p is constitutive, whereas that of Pex31p is oleate-induced but at a much lower level relative to Pex30p. Pex30p interacts with Pex31p and is required for its stability. At steady state, both Pex30p and Pex31p exhibit a dual localization to the endoplasmic reticulum (ER) and peroxisomes. However, Pex30p is localized mostly to the ER, whereas Pex31p is predominantly on peroxisomes. Consistent with ER-to-peroxisome trafficking of these proteins, Pex30p accumulates on peroxisomes upon overexpression of Pex31p. Additionally, Pex31p colocalizes with Pex30p at the ER in pex19Δ cells and can be chased from the ER to peroxisomes in a Pex19p-dependent manner. The dysferlin domains of Pex30p and Pex31p, which are dispensable for their interaction, stability, and subcellular localization, are essential for normal peroxisome number and size. The growth environment-specific role of these peroxins, their dual localization, and the function of their dysferlin domains provide novel insights into peroxisome morphogenesis.
INTRODUCTION
Peroxisomes are highly dynamic because their size, number, protein composition, and biochemical functions vary dramatically depending on the organism, cell type, and environmental milieu. They divide both by growth and fission of pre-existing peroxisomes and by biogenesis from the endoplasmic reticulum (ER). They comprise an essential subcellular compartment that is necessary for proper development, differentiation, and human survival, playing central roles in lipid metabolism, decomposition of toxic hydrogen peroxide, and, depending on the organism, in the metabolism of amino acids, purines, methanol, bile acids, cholesterol, and fatty acids (Wanders, 2004). Dysfunctional peroxisome assembly causes a group of lethal, autosomal recessive peroxisome biogenesis disorders in humans (Steinberg et al., 2006). In yeasts, the peroxisome is the only compartment in which the β-oxidation of fatty acids occurs so that peroxisomes are required for growth on medium containing fatty acids, such as oleic acid, as the sole carbon source (Poirier et al., 2006). Methylotrophic yeasts such as Pichia pastoris (Pp) also use peroxisomes to oxidize methanol (van der Klei et al., 2006).
All peroxisomal membrane proteins (PMP) and metabolic enzymes in the peroxisome matrix are encoded by nuclear genes and are post-translationally targeted to peroxisomes. Like the sorting of proteins to other subcellular compartments, protein targeting to peroxisomes is signal-dependent (Subramani et al., 2000). Most peroxisomal matrix proteins possess a peroxisome-targeting signal (PTS) 1, a C-terminal tripeptide, recognized by the PTS1 receptor, Pex5p. A few matrix proteins are targeted using a PTS2 sequence, a N-terminal nonapeptide, and its cognate PTS2 receptor, Pex7p. These receptors, when bound to cargoes, dock at the peroxisome membrane, and cargoes are imported into the matrix through the importomer complex (Rayapuram and Subramani, 2006). After releasing their cargoes, PTS receptors recycle to the cytosol (Dammai and Subramani, 2001; Nair et al., 2004; Leon et al., 2006). The sorting of PMPs is less well understood. Pex19p may act as a shuttling receptor for a subset of PMPs and/or as a chaperone assisting the assembly of multimeric complexes after their docking at the peroxisome membrane. Pex3p serves as the membrane-anchoring site on the peroxisomal membrane for Pex19p (Fujiki et al., 2006). Recent evidence shows that Pex3p appears at the ER transiently after synthesis and then appears, in a Pex19p-dependent manner, on mature peroxisomes, suggesting an involvement of the ER in peroxisomes biogenesis (Hoepfner et al., 2005).
The PTS receptors, importomer and components of the PTS receptor recycling complex, which are essential for the assembly of functional peroxisomes, form a “classical” peroxin group. A deficiency in any member of this group blocks peroxisome biogenesis, resulting in empty peroxisomes (“ghosts”), small peroxisome remnants or, rarely, no detectable peroxisomes. In contrast, a “nonclassical” peroxin group is involved in peroxisome proliferation. Peroxisomes in cells lacking components of this group may still be functional but are of abnormal number and size. Pex11p was the first member of this group to be described (Erdmann and Blobel, 1995). Overexpression of Pex11p leads to the formation of peroxisomes that are more abundant but smaller than those of wild-type cells, whereas cells lacking Pex11p have fewer but larger peroxisomes than normal. Similar phenotypes are also found in Saccharomyces cerevisiae (Sc) cells lacking Pex25p or Pex27p, which actually share extensive sequence similarity with Pex11p (Smith et al., 2002; Rottensteiner et al., 2003; Tam et al., 2003). In contrast, the absence of ScPex28p or ScPex29p, which are homologues of Yarrowia lipolytica (Yl) Pex24p (Tam and Rachubinski, 2002) leads to more, but smaller and clustered, peroxisomes (Vizeacoumar et al., 2003). The most recently identified peroxins of S. cerevisiae, ScPex30p, ScPex31p, and ScPex32p, which are homologues of YlPex23p (Brown et al., 2000), form another family controlling peroxisome morphogenesis. Cells without members of this family usually exhibit either a greater number of normal-size peroxisomes (Scpex30Δ) or a somewhat normal number of enlarged peroxisomes (Scpex31Δ or Scpex32Δ; Vizeacoumar et al., 2004). In addition, dynamin-related proteins and their peroxisomal receptors are also involved in peroxisome division and thus affect the size and number of peroxisomes (Yan et al., 2005). Recently identified peroxisomal membrane proteins, Inp1p and Inp2p, which perform antagonistic functions in regulating peroxisome inheritance in S. cerevisiae, control the distribution of peroxisomes during mitosis and influence the number of peroxisomes in mother and daughter cells (Fagarasanu et al., 2007).
A search of the P. pastoris genome database revealed two proteins homologous to YlPex23p, ScPex30p, ScPex31p, and ScPex32p. We designated these proteins as P. pastoris Pex30p and Pex31p, and, unless stated explicitly by designations for other species, we refer exclusively to the P. pastoris homologues of these proteins in this article. We describe here the function, inducibility, interactions, stability, and dynamic subcellular localization of Pex30p and Pex31p. We also provide evidence for a novel function for the conserved dysferlin domain in this peroxin family and uncover an unappreciated role for both ER- and peroxisomally localized peroxins in the control of peroxisome number and size.
MATERIALS AND METHODS
Strains and Culture Conditions
P. pastoris strains used in this study are listed in Table 1. All strains were cultured at 30°C. Media components were as follows: YPD, 1% yeast extract, 2% peptone, 2% glucose; YYHR, 0.67% yeast nitrogen base without amino acids, 0.1% yeast extract, 0.02 g l-histidine/l, 0.02 g l-arginine/l; YYHRM, YYHR supplemented with 0.5% methanol; YYHROT, YYHR supplemented with 0.2% oleic acid and 0.02% Tween 40; YPD plates, YPD, 2% agar, 0.1 g zeocin, 0.05 g G418 or 0.15 g hygromycin B/l for drug-resistant selection; and SD plates, 0.67% yeast nitrogen base without amino acids, 2% glucose, 2% agar, 0.02 g l-histidine or l-arginine/l for auxotrophic selection.
Table 1.
P. pastoris strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| PPY12 | arg4, his4 | Gould et al. (1992) |
| SJS47 | pex19Δ::ZeoR, AOX1::BFP-SKL(HygR), ARG4::PAOX1-PEX19, HIS4::PACOX-YFP-PEX31 | This study |
| SJS49 | pex19Δ::ZeoR, SEC61::SEC61-RFP(HygR), ARG4::PAOX1-PEX19, HIS4::PACOX-YFP-PEX31 | This study |
| SKF13 | pex19Δ::ZeoR, arg4, his4 | Snyder et al. (1999) |
| SMY142 | pex31Δ::ZeoR, arg4, his4 | This study |
| SMY169 | PEX31::PEX31-HA(ZeoR), arg4, his4 | This study |
| SMY235 | pex19Δ::ZeoR, PEX30::PEX30-GFP(ARG4), HIS4::PGAP-PEX31-RFP | This study |
| SMY240 | PEX30::PEX30-GFP(ARG4), HIS4::PGAP-PEX31-RFP | This study |
| SMY275 | pex30Δ::ZeoR, PEX31::PEX31-HA(KanR), HIS4::PGAP-GFP-PEX30ΔC, arg4 | This study |
| SMY283 | pex30Δ::ZeoR, arg4, his4 | This study |
| SMY300 | PEX30::PEX30-HA(ZeoR), arg4, his4 | This study |
| SMY376 | pex31Δ::ZeoR, PEX30::PEX30-HA(KanR), arg4, his4 | This study |
| SMY377 | pex30Δ::ZeoR, PEX31::PEX31-HA(KanR), arg4, his4 | This study |
| SMY382 | pex30Δ::ZeoR, PEX31::PEX31-HA(KanR), HIS4::PGAP-GFP-PEX30, arg4 | This study |
| SMY391 | pex31Δ::ZeoR, HIS4::PGAP-PEX31-FLAG, PEX30::PEX30-HA(KanR), arg4 | This study |
| SMY393 | PEX30::PEX30-GFP(ARG4), SEC61::SEC61-RFP(HygR), his4 | This study |
| SMY394 | pex31Δ::ZeoR, HIS4::PGAP-PEX31-FLAG, arg4 | This study |
| SMY395 | pex31Δ::ZeoR, HIS4::PGAP-PEX31ΔC-FLAG, arg4 | This study |
| SMY404 | PEX31::PEX31-GFP(ARG4), SEC61::SEC61-RFP(HygR), his4 | This study |
| SMY405 | PEX31::PEX31-GFP(ARG4), HIS4::PEX3-RFP | This study |
| SMY406 | PEX30::PEX30-GFP(ARG4), HIS4::PEX3-RFP | This study |
| SMY411 | HIS4::PGAP-GFP-PEX31, SEC61::SEC61-RFP(HygR), arg4 | This study |
| SMY419 | HIS4::PGAP-GFP-PEX31, ARG4::PEX3-RFP | This study |
| SMY420 | PEX30::PEX30ΔC-GFP(ARG4), HIS4::PEX3-RFP | This study |
| SMY421 | HIS4::PGAP-GFP-PEX31ΔC, ARG4::PEX3-RFP | This study |
| SMY440 | PEX30::PEX30ΔC-GFP(ARG4), SEC61::SEC61-RFP(HygR), his4 | This study |
| SMY442 | HIS4::PGAP-GFP-PEX31ΔC, SEC61::SEC61-RFP(HygR), arg4 | This study |
Glycerol stock cells were first streaked onto YPD plates. Colonies from a YPD plate were grown overnight in YPD medium to stationary phase (preculture). An appropriate volume of preculture was used to inoculate fresh YPD medium to achieve an initial optical density at 600 nm (OD600) of 0.2. Once the OD600 of a culture reached 1, cells were subjected to centrifugation, washed with water, introduced into YYHRM or YYHROT at an OD600 of 0.75, and incubated for the times indicated to induce peroxisomes.
Cloning of PpPEX30 and PpPEX31
Raw data from the ERGO database (courtesy of Integrated Genomics, Chicago, IL) showed two homologues, RPPA06010 and RPPA09211, of YlPEX23. These open reading frames (ORFs) were amplified using PCR from genomic DNA of P. pastoris strain PPY12, and sequences were corrected according to DNA sequencing data. Based on their homology to known genes and their functions (see Results), the ORF in RPPA06010 was designated as PpPEX30 and that for RPPA09211 as PpPEX31.
Construction of Deletion Strains
The 0.6-kbp 5′ flanking region of PEX30 was amplified from PPY12 genomic DNA using oligonucleotides OMY233 and OMY234 (Table 2). The 0.7-kbp 3′ flanking region of PEX30 was amplified by using the oligonucleotides OMY236 and OMY237. These two fragments were cloned into a vector encoding zeocin resistance, pMYZeo, to obtain pMYΔ30. pMYΔ30 was digested with EcoRI and NheI and transformed into PPY12 cells to make the pex30Δ strain, SMY283.
Table 2.
Oligonucleotide primers used for PCR in this study
| Primer | Sequence (5′ to 3′) |
|---|---|
| JS6 | AATGCTCGAGATGAGTAGCGAGGACGAACTAG-ATGATC |
| JS7 | CTATAAGCTTTCACTGCTGTTTACAAGTCTCAT-CCAATTC |
| JS11 | AATGCTCGAGATGGTGAGCAAGGGCGAGGA |
| JS18 | CTATAAGCTTTTAGATACTTCCCTGAACTGTCA-GTTTG |
| OMY223 | GTCCCTTAAGGTCAGTTTTGGCTTTACTTTTATC-TTTTTCTATGGC |
| OMY224 | CAGGTCTAGATCTCAATGGCGCACATTCCAAC |
| OMY233 | CGTATCTAGAATTCGACCTTCTTACGTTCAGAA-AAGA |
| OMY234 | GGAACTCGAGATTCTGGTGACTTACACGGTGA-TAGTAATTGA |
| OMY236 | AAAAAGATCTAAAGTGGACATCTCTCATCTCTC-ATCTCTC |
| OMY237 | TTATTCTAGATGCTAGCTAACTCTCCCTCGCCT-ATATG |
| OMY309 | AATCGGATCCATGGCTTCCATCAATTCGCAGCCA |
| OMY310 | CGAGGTCGACTTAGTCAGTTTTGGCTTTACTTT-TATCTTTTTC |
| OMY319 | TGGTCCTGCTGGAGTTTGTGAC |
| OMY344 | CAGCTCTCATCCCAAAGATGCTCTTGT |
| OMY345 | GATCCTCGAGTGGTTAGCTCGTTAAAAAATCAG-ACGAA |
| OMY346 | GGAAAGATCTAATATAGTCACGATTACATAAGC-ATATCT |
| OMY347 | TCATCTTAGATTCCTTATGGGTTGCTTTCTGT |
| OMY348 | GAGCGAATTCATGGCAGATCCACCTAGTAAAG-ATGGC |
| OMY349 | TAATGTCGACTATATTAGATACTTCCCTGAACT-GTCAGT |
| OMY350 | TTACATCGATGGTTTATCGTGACTCAAAAGAA-CCG |
| OMY351 | GACTCTTAAGGATACTTCCCTGAACTGTCAGT-TTGGCTT |
| OMY380 | AAGCAAGCTTAGACGTCTGAACCATTTCCTCCG |
| OMY382 | ACAAAAGCTTACTGCAATGACATGTCTGCTGT-GTG |
| OMY383 | CTAGGTCGACCGGTCTTCTCGTAAGTGCCCA |
| OMY518 | CAATCTTAAGCTGCAATGACATGTCTGCTGT-GTGTTC |
| OMY550 | AATCCCCGGGGATACTTCCCTGAACTGTCAGT-TTGGCT |
| OMY553 | ACCAATCGATGCTTCCATCAATTCGCAGCCA |
| OMY554 | AGCGCTTAAGGACGTCTGAACCATTTCCTCC |
The 0.4 kbp of 5′ flanking region of PEX31 was amplified using the oligonucleotides OMY344 and OMY345. The 0.6 kbp of 3′ flanking region of PEX31 was amplified using the oligonucleotides OMY346 and OMY347. These two fragments were cloned into pMYZeo to obtain pMY94. pMY94 was linearized with PvuII and transformed into PPY12 to make the pex31Δ strain, SMY142.
Plasmids (Table 3) were prepared with a miniprep kit (Promega, Madison, WI) and manipulated with restriction enzymes or T4 DNA ligase (New England Biolabs, Ipswich, MA). PPY12 was transformed by electrotransformation (Cregg and Russell, 1998). Transformants were selected on zeocin-containing YPD plates, and the knock out of a given locus was confirmed by PCR.
Table 3.
Plasmids used in this study
| Plasmid | Characteristics | Source |
|---|---|---|
| pJCF235 | PEX3-RFP*(mRFP1), ARG4 | Laboratory stock |
| pJS15 | PAOX1-PEX19, ARG4 | This study |
| pJS16 | PACOX-YFP-PEX31, HIS4 | This study |
| pKNSD52 | VP16 yeast two-hybrid vector | Faber et al. (1998) |
| pKNSD55 | LexA yeast two-hybrid vector | Faber et al. (1998) |
| pKSN183 | PEX3-RFP(mRFP1), HIS4 | Laboratory stock |
| pKSN256 | SEC61 3′-RFP(mCherry) for genomic tag, HygR | Laboratory stock |
| pMY46 | LexA-PEX30 | This study |
| pMY47 | VP16-PEX30 | This study |
| pMY52 | PGAP-GFP-PEX30, HIS4 | This study |
| pMY94 | PEX31 knock out construct, ZeoR | This study |
| pMY96 | PEX31 3′-HA for genomic tag, ZeoR | This study |
| pMY98 | LexA-PEX31 | This study |
| pMY99 | VP16-PEX31 | This study |
| pMY101 | PGAP-GFP-PEX31, HIS4 | This study |
| pMY104 | PEX30 3′-HA for genomic tag, KanR | This study |
| pMY105 | PEX31 3′-HA for genomic tag, KanR | This study |
| pMY201 | PEX30 3′-HA for genomic tag, ZeoR | This study |
| pMY219 | PGAP-PEX31-FLAG, HIS4 | This study |
| pMY255 | PGAP-GFP-PEX30ΔC, HIS4 | This study |
| pMY257 | PGAP-GFP-PEX31ΔC, HIS4 | This study |
| pMY263 | LexA-PEX30ΔC | This study |
| pMY265 | LexA-PEX31ΔC | This study |
| pMY268 | VP16-PEX30ΔC | This study |
| pMY270 | VP16-PEX31ΔC | This study |
| pMY333 | PGAP-PEX31ΔC-FLAG, HIS4 | This study |
| pMY356 | PEX30 3′-GFP for genomic tag, ARG4 | This study |
| pMY364 | PGAP-PEX31-RFP(mCherry), HIS4 | This study |
| pMY366 | pex30ΔC 3′-GFP for genomic tag, ARG4 | This study |
| pMY370 | PEX31 3′-GFP for genomic tag, ARG4 | This study |
| pMYΔ30 | PEX30 knockout construct, ZeoR | This study |
*Note that for simplicity, in the figures and text we used the generic label RFP for both mRFP1 and mCherry, but the specific gene used is shown in this table.
HA Tags at Genomic Loci
A previously published strategy developed for TAP-tags at genomic loci was used to introduce HA-tags at genomic loci (Leon et al., 2007). A 0.8-kbp fragment of the PEX30 ORF without its stop codon was amplified using oligonucleotides OMY223 and OMY224 and cloned into a C-terminal 2×HA-tag vector, pMY155. The resulting construct, pMY201, was linearized with SacI and transformed into PPY12 to make strain SMY300. Transformants were selected for zeocin resistance, and correct integration at the endogenous PEX30 locus was confirmed by PCR. Another kanamycin/G418 construct, pMY104, was made by replacing the resistance cassette in pMY201. pMY104 was linearized with SacI and transformed into SMY142 to yield strain SMY376. Transformants were selected for G418 resistance and confirmed by PCR.
A 0.8-kbp fragment of the PEX31 ORF without its stop codon was amplified using oligonucleotides OMY350 and OMY351 and cloned into a C-terminal 2×HA-tag vector, pMY69′. The resulting construct, pMY96, was linearized with PstI and transformed into PPY12 to make strain SMY169. Transformants were selected for zeocin resistance, and correct integration at the endogenous PEX31 locus was confirmed by PCR. Another kanamycin/G418 construct, pMY105, was made by replacing the resistance cassette in pMY96. pMY105 was linearized with PstI and transformed into SMY283 to produce strain SMY377. Transformants were selected for G418 resistance and confirmed by PCR.
To visualize the synthesis of HA-fusion proteins, lysates were prepared by an alkaline treatment of cells (Kushnirov, 2000) and analyzed by Western blotting with anti-HA antibody (Covance, Berkeley, CA).
GFP Tags at Genomic Loci
The PEX30 ORF without its stop codon in pMY201 was subcloned into a C-terminal GFP vector, pMY328. The resulting construct, pMY356, was linearized with BstZ17I and transformed to integrate at the endogenous PEX30 locus in appropriate strains to produce SMY235, SMY240, SMY393, and SMY406. The pex30ΔC (deletion of the C-terminal region of Pex30p beginning at the dysferlin domain) partial fragment was amplified using oligonucleotides OMY553 and OMY554 and cloned into pMY328. The resulting construct, pMY366, was linearized with HpaI and used to make strains SMY420 and SMY440. The PEX31 ORF without its stop codon was amplified using oligonucleotides OMY350 and OMY351 and cloned into pMY328. The resulting plasmid, pMY370, was linearized with AccI and transformed to yield strains SMY404 and SMY405. Transformants were selected by arginine prototrophy and confirmed by fluorescence microscopy.
Ectopic Expression
The full-length PEX30 ORF was amplified using oligonucleotides OMY309 and OMY310 to make a fusion protein with N-terminal GFP expressed from the glyceraldehyde-3-phosphate (GAP) promoter in the vector pPS55. The resulting construct, pMY52, was linearized with StuI and transformed to integrate at the HIS4 locus for ectopic expression of GFP-Pex30p driven by the GAP promoter. SMY377 transformed with pMY52 was designated as strain SMY382. The pex30ΔC fragment was amplified from pMY52 with oligonucleotides OMY319 and OMY380 and cloned into pPS55. The resulting construct, pMY255, was linearized with StuI and transformed into SMY377 to make strain SMY275. Transformants were selected by histidine prototrophy and confirmed by fluorescence microscopy.
The full-length PEX31 ORF was amplified using oligonucleotides OMY348 and OMY349 and cloned into pPS55. The resulting construct, pMY101, was linearized with StuI and transformed into appropriate strains to produce SMY411 and SMY419. The pex31ΔC fragment was amplified from pMY101 with oligonucleotides OMY319 and OMY382 and cloned into pPS55. The resulting pMY257 was linearized with StuI and transformed into appropriate strains to yield SMY421 and SMY442. Transformants were selected by histidine prototrophy and confirmed by fluorescence microscopy.
The full-length PEX31 ORF without its stop codon was amplified using oligonucleotides OMY348 and OMY351 to create a fusion protein with a C-terminal 3×FLAG tag expressed from the GAP promoter in a vector derived from pIB2 (Sears et al., 1998). The resulting plasmid, pMY219, was linearized with StuI and transformed into cells to integrate at the HIS4 locus for ectopic expression of PEX31-FLAG from the GAP promoter. pMY219 was transformed into SMY142 to make SMY394 and into SMY376 to make SMY391. The pex31ΔC fragment without its stop codon was amplified using oligonucleotides OMY348 and OMY518 to fuse this ORF to the C-terminal 3×FLAG tag expressed from the GAP promoter. The resulting plasmid, pMY333, was linearized with StuI and transformed into SMY142 to make strain SMY395. Transformants were selected by histidine prototrophy and confirmed by Western blotting with anti-FLAG antibody (Covance).
The full-length PEX31 ORF without its stop codon was amplified using oligonucleotides OMY348 and OMY550 to fuse it to the C-terminal RFP (mCherry) expressed from the GAP promoter in a vector pMY363. The resulting plasmid, pMY364, was linearized with SalI and transformed into appropriate strains to produce SMY235 and SMY240. Transformants were selected by histidine prototrophy and confirmed by fluorescence microscopy.
The full-length PEX19 ORF was amplified using oligonucleotides JS6 and JS7 and cloned into an alcohol oxidase 1 (AOX1) promoter–containing vector, pJCF230, resulting in pJS15. The ORF for the YFP-PEX31 fusion in pMY368 was amplified using oligonucleotides JS11 and JS18 and subcloned into a pIB1-based, acyl-CoA oxidase (ACOX) promoter–containing vector, resulting in pJS16. pJS15 and pJS16 linearized with NruII and StuI, respectively, were transformed into appropriate strains to produce SJS47 and SJS49. Transformants were selected by arginine/histidine prototrophy as well as hygromycin B resistance and confirmed by fluorescence microscopy.
Fluorescence Microscopy
For colocalization studies, a peroxisomal membrane marker, Pex3p-RFP (pJCF235, courtesy of Dr. Jean-Claude Farré; pKSN183, courtesy of Dr. Kanae Noda), and an ER membrane marker, Sec61p-RFP (pKSN256, courtesy of Dr. Kanae Noda, both from the University of California, San Diego), were used together with GFP-Pex30p or -Pex31p constructs. Observations were made using an Axioscope 2 MOT fluorescence microscope (Carl Zeiss, Thornwood, NY). Images were captured using an AxioCam MR camera and analyzed using AxioVision software.
For pulse-chase experiments, cells were first induced in the oleate medium for the expression of yellow fluorescent protein (YFP)-Pex31p. After 6-h induction, cells were imaged by Axioscope 2 MOT fluorescence microscope. Cells were then washed with water to turn off the expression of YFP-Pex31p and shifted to the methanol medium for the expression of Pex19p and induction of functional peroxisomes. After overnight growth, cells were analyzed with fluorescence microscopy again.
For immunofluorescence, samples were prepared as described previously (Rossanese et al., 1999), with some modifications. Briefly, cells were fixed with 4% paraformaldehyde, spheroplasted with Zymolyase 20T, and then adhered to a poly-lysine–coated glass slide followed by acetone postfixation at −20°C. Samples were rehydrated in phosphate-buffered saline (PBS), blocked with 4.3 mM Na2HPO4, 1.4 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl, pH 7.4, 1% skim milk, 0.1% bovine serum albumin (BSA), 0.1% n-octyl glucoside for 30 min and then incubated with primary anti-Pex3p antibody (Wiemer et al., 1996) and/or anti-HA antibody (1:2000 dilution in PBS blocking buffer). After incubation overnight at 4°C, samples were washed with PBS blocking buffer and incubated with secondary Cy3-conjugated anti-rabbit IgG antibody (Jackson ImmunoResearch, West Grove, PA) and/or Alexa Fluor 488–conjugated anti-mouse IgG antibody (Molecular Probes, Eugene, OR; diluted 1:200 in PBS blocking buffer). After a 1-h incubation in the dark, samples were washed with PBS blocking buffer. A drop of mounting medium (95% glycerol, 0.1% p-phenylenediamine) was added, and a coverslip was then placed over the sample.
Morphometric Analysis of Peroxisomes by Electron Microscopy
For each strain analyzed, electron micrographs of 50 randomly selected cells at a magnification of 17,000 were scanned, and the areas of individual cells and of individual peroxisomes were determined by counting the number of individual pixels in a particular cell or peroxisome with Image Tool for Windows (University of Texas Health Science Center, San Antonio, TX). To determine the average area of a peroxisome, the total peroxisome area was calculated and divided by the total number of peroxisomes counted. To quantify peroxisome number, the numerical density of peroxisomes (number of peroxisomes per μm3 of cell volume) was calculated by a method for spherical organelles (Weibel and Bolender, 1973). Statistical data were processed by Excel software (Microsoft, Redmond, WA).
Yeast Two-Hybrid Analysis
The full-length PEX30 ORF was amplified from PPY12 genomic DNA with oligonucleotides OMY309 and OMY310 and cloned into the LexA DNA-binding domain vector, pKNSD55, and the VP16 transactivation domain vector, pKNSD52 (Faber et al., 1998), resulting in pMY46 and pMY47, respectively. The full-length PEX31 ORF was amplified from PPY12 genomic DNA with oligonucleotides OMY348 and OMY349 and cloned into pKNSD55 and pKNSD52, resulting in pMY98 and pMY99, respectively. pex30ΔC was amplified from pMY255 with oligonucleotides OMY319 and OMY383 and cloned into pKNSD55 and pKNSD52, resulting in pMY263 and pMY268, respectively. pex31ΔC was amplified from pMY257 with oligonucleotides OMY319 and OMY383 and cloned into pKNSD55 and pKNSD52, resulting in pMY265 and pMY270, respectively. Construct combinations were transformed into S. cerevisiae strain L40 and selected on synthetic medium lacking tryptophan and leucine. Transformants were tested for the activation of the integrated lacZ reporter construct using a β-galactosidase filter assay.
Immunoprecipitation
Twenty-five OD600 of cells were resuspended in 2.5 ml immunoprecipitation buffer (IP buffer; 20 mM Tris-HCl, pH 7.5, 0.3 M NaCl, 0.3% NP-40, 1 mM EDTA, protease inhibitor cocktail; Sigma-Aldrich, St. Louis, MO) supplied with 1% digitonin and lysed by vortexing the cells with acid-washed glass beads. The lysate was subjected to centrifugation at 20,000 × g for 3 min to eliminate debris. One milliliter of lysate was incubated with 5 μl of the desired antibody (anti-HA mouse mAb or anti-FLAG rabbit polyclonal antibody; Covance) for 4–5 h at 4°C. Fifty microliters of Gamma-Bind G Sepharose beads (GE Healthcare, Piscataway, NJ) prewashed in IP buffer were added to the lysate and incubated further for 1 h. The beads were then washed with 10 ml of IP buffer three times and boiled in 100 μl of SDS loading buffer. Samples free of beads were analyzed by a standard Western blotting procedure.
RESULTS
P. pastoris Pex30p and Pex31p Share Extensive Similarity with YlPex23p, ScPex30p, ScPex31p, and ScPex32p
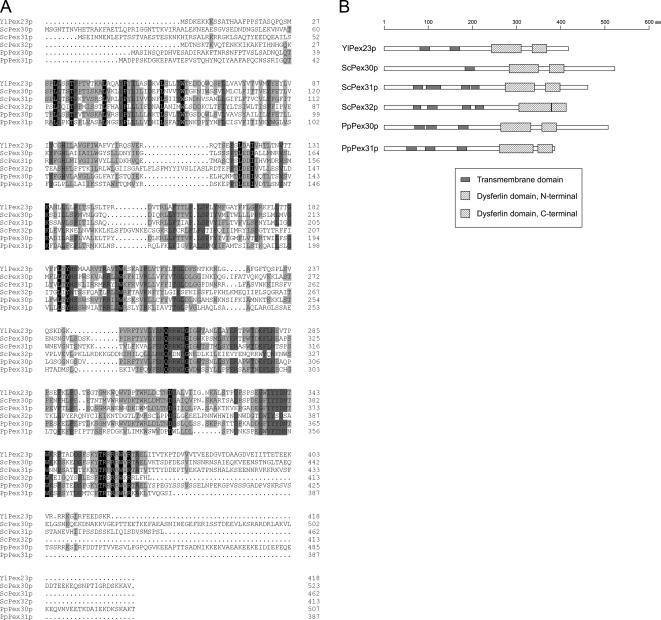
YlPex23p is an integral PMP required for peroxisome assembly in Y. lipolytica (Brown et al., 2000). A search for homologues in S. cerevisiae identified three integral PMPs, ScPex30p, ScPex31p, and ScPex32p, that are involved in the regulation of peroxisome size and number (Vizeacoumar et al., 2004). Raw data from the P. pastoris genome sequencing project showed two potential ORFs whose products share extensive similarity with these proteins (Figure 1A). Because they function in the regulation of peroxisome size and number (see below), we have named these proteins PpPex30p and PpPex31p (GenBank accession numbers EF619963 and EF619964) after their most closely related S. cerevisiae counterparts. PpPex30p is predicted to be a protein of 508 amino acids with a molecular weight of 57,355 Da. PpPex31p is predicted to be a protein of 387 amino acids with a molecular weight of 44,087 Da. PpPex30p and YlPex23p exhibit 53% amino acid identity and 17% amino acid similarity (at positions of nonidentity), and PpPex31p and YlPex23p exhibit 32% amino acid identity and 18% amino acid similarity, whereas PpPex30p and PpPex31p exhibit 34% amino acid identity and 19% amino acid similarity between themselves. These six proteins show similar domain architecture as predicted by the SMART program (Letunic et al., 2006), i.e., several transmembrane domains followed by dysferlin domains (Figure 1B). For Pex30p, three transmembrane domains, as well as N- and C-terminal dysferlin domains, are predicted at amino acids 70–92, 97–119, 169–191, 265–333, and 358–391. For Pex31p, similar domains are predicted at amino acids 52–74, 94–116, 166–188, 262–338, and 349–382.
Figure 1.
Sequence alignment and domain architecture of P. pastoris Pex30p and Pex31p with their homologues from Y. lipolytica and S. cerevisiae. (A) Alignment of the deduced amino acid sequences of YlPex23p, ScPex30p, ScPex31p, ScPex32p, PpPex30p, and PpPex31p was performed by ClustalW. Identical residues in all six or five of the proteins are shaded black or dark gray, respectively, whereas those present in at least three of the proteins are shaded light gray. Similarity rules are applied. Dots represent gaps. (B) Schematic presentation of the domain architecture predicted by the SMART program.
Deletion of the PEX30 or PEX31 Gene Has No Effect on Methanol-induced Peroxisomes But Causes Fewer and Enlarged Oleate-induced Peroxisomes
The pex30Δ and pex31Δ strains grew as well as the wild-type strain in all media tested, including methanol and oleate in which functional peroxisomes are required for cell growth (our unpublished data), suggesting that Pex30p and Pex31p are not directly involved in peroxisome assembly.
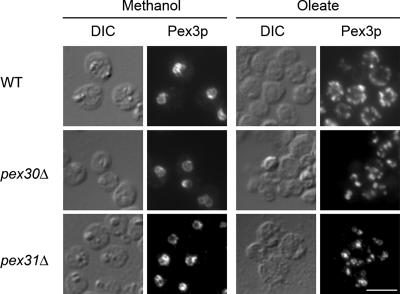
Immunofluorescence analysis of methanol- or oleate-induced cells with an antibody against the PMP, Pex3p, was performed to examine peroxisome morphology. Methanol-induced pex30Δ or pex31Δ cells showed typical peroxisome clusters as in wild-type cells (Figure 2, left panels). In contrast, when compared with numerous peroxisomes distributing evenly in wild-type cells, peroxisomes in oleate-grown pex30Δ and pex31Δ cells were fewer in number and tended to cluster together (Figure 2, right panels).
Figure 2.
pex30Δ and pex31Δ cells have normal methanol-induced peroxisomes but abnormal oleate-induced peroxisomes. Wild-type (WT, PPY12), pex30Δ (SMY283), or pex31Δ (SMY142) cells were induced in methanol or oleate medium overnight for peroxisome proliferation and analyzed by indirect immunofluorescence after labeling with anti-Pex3p antibody and Alexa Fluor 488–conjugated secondary antibody. DIC, differential interference contrast. Scale bar, 5 μm.
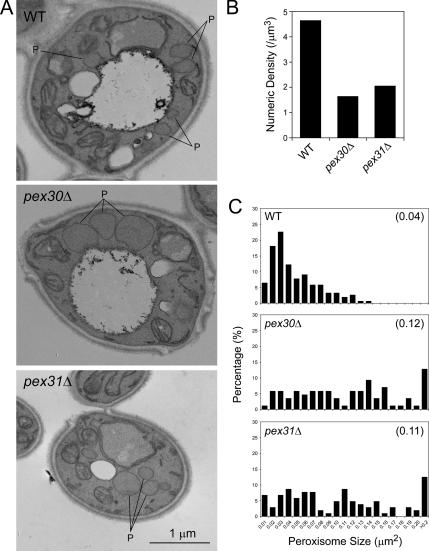
In electron micrographs (Figure 3A), oleate-induced peroxisomes in pex30Δ or pex31Δ cells were noticeably larger than those of wild-type cells and exhibited the characteristics of reduced numbers and clustering also observed by immunofluorescence. Fifty randomly selected cell images were quantified for cell area, peroxisome number and individual peroxisome area. The calculated numerical density of peroxisomes in deletion strains (1.64 peroxisomes/μm3 in pex30Δ, 2.05 peroxisomes/μm3 in pex31Δ) was about a third to half of that in wild-type cells (4.64 peroxisomes/μm3; Figure 3B). Histograms were generated to depict the percentage of total peroxisomes occupied by the peroxisomes of each size category (Figure 3C). The area of peroxisomes in wild-type cells was in a range of 0–0.14 μm2, with a peak at 0.02–0.03 μm2. In contrast, pex30Δ or pex31Δ cells showed a wider distribution of various peroxisome areas, with a higher percentage of bigger peroxisomes that were rarely seen or were absent in wild-type cells. Cells of the double deletion strain, pex30Δ pex31Δ, did not exhibit any synergistic effects with respect to peroxisome size and number (numerical density of 2.12 peroxisomes/μm3 and average peroxisome area of 0.10 μm2), and behaved essentially like pex31Δ cells, suggesting that Pex30p and Pex31p likely act in the same pathway. Pex30p could not complement pex31Δ cells and vice versa (our unpublished data), showing that Pex30p and Pex31p are not redundant proteins.
Figure 3.
Oleate-induced peroxisomes in pex30Δ or pex31Δ cells are fewer and larger in area than peroxisomes of wild-type cells. Wild-type (WT, PPY12), pex30Δ (SMY283), and pex31Δ (SMY142) cells were induced in oleate medium overnight and analyzed by EM at a magnification of 17,000. Fifty randomly selected cell images were scanned and quantified for cell area, peroxisome number, and individual peroxisome area. (A) Representative micrographs. P, peroxisome. (B) Numerical density of peroxisomes. (C) Histograms depicting the percentage of total peroxisomes occupied by peroxisomes of each size category. The numbers in parenthesis indicate the mean areas of peroxisomes.
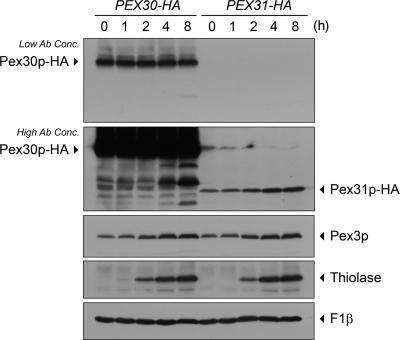
Synthesis of Pex30p Remains Constant, Whereas Pex31p Is Induced at Low Levels by Oleate
To trace the expression profiles, the sequence encoding a 2×HA-tag was genomically fused to the 3′-terminus of the ORF at the endogenous PEX30 or PEX31 locus. Pex30p-HA and Pex31p-HA fusion proteins were thus synthesized under the control of their native gene promoters. Cells were grown in YPD medium and then shifted to oleate medium. Aliquots of cells were removed at various times, and their lysates were analyzed by SDS-PAGE and Western blotting (Figure 4). Pex30p-HA and Pex31p-HA were detected at the expected molecular weights. The level of Pex30p-HA remained unchanged during oleate induction. The level of Pex31p-HA was much lower than that of Pex30p-HA but increased with time of incubation of cells in oleate-medium. The level of induction of Pex31p was not as great as that of the peroxisomal matrix protein, thiolase, but was comparable to that seen for the PMP, Pex3p. Mitochondrial F1-ATPase β subunit (F1β) served as a control for equal loading in each lane.
Figure 4.
Pex30p remains at a constant level, whereas Pex31p is inducible, in oleate. Genomically tagged PEX30-HA (SMY300) and PEX31-HA (SMY169) strains were precultured in YPD medium to log phase and then switched to oleate medium. Aliquots of cells were collected at the times indicated, and total cell lysates were prepared. Equal amounts of proteins were analyzed by Western blotting and decorated with antibodies against HA, Pex3p, thiolase, or F1β. Low and high concentrations of anti-HA antibody were used for appropriate visualization of Pex30p-HA and Pex31p-HA, respectively.
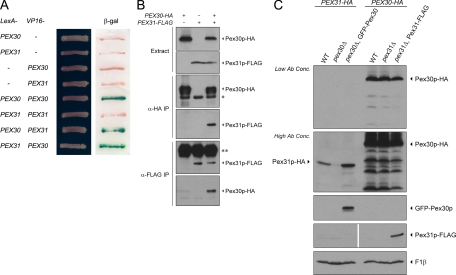
Pex30p Interacts with Pex31p and Is Required for Its Stability
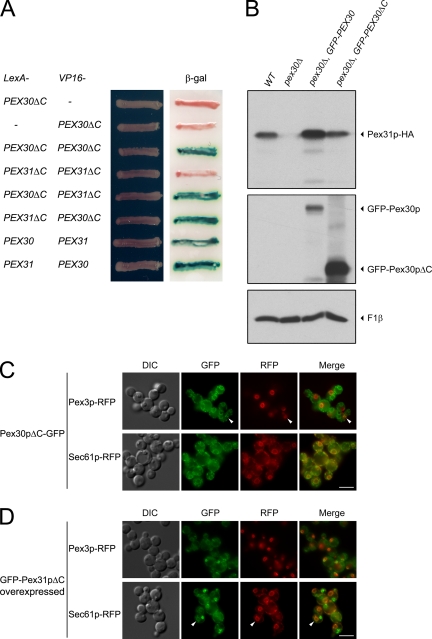
Interaction between Pex30p and Pex31p was tested using the yeast two-hybrid system. Pex30p-Pex30p and Pex30p-Pex31p positive interactions were detected by a β-galactosidase activity assay (Figure 5A). To confirm these interactions between Pex30p and Pex31p, PEX30 and PEX31 were tagged with HA and FLAG epitopes, respectively. In a strain expressing Pex30p-HA and Pex31p-FLAG, immunoprecipitation of Pex30p-HA coimmunoprecipitated Pex31p-FLAG, and immunoprecipitation of Pex31p-FLAG coimmunoprecipitated Pex30p-HA (Figure 5B). Immunoprecipitations of strains expressing only one fusion protein served as negative controls.
Figure 5.
Pex30p interacts with Pex31p and influences its stability. (A) Interactions were tested by yeast two-hybrid assay. Cells of S. cerevisiae strain L40 were transformed simultaneously with a LexA-containing plasmid and a VP16-containing plasmid. Transformants were grown on synthetic medium lacking tryptophan and leucine and tested for activation of the integrated lacZ reporter construct using a β-galactosidase (β-gal) filter assay. (B) PEX30 was tagged with HA (+) or not (−), whereas PEX31 was tagged with FLAG (+) or not (−). Strains SMY300 for PEX30-HA, SMY394 for PEX31-FLAG, and SMY391 for both were induced in oleate medium overnight and analyzed by immunoprecipitation (IP) with antibody against HA or FLAG. Pex30p-HA and Pex31p-FLAG were visualized by Western blotting. Asterisk, IgG heavy chain of mouse anti-HA; double asterisk, IgG heavy chain of rabbit anti-FLAG. (C) PEX31 was genomically tagged with HA in wild-type (WT, SMY169) and pex30Δ (SMY377) strains. The latter was transformed to ectopically express GFP-Pex30p (SMY382). Similarly, PEX30 was genomically tagged with HA in wild-type (WT, SMY300) and pex31Δ (SMY376) strains. The latter was transformed to ectopically express Pex31p-FLAG (SMY391). Cells were induced in oleate medium overnight, analyzed by Western blotting, and decorated with antibodies against HA (for Pex30p-HA and Pex31p-HA), GFP (for GFP-Pex30p), FLAG (for Pex31p-FLAG), and F1β.
Interaction between proteins may affect their stabilities. To investigate the protein stability of Pex31p, levels of genomically tagged Pex31p-HA were compared in wild-type, pex30Δ, and complemented pex30Δ strains (Figure 5C). Pex31p-HA became unstable in the absence of Pex30p and more stable in the presence of excess Pex30p caused by overexpression of GFP-Pex30p. On the other hand, the level of Pex30p-HA did not apparently change in the pex31Δ strain or in cells overexpressing Pex31p-FLAG.
Taken together, yeast two-hybrid and coimmunoprecipitation analyses, as well as the role of Pex30p in stabilizing Pex31p, provide strong evidence that these two proteins interact physiologically.
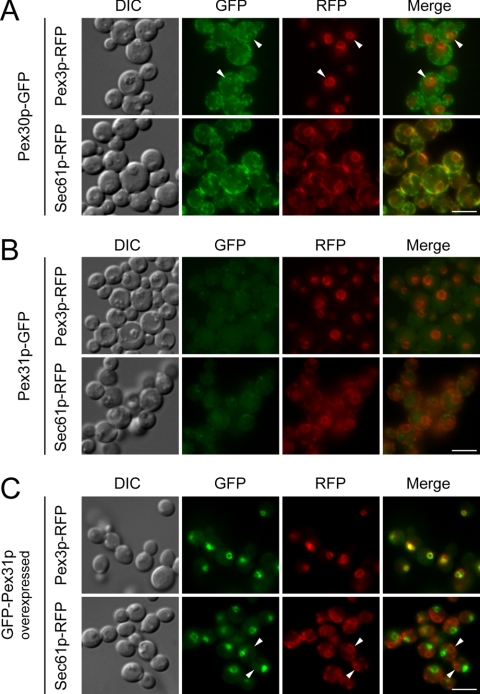
Pex30p and Pex31p Localize to the ER and Peroxisomes at Steady State
To visualize Pex30p or Pex31p in living cells at their native levels, the DNA encoding GFP was fused to the 3′-terminus of the ORF at the endogenous PEX30 or PEX31 locus. Control experiments showed that these tags do not affect the localization of these proteins. Constitutively expressed Pex30p-GFP localized primarily to the ER as shown by colocalization with the ER marker, Sec61p-RFP, in every medium tested (glucose or oleate: Supplementary Figure 1A; methanol: Figure 6A). A small portion of Pex30p-GFP could also be found on peroxisomes as shown by colocalization with the peroxisome marker, Pex3p-RFP. This partial colocalization was much clearer on big clustered peroxisomes induced by methanol (arrowheads in Figure 6A).
Figure 6.
Pex30p localizes primarily to the ER with a small amount on peroxisomes, whereas Pex31p localizes primarily to peroxisomes with a small amount at the ER. (A) Pex30p-GFP expressed from a genomically tagged locus was colocalized with a peroxisome marker, Pex3p-RFP (SMY406), or an ER marker, Sec61p-RFP (SMY393), in methanol-induced cells. Arrowheads indicate partial colocalization on peroxisomes. (B) Pex31p-GFP expressed from its genomically tagged locus was colocalized with Pex3p-RFP (SMY405) or Sec61p-RFP (SMY404) in methanol-induced cells. (C) Pex31p-GFP ectopically expressed from the GAP promoter was colocalized with Pex3p-RFP (SMY419) or Sec61p-RFP (SMY411) in methanol-induced cells. Arrowheads indicate small amounts of colocalization at the ER. DIC, differential interference contrast. Scale bar, 5 μm.
Consistent with the results of Western blotting, only a very weak signal of Pex31p-GFP expressed from its endogenous promoter could be detected. This weak signal showed colocalization with Pex3p-RFP, indicating that Pex31p is localized primarily to peroxisomes (Figure 6B and Supplementary Figure 1B). To enhance its visualization, Pex31p-GFP was ectopically expressed from the GAP promoter. Overexpressed Pex31p-GFP clearly showed a predominantly peroxisomal localization (Figure 6C and Supplementary Figure 1C). A certain amount of Pex31p-GFP could also be found at the ER. This partial colocalization is obvious in glucose medium where only small peroxisomes were present in the cells (Supplementary Figure 1C, left panels). When peroxisomes were induced by methanol or oleate, the level of partial colocalization at ER was reduced (arrowheads in Figure 6C and Supplementary Figure 1C, right panels).
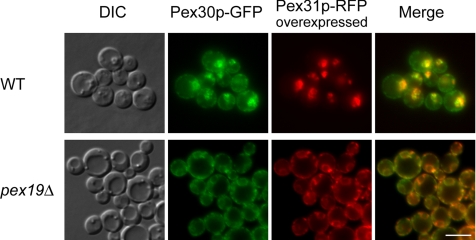
Perturbation of the Steady-State Pools of Pex30p and Pex31p
Pex31p-RFP was overexpressed in cells expressing Pex30p-GFP. Interestingly, upon methanol induction, most of the Pex30p-GFP colocalized with Pex31p-RFP (Figure 7, top panels). In comparison with the predominantly ER localization of Pex30p-GFP (Figure 6A), excess Pex31p on peroxisomes increased the peroxisome/ER distribution ratio of Pex30p-GFP.
Figure 7.
Colocalization of Pex30p and overexpressed Pex31p in wild-type and pex19Δ cells. Pex30p-GFP expressed from a genomically tagged locus and Pex31p-RFP expressed ectopically from the GAP promoter were colocalized in wild-type (WT, SMY240) and pex19Δ cells (SMY235). DIC, differential interference contrast. Scale bar, 5 μm.
The localization of Pex30p and Pex31p was also investigated in pex19Δ cells, which are deficient in peroxisome membrane and matrix protein biogenesis. Pex30p-GFP and Pex31p-RFP colocalized with an apparent ER pattern (Figure 7, bottom panels).
These data on the dual localization of these proteins demonstrate that at steady state, Pex30p resides mostly at the ER, whereas Pex31p is primarily peroxisomal. Additionally, the pool of each protein can be redistributed from one compartment to the other by specific manipulations.
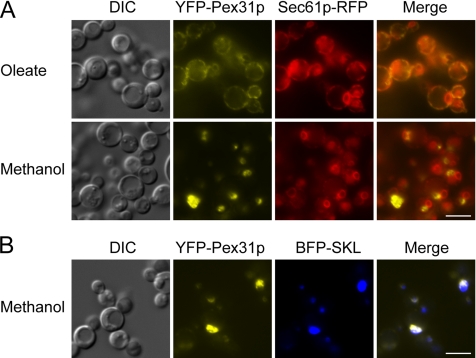
Pex31p Can Be Accumulated at the ER and Chased to Peroxisomes in a Pex19p-dependent Manner
To determine whether Pex31p traffics from the ER to peroxisomes, we expressed YFP-Pex31p from the oleate-inducible acyl-CoA oxidase promoter in pex19Δ cells to accumulate it at the ER, where it colocalized with Sec61p-RFP (Figure 8A, top panel). These cells were then shifted to methanol, where the expression of YFP-Pex31p was turned off and that of Pex19p was induced from the methanol-inducible alcohol oxidase promoter. Under these conditions, the ER-localized YFP-Pex31p disappeared and reappeared at punctate structures (Figure 8A, bottom panel), which were shown to be peroxisomes by colocalization of YFP-Pex31p with BFP-SKL (Figure 8B). As expected, no newly synthesized YFP-Pex31p appeared at the ER because, as proved by this and independent control experiments, the acyl-CoA oxidase promoter was inactive in methanol. These data demonstrate that Pex31p can be chased from the ER to peroxisomes.
Figure 8.
ER to peroxisome traffic of Pex31p in a Pex19-dependent manner. (A) Cells (SJS49) expressing YFP-Pex31p driven by the acyl-CoA oxidase (ACOX) promoter was first induced in oleate medium and colocalized with Sec61p-RFP in pex19Δ cells. The cells were then shifted to methanol medium to turn off the expression of YFP-Pex31p and induce that of Pex19p driven by the alcohol oxidase 1 (AOX) promoter. (B) YFP-Pex31p in pex19Δ cells (SJS47) was first induced in oleate medium. Cells were then shifted to methanol medium to turn off the expression of YFP-Pex31p and induce that of Pex19p. YFP-Pex31p was colocalized with BFP-SKL. DIC, differential interference contrast. Scale bar, 5 μm.
Because Pex31p was peroxisomal at steady-state and its transport from the ER to peroxisomes required Pex19p, we tested whether it interacts with Pex19p. In coimmunoprecipitates of cells overexpressing Pex31p-HA, Pex19p was coimmunoprecipitated (Supplementary Figure 2). This result is analogous to previous studies in S. cerevisiae reporting interactions between ScPex19p and peroxisomal ScPex30p and ScPex32p (Vizeacoumar et al., 2006).
The Dysferlin Domains of Pex30p and Pex31p Are Not Essential for Their Interactions, Stability, or Localization
The dysferlin domain comprises two motifs, DysFN and DysFC, in dysferlin protein, which is mutated in limb girdle muscular dystrophy (Ponting et al., 2001). The function of the dysferlin domain is unknown. ScPex30p, ScPex31p, and ScPex32p are the only proteins in S. cerevisiae that contain a complete dysferlin domain. Genome sequencing data indicate that Pex30p and Pex31p are the only dysferlin domain-containing proteins in P. pastoris.
To investigate the function of this domain, we made constructs of PEX30 and PEX31 that lack their C-terminal dysferlin domains (ΔC). Yeast two-hybrid analysis showed that the interactions of Pex30p-Pex30p and Pex30p-Pex31p were not affected by the absence of the dysferlin domains (Figure 9A). Western blotting was done to analyze the levels of genomically tagged endogenous Pex31p in pex30Δ cells upon expression of either full-length or dysferlin domain–deleted Pex30p. Although not as efficient as full-length GFP-Pex30p, GFP-Pex30pΔC partially restored Pex31p levels, suggesting that the requirement of Pex30p for the stabilization of Pex31p did not require its dysferlin domain (Figure 9B). However, the dysferlin domain of Pex30p may enhance the stability of Pex31p, although it is not essential for this role. Furthermore, GFP-Pex30pΔC showed exactly the same subcellular distribution (Figure 9C) as full-length Pex30p-GFP (Figure 6A). As for Pex31pΔC, although absence of its dysferlin domain increased its ER-localized pool slightly, the majority of Pex31pΔC still localized to peroxisomes (Figure 9D).
Figure 9.
The dysferlin domains of Pex30p and Pex31p are not necessary for their interaction, stability and localization. (A) Interactions were tested by the yeast two-hybrid assay in S. cerevisiae. Transformants were grown on synthetic medium lacking tryptophan and leucine and tested for the activation of the integrated lacZ reporter construct using a β-galactosidase (β-gal) filter assay. (B) PEX31 was genomically tagged with HA in wild-type (WT, SMY169) and pex30Δ (SMY377) cells. The latter was transformed to ectopically express GFP-PEX30 (SMY382) or GFP-PEX30ΔC (SMY275). Cells were induced in oleate medium overnight, analyzed by Western blotting, and decorated with antibodies against HA, GFP, and F1β. (C) Pex30pΔC-GFP expressed from its genomically tagged locus was colocalized with the peroxisome marker, Pex3p-RFP (SMY420), or the ER marker, Sec61p-RFP (SMY440), in methanol-induced cells. Arrowheads indicate small amounts of colocalization at peroxisomes. (D) GFP-Pex31pΔC ectopically expressed from the GAP promoter was colocalized with Pex3p-RFP (SMY421) or Sec61p-RFP (SMY442) in methanol-induced cells. Arrowheads indicate a small level of colocalization at the ER. DIC, differential interference contrast. Scale bar, 5 μm.
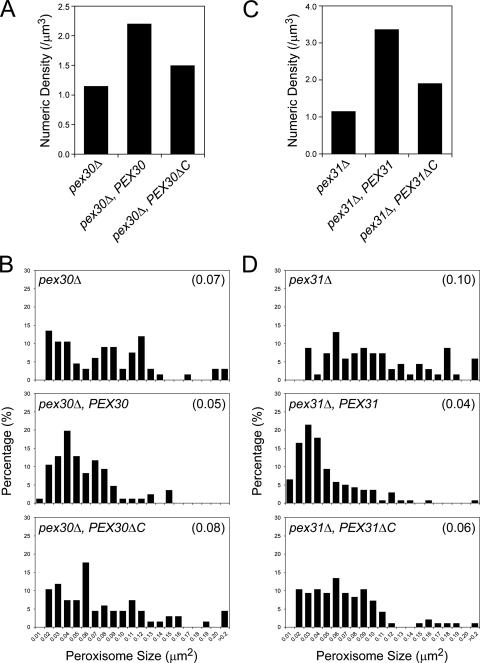
The Dysferlin Domains of Pex30p and Pex31p Are Essential for Their Control of Peroxisome Number and Size
To investigate the function of the dysferlin domain in peroxisome morphogenesis, full-length or dysferlin domain-deleted ΔC truncations of Pex30p or Pex31p were reintroduced into the respective knockout strains. Cells were induced in oleate overnight and analyzed by electron microscopy (EM) for a quantitative assessment of peroxisome morphology. Full-length PEX30 increased the numerical density of peroxisomes twofold in pex30Δ cells, indicating full complementation (Figure 10A). However, pex30ΔC could not restore peroxisome number to wild-type levels. Histograms of peroxisome size distributions also showed that pex30ΔC was impaired in its capacity to maintain peroxisomes of normal area (Figure 10B). Similarly, full-length PEX31 fully complemented pex31Δ, as expected, but pex31ΔC could not completely restore normal peroxisome number and size in pex31Δ cells (Figure 10, C and D).
Figure 10.
The dysferlin domains of Pex30p and Pex31p are required for the control of peroxisome number and size. Cells of the pex30Δ (SMY377) strain and strains containing full-length PEX30 (SMY382) or dysferlin domain–deleted pex30ΔC (SMY275) were induced in oleate medium overnight and analyzed by EM. Fifty randomly selected cell images were scanned and quantified for cell area, peroxisome number, and individual peroxisome area. (A) Numerical densities of peroxisomes. (B) Histograms depicting the percentage of total peroxisomes occupied by the peroxisomes of each size category. Average size is indicated in parenthesis. Similarly, cells of the pex31Δ (SMY142) strain and strains containing full-length PEX31 (SMY394) or dysferlin domain–deleted pex31ΔC (SMY395) were induced in oleate medium overnight and analyzed by EM for the numerical densities of peroxisomes (C), and the percentage of total peroxisomes occupied by the peroxisomes of each size category (D).
DISCUSSION
This article sheds new light on a number of aspects of the subcellular location and function of Pex30p and Pex31p that go beyond previously published results regarding these peroxins.
First, though this protein family is known to be involved in the regulation of peroxisome size and number, the precise functions of this family vary from species to species. For example, YlPex23p is required for peroxisome biogenesis (Brown et al., 2000), but ScPex30p-ScPex32p, as well as PpPex30p and PpPex31p, are not. Also, ScPex31p and ScPex32p regulate mostly peroxisome size and ScPex30p peroxisome number (Vizeacoumar et al., 2004), whereas both PpPex30p and PpPex31p affect peroxisome size and number similarly. Moreover, previous studies had used nonmethylotrophic yeasts and had therefore studied the functions of these proteins only in the presence of oleate, leaving the impression that these proteins control peroxisome size and number under all conditions. However, our studies show for the first time that these proteins control peroxisome size and number only on oleate and not on methanol, which suggests that peroxisome size and number are controlled by different mechanisms when cells are exposed to different conditions.
Second, previous studies had localized these peroxins only to peroxisomes in other yeasts, whereas our studies reveal a dual localization of this class of peroxins to the ER and peroxisomes. Moreover, we show that PpPex31p traffics to the peroxisomes via the ER in a Pex19p-dependent manner.
Third, our report of the dual and exchangeable localization of these proteins on the ER and peroxisomes suggests not only that peroxisome biogenesis can originate from the ER, but also that the ER plays a role in the control of peroxisome size and number.
Fourth, our study provides a working model for how Pex30p and Pex31p function, which was missing in previous reports.
A Family of Dysferlin Domain Proteins Involved in the Control of Peroxisome Morphogenesis
The discoveries of YlPex23p (Brown et al., 2000); ScPex30p, ScPex31p, and ScPex32p (Vizeacoumar et al., 2004); and PpPex30p and PpPex31p reported in this study establish a novel dysferlin domain–containing protein family in yeast (Figure 1). Currently, there are no significant homologues of these proteins in higher eukaryotic organisms. However, proteins containing the dysferlin domain are present in higher eukaryotes, including humans, where they are believed to play a role in the repair and resealing of membranes damaged by mechanical stress (Glover and Brown, 2007). In humans, mutation of dysferlin causes dysferlinopathies, including limb girdle muscular dystrophy and Miyoshi myopathy (Ponting et al., 2001; Bansal et al., 2003). The C. elegans homolog of human dysferlin, fer-1, is responsible for the fusion of multiple intracellular vesicles called “membranous organelles” to the spermatid plasma membrane (Washington and Ward, 2006). Another related protein with the dysferlin domain is myoferlin, required for myoblast fusion and myotube formation (Doherty et al., 2005).
Our work provides also new insight into the role of the dysferlin domain in yeasts. Although all yeast dysferlin domain proteins are peroxins involved primarily in the control of peroxisome number and size, their precise functions vary in the different yeast species. YlPex23p is required for the assembly of functional peroxisomes (Brown et al., 2000). Cells deleted for YlPEX23 cannot grow on medium containing oleate as the sole carbon source. Therefore, YlPex23p may be classified as a member of the “classical” peroxin group responsible for the assembly of functional peroxisomes. In contrast, ScPex30p, ScPex31p, ScPex32p, PpPex30p, and PpPex31p are members of the “nonclassical” peroxin group, which are not required for peroxisome assembly. Cells harboring deletions of genes of this latter group are able to grow in oleate-containing medium with essentially the same kinetics as wild-type cells. The only abnormality in cells mutant for these genes is seen in the number and size of peroxisomes, which explains why these genes were not identified in screens for mutants affected in peroxisome biogenesis per se.
Immunofluorescence analyses of P. pastoris pex30Δ and pex31Δ cells using the peroxisome marker, Pex3p, revealed that they have normal methanol-induced peroxisomes but abnormal oleate-induced peroxisomes (Figure 2). A consideration of the different contents and morphology of peroxisomes in these two carbon sources suggests that different mechanisms are used to control the number and size of peroxisomes depending on their growth environment. This interesting result is the first indication that the functions of Pex30p and Pex31p depend strictly on the carbon source in which P. pastoris grows and raises the possibility that other proteins may perform the same or similar functions in cells grown in other carbon sources.
Quantification of EM data showed that in oleate-induced P. pastoris pex30Δ or pex31Δ cells, peroxisomes are fewer and larger than in wild-type cells (Figure 3). Therefore, these proteins appear to be positive regulators of peroxisome number and negative regulators of peroxisome size. Absence of ScPex31p or ScPex32p causes enlarged peroxisomes and somewhat increased numbers of peroxisomes, suggesting that these proteins act mostly as negative regulators of peroxisome size. Scpex30Δ cells exhibit only increased number of peroxisomes, leading to the conclusion that it is primarily a negative regulator of peroxisome number (Vizeacoumar et al., 2004). Not surprisingly, ScPex30p, ScPex31p, and ScPex32p could not complement P. pastoris pex30Δ or pex31Δ cells (our unpublished data). These differences between P. pastoris and S. cerevisiae could also be influenced by the complex interplay among these proteins and others to tailor peroxisome growth and division, specifically for each yeast.
ScPex31p is not induced by oleate, but YlPex23p, ScPex30p, and ScPex32p are (Brown et al., 2000; Vizeacoumar et al., 2004). In contrast, PpPex30p is constitutively expressed in either glucose medium or oleate medium, whereas PpPex31p is induced by oleate (Figure 4). Considering the relatively high level of Pex30p (Figure 4) and its primary localization to the ER (Figure 6A), the proliferation of peroxisomes upon oleate induction does not require additional synthesis of Pex30p. On the other hand, given the low level of Pex31p and its primarily peroxisomal localization, the induction of Pex31p synthesis is a logical response to peroxisome proliferation in oleate medium.
Interactions of PpPex30p and PpPex31p and Their Dynamic, Dual Subcellular Localization
Yeast two-hybrid and coimmunoprecipitation analyses demonstrated an interaction between Pex30p and Pex31p (Figure 5, A and B), consistent with data from S. cerevisiae in which a strong interaction between ScPex30p and ScPex31p was observed (Vizeacoumar et al., 2004). The presence of Pex30p is essential for the stability of Pex31p (Figure 5C). Likely, this stabilization is mediated by that portion of Pex30p that physically interacts with Pex31p and is independent of the portion of Pex30p that regulates peroxisome size and number. C-terminally truncated Pex30pΔC, which is nonfunctional for the regulation of peroxisome size and number (Figure 10, A and B), maintains its interaction with Pex31p and can still stabilize Pex31p (Figure 9, A and B). Our unpublished data also show that ScPex30p can substitute for PpPex30p in its interaction with, and stabilization of, PpPex31p.
Fluorescence microscopy of GFP-fusions of Pex30p and Pex31p shows their dual, steady-state localization to two different organelles (Figure 6). Pex30p was found primarily at the ER, with a small amount on peroxisomes. Pex31p shows the opposite distribution, with most of it on peroxisomes and only a small fraction that is ER-associated. In contrast, YlPex23p, ScPex30p, ScPex31p, and ScPex32p are reported to be peroxisomal (Brown et al., 2000; Vizeacoumar et al., 2004).
The differing locations of Pex30p and Pex31p may reflect a dynamic relocation of these proteins depending on the state of the cells rather than differences in yeast species used or in methods of protein detection. In particular, ScPex30p and ScPex31p were found by subcellular fractionation in fractions less dense than those containing mature peroxisomes, suggesting that some portion of these proteins may be localized to some compartment other than mature peroxisomes (Vizeacoumar et al., 2004).
Our data show that both Pex30p and Pex31p are localized to the ER, as well as to peroxisomes, suggesting that they probably shuttle between these two compartments. The pulse-chase experiment showing trafficking of Pex31p from the ER to peroxisomes in a Pex19p-dependent manner, the interactions between Pex30p and Pex31p, the requirement of Pex30p for the stability of Pex31p, and the fact that we can redistribute the pools of each of these proteins between these compartments is certainly consistent with such shuttling. Overexpression of Pex31p dramatically increases the peroxisomal pool of Pex30p (Figure 7, top panels). Conversely, in the absence of Pex19p, Pex31p relocates to the ER, where it colocalizes with Pex30p (Figure 7, bottom panels). Because the ER and peroxisomes are not directly contiguous compartments, the most plausible mechanism by which the two proteins could find each other would be if there is a vesicle-mediated trafficking system, between the ER and peroxisomes. Recent work has provided evidence for such a vesicle-mediated ER-to-peroxisome transport system that contributes to the growth of pre-existing peroxisomes (Mullen and Trelease, 2006; Motley and Hettema, 2007). Finally, for two interacting proteins to be localized to different compartments, there is likely to be both an anterograde and retrograde trafficking system (Mullen and Trelease, 2006) or, alternatively, there must be a mechanism to degrade Pex30p at peroxisomes.
Like Pex31p, there are other peroxins, including YlPex2p, ScPex3p, and Pex16p in Y. lipolytica and mammals, whose initial sorting site is at the ER before they reach the peroxisome membrane (Titorenko and Rachubinski, 1998; Hoepfner et al., 2005; Kim et al., 2006). The translocation of Pex31p from the ER to the peroxisome does not depend on Pex30p, because overexpressed Pex31p in pex30Δ cells was still primarily localized to peroxisomes (our unpublished data). Additionally, Pex31p is retained at the ER in pex19Δ cells (Figure 7). Like Pex31p, ScPex3p also traffics to peroxisomes via the ER, accumulating at the ER in pex19Δ cells and moving to the peroxisomes in a Pex19p-dependent manner (Hoepfner et al., 2005). The dual localization of PpPex30p and PpPex31p also implicates the de novo peroxisome assembly pathway from the ER in the morphogenesis of oleate-induced peroxisomes.
A Working Model for the Role of PpPex30p and PpPex31p in Controlling Peroxisome Division
Our studies show that both Pex30p and Pex31p are needed for proper peroxisome size and number and that in their absence, peroxisome division, but not growth, is affected. This would yield larger and fewer peroxisomes in pex30Δ and pex31Δ cells, as observed. Previous work in many model organisms, confirmed by our observations in P. pastoris, has shown that Pex11p is required for peroxisome division. Interestingly, like Pex30p and Pex31p, P. pastoris Pex11p only affects oleate-induced peroxisomes but not methanol-induced peroxisomes (Supplementary Figure 3A). The absence of Pex11p causes fewer and larger peroxisomes (Supplementary Figure 3A, right panel, and Figure 3B), a phenotype similar to, but more severe than, that seen in pex30Δ and pex31Δ cells. It has been reported that the multimerization state of Pex11p controls its activity, with the oligomeric state being inactive and the monomeric state being the active form (Marshall et al., 1996). But how this switch is controlled and what signals the transition from the inactive to the active state of Pex11p is unknown. Our working model is that peroxisomes grow by vesicle-mediated delivery of lipids (and certain proteins such as Pex3p) from the ER. If Pex30p and Pex31p accompany these vesicles, but are not necessary for their formation, they could transmit a signal to Pex11p as to when sufficient lipid has been delivered. Consistent with this idea, we found an interaction between Pex11p and Pex31p. The interaction is weak or conditional so that it could not be shown by yeast two-hybrid (our unpublished data), but coimmunoprecipitation clearly shows that Pex11p pulls down Pex31p (Supplementary Figure 3C). Weak interaction between Pex11p and Pex30p also exists (our unpublished data), probably mediated by Pex31p. It is possible that the dysferlin domains have some role to play in relaying this signal, because in their absence we observe larger and fewer peroxisomes despite the fact that Pex11p is still on peroxisomes (our unpublished data) and the mutant proteins Pex30pΔC or Pex31pΔC still interact with each other and are properly localized (Figure 9). Once Pex11p-mediated peroxisome division is complete, the signal would need to be attenuated. This might be achieved either by retrieval of Pex30p to the ER or by local degradation of Pex30p such that it never accumulates to a large extent at peroxisomes. Experiments are underway to test this model.
Recent studies have shown that in Y. lipolytica, a signal mediated by Pex16p and acyl-CoA oxidase from within the peroxisome matrix can also initiate peroxisome division (Guo et al., 2007). It is likely that there may be redundant signals for peroxisome division that do not need to emanate from the peroxisome matrix, because in pex5-deficient cells that are incapable of all matrix protein import, Pex11p overexpression still promotes the division of empty peroxisome ghosts (Li and Gould, 2002). This study also showed that in S. cerevisiae, metabolic flux through the fatty acid β-oxidation pathway is not required for Pex11p-mediated peroxisome division. The mechanism by which such redundant peroxisome division pathways communicate with each other is an interesting unsolved puzzle.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. James Cregg (Keck Graduate Institute, Claremont, CA) and Integrated Genomics for access to the ERGO database, Drs. Jean-Claude Farré and Kanae Noda for plasmids, and Dr. Naganand Rayapuram for advice and input on this project. R.R. is Canada Research Chair in Cell Biology and an International Research Scholar of the Howard Hughes Medical Institute. This work was supported by a grant to S.S. from the National Institutes of Health (DK41737).
Abbreviations used:
- BFP
blue fluorescent protein
- BSA
bovine serum albumin
- EM
electron microscopy
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
- HA
hemagglutinin
- PMP
peroxisomal membrane protein
- Pp
Pichia pastoris
- PTS
peroxisome targeting signal
- RFP
red fluorescent protein
- Sc
Saccharomyces cerevisiae
- YFP
yellow fluorescent protein
- Yl
Yarrowia lipolytica.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-10-1042) on December 19, 2007.
REFERENCES
- Bansal D., Miyake K., Vogel S. S., Groh S., Chen C. C., Williamson R., McNeil P. L., Campbell K. P. Defective membrane repair in dysferlin-deficient muscular dystrophy. Nature. 2003;423:168–172. doi: 10.1038/nature01573. [DOI] [PubMed] [Google Scholar]
- Brown T. W., Titorenko V. I., Rachubinski R. A. Mutants of the Yarrowia lipolytica PEX23 gene encoding an integral peroxisomal membrane peroxin mislocalize matrix proteins and accumulate vesicles containing peroxisomal matrix and membrane proteins. Mol. Biol. Cell. 2000;11:141–152. doi: 10.1091/mbc.11.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cregg J. M., Russell K. A. Transformation. In: Higgins D. R., Cregg J. M., editors. Pichia Protocols. vol. 103. Totowa, NJ: Humana Press; 1998. pp. 27–39. [Google Scholar]
- Dammai V., Subramani S. The human peroxisomal targeting signal receptor, Pex5p, is translocated into the peroxisomal matrix and recycled to the cytosol. Cell. 2001;105:187–196. doi: 10.1016/s0092-8674(01)00310-5. [DOI] [PubMed] [Google Scholar]
- Doherty K. R., Cave A., Davis D. B., Delmonte A. J., Posey A., Earley J. U., Hadhazy M., McNally E. M. Normal myoblast fusion requires myoferlin. Development. 2005;132:5565–5575. doi: 10.1242/dev.02155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann R., Blobel G. Giant peroxisomes in oleic acid-induced Saccharomyces cerevisiae lacking the peroxisomal membrane protein Pmp27p. J. Cell Biol. 1995;128:509–523. doi: 10.1083/jcb.128.4.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber K. N., Heyman J. A., Subramani S. Two AAA family peroxins, PpPex1p and PpPex6p, interact with each other in an ATP-dependent manner and are associated with different subcellular membranous structures distinct from peroxisomes. Mol. Cell. Biol. 1998;18:936–943. doi: 10.1128/mcb.18.2.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagarasanu A., Fagarasanu M., Rachubinski R. A. Maintaining peroxisome populations: a story of division and inheritance. Annu. Rev. Cell Dev. Biol. 2007;23:321–344. doi: 10.1146/annurev.cellbio.23.090506.123456. [DOI] [PubMed] [Google Scholar]
- Fujiki Y., Matsuzono Y., Matsuzaki T., Fransen M. Import of peroxisomal membrane proteins: the interplay of Pex3p- and Pex19p-mediated interactions. Biochim. Biophys. Acta. 2006;1763:1639–1646. doi: 10.1016/j.bbamcr.2006.09.030. [DOI] [PubMed] [Google Scholar]
- Glover L., Brown R. H., Jr Dysferlin in membrane trafficking and patch repair. Traffic. 2007;8:785–794. doi: 10.1111/j.1600-0854.2007.00573.x. [DOI] [PubMed] [Google Scholar]
- Gould S. J., McCollum D., Spong A. P., Heyman J. A., Subramani S. Development of the yeast Pichia pastoris as a model organism for a genetic and molecular analysis of peroxisome assembly. Yeast. 1992;8:613–628. doi: 10.1002/yea.320080805. [DOI] [PubMed] [Google Scholar]
- Guo T., et al. A signal from inside the peroxisome initiates its division by promoting the remodeling of the peroxisomal membrane. J. Cell Biol. 2007;177:289–303. doi: 10.1083/jcb.200609072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoepfner D., Schildknegt D., Braakman I., Philippsen P., Tabak H. F. Contribution of the endoplasmic reticulum to peroxisome formation. Cell. 2005;122:85–95. doi: 10.1016/j.cell.2005.04.025. [DOI] [PubMed] [Google Scholar]
- Kim P. K., Mullen R. T., Schumann U., Lippincott-Schwartz J. The origin and maintenance of mammalian peroxisomes involves a de novo PEX16-dependent pathway from the ER. J. Cell Biol. 2006;173:521–532. doi: 10.1083/jcb.200601036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnirov V. V. Rapid and reliable protein extraction from yeast. Yeast. 2000;16:857–860. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Leon S., Suriapranata I., Yan M., Rayapuram N., Patel A., Subramani S. Characterization of protein-protein interactions: application to the understanding of peroxisome biogenesis. Methods Mol. Biol. 2007;389:219–238. doi: 10.1007/978-1-59745-456-8_16. [DOI] [PubMed] [Google Scholar]
- Leon S., Zhang L., McDonald W. H., Yates J., 3rd, Cregg J. M., Subramani S. Dynamics of the peroxisomal import cycle of PpPex20p: ubiquitin-dependent localization and regulation. J. Cell Biol. 2006;172:67–78. doi: 10.1083/jcb.200508096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Copley R. R., Pils B., Pinkert S., Schultz J., Bork P. SMART5, domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Gould S. J. PEX11 promotes peroxisome division independently of peroxisome metabolism. J. Cell Biol. 2002;156:643–651. doi: 10.1083/jcb.200112028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall P. A., Dyer J. M., Quick M. E., Goodman J. M. Redox-sensitive homodimerization of Pex11p: a proposed mechanism to regulate peroxisomal division. J. Cell Biol. 1996;135:123–137. doi: 10.1083/jcb.135.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley A. M., Hettema E. H. Yeast peroxisomes multiply by growth and division. J. Cell Biol. 2007;178:399–410. doi: 10.1083/jcb.200702167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen R. T., Trelease R. N. The ER-peroxisome connection in plants: development of the “ER semi-autonomous peroxisome maturation and replication” model for plant peroxisome biogenesis. Biochim. Biophys. Acta. 2006;1763:1655–1668. doi: 10.1016/j.bbamcr.2006.09.011. [DOI] [PubMed] [Google Scholar]
- Nair D. M., Purdue P. E., Lazarow P. B. Pex7p translocates in and out of peroxisomes in Saccharomyces cerevisiae. J. Cell Biol. 2004;167:599–604. doi: 10.1083/jcb.200407119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier Y., Antonenkov V. D., Glumoff T., Hiltunen J. K. Peroxisomal beta-oxidation—a metabolic pathway with multiple functions. Biochim. Biophys. Acta. 2006;1763:1413–1426. doi: 10.1016/j.bbamcr.2006.08.034. [DOI] [PubMed] [Google Scholar]
- Ponting C. P., Mott R., Bork P., Copley R. R. Novel protein domains and repeats in Drosophila melanogaster: insights into structure, function, and evolution. Genome Res. 2001;11:1996–2008. doi: 10.1101/gr.198701. [DOI] [PubMed] [Google Scholar]
- Rayapuram N., Subramani S. The importomer—a peroxisomal membrane complex involved in protein translocation into the peroxisome matrix. Biochim. Biophys. Acta. 2006;1763:1613–1619. doi: 10.1016/j.bbamcr.2006.08.035. [DOI] [PubMed] [Google Scholar]
- Rossanese O. W., Soderholm J., Bevis B. J., Sears I. B., O'Connor J., Williamson E. K., Glick B. S. Golgi structure correlates with transitional endoplasmic reticulum organization in Pichia pastoris and Saccharomyces cerevisiae. J. Cell Biol. 1999;145:69–81. doi: 10.1083/jcb.145.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottensteiner H., Stein K., Sonnenhol E., Erdmann R. Conserved function of Pex11p and the novel Pex25p and Pex27p in peroxisome biogenesis. Mol. Biol. Cell. 2003;14:4316–4328. doi: 10.1091/mbc.E03-03-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears I. B., O'Connor J., Rossanese O. W., Glick B. S. A versatile set of vectors for constitutive and regulated gene expression in Pichia pastoris. Yeast. 1998;14:783–790. doi: 10.1002/(SICI)1097-0061(19980615)14:8<783::AID-YEA272>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Smith J. J., Marelli M., Christmas R. H., Vizeacoumar F. J., Dilworth D. J., Ideker T., Galitski T., Dimitrov K., Rachubinski R. A., Aitchison J. D. Transcriptome profiling to identify genes involved in peroxisome assembly and function. J. Cell Biol. 2002;158:259–271. doi: 10.1083/jcb.200204059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder W. B., Faber K. N., Wenzel T. J., Koller A., Luers G. H., Rangell L., Keller G. A., Subramani S. Pex19p interacts with Pex3p and Pex10p and is essential for peroxisome biogenesis in Pichia pastoris. Mol. Biol. Cell. 1999;10:1745–1761. doi: 10.1091/mbc.10.6.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg S. J., Dodt G., Raymond G. V., Braverman N. E., Moser A. B., Moser H. W. Peroxisome biogenesis disorders. Biochim. Biophys. Acta. 2006;1763:1733–1748. doi: 10.1016/j.bbamcr.2006.09.010. [DOI] [PubMed] [Google Scholar]
- Subramani S., Koller A., Snyder W. B. Import of peroxisomal matrix and membrane proteins. Annu. Rev. Biochem. 2000;69:399–418. doi: 10.1146/annurev.biochem.69.1.399. [DOI] [PubMed] [Google Scholar]
- Tam Y.Y., Rachubinski R.A. Yarrowia lipolytica cells mutant for the PEX24 gene encoding a peroxisomal membrane peroxin mislocalize peroxisomal proteins and accumulate membrane structures containing both peroxisomal matrix and membrane proteins. Mol. Biol. Cell. 2002;13:2681–2691. doi: 10.1091/mbc.E02-02-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam Y. Y., Torres-Guzman J. C., Vizeacoumar F. J., Smith J. J., Marelli M., Aitchison J. D., Rachubinski R. A. Pex11-related proteins in peroxisome dynamics: a role for the novel peroxin Pex27p in controlling peroxisome size and number in Saccharomyces cerevisiae. Mol. Biol. Cell. 2003;14:4089–4102. doi: 10.1091/mbc.E03-03-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titorenko V. I., Rachubinski R. A. Mutants of the yeast Yarrowia lipolytica defective in protein exit from the endoplasmic reticulum are also defective in peroxisome biogenesis. Mol. Cell. Biol. 1998;18:2789–2803. doi: 10.1128/mcb.18.5.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Klei I. J., Yurimoto H., Sakai Y., Veenhuis M. The significance of peroxisomes in methanol metabolism in methylotrophic yeast. Biochim. Biophys. Acta. 2006;1763:1453–1462. doi: 10.1016/j.bbamcr.2006.07.016. [DOI] [PubMed] [Google Scholar]
- Vizeacoumar F. J., Torres-Guzman J. C., Bouard D., Aitchison J. D., Rachubinski R. A. Pex30p, Pex31p, and Pex32p form a family of peroxisomal integral membrane proteins regulating peroxisome size and number in Saccharomyces cerevisiae. Mol. Biol. Cell. 2004;15:665–677. doi: 10.1091/mbc.E03-09-0681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizeacoumar F. J., Torres-Guzman J. C., Tam Y. Y., Aitchison J. D., Rachubinski R. A. YHR150w and YDR479c encode peroxisomal integral membrane proteins involved in the regulation of peroxisome number, size, and distribution in Saccharomyces cerevisiae. J. Cell Biol. 2003;161:321–332. doi: 10.1083/jcb.200210130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizeacoumar F. J., Vreden W. N., Aitchison J. D., Rachubinski R. A. Pex19p binds Pex30p and Pex32p at regions required for their peroxisomal localization but separate from their peroxisomal targeting signals. J. Biol. Chem. 2006;281:14805–14812. doi: 10.1074/jbc.M601808200. [DOI] [PubMed] [Google Scholar]
- Wanders R. J. Metabolic and molecular basis of peroxisomal disorders: a review. Am. J. Med. Genet. A. 2004;126:355–375. doi: 10.1002/ajmg.a.20661. [DOI] [PubMed] [Google Scholar]
- Washington N. L., Ward S. FER-1 regulates Ca2+-mediated membrane fusion during C. elegans spermatogenesis. J. Cell Sci. 2006;119:2552–2562. doi: 10.1242/jcs.02980. [DOI] [PubMed] [Google Scholar]
- Weibel E. R., Bolender P. Stereological techniques for electron microscopic morphometry. In: Hayat M. A., editor. Principles and Techniques of Electron Microscopy. Vol. 3. New York: Van Nostrand Reinhold; 1973. pp. 237–296. [Google Scholar]
- Wiemer E. A., Luers G. H., Faber K. N., Wenzel T., Veenhuis M., Subramani S. Isolation and characterization of Pas2p, a peroxisomal membrane protein essential for peroxisome biogenesis in the methylotrophic yeast Pichia pastoris. J. Biol. Chem. 1996;271:18973–18980. doi: 10.1074/jbc.271.31.18973. [DOI] [PubMed] [Google Scholar]
- Yan M., Rayapuram N., Subramani S. The control of peroxisome number and size during division and proliferation. Curr. Opin. Cell Biol. 2005;17:376–383. doi: 10.1016/j.ceb.2005.06.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.