Abstract
The lipid polyunsaturated fatty acids are highly enriched in synaptic membranes, including synaptic vesicles, but their precise function there is unknown. Caenorhabditis elegans fat-3 mutants lack long-chain polyunsaturated fatty acids (LC-PUFAs); they release abnormally low levels of serotonin and acetylcholine and are depleted of synaptic vesicles, but the mechanistic basis of these defects is unclear. Here we demonstrate that synaptic vesicle endocytosis is impaired in the mutants: the synaptic vesicle protein synaptobrevin is not efficiently retrieved after synaptic vesicles fuse with the presynaptic membrane, and the presynaptic terminals contain abnormally large endosomal-like compartments and synaptic vesicles. Moreover, the mutants have abnormally low levels of the phosphoinositide phosphatase synaptojanin at release sites and accumulate the main synaptojanin substrate phosphatidylinositol 4,5-bisphosphate at these sites. Both synaptobrevin and synaptojanin mislocalization can be rescued by providing exogenous arachidonic acid, an LC-PUFA, suggesting that the endocytosis defect is caused by LC-PUFA depletion. By showing that the genes fat-3 and synaptojanin act in the same endocytic pathway at synapses, our findings suggest that LC-PUFAs are required for efficient synaptic vesicle recycling, probably by modulating synaptojanin localization at synapses.
INTRODUCTION
Neurons contain an enormous variety of complex lipids, the functions of which are largely unknown. Polyunsaturated fatty acids (PUFAs) contain at least two double bonds and are mostly located in neuronal cell membranes as phospholipid esters. Certain PUFAs, particularly those with long acyl chains (for example, docosahexaenoic acic [DHA]) are highly enriched in synaptic membranes, including synaptic vesicles (SVs; Cotman et al., 1969; Deutsch and Kelly, 1981; Takamori et al., 2006), suggesting that PUFAs have important synaptic functions (Lauritzen et al., 2001). Mutations in enzymes involved in the regulation of PUFA metabolism cause serious neuronal disorders and behavioral defects in humans (Zhang et al., 2001; Meloni et al., 2002). Exogenous PUFAs also stimulate neurite outgrowth (Darios and Davletov, 2006) and modulate the activity of various ion channels, including TRP channels (Vreugdenhil et al., 1996; Chyb et al., 1999; Kahn-Kirby et al., 2004). Although these findings demonstrate that PUFAs have important roles in neurons, it is still unclear which synaptic function(s) they modulate and how they do it.
The nematode Caenorhabditis elegans synthesizes many of the PUFAs present in humans, suggesting that these molecules have evolutionarily conserved functions. To investigate the roles of PUFAs in neurons, we generated PUFA-deficient C. elegans mutants by mutating the gene fat-3, which encodes a Δ6-desaturase (Watts and Browse, 2002; Lesa et al., 2003). fat-3 mutants are depleted of long-chain (LC) PUFAs, including arachidonic acid (AA, C20) and DHA (C22), but not of short-chain PUFAs (≤C18) like linoleic acid (LIN) and α-linolenic acid (ALA), and they exhibit abnormal neuronal function. The neuronal defects observed in fat-3 mutants are likely to be caused by depletion of LC-PUFAs because exogenous AA or DHA rescue them (Watts and Browse, 2002; Lesa et al., 2003).
We previously showed that, although fat-3 mutants cannot make LC-PUFAs, they undergo normal neuronal development and display normal neuronal morphology. The mutants, however, have reduced synaptic function, as indicated by the abnormally low levels of neurotransmitter they release at cholinergic and serotonergic neuromuscular junctions. This decrease in neurotransmitter release is caused by a severe depletion of SVs at release sites (Lesa et al., 2003), suggesting that LC-PUFAs are required to maintain a normal pool of SVs.
SVs are initially generated in neuronal cell bodies as precursor vesicles, which are transported along the axon to the nerve terminals (Hannah et al., 1999). After fusion with the presynaptic plasma membrane, SV proteins and lipids are rapidly endocytosed via a clathrin-mediated mechanism and are reused to form new SVs (Sudhof, 2004). Although there has been great progress in recent years elucidating the role of specific proteins in endocytosis (Sudhof, 2004), much less is known about the role of lipids in the SV cycle (Rohrbough and Broadie, 2005). Among the best characterized lipids involved in SV recycling are the phosphoinositides, which are phospholipids stably anchored to the plasma membrane via two long-chain fatty acid chains, one of which is always a LC-PUFA (Cremona and De Camilli, 2001; Wenk and De Camilli, 2004).
At early stages of endocytosis in synapses, the phosphoinositide phosphatidylinositol 4,5-bisphosphate [PtdIns(4,5)P2] recruits several essential proteins to synaptic membranes, including dynamin and the clathrin adaptor proteins AP2 and AP180. At later stages, PtdIns(4,5)P2 is dephosphorylated by synaptojanin-1, decreasing the affinity of the clathrin adaptor proteins for the SV membrane (Cremona and De Camilli, 2001; Wenk and De Camilli, 2004). Studies in C. elegans and Drosophila have shown that endophilin A is required for synaptojanin recruitment to sites of neurotransmitter release (Schuske et al., 2003; Verstreken et al., 2003). The C. elegans genome encodes a single synaptojanin, UNC-26, and unc-26-synaptojanin mutants exhibit a phenotype similar to that of fat-3 and unc-57-endophilin A mutants, with a depletion of SVs at presynaptic sites (Lesa et al., 2003; Schuske et al., 2003).
SVs are very small (diameter 30–40 nm), and so their membrane is highly curved. The degree of membrane curvature is greatly influenced by the composition and orientation of the lipids (Hammond et al., 1984), as well as by membrane-bending proteins (McMahon and Gallop, 2005; Dawson et al., 2006). SV recycling is critically dependent on the ability of the membrane to become curved, and so it is not surprising that it is influenced by both membrane lipids (Rohrbough and Broadie, 2005) and the membrane-bending protein endophilin (Gallop et al., 2006; Masuda et al., 2006).
The reduced number of SVs at synapses and the impaired neurotransmission observed in animals depleted of LC-PUFAs are consistent with a role for LC-PUFAs in vesicle recycling, but other roles, including a role in SV formation, are possible. The parallel decrease in neurotransmitter release and SV number makes a defect in SV fusion unlikely. To clarify the role of LC-PUFAs at the synapse, we have investigated the mechanisms underlying the depletion of SVs observed in LC-PUFA–deficient fat-3 mutants by determining the subcellular distribution of SVs and their components at presynaptic sites. We have analyzed two fat-3 alleles, a reduction-of-function allele called wa22 and a null allele called lg8101. Our results suggest that reducing the activity of the fat-3 gene causes defects in SV recycling. First, the SV component synaptobrevin is partially mislocalized at sites of release in the mutants. Second, SVs are enlarged in the mutants. Third, fat-3 acts in the same genetic pathway as synaptojanin at release sites. Fourth, inactivation of fat-3 results in synaptojanin depletion and in accumulation of PtdIns(4,5)P2 at synapses. Fifth, exogenous LC-PUFAs largely rescue the mislocalization of synaptobrevin and synaptojanin and the functional deficits. We therefore propose that LC-PUFAs are essential for efficient SV recycling by influencing the localization of synaptojanin at sites of neurotransmitter release.
MATERIALS AND METHODS
C. elegans Strains
Strains were cultured at 20°C as described (Brenner, 1974). fat-3(wa22) and snb-1(md247) were obtained from the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis, MN). The allele fat-3(lg8101) was isolated by us (Lesa et al., 2003). The strains lin-15(n767ts) nIs52[Punc-25::snb-1::gfp] (Jorgensen et al., 1995), unc-57(ok310); nIs52 (Schuske et al., 2003), and unc-26(s1710); oxEx749[Prab-3::gfp::unc-26] were kindly provided by Erik Jorgensen (University of Utah, Salt Lake City). The strain hpIs76[lin-15(+); Punc-25::syd-2::rfp]; lin-15(n765), which expresses SYD-2::RFP was a gift from Mei Zhen (University of Toronto, Canada). The strains fat-3(wa22); nIs52, unc-57(ok310); fat-3(wa22); nIs52, fat-3(wa22)unc-26(s1710); nIs52, fat-3(wa22) unc-26(s1710); oxEx749[Prab-3::gfp::unc-26], unc-26(s1710); hpIs76; oxEx749, dpy-20(e1282); gxEx151[dpy(+); Punc-25::unc-57::gfp], fat-3(wa22)dpy-20(e1282); gxEx151, dpy-20(e1282); gxEx152[dpy(+); Punc-25::amph-1::gfp], fat-3(wa22)dpy-20(e1282); gxEx152, Ex108[Prab-3::phplc-δ1::gfp; Pttx-3::rfp; rol-6(su1006)] and fat-3(wa22); Ex108 were obtained by conventional genetic methods. The unc-26(s1710) background was used to avoid Prab-3::GFP::UNC-26 mislocalization (Kim Schuske and Erik Jorgensen, personal communication). The presence of nIs52, fat-3(wa22), unc-57(ok310), and oxEx749 were verified by PCR or by DNA sequencing.
To construct the strain expressing PHPLC-∂1::GFP, DNA coding for PHPLC-∂1 was excised as a KpnI fragment from pHD86 (Patton et al., 2005), subcloned into the green fluorescent protein (GFP) vector pPD115.62 (a gift from Andy Fire, Stanford University, Stanford, CA) and placed under the control of a 1.2-kb rab-3 promoter fragment. Transgenic strains were generated by microinjection (Mello and Fire, 1995) of the plasmid Prab-3::phplc-δ1::gfp (10 ng/μl) along with the coinjection markers Pttx-3::rfp (10 ng/μl) and pRF4 [rol-6(su1006), 40 ng/μl]. The injection mix was adjusted to a total DNA concentration of 100 ng/μl by the addition of pBlueScript.
Analysis of Locomotion
The number of body bends per second was assayed by visual inspection in the presence of food on Nematode Growth Medium culture plates according to McMullan et al. (2006). For each tested strain, at least 25 L4 hermaphrodites were assayed twice for 1 min.
Arachidonic Acid Supplementation
AA sodium salt (Sigma, Poole, United Kingdom) was dissolved in water and added with 0.1% NP-40 (Merck Biosciences, Nottingham, United Kingdom) to Nematode Growth Medium as described (Watts et al., 2003) so that the final concentration of AA was ∼0.75 mM. Plates were poured, seeded with Escherichia coli OP50, and allowed to dry for 2 d at room temperature. L4 hermaphrodites were transferred to the plates and allowed to grow for 4 d, and their L4 progeny were then used for analysis.
Immunohistochemistry
Immunohistochemistry was carried out as described in (Nonet et al., 1999). We used antibodies against the following proteins: SNB-1 (SB1, Developmental Studies Hybridoma Bank, University of Iowa), UNC-11 (kindly provided by Aixa Alfonso, University of Illinois, Chicago, IL), SNT-1 and RAB-3 (AB1095 and AB326, respectively, kindly provided by Michael Nonet, Washington University, St. Louis, MO) and performed immunohistochemistry as described in Nonet et al. (1999). Worms were fixed in Bouin's buffer for RAB-3 and UNC-11 detection or with 4% paraformaldehyde for SNB-1 and SNT-1 detection. Anti-RAB-3, anti-SNT-1, anti-SNB-1, and anti-UNC-11 antibodies and Alexa fluor 488 (Invitrogen Ltd, Paisley, United Kingdom) secondary antibody were used at a dilution of 1:2000, 1:5000, 1:150, 1:50, and 1:500, respectively.
Protein Extraction and Western Blotting
Mixed-stage populations of worms were suspended in 5 volumes of ice-cold lysis buffer (0.36 M sucrose, 12 mM HEPES, Complete Mini Protease inhibitor cocktail [Roche, Lewes, United Kingdom], 1 μg/ml pepstatin A [Sigma]) and sonicated on ice with four 5-s bursts using a 1/8 microtip at full power. Worm debris was removed by centrifugation at 2000 × g for 15 min, and the supernatant was centrifuged at 100,000 × g for 45 min at 4°C. The membrane pellet proteins (45 μg) were separated by SDS-PAGE and blotted onto a PVDF membrane (Bio-Rad, Hemel Hempstead, Herts, United Kingdom) and assayed with anti-SNB-1 (SB-1, 1:1000) and anti-syntaxin (kindly provided by Vincent O'Connor, University of Southampton, UK, 1:500) antibodies. Peroxidase-conjugated goat anti-mouse secondary antibody (Perbio, Cramlington, United Kingdom) was used at a dilution of 1:10,000 and revealed by electrochemiluminescence (Amersham Biosciences, Little Chalfont, United Kingdom).
Biochemical Quantification of Total PtdIns(4,5)P2 Levels
Worms were grown in liquid culture in the presence of 10 μCi/ml myo[1,2-3H]inositol (American Radiolabeled Chemicals, St. Louis, MO) for 4 to 5 d. Lipids were extracted from mixed-stage populations of wild-type (WT), fat-3(wa22), and unc-26(s1710) worms in a chloroform solution containing 0.02% of Folch type I brain extract (Sigma). These lipids were extracted twice, deacylated, resuspended in 1 mM EDTA, and resolved using anion exchange high-performance liquid chromatography (HPLC) with a Whatman PartiSphere SAX (4.6 × 250 mm) column (Whatman plc, Maidstone, Kent, UK). Three independent measurements were performed for each strain tested.
Quantitative RT-PCR
Total RNA was extracted from mixed-stage populations of worms using the RNeasy kit (Qiagen, Crawley, United Kingdom). Total RNA, 1.5 μg per sample, was reverse-transcribed with Superscript II Reverse Transcriptase (Invitrogen) and hexamer random primers (Promega, Southampton, United Kingdom). Quantitative PCR was carried out in duplicates of 30 μl volume, on a Mx3000P QPCR system (Stratagene, Amsterdam, The Netherlands). We used 2× iQ SYBR Green Supermix (Bio-Rad), 2 μl of 1/10 diluted cDNA and primers to a final concentration of 0.3 μM each. Primers used to amplify a 137-base pair fragment of unc-26-synaptojanin cDNA (sense GCAACGTTAGATTCGGATTC and antisense TCAGCTCCATAAAGTTGTCC), a 180-base pair fragment of nhr-23 cDNA (sense GATGCCGAGATGCAATTGATC and antisense GAAGAGCCTCTTGATGCAAAC), and a 130-base pair fragment of β-actin (sense GCCCCATCAACCATGAAG and antisense CCGGACTCGTCGTATTCTTG) were designed on exon boundaries. The PCR profile consisted of an initial denaturation step at 95°C for 10 min followed by 30 cycles of denaturation at 95°C for 20 s, annealing at 56°C for 20 s, and extension at 72°C for 20 s. Standard curves were generated using serial dilutions of the purified amplicons from the target and the control genes. No amplification was detected for reverse-transcription–negative controls or for controls with no template. Three independent samples were analyzed three independent times.
Fluorescence Microscopy and Quantitative Analyses
Live animals were immobilized in 10 mM levamisole (Sigma). Images were captured from portions of the dorsal or ventral nerve cord (see below) with a 40× 1.30 NA objective (1 pixel = 400 nm) for RAB-3, SNT-1, SNB-1::GFP, and UNC-57::GFP or a 60× 1.40 NA objective (1 pixel = 130 nm) for UNC-11, SNB-1, SNB-1::GFP, PHPLC-∂1::GFP, and AMPH-1::GFP on a Bio-Rad Radiance 2100 Multiphoton Confocal microscope using LaserSharp software. Images showed a similar pattern of fluorescence independently of region of the nerve cord from which they were collected.
For SNB-1::GFP expressing animals and for anti-UNC-11 and anti-RAB-3 immunostained animals, all the images were taken from the same 50 μm region of the dorsal nerve cord, located 28 μm posterior to the motor neuron cell body RMED (Figure 1A). A corresponding 50 μm region in the ventral nerve cord was used to image anti-SNT-1–immunostained worms (Supplementary Figure S2C). For anti-SNB-1 immunostaining, images were taken from a 50 μm region of the dorsal nerve cord just above the vulva (Supplementary Figure S2C). For worms expressing GFP::UNC-26, SYD-2::RFP, UNC-57::GFP, AMPH-1::GFP, and/or PHPLC-∂1::GFP, images were collected from a 50 μm stretch of the dorsal nerve cord lying 34 μm anterior to the anus (see Figure 4F).
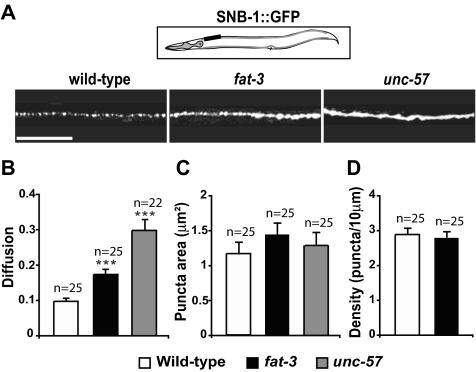
Figure 1.
Synaptobrevin is partially mislocalized in fat-3 mutants. (A) Localization of SV proteins at sites of release in WT (first column), fat-3(wa22) (second column), and unc-57(ok310)-endophilin animals (third column). GFP-tagged synaptobrevin (SNB-1::GFP) exhibited a punctate pattern in the dorsal nerve cord of WT animals. SNB-1::GFP was partially diffused along the axons in fat-3(wa22) mutants and was even more diffused in unc-57-endophilin mutants. The black box in the schematic shows the portion of the nerve cord imaged. The fluorescent pattern was similar for any portion of the nerve cord we analyzed. Anterior, left; dorsal, up. Scale bar, 30 μm. (B) Diffusion of SNB-1::GFP was significantly increased in fat-3(wa22) mutants. Diffusion was defined as [Σ(fa/fp)]/n. fp is the fluorescence intensity of each individual synapse (punctum) and fa is the fluorescence intensity of each axonal membrane between synapses. For further details see Materials and Methods. The average fluorescence intensities detected in puncta and in axonal membranes between synapses are reported in Supplementary Figure S1. (C and D) Area (C) and density (D) of the puncta were similar in WT animals and fat-3(wa22) mutants. In unc-57(ok310)-endophilin mutants, the high level of diffusion did not allow us to accurately count all the puncta. Data are plotted as mean ± SEM. ***p < 0.0001.
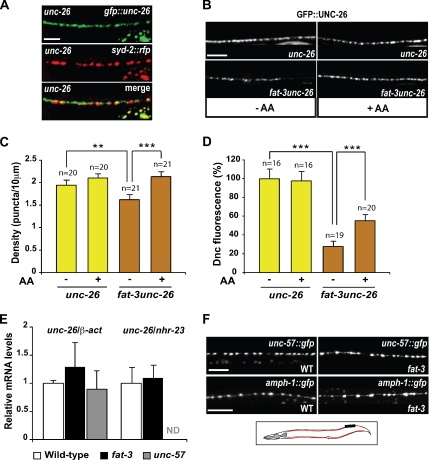
Figure 4.
Synaptojanin levels at sites of release are severely reduced in fat-3 mutants. All images were taken from the same 50 μm portion of the dorsal nerve cord (in black in the schematic on panel F). Anterior, left; dorsal, up. The nerve cord is shown in red. (A) Colocalization of GFP::UNC-26 with the presynaptic marker SYD-2::RFP in unc-26-synaptojanin animals. (B and C) Effect of the LC-PUFA AA on synaptojanin localization at release sites. (B) In unc-26-synaptojanin animals, which have a fully functional FAT-3 protein, GFP::UNC-26 is localized along the dorsal nerve cord and accumulates in synaptic varicosities exhibiting a punctate pattern. In fat-3unc-26-synaptojanin mutants, which have a nonfunctional FAT-3 protein, GFP::UNC-26 fluorescence is reduced at sites of release and often even puncta are hardly visible. Exogenous AA restores the WT punctate pattern in fat-3unc-26-synaptojanin mutants. (C) Quantification of the effect of AA on GFP::UNC-26 puncta density. (D) Quantification of GFP::UNC-26 fluorescence at release sites. (E) Quantitative RT-PCR analysis of unc-26-synaptojanin mRNA levels. Two different control genes, nhr-23 and β-actin, were used to normalize for the levels of unc-26-synaptojanin mRNA. No significant difference was detected in the mRNA levels of fat-3 mutants and unc-57-endophilin mutants compared with WT animals. Values are represented as the ratio between the mRNA copy number of unc-26-synaptojanin and that of either β-actin or nhr-23, and are normalized to WT. (F) UNC-57:: GFP and AMPH-1::GFP localizations appear normal in fat-3 mutants. In A, B, and F the fluorescence detected outside the nerve cord is gut autofluorescence. In C, D, and E data were plotted as mean ± SEM. Scale bars, 10 μm. **p = 0.026; ***p < 0.001. Dnc, dorsal nerve cord. Alleles: fat-3(wa22), unc-57(ok310) and unc-26(s1710).
Images were quantitatively analyzed using ImageJ software (http://rsb.info.nih.gov/ij/; National Institutes of Health, Bethesda, MD). The average number of GABAergic cell bodies per animal expressing SNB-1::GFP was determined by counting the number of fluorescent DD1, VD1, and VD2 cell bodies in 21 N2 animals and in 19 fat-3(wa22) mutants. Fluorescence intensity of SNB-1::GFP in the DD1, VD1, and VD2 neuronal cell bodies was calculated as the mean of the fluorescence intensity (F) of each single neuron divided by the fluorescence background (F0). The total fluorescence represents the sum of the fluorescence intensity in neuronal cell bodies and the fluorescence intensity within the dorsal nerve cord.
SNB-1::GFP, GFP::UNC-26, and PHPLC-∂1::GFP fluorescence in the dorsal nerve cord was measured as the fluorescence intensity within a 50 μm fragment of the nerve cord (F) divided by the fluorescence background (F0). The mean fluorescence intensities of each individual punctum (fp) and of the axonal membrane between each individual synapse (fa) were measured. Diffusion was defined as [Σ(fa/fp)]/n, where n is the number of puncta. In Supplementary Figure S1 we also show the values of the fluorescence intensity of axonal membranes (Fa), which reflects the average level of mislocalized SNB-1::GFP and of puncta (Fp). Fa = (Σfa)/n; Fp = (Σfp)/n. The area of the puncta was measured in an automated manner using ImageJ software. Puncta density is expressed as the average number of puncta contained in a 10 μm segment of the dorsal nerve cord.
Electron Microscopy
Young adult nematodes were immersed in fixatives containing buffered aldehydes and osmium tetroxide (Sigma; Hall, 1995). Postfixation steps contained 0.5% of potassium ferrocyanide (Sigma). Samples were embedded into plastic resin, and serial ultrathin sections were performed with a diamond knife and examined on a Philips CM10 transmission electron microscope (Philips Electron Optics, Eindhoven, The Netherlands). In parallel, young adult nematodes were fast-frozen by high-pressure freezing, stained in osmium tetroxide and uranyl acetate in acetone at low temperature, embedded in Epon resin, and cured into plastic (McDonald, 1999). For quantitative measurements of vesicle populations at synapses, WT and fat-3 samples were fixed by high-pressure freezing. For quantitative measurements in neuron cell bodies, chemically fixed samples were used. We used two fixation conditions because fat-3 mutants are, for some unknown reason, difficult to fix well, and the quality of the images obtained was not optimal. However, we found similar results with either fixation method.
To quantify SV diameter in the hypomorph fat-3(wa22) allele, we measured the diameter of 497 SVs from four mutants and 511 SVs from six WT animals. The average measurement for each synapse on each negative was calculated. These averages were used for statistical analysis. To quantify SV diameter in the null fat-3(lg8101) allele, we measured the diameter of 781 SVs and round membrane-bounded structures from six mutants and 624 SVs from two WT animals. For SVs, the average measurement of round structures up to 56 nm was considered, whereas for endocytotic structures, the diameter 71–96 nm was considered. The average measurements were used for statistical analysis.
Statistical Analyses
Statistical analyses were performed using SAS/STAT software (SAS Institute, Cary, NC) or InStat software (GraphPad Software, San Diego, CA). Significance was tested using an unpaired two-tailed Student's t test or the Mann-Whitney nonparametric test, depending on the experiment. Throughout the article data are expressed as mean ± SEM.
RESULTS
Production and Transport of SV Precursors Appear Normal in fat-3 Mutants
To examine whether the decrease in SV number in fat-3 mutants is caused by defects in the biogenesis or the transport of SV precursors, we visualized and quantified the endogenous SV protein synaptobrevin (SNB-1), as well as SNB-1 tagged with GFP (SNB-1::GFP; Jorgensen et al., 1995; Supplementary Figure S1). Because the vast majority of SNB-1 is tightly associated with the membrane of SVs and their precursor vesicles, its distribution faithfully reflects the distribution of these vesicles (Trimble et al., 1988; Dittman and Kaplan, 2006). We found that the total levels of endogenous SNB-1 and the total fluorescence intensity of SNB-1::GFP were similar in fat-3(wa22) mutants and in WT animals (Supplementary Figure S1, A and B), suggesting that normal levels of SNB-1 are synthesized in fat-3(wa22) mutants. The levels were also normal in the endocytosis mutant unc-57-endophilin.
It has been shown that mutants defective in the transport of SV precursors to sites of release accumulate an abnormally high number of small precursor vesicles (30–50 nm) in their neuronal cell bodies (Hall and Hedgecock, 1991; Jorgensen et al., 1995). To test whether SVs are mobilized correctly to sites of release in fat-3 mutants, we quantified the number of small vesicles in neuronal cell bodies. Although we found a small increase in the average number of these vesicles in the null fat-3(lg8101) mutants, compared with WT animals, the increase was not statistically significant (p = 0.43; Supplementary Figure S1, C–E), suggesting that the transport of SV precursors to sites of release is either normal or minimally affected in fat-3 mutants. In agreement with this conclusion, we could not detect accumulation of SNB-1::GFP in GABAergic neuronal cell bodies or an increase in the number of neuronal cell bodies with detectable SNB-1::GFP (Supplementary Figure S1, F–H) in fat-3(wa22) mutants. In addition, we found that normal levels of SNB-1::GFP reach sites of release (Supplementary Figure S1I) in fat-3(wa22) mutants, suggesting that normal amounts of SV precursors reach release sites in the mutants. Because both the formation and transport of SV precursors were apparently normal in fat-3 mutants, we went on to determine whether the recycling of SVs was impaired in the mutants.
SV Recycling Is Defective in fat-3 Mutants
Mutants defective in the recycling of SV components, such as strains carrying a mutation in synaptojanin (unc-26), endophilin A (unc-57), or the clathrin adaptor AP180 (unc-11) are unable to efficiently retrieve SV components, including SNB-1::GFP, from the presynaptic plasma membrane. As a result, SNB-1::GFP diffuses along the axonal plasma membrane and is visible between release sites. By contrast, mutants with impaired SV fusion show the opposite phenotype with respect to SNB-1::GFP localization: they have less SNB-1::GFP than WT animals along the axonal plasma membrane (Nonet et al., 1999; Harris et al., 2000; Schuske et al., 2003; Dittman and Kaplan, 2006). Therefore, diffusion of SNB-1::GFP in the plasma membrane away from release sites is indicative of a defect in SV recycling.
We assessed the distribution of SNB-1::GFP in the nerve cord of fat-3(wa22) mutants, WT animals, and the endocytosis mutant unc-57(ok310)-endophilin (Figure 1). In WT animals, SNB-1::GFP was distributed in puncta, corresponding to clusters of SVs at neuromuscular junctions. By contrast, SNB-1::GFP was mislocalized in the dorsal nerve cord of both unc-57-endophilin and fat-3 mutants (Figure 1A and Supplementary Figure S2A). To quantify SNB-1::GFP mislocalization, we calculated protein diffusion values (Figure 1B; for the definition of these diffusion values, see Materials and Methods). The diffusion values obtained for fat-3(wa22) mutants (0.174 ± 0.013, n = 25) and unc-57-endophilin mutants (0.299 ± 0.029, n = 22) were 1.7-fold (p < 0.0001) and 3-fold (p < 0.0001) greater than those of WT animals (0.098 ± 0.007, n = 25), respectively. Similar diffusion values were obtained from images captured at very high resolution (Supplementary Figure S2B). Endogenous SNB-1 also appeared to be partially mislocalized in the fat-3(wa22) mutant, whereas the localizations of other SV proteins including RAB-3, SNT-1, and UNC-11 were not affected (Supplementary Figure S2, C and D).
The sizes of the SNB-1::GFP puncta were similar in the fat-3(wa22) mutant (1.42 ± 0.11 μm2, n = 25; p = 0.11), WT animals (1.18 ± 0.10 μm2, n = 25), and the unc-57(ok310)-endophilin mutant (1.29 ± 0.18 μm2, n = 25; p = 0.81; Figure 1C), suggesting that synaptic size was normal in the mutants. The density of puncta along the dorsal nerve cord was normal in the fat-3(wa22) mutant when compared with WT worms (2.8 ± 0.1 vs. 2.9 ± 0.1 puncta per 10 μm, n = 25; p = 0.55; Figure 1D), suggesting that the number of synapses along the dorsal nerve cord segment is normal in the fat-3(wa22) mutants, in agreement with our previous findings that synaptic and neuronal morphologies are normal in the mutants (Lesa et al., 2003). The partial mislocalization of SNB-1, however, suggests that SV recycling is inefficient in fat-3 mutants, consistent with a defective retrieval of SV components from the presynaptic plasma membrane.
fat-3 Mutants Have Abnormally Large SVs
Defects in endocytosis are often associated with an increase in SV size and the appearance of abnormal, intracellular, membrane-bounded structures at synapses (Zhang et al., 1998; Fergestad et al., 1999; Nonet et al., 1999; Verstreken et al., 2003). We therefore examined the ultrastructure of synapses in both the reduction-of-function wa22 and null lg8101 fat-3 mutants (Figure 2).
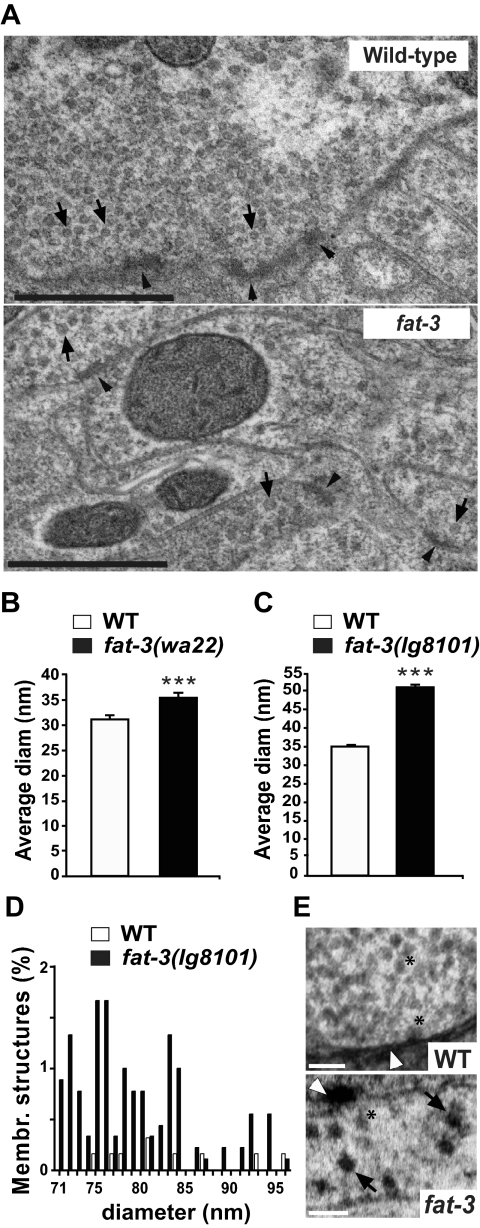
Figure 2.
SVs and membrane-bounded structures at synapses. (A) Electron micrographs of WT and fat-3 mutant synapses fixed by high-pressure freezing. In fat-3(wa22) mutants, SVs (black arrows) appear larger and more heterogeneous than in WT animals. The presynaptic dense bar is indicated by black arrowheads. Scale bar, 0.5 μm. (B) Average SV diameter in fat-3(wa22) mutants. The average diameter was significantly higher in the mutants than in WT animals (35.13 ± 0.86 vs. 31.06 ± 0.94 nm, p = 0.004). (C) Average SV diameter in fat-3(lg8101) mutants. To confirm that SV had a larger diameter in fat-3 mutants, we measured SV size in the fat-3(lg8101) null mutant (C) and found that the SV diameter was significantly higher than in WT animals (50.37 ± 0.80 vs. 34.59 ± 0.46 nm, p < 0.0001). (D) Diameter distribution of round membrane-bounded structures found at presynaptic sites in the fat-3(lg8101) mutant. These structures are distinctly larger in diameter than SVs and are likely to be generated by defective endocytosis. (E) Electron micrographs showing WT and fat-3(lg8101) synapses at high magnification. Round membrane-bounded structures are visible in fat-3(lg8101) mutants (black arrows), but not in WT animals.
The number of SVs in fat-3(lg8101) was reduced to 37% of that in WT animals (data not shown), similar to that detected in our previous study (Lesa et al., 2003). This reduction was virtually identical to that reported in unc-26-synaptojanin mutants (38%; Harris et al., 2000) and in unc-57-endophilin mutants (35%; Schuske et al., 2003). Enlarged SVs were visible in both fat-3(wa22) (Figure 2A) and fat-3(lg8101) (Figure 2E) mutants. We also determined the diameter of SVs in the mutants (Figure 2, B and C) and found that the average SV diameter was significantly increased 1.13-fold in the fat-3(wa22) mutant and 1.46-fold in the fat-3(lg8101) mutant. We also detected a significant number of larger membrane-bounded structures (diameter 71–96 nm) in the fat-3(lg8101) mutant that were not detected in WT animals (Figure 2, D and E) and were only rarely visible in the mild fat-3(wa22) mutant, suggesting that their formation is inversely proportional to fat-3 activity. These structures presumably represent abnormal endocytic structures generated by aberrant SV recycling, rather than functional SVs. These results are consistent with a defect in synaptic vesicle recycling in fat-3 mutants.
Exogenous AA Rescues the SV Recycling Defect in fat-3 Mutants
To test whether the defect in SV recycling observed in fat-3 mutants is dependent on LC-PUFA depletion, we analyzed the localization of SNB-1::GFP in fat-3(wa22) mutants grown in medium supplemented with the LC-PUFA AA (Figure 3). We and others had previously shown that providing exogenous LC-PUFAs, including AA, could rescue the functional defects associated with the fat-3 mutation (Lesa et al., 2003; Watts et al., 2003; Kahn-Kirby et al., 2004).
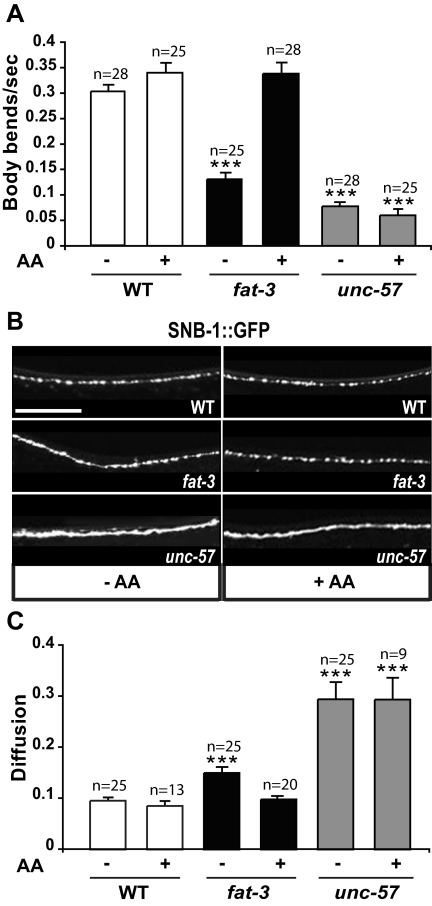
Figure 3.
The LC-PUFA arachidonic acid rescues the locomotion defect and the mislocalization of synaptobrevin in fat-3 mutants. (A) Exogenous AA rescues the locomotion defect of fat-3 mutants but not of unc-57-endophilin mutants. (B) Exogenous AA restores WT localization (top) of SNB-1::GFP in fat-3 mutants (middle) but not in unc-57-endophilin mutants (bottom panel). Scale bar, 30 μm. (C) AA restores a diffusion value similar to that observed in WT animals in fat-3 but not in unc-57-endophilin mutants. The quantification was performed on a 50 μm portion 28 μm posterior to the ring motor neuron RMED (schematic in Figure 1A). In A and C, data are plotted as mean ± SEM. ***p < 0.0001, versus WT. Alleles: fat-3(wa22) and unc-57(ok310).
We first verified that exposure to exogenous AA rescued the locomotion defect observed in fat-3(wa22) mutants (Figure 3A). We calculated the locomotion rate by counting the number of body bends per second. In the absence of AA, fat-3(wa22) mutants moved much more slowly than WT animals (0.13 ± 0.01, n = 25, vs. 0.30 ± 0.01, n = 28; p = 0.0001). When these mutants were grown in media supplemented with AA, the locomotion rate was indistinguishable from that of WT animals exposed to the LC-PUFA (0.34 ± 0.02, n = 25, vs. 0.34 ± 0.01, n = 28; p = 0.65). The effect of exogenous AA was specific to fat-3(wa22) mutants, because unc-57(ok310)-endophilin mutants did not move faster when they were grown in the presence of AA (0.08 ± 0.01, n = 28, vs. 0.06 ± 0.01, n = 25).
The defect in the localization of SNB-1::GFP in the axonal plasma membrane was also largely rescued by providing exogenous AA (Figure 3B). fat-3(wa22) mutants exposed to AA showed the same punctate SNB-1::GFP fluorescence observed in WT animals, and the diffusion value, which was 0.174 ± 0.013 in fat-3(wa22) mutants grown in the absence of AA, was virtually identical to that in WT animals (0.098 ± 0.005, n = 20 vs. 0.096 ± 0.006, n = 25; Figure 3C). Conversely, WT animals and unc-57-endophilin mutants exposed to exogenous AA showed no change in the pattern of SNB-1::GFP fluorescence (Figure 3B) or in the diffusion value (Figure 3C), which went from 0.096 ± 0.006 (n = 25) to 0.085 ± 0.008 (n = 13) in WT animals and from 0.293 ± 0.032 (n = 25) to 0.294 ± 0.043 (n = 9) in unc-57-endophilin mutants.
These experiments suggest that the defect in SV recycling observed in fat-3 mutants is caused by LC-PUFA depletion. Therefore, normal levels of LC-PUFAs are required for normal recycling of SV components at synapses.
fat-3 Is Required for Normal Synaptojanin Localization at Sites of Release
We observed that the behavioral phenotype of fat-3(wa22) mutants is qualitatively similar, although weaker, to that of the endocytosis mutants unc-26-synaptojanin, raising the possibility that the normal function of synaptojanin might require LC-PUFAs. We examined the localization of the fully functional, GFP-tagged synaptojanin protein (GFP::UNC-26, Figure 4, A–D) at release sites of unc-26(s1710)-synaptojanin mutants. We used the unc-26 mutant background because GFP::UNC-26 is mislocalized in a WT unc-26 background (Kim Schuske and Erik Jorgensen, personal communication). We confirmed that the puncta observed with GFP::UNC-26 were release sites by showing that they colocalized with the active zone marker SYD-2 (Zhen and Jin, 1999; Figure 4A). In fat-3(wa22)unc-26(s1710)-synaptojanin mutants, GFP::UNC-26 fluorescence in the dorsal nerve cord was greatly reduced, such that most fluorescent puncta were hardly detectable, and there were fewer of them than in WT animals (Figure 4B). The puncta density was decreased from 1.95 ± 0.09 puncta/10 μm in the presence of the WT fat-3 gene (unc-26 animals, n = 20) to 1.63 ± 0.10 in fat-3(wa22)unc-26(s1710)-synaptojanin mutants (p = 0.026, n = 21; Figure 4C). The number of puncta was restored to normal when fat-3(wa22)unc-26-synaptojanin mutants were grown in the presence of exogenous AA (2.14 ± 0.09, n = 21, vs. 2.11 ± 0.08, n = 20). The decrease in puncta in the fat-3 mutants was not caused by a reduction in synapse number along the nerve cord, because the density of SNB-1::GFP fluorescent puncta was virtually identical in fat-3(wa22) mutants and WT animals (see Figure 1D); it reflected a decrease in GFP::UNC-26 fluorescence of individual puncta. The levels of GFP::UNC-26 fluorescence detected in the dorsal nerve cord were drastically reduced in fat-3(wa22) mutants in comparison to WT animals (26.9 ± 2.9%, n = 19 vs. 100 ± 13.4%, n = 16, p = 0.0004; Figure 4D). These very low levels of GFP::UNC-26 fluorescence were partially rescued by providing exogenous AA (from 26.9 ± 2.9%, n = 19, to 57.2 ± 6.2%, n = 20; p < 0.001), suggesting that increased levels of LC-PUFAs increase synaptojanin localization or stabilization at release sites.
It has been shown that LC-PUFAs regulate the transcription of certain genes (Sampath and Ntambi, 2004). Therefore, we investigated whether the depletion of UNC-26-synaptojanin observed in fat-3(wa22) mutants is caused by the down-regulation of unc-26-synaptojanin transcription by performing real-time quantitative RT-PCR (Figure 4E). The ratio between the unc-26-synaptojanin and the β-actin mRNA levels was not significantly different in fat-3(wa22) mutants (1.29 ± 0.42, p = 0.54) or unc-57-endophilin mutants (0.90 ± 0.32, p = 0.76), when compared with WT animals (1.00 ± 0.04). We confirmed these results using the nuclear hormone receptor gene nhr-23 (Van Gilst et al., 2005) to normalize for unc-26-synaptojanin mRNA. We found that the ratio between unc-26-synaptojanin and nhr-23 mRNA levels in fat-3(wa22) mutants (1.09 ± 0.04) was similar to that obtained in WT animals (1.00 ± 0.27). These data suggest that the transcription of the unc-26-synaptojanin gene is normal in fat-3(wa22) mutants, making it unlikely that the synaptojanin depletion at release sites is caused by a decrease in synaptojanin mRNA.
In principle, the degradation of synaptojanin at release sites in fat-3(wa22) mutants could be caused by the mislocalization of synaptojanin-localizing factors in the absence of LC-PUFAs. To begin to test this possibility, we followed the distribution of GFP-tagged forms of two proteins that localize and/or stabilize synaptojanin at sites of neurotransmitter release: UNC-57-endophilin (Harris et al., 2000; Schuske et al., 2003; Verstreken et al., 2003) and amphiphysin (AMPH-1). Amphiphysin has many common features with endophilin, associates and colocalizes with synaptojanin at synapses, and is required in vitro for synaptojanin binding to liposomes composed of total brain lipids (McPherson et al., 1996; Micheva et al., 1997; Di Paolo et al., 2002; Dawson et al., 2006). We could not detect differences between the distribution patterns of these proteins in fat-3(wa22) mutants and in WT animals (Figure 4F), suggesting that the localization and abundance of endophilin and amphiphysin at sites of release are not dependent on LC-PUFAs.
Decreased synaptojanin function is predicted to impair SV uncoating and thereby lead to defective SV recycling. Synaptojanin depletion from fat-3(wa22) mutants release sites could therefore explain the defects in SV recycling observed in LC-PUFA–deficient animals.
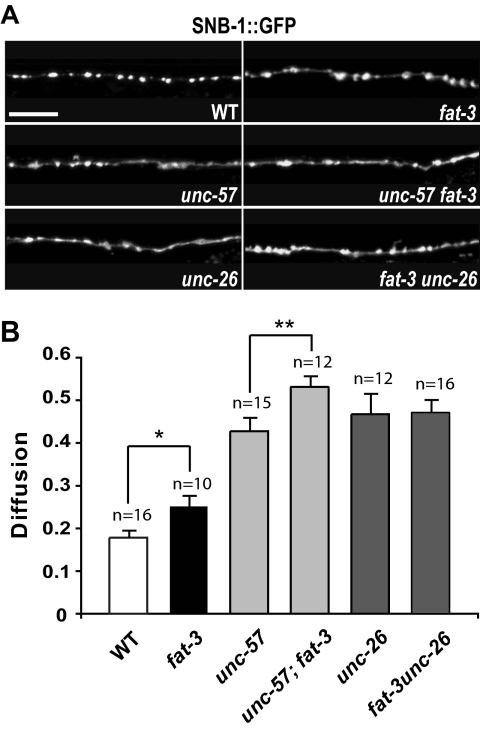
fat-3 Acts in the Same Genetic Pathway as synaptojanin and Independently of endophilin to Retrieve Synaptobrevin from the Plasma Membrane
Because the fat-3(wa22) mutation causes depletion of synaptojanin from release sites, we tested whether fat-3 acts in the same genetic pathway as unc-26-synaptojanin and unc-57-endophilin. We constructed double mutant strains expressing SNB-1::GFP in GABAergic neurons and carrying both the fat-3(wa22) mutation and either the unc-57(ok310)-endophilin or unc-26(s1710)-synaptojanin mutations. We reasoned that if fat-3 acts in the same biological process as unc-57-endophilin or unc-26-synaptojanin, the double mutants should show a mislocalization of SNB-1::GFP at release sites similar to—but not higher than—that of the most severe single mutant. We therefore quantified SNB-1::GFP fluorescence at release sites in the dorsal nerve cord at high resolution (Figure 5 and Supplementary Figure S3). SNB-1::GFP diffusion values obtained for the single mutants fat-3(wa22) (0.243 ± 0.023, n = 10), unc-57(ok310)-endophilin (0.428 ± 0.030, n = 15), and unc-26(s1710)-synaptojanin (0.468 ± 0.046, n = 12) were 1.36-fold (p = 0.027), 2.4-fold (p < 0.0001), and 2.63-fold (p < 0.0001), respectively, greater than those of WT animals (0.178 ± 0.015, n = 16). We found that SNB-1::GFP diffusion was significantly increased in unc-57-endophilin; fat-3(wa22) double mutants (0.531 ± 0.024, n = 12) compared with that of the most severe single mutant, unc-57-endophilin (0.428 ± 0.030, n = 15; p = 0.026), suggesting that fat-3 and endophilin operate in parallel biological processes. We also found that fat-3(wa22)unc-26-synaptojanin double mutants and unc-26-synaptojanin single mutants had almost identical SNB-1::GFP diffusion values (0.471 ± 0.028, n = 16 vs. 0.468 ± 0.046, n = 12; p = 0.94), suggesting that fat-3 and synaptojanin operate in the same genetic pathway to regulate SNB-1::GFP retrieval from the plasma membrane. Our genetic data, along with our UNC-57-endophilin localization data, and together with our finding that inactivation of fat-3 causes synaptojanin mislocalization at release sites, suggest that FAT-3 acts independently of endophilin to influence synaptojanin localization at these sites.
Figure 5.
fat-3 acts in the same genetic pathway as synaptojanin to retrieve synaptobrevin from the plasma membrane. (A) Localization of SNB-1::GFP at sites of release in WT, fat-3, unc-57-endophilin, unc-57-endophilin; fat-3, unc-26-synaptojanin, and fat-3unc-26-synaptojanin animals. SNB-1::GFP exhibits a punctate pattern in WT animals, whereas it is mislocalized in all the other strains. All images were taken in the same 50 μm region of the dorsal nerve cord, 28 μm posterior to the nerve ring motor neuron RMED (schematic in Figure 1A). Scale bar, 10 μm. (B) Quantification of SNB-1::GFP diffusion. Inactivation of fat-3 increases significantly SNB-1::GFP diffusion in unc-57-endophilin; fat-3 animals in comparison to the most severe single mutant, unc-57-endophilin, whereas diffusion values are virtually unchanged in fat-3unc-26-synaptojanin and unc-26-synaptojanin animals. The average fluorescence intensities detected in puncta and in axonal membranes between synapses are reported in Supplementary Figure S3. Data were plotted as mean ± SEM. *p = 0.027; **p = 0.026. Alleles: fat-3(wa22), unc-57(ok310), unc-26(s1710).
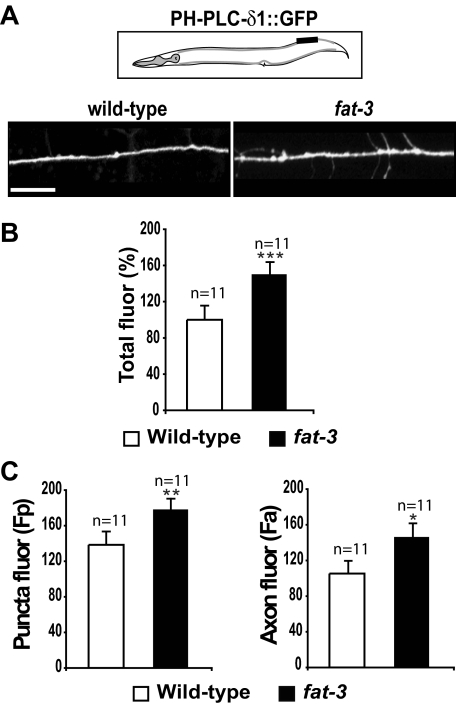
fat-3 Influences the Levels of PtdIns(4,5)P2 at Sites of Release
LC-PUFAs, and particularly AA, are almost invariably esterified at the position sn2 of the phosphoinositide PtdIns(4,5)P2 (Wenk and De Camilli, 2004), which is the main substrate of synaptojanin in the brain. In principle, therefore, the depletion of LC-PUFAs in fat-3 mutants could cause a decrease of PtdIns(4,5)P2 synthesis, resulting in inefficient binding or stabilization of synaptojanin at release sites. To test this possibility, we used a PtdIns(4,5)P2 binding domain. The pleckstrin homology (PH) domain of the phospholipase C ∂1 (PHPLC-∂1) binds selectively to PtdIns(4,5)P2 (Varnai and Balla, 1998;Patton et al., 2005). To follow the levels of PtdIns(4,5)P2 at sites of release, we expressed PHPLC-∂1 tagged with GFP (PHPLC-∂1::GFP) in neurons of WT and fat-3(wa22) mutants (Figure 6). In WT animals, PHPLC-∂1::GFP was distributed in a punctate pattern in the sublateral dorsal nerve cords, as expected for sites of release (Figure 6A). In fat-3(wa22) mutants, we observed a similar distribution of PHPLC-∂1::GFP, but we detected higher levels of fluorescence. We quantified the total levels of PHPLC-∂1::GFP fluorescence in the sublateral nerve cord and found a significant increase in fat-3(wa22) mutants (150.2 ± 13.2%), compared with WT animals (100.0 ± 15.2%, p = 0.021; Figure 6B). We detected a significant increase in PHPLC-∂1::GFP levels both in the puncta [fat-3(wa22) mutants: 178.2 ± 11.8 compared with WT animals: 138.5 ± 14.5, p = 0.028] and in axonal plasma membranes [fat-3(wa22) mutants: 146.4 ± 14.8 compared with WT animals: 105.4 ± 13.8, p = 0.036; Figure 6C], suggesting that in the mutant PtdIns(4,5)P2 levels are higher at sites of release as well as in axonal plasma membranes. The increase in PtdIns(4,5)P2 in fat-3(wa22) mutants was neuronal specific, because the total levels of PtdIns(4,5)P2 measured by HPLC in whole worms were slightly decreased in fat-3(wa22) mutants compared with WT animals (Supplementary Figure S4). These results suggest that synaptojanin mislocalization is not caused by low levels of PtdIns(4,5)P2 at release sites in fat-3 mutants.
Figure 6.
PtdIns(4,5)P2 levels at sites of release are increased in fat-3 mutants. (A) Distribution of the PtdIns(4,5)P2 reporter PHPLC-∂1::GFP in WT animals and fat-3(wa22) mutants. PHPLC-∂1:: GFP exhibits a punctate pattern in the sublateral dorsal nerve cord of WT animals and fat-3(wa22) mutants, as expected for sites of release. In fat-3(wa22) mutants, the level of PHPLC-∂1::GFP fluorescence is higher than in WT animals, suggesting that PtdIns(4,5)P2 levels are increased. Images were taken from the same 50-μm portion of the sublateral nerve cord, which is indicated in the schematic (black box). Anterior, left; dorsal, up. Scale bar, 10 μm. (B) Quantification of PHPLC-∂1::GFP fluorescence in the sublateral dorsal nerve cord. The PHPLC-∂1::GFP fluorescence measured in fat-3(wa22) mutants is significantly higher than in WT animals. ***p = 0.021. (C) Quantification of PHPLC-∂1::GFP fluorescence at sites of release and in the axonal membrane. fat-3(wa22) mutants display significantly higher levels of PHPLC-∂1::GFP fluorescence than WT animals at sites of release (puncta fluor, left graph) as well as in the axonal membrane (axon fluor, right graph). Data are plotted as mean ± SEM. **p = 0.028; *p = 0.036.
DISCUSSION
We had previously shown that fat-3 mutants, which are depleted of LC-PUFAs, have a reduction in the number of SVs at release sites and a corresponding decrease in neurotransmitter release (Lesa et al., 2003). Here, we have investigated the reasons for the SV depletion in fat-3 mutants. We find that, although we cannot detect defects in the biosynthesis and transport of SV components from neuronal cell bodies to presynaptic sites, the mutants show a variety of defects that are consistent with an impairment in SV recycling. First, the SV protein synaptobrevin is mislocalized in a way that suggests that it is not efficiently retrieved from the presynaptic plasma membrane. Second, we find that fat-3 acts in the same biological process as unc-26-synaptojanin, a gene involved in the endocytic retrieval of SV components, implying that the FAT-3 protein is also involved in this process. Third, fat-3 mutants have abnormally low levels of synaptojanin at sites of neurotransmitter release and have increased levels of the main synaptojanin substrate PtdIns(4,5)P2 at these sites, suggesting that LC-PUFAs are required for correct synaptojanin localization and function at synapses. Together, our findings suggest that FAT-3 is required upstream of synaptojanin at release sites to regulate the localization of synaptojanin and allow SV recycling to proceed efficiently.
LC-PUFAs Are Required for Efficient SV Recycling
The synaptobrevin mislocalization at synaptic sites in fat-3(wa22) mutants is similar to that observed in C. elegans and Drosophila endocytic mutants, including mutants in AP180 (Nonet et al., 1999; Bao et al., 2005), endophilin (Schuske et al., 2003), synaptojanin (Harris et al., 2000), and dynamin (Dittman and Kaplan, 2006). In all of these endocytic mutants, synaptobrevin is not retrieved efficiently after SV fusion and diffuses back along the axonal plasma membrane. In principle, synaptobrevin mislocalization could also be caused by an abnormally high number of SVs fusing with the presynaptic membrane, as reported for the tomosyn mutant (Dittman and Kaplan, 2006; McEwen et al., 2006). This cannot be the explanation for synaptobrevin mislocalization in fat-3 mutants, however, because the number of SVs fusing with the presynaptic membrane at release sites is abnormally low in these animals. Indeed, compared with WT animals, fat-3(wa22) mutants display decreased evoked excitatory postsynaptic currents, as well as a lower frequency of miniature postsynaptic currents (Lesa et al., 2003). Therefore, we conclude that synaptobrevin mislocalization in fat-3 mutants reflects deficits in SV recycling. Consistent with this conclusion, we find that fat-3 and unc-26-synaptojanin operate in the same biological process to control synaptobrevin retrieval from the plasma membrane, suggesting that FAT-3 functions in the endocytic part of the SV recycling process. The finding that an exogenous LC-PUFA largely rescues both synaptobrevin and synaptojanin mislocalizations in fat-3 mutants argues that the defect in endocytosis in the mutant is caused by LC-PUFA depletion.
Recent studies have shown that the PUFAs ALA, LIN, AA, and DHA induce SNARE complex formation, thus promoting vesicle docking and/or fusion (Darios and Davletov, 2006). Could inefficient formation of SNARE complexes be the cause of the SV recycling defects in fat-3 mutants? This seems unlikely, as fat-3 mutants are only depleted of LC-PUFAs and contain high levels of other PUFAs including ALA and LIN, which are both able to induce SNARE complex formation (Darios and Davletov, 2006). In addition, exogenous LIN does not rescue the inefficient neurotransmitter release associated with loss of fat-3 function (Lesa et al., 2003). Moreover, it is unlikely that fat-3 mutants are defective in SV docking or fusion, as this should result in an increased number of SVs docked at the presynaptic membrane, and fat-3 mutants are depleted of these vesicles (Lesa et al., 2003).
The SV Recycling Defect Observed in fat-3 Mutants Is Caused by Reduced Synaptojanin Function
Animals depleted of LC-PUFAs have abnormally low levels of synaptojanin at release sites. Because synaptojanin mRNA levels in these animals are similar to those in WT animals, it is unlikely that the depletion of synaptojanin at release sites is caused by a decrease in synaptojanin mRNA. The simplest possibility is that synaptojanin fails to bind efficiently at these sites when LC-PUFAs are depleted. Consistent with this possibility, we find that overexpression of synaptojanin fails to rescue the neuronal phenotypes in fat-3(wa22) mutants (data not shown), suggesting that FAT-3 may act on one or more factors important for synaptojanin localization and/or stabilization at release sites.
Synaptojanin dephosphorylates the phosphoinositide PtdIns (4,5)P2 (McPherson et al., 1996), thereby promoting the uncoating of presynaptic endocytic vesicles (Cremona et al., 1999; Cremona and De Camilli, 2001; Wenk and De Camilli, 2004). C. elegans and Drosophila synaptojanin mutants are defective in SV recycling, as well as in neurotransmitter release; they have a depleted SV pool and the SVs are larger than normal (Harris et al., 2000; Verstreken et al., 2003; Dickman et al., 2005). fat-3 mutants also show all of these defects, consistent with the proposal that FAT-3 and LC-PUFAs promote endocytosis at synapses by regulating synaptojanin localization or stabilization at sites of release. In addition, our epistasis analysis indicates that FAT-3 acts upstream of and in the same genetic pathway as synaptojanin to retrieve SNB-1::GFP from the presynaptic plasma membrane. Because fat-3 mutants retrieve SNB-1::GFP from these sites only poorly and inefficient SNB-1::GFP retrieval directly correlates with defects in endocytosis, our findings suggest that the endocytosis defects observed in fat-3 mutants are caused by a reduction of synaptojanin function at release sites. The local increase in PtdIns(4,5)P2 that we demonstrate at release sites in fat-3 mutants is presumably a consequence of the decreased synaptojanin function at these sites (Cremona et al., 1999; Stefan et al., 2002).
How Does a Lack of LC-PUFAs Affect Synaptojanin Localization?
We observe a severe depletion of synaptojanin at synapses in LC-PUFA-depleted animals. By contrast, two other proteins that are concentrated at release sites and bind synaptojanin—endophilin and amphiphysin (Micheva et al., 1997; Di Paolo et al., 2002; Schuske et al., 2003; Verstreken et al., 2003)—do not appear to be mislocalized in fat-3 mutants, suggesting that these proteins do not require LC-PUFAs to bind to release sites and the decreased recruitment of synaptojanin at release sites is not caused by a decrease in either endophilin or amphiphysin. Indeed, our genetic and GFP:: UNC-26 localization findings show that FAT-3 influences synaptojanin recruitment at release sites independently of endophilin.
It is unclear why LC-PUFAs are required for the normal recruitment of synaptojanin at release sites. Intuitively, it seems unlikely that the protein binds directly to LC-PUFAs, as their fatty acid chains are buried in the interior of the lipid bilayer. On the other hand, synaptojanin must bind PtdIns(4,5)P2 to dephosphorylate it (Tsujishita et al., 2001) and experiments in vitro have demonstrated that the catalytic domain of synaptojanin has a substrate preference for membranes enriched in a natural PtdIns(4,5)P2 carrying an LC-PUFA compared with membranes containing a synthetic PtdIns(4,5)P2 with two saturated fatty acids (Schmid et al., 2004). fat-3 mutants are devoid of LC-PUFAs, and therefore it is likely that most of their PtdIns(4,5)P2 does not contain LC-PUFAs. It seems therefore possible that the substrate preference of synaptojanin for the PtdIns(4,5)P2 present in fat-3 membranes decreases, resulting in synaptojanin being unable to localize efficiently to these membranes and effectively resulting in decreased synaptojanin function.
Supplementary Material
ACKNOWLEDGMENTS
We thank Michael Nonet, Aixa Alfonso, and Vincent O'Connor for antibodies; Erik Jorgensen and Mei Zhen for strains; Andy Fire for plasmids; Andrew Vaughan, Suzumi Tokuoka, and Remi Fronzes for technical help and advice; and Martin Raff for discussions. We also thank Yukiko Goda, Neil Hopper, Giovanna Lalli, Stephen Nurrish, Martin Raff, and Giampietro Schiavo for critically reading the manuscript. Some nematode strains were supplied by the Caenorhabditis Genetics Center, which is supported by the National Institutes of Health (NIH) and the University of Minnesota. This work was supported by the Royal Society (G.M.L.), by the Medical Research Council (G.M.L. and E.M.) and by the NIH Center for Research Resources Grant RR12596 (D.H.H.).
Abbreviations used:
- AA
arachidonic acid
- ALA
α-linolenic acid
- DHA
docosahexaenoic acid
- GABA
γ-aminobutyric acid
- LIN
linoleic acid
- LC
long chain
- PtdIns
phosphatidylinositol
- PUFA
polyunsaturated fatty acid
- SV
synaptic vesicle
- WT
wild type.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-07-0719) on December 19, 2007.
REFERENCES
- Bao H., Daniels R. W., MacLeod G. T., Charlton M. P., Atwood H. L., Zhang B. AP180 maintains the distribution of synaptic and vesicle proteins in the nerve terminal and indirectly regulates the efficacy of Ca2+-triggered exocytosis. J. Neurophysiol. 2005;94:1888–1903. doi: 10.1152/jn.00080.2005. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chyb S., Raghu P., Hardie R. C. Polyunsaturated fatty acids activate the Drosophila light-sensitive channels TRP and TRPL. Nature. 1999;397:255–259. doi: 10.1038/16703. [DOI] [PubMed] [Google Scholar]
- Cotman C., Blank M. L., Moehl A., Snyder F. Lipid composition of synaptic plasma membranes isolated from rat brain by zonal centrifugation. Biochemistry. 1969;8:4606–4612. doi: 10.1021/bi00839a056. [DOI] [PubMed] [Google Scholar]
- Cremona O., De Camilli P. Phosphoinositides in membrane traffic at the synapse. J. Cell Sci. 2001;114:1041–1052. doi: 10.1242/jcs.114.6.1041. [DOI] [PubMed] [Google Scholar]
- Cremona O., et al. Essential role of phosphoinositide metabolism in synaptic vesicle recycling. Cell. 1999;99:179–188. doi: 10.1016/s0092-8674(00)81649-9. [DOI] [PubMed] [Google Scholar]
- Darios F., Davletov B. Omega-3 and omega-6 fatty acids stimulate cell membrane expansion by acting on syntaxin 3. Nature. 2006;440:813–817. doi: 10.1038/nature04598. [DOI] [PubMed] [Google Scholar]
- Dawson J. C., Legg J. A., Machesky L. M. Bar domain proteins: a role in tubulation, scission and actin assembly in clathrin-mediated endocytosis. Trends Cell Biol. 2006;16:493–498. doi: 10.1016/j.tcb.2006.08.004. [DOI] [PubMed] [Google Scholar]
- Deutsch J. W., Kelly R. B. Lipids of synaptic vesicles: relevance to the mechanism of membrane fusion. Biochemistry. 1981;20:378–385. doi: 10.1021/bi00505a024. [DOI] [PubMed] [Google Scholar]
- Di Paolo G., et al. Decreased synaptic vesicle recycling efficiency and cognitive deficits in amphiphysin 1 knockout mice. Neuron. 2002;33:789–804. doi: 10.1016/s0896-6273(02)00601-3. [DOI] [PubMed] [Google Scholar]
- Dickman D. K., Horne J. A., Meinertzhagen I. A., Schwarz T. L. A slowed classical pathway rather than kiss-and-run mediates endocytosis at synapses lacking synaptojanin and endophilin. Cell. 2005;123:521–533. doi: 10.1016/j.cell.2005.09.026. [DOI] [PubMed] [Google Scholar]
- Dittman J. S., Kaplan J. M. Factors regulating the abundance and localization of synaptobrevin in the plasma membrane. Proc. Natl. Acad. Sci. USA. 2006;103:11399–11404. doi: 10.1073/pnas.0600784103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fergestad T., Davis W. S., Broadie K. The stoned proteins regulate synaptic vesicle recycling in the presynaptic terminal. J. Neurosci. 1999;19:5847–5860. doi: 10.1523/JNEUROSCI.19-14-05847.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallop J. L., Jao C. C., Kent H. M., Butler P. J., Evans P. R., Langen R., McMahon H. T. Mechanism of endophilin N-BAR domain-mediated membrane curvature. EMBO J. 2006;25:2898–2910. doi: 10.1038/sj.emboj.7601174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall D. H. Electron microscopy and three-dimensional image reconstruction. Methods Cell Biol. 1995;48:395–436. doi: 10.1016/s0091-679x(08)61397-7. [DOI] [PubMed] [Google Scholar]
- Hall D. H., Hedgecock E. M. Kinesin-related gene unc-104 is required for axonal transport of synaptic vesicles in C. elegans. Cell. 1991;65:837–847. doi: 10.1016/0092-8674(91)90391-b. [DOI] [PubMed] [Google Scholar]
- Hammond K., Reboiras M. D., Lyle I. G., Jones M. N. Characterisation of phosphatidylcholine/phosphatidylinositol sonicated vesicles. Effects of phospholipid composition on vesicle size. Biochim. Biophys. Acta. 1984;774:19–25. doi: 10.1016/0005-2736(84)90269-4. [DOI] [PubMed] [Google Scholar]
- Hannah M. J., Schmidt A. A., Huttner W. B. Synaptic vesicle biogenesis. Annu. Rev. Cell Dev. Biol. 1999;15:733–798. doi: 10.1146/annurev.cellbio.15.1.733. [DOI] [PubMed] [Google Scholar]
- Harris T. W., Hartwieg E., Horvitz H. R., Jorgensen E. M. Mutations in synaptojanin disrupt synaptic vesicle recycling. J. Cell Biol. 2000;150:589–600. doi: 10.1083/jcb.150.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen E. M., Hartwieg E., Schuske K., Nonet M. L., Jin Y., Horvitz H. R. Defective recycling of synaptic vesicles in synaptotagmin mutants of Caenorhabditis elegans. Nature. 1995;378:196–199. doi: 10.1038/378196a0. [DOI] [PubMed] [Google Scholar]
- Kahn-Kirby A. H., Dantzker J. L., Apicella A. J., Schafer W. R., Browse J., Bargmann C. I., Watts J. L. Specific polyunsaturated fatty acids drive TRPV-dependent sensory signaling in vivo. Cell. 2004;119:889–900. doi: 10.1016/j.cell.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Lauritzen L., Hansen H. S., Jorgensen M. H., Michaelsen K. F. The essentiality of long chain n-3 fatty acids in relation to development and function of the brain and retina. Prog. Lipid Res. 2001;40:1–94. doi: 10.1016/s0163-7827(00)00017-5. [DOI] [PubMed] [Google Scholar]
- Lesa G. M., Palfreyman M., Hall D. H., Clandinin M. T., Rudolph C., Jorgensen E. M., Schiavo G. Long chain polyunsaturated fatty acids are required for efficient neurotransmission in C. elegans. J. Cell Sci. 2003;116:4965–4975. doi: 10.1242/jcs.00918. [DOI] [PubMed] [Google Scholar]
- Masuda M., Takeda S., Sone M., Ohki T., Mori H., Kamioka Y., Mochizuki N. Endophilin BAR domain drives membrane curvature by two newly identified structure-based mechanisms. EMBO J. 2006;25:2889–2897. doi: 10.1038/sj.emboj.7601176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald K. High-pressure freezing for preservation of high resolution fine structure and antigenicity for immunolabeling. Methods Mol. Biol. 1999;117:77–97. doi: 10.1385/1-59259-201-5:77. [DOI] [PubMed] [Google Scholar]
- McEwen J. M., Madison J. M., Dybbs M., Kaplan J. M. Antagonistic regulation of synaptic vesicle priming by Tomosyn and UNC-13. Neuron. 2006;51:303–315. doi: 10.1016/j.neuron.2006.06.025. [DOI] [PubMed] [Google Scholar]
- McMahon H. T., Gallop J. L. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- McMullan R., Hiley E., Morrison P., Nurrish S. J. Rho is a presynaptic activator of neurotransmitter release at pre-existing synapses in C. elegans. Genes Dev. 2006;20:65–76. doi: 10.1101/gad.359706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson P. S., Garcia E. P., Slepnev V. I., David C., Zhang X., Grabs D., Sossin W. S., Bauerfeind R., Nemoto Y., De Camilli P. A presynaptic inositol-5-phosphatase. Nature. 1996;379:353–357. doi: 10.1038/379353a0. [DOI] [PubMed] [Google Scholar]
- Mello C., Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- Meloni I., et al. FACL4, encoding fatty acid-CoA ligase 4, is mutated in nonspecific X-linked mental retardation. Nat. Genet. 2002;30:436–440. doi: 10.1038/ng857. [DOI] [PubMed] [Google Scholar]
- Micheva K. D., Kay B. K., McPherson P. S. Synaptojanin forms two separate complexes in the nerve terminal. Interactions with endophilin and amphiphysin. J. Biol. Chem. 1997;272:27239–27245. doi: 10.1074/jbc.272.43.27239. [DOI] [PubMed] [Google Scholar]
- Nonet M. L., Holgado A. M., Brewer F., Serpe C. J., Norbeck B. A., Holleran J., Wei L., Hartwieg E., Jorgensen E. M., Alfonso A. UNC-11, a Caenorhabditis elegans AP180 homologue, regulates the size and protein composition of synaptic vesicles. Mol. Biol. Cell. 1999;10:2343–2360. doi: 10.1091/mbc.10.7.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton A., Knuth S., Schaheen B., Dang H., Greenwald I., Fares H. Endocytosis function of a ligand-gated ion channel homolog in Caenorhabditis elegans. Curr. Biol. 2005;15:1045–1050. doi: 10.1016/j.cub.2005.04.057. [DOI] [PubMed] [Google Scholar]
- Rohrbough J., Broadie K. Lipid regulation of the synaptic vesicle cycle. Nat. Rev. Neurosci. 2005;6:139–150. doi: 10.1038/nrn1608. [DOI] [PubMed] [Google Scholar]
- Sampath H., Ntambi J. M. Polyunsaturated fatty acid regulation of gene expression. Nutr. Rev. 2004;62:333–339. doi: 10.1111/j.1753-4887.2004.tb00058.x. [DOI] [PubMed] [Google Scholar]
- Schmid A. C., Wise H. M., Mitchell C. A., Nussbaum R., Woscholski R. Type II phosphoinositide 5-phosphatases have unique sensitivities towards fatty acid composition and head group phosphorylation. FEBS Lett. 2004;576:9–13. doi: 10.1016/j.febslet.2004.08.052. [DOI] [PubMed] [Google Scholar]
- Schuske K. R., Richmond J. E., Matthies D. S., Davis W. S., Runz S., Rube D. A., van der Bliek A. M., Jorgensen E. M. Endophilin is required for synaptic vesicle endocytosis by localizing synaptojanin. Neuron. 2003;40:749–762. doi: 10.1016/s0896-6273(03)00667-6. [DOI] [PubMed] [Google Scholar]
- Stefan C. J., Audhya A., Emr S. D. The yeast synaptojanin-like proteins control the cellular distribution of phosphatidylinositol (4,5)-bisphosphate. Mol. Biol. Cell. 2002;13:542–557. doi: 10.1091/mbc.01-10-0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhof T. C. The synaptic vesicle cycle. Annu. Rev. Neurosci. 2004;27:509–547. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- Takamori S., et al. Molecular anatomy of a trafficking organelle. Cell. 2006;127:831–846. doi: 10.1016/j.cell.2006.10.030. [DOI] [PubMed] [Google Scholar]
- Trimble W. S., Cowan D. M., Scheller R. H. VAMP- 1, a synaptic vesicle-associated integral membrane protein. Proc. Natl. Acad. Sci. USA. 1988;85:4538–4542. doi: 10.1073/pnas.85.12.4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujishita Y., Guo S., Stolz L. E., York J. D., Hurley J. H. Specificity determinants in phosphoinositide dephosphorylation: crystal structure of an archetypal inositol polyphosphate 5-phosphatase. Cell. 2001;105:379–389. doi: 10.1016/s0092-8674(01)00326-9. [DOI] [PubMed] [Google Scholar]
- Van Gilst M. R., Hadjivassiliou H., Yamamoto K. R. A Caenorhabditis elegans nutrient response system partially dependent on nuclear receptor NHR-49. Proc. Natl. Acad. Sci. USA. 2005;102:13496–13501. doi: 10.1073/pnas.0506234102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnai P., Balla T. Visualization of phosphoinositides that bind pleckstrin homology domains: calcium- and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J. Cell Biol. 1998;143:501–510. doi: 10.1083/jcb.143.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstreken P., Koh T. W., Schulze K. L., Zhai R. G., Hiesinger P. R., Zhou Y., Mehta S. Q., Cao Y., Roos J., Bellen H. J. Synaptojanin is recruited by endophilin to promote synaptic vesicle uncoating. Neuron. 2003;40:733–748. doi: 10.1016/s0896-6273(03)00644-5. [DOI] [PubMed] [Google Scholar]
- Vreugdenhil M., Bruehl C., Voskuyl R. A., Kang J. X., Leaf A., Wadman W. J. Polyunsaturated fatty acids modulate sodium and calcium currents in CA1 neurons. Proc. Natl. Acad Sci. USA. 1996;93:12559–12563. doi: 10.1073/pnas.93.22.12559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts J. L., Browse J. Genetic dissection of polyunsaturated fatty acid synthesis in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2002;99:5854–5859. doi: 10.1073/pnas.092064799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts J. L., Phillips E., Griffing K. R., Browse J. Deficiencies in C20 polyunsaturated fatty acids cause behavioral and developmental defects in Caenorhabditis elegans fat-3 mutants. Genetics. 2003;163:581–589. doi: 10.1093/genetics/163.2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenk M. R., De Camilli P. Protein-lipid interactions and phosphoinositide metabolism in membrane traffic: insights from vesicle recycling in nerve terminals. Proc. Natl. Acad. Sci. USA. 2004;101:8262–8269. doi: 10.1073/pnas.0401874101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Koh Y. H., Beckstead R. B., Budnik V., Ganetzky B., Bellen H. J. Synaptic vesicle size and number are regulated by a clathrin adaptor protein required for endocytosis. Neuron. 1998;21:1465–1475. doi: 10.1016/s0896-6273(00)80664-9. [DOI] [PubMed] [Google Scholar]
- Zhang K., et al. A 5-bp deletion in ELOVL4 is associated with two related forms of autosomal dominant macular dystrophy. Nat. Genet. 2001;27:89–93. doi: 10.1038/83817. [DOI] [PubMed] [Google Scholar]
- Zhen M., Jin Y. The liprin protein SYD-2 regulates the differentiation of presynaptic termini in C. elegans. Nature. 1999;401:371–375. doi: 10.1038/43886. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.