Abstract
McKusick–Kaufman syndrome (MKKS) is a recessively inherited human genetic disease characterized by several developmental anomalies. Mutations in the MKKS gene also cause Bardet–Biedl syndrome (BBS), a genetically heterogeneous disorder with pleiotropic symptoms. However, little is known about how MKKS mutations lead to disease. Here, we show that disease-causing mutants of MKKS are rapidly degraded via the ubiquitin–proteasome pathway in a manner dependent on HSC70 interacting protein (CHIP), a chaperone-dependent ubiquitin ligase. Although wild-type MKKS quickly shuttles between the centrosome and cytosol in living cells, the rapidly degraded mutants often fail to localize to the centrosome. Inhibition of proteasome functions causes MKKS mutants to form insoluble structures at the centrosome. CHIP and partner chaperones, including heat-shock protein (HSP)70/heat-shock cognate 70 and HSP90, strongly recognize MKKS mutants. Modest knockdown of CHIP by RNA interference moderately inhibited the degradation of MKKS mutants. These results indicate that the MKKS mutants have an abnormal conformation and that chaperone-dependent degradation mediated by CHIP is a key feature of MKKS/BBS diseases.
INTRODUCTION
McKusick–Kaufman syndrome (MKKS) is a recessively inherited human genetic disease predominantly characterized by developmental anomalies, including vaginal atresia with hydrometrocolpos, polydactyly, and congenital heart defects (McKusick et al., 1964). The MKKS gene encodes a 570-amino acid polypeptide with weak but significant similarity to group II chaperonins (Stone et al., 2000). Mutations in the MKKS gene also cause Bardet–Biedl syndrome (BBS), a genetically heterogeneous disorder characterized by obesity, retinal dystrophy, polydactyly, mental retardation, renal malformation, and hypogenitalism (Beales et al., 1999), and thus MKKS is also called BBS6. The MKKS mRNA is widely expressed in various tissues, including those affected by MKKS and BBS diseases (Slavotinek et al., 2000; Stone et al., 2000). Although more than 10 mutations in the MKKS gene have been identified in patients (Beales et al., 2001; Slavotinek et al., 2002), little is known about how they cause MKKS and BBS.
Twelve of the genes that are responsible for BBS (BBS1-12) have been identified so far, and the products of several of these genes have been suggested to be involved in intraflagellar transport in cilia and intracellular trafficking in the cytosol (Mykytyn and Sheffield, 2004; Badano et al., 2005; Beales, 2005; Blacque and Leroux, 2006). Cargo-carrying motors play crucial roles in intraflagellar and intracellular transport systems by bidirectionally moving on microtubules, and BBS4 interacts with the dynactin complex, which mediates interactions between cargoes and the dynein motor (Kim et al., 2004). Retrograde transport of melanosomes in zebrafish is slowed by knockdown of BBS2 or BBS4-8 (Yen et al., 2006), whereas mutations in Caenorhabditis elegans BBS7 and BBS8 cause delays in intraflagellar transport driven by kinesin motors (Ou et al., 2005). The Chlamydomonas homologue of BBS5 is essential for flagellum formation (Li et al., 2004), and BBS3 shows significant similarities to the ADP ribosylation factor family of small guanosine triphosphate-binding proteins, including those involved in vesicular trafficking (Chiang et al., 2004). BBS1, BBS2, and BBS4-8 are concentrated in centrosome/basal bodies, which act as microtubule-organizing centers (MTOCs) (Ansley et al., 2003; Blacque et al., 2004; Kim et al., 2004, 2005; Li et al., 2004; Badano et al., 2006). Whereas BBS4 and BBS8 are tetratricopeptide repeat (TPR) domain proteins (Ansley et al., 2003), BBS1, BBS2, and BBS7 are putative β-propeller proteins (Badano et al., 2003). These distinctive domains of these proteins have been suggested to be involved in protein interactions, although BBS5 (Li et al., 2004) and BBS9 (Nishimura et al., 2005) have no obvious motifs. Thus, BBS genes are suggested to be involved in motor-driven intraflagellar and intracellular transport through their ability to mediate protein interactions. A large complex (BBSome) containing BBS1, BBS2, BBS4, BBS5, and BBS7-9 associates Rab8, a GTPase required for ciliary membrane growth, suggesting that this complex is involved in vesicular trafficking to the cilium (Nachury et al., 2007). However, the molecular mechanisms of BBS protein-assisted trafficking are still largely unknown.
Amino acid sequence similarity between MKKS protein and group II chaperonins suggests that MKKS function may have evolved from the chaperonin family (Stone et al., 2000). In addition to MKKS, two other BBS proteins, BBS10 (Stoetzel et al., 2006) and BBS12 (Stoetzel et al., 2007), were recently shown to share weak but significant sequence similarities with group II chaperonins. Interestingly, none of the three chapeonin-like BBS proteins were found in the BBSome (Nachury et al., 2007), suggesting that these chaperonin like proteins have a distinctive role in BBS-associated functions. Taken together with a recent report indicating that BBS11 encodes an E3 ubiquitin ligase (Chiang et al., 2006), these observations suggest that the functions of BBS family proteins may be related to protein quality control systems and protein transport.
Molecular chaperones play essential roles in assisting protein folding and preventing aggregation of misfolded proteins. HSP70 and chaperonin (HSP60) family proteins, for example, assist in protein folding through distinctive mechanisms (Nollen and Morimoto, 2002; Young et al., 2004; Horwich et al., 2007). Whereas HSP70 proteins interact with the exposed hydrophobic surfaces of unfolded and partially folded substrate proteins in concert with J-domain proteins, chaperonins sequester nonnative state proteins in cage-like structures. A group II chaperonin, chaperonin containing t-complex polypeptide 1 (CCT, also called TRiC), has been identified in the eukaryotic cytosol (Kubota, 2002; Spiess et al., 2004). CCT is a hexadecameric complex composed of eight different subunits (Kubota et al., 1994; Llorca et al., 1999) and facilitates protein folding (Tian et al., 1995; Farr et al., 1997; Meyer et al., 2003; Kubota et al., 2006). Recently, CCT was shown to inhibit protein aggregation and counteract the cytotoxicity of misfolded proteins (Behrends et al., 2006; Kitamura et al., 2006; Kubota et al., 2006; Tam et al., 2006). These observations indicate that molecular chaperones are indispensable for protecting cells against the pernicious effects of misfolded proteins.
Irreversibly misfolded proteins are toxic to the cell because they trap normal proteins in aggregates and they inhibit the proteasome-dependent protein degradation pathway (Holmberg et al., 2004; Schaffar et al., 2004; Bennett et al., 2005). Mutations that alter amino acid residues often cause protein misfolding, which leads to various diseases, including neurodegenerative disease (Barral et al., 2004). Thus, the ubiquitin-proteasome system plays a crucial role in protecting cells by degrading misfolded proteins (Ciechanover and Brundin, 2003). The ubiquitin ligating enzyme E3 is a key factor in the specific recognition of target proteins (Pickart, 2004), and the carboxy terminus of heat-shock cognate (HSC)70 interacting protein (CHIP) is an E3 enzyme that polyubiquitinates misfolded proteins with the assistance of the molecular chaperones HSC70/HSP70 or HSP90 (Connell et al., 2001; Meacham et al., 2001; Murata et al., 2001). CHIP interacts with HSC70 and HSP90 through its amino-terminal TPR domain, and the ubiquitin ligating reaction is performed by a U-box motif in its carboxy terminus. In association with these chaperones, CHIP plays an essential role in the quality control of misfolded proteins (Tateishi et al., 2004; Younger et al., 2004, 2006; Qian et al., 2006).
Here, we report that many of the MKKS mutants that are responsible for the MKKS/BBS diseases are degraded in the cell much more rapidly than the wild-type protein and that a CHIP-dependent ubiqutin-proteasome pathway plays a crucial role in this rapid degradation. Although the wild-type MKKS rapidly shuttles between the centrosome and cytosol, rapidly degraded MKKS mutants often fail to localize to the centrosome. Despite that MKKS mutants tend to from insoluble structure at the centrosome, CHIP effectively degrades these mutants through preferential recognition of these mutants by molecular chaperones. We discuss how the MKKS mutations cause the diseases MKKS and BBS with particular emphasis on the role played by CHIP-dependent degradation.
MATERIALS AND METHODS
MKKS and Mutant cDNAs
Human and mouse cDNAs encoding MKKS were amplified from cDNA libraries (BD Biosciences Clontech, Palo Alto, CA) by polymerase chain reaction (PCR). Human MKKS cDNAs carrying disease-causing mutations were produced by site-directed mutagenesis (Sawano and Miyawaki, 2000).
Expression of MKKS in Cultured Cells
Human MKKS cDNA or FLAG-tagged MKKS was subcloned into the mammalian expression vector pCAGGS (Niwa et al., 1991). Human embryonic kidney (HEK)293 cells (3.5-mm dish) were transfected with 0.3 μg of the expression vector by using 3 μl of FuGENE 6 (Roche Diagnostics, Indianapolis, IN), unless otherwise indicated. Transfected cells were cultured for 24 h before analysis. Stable cell lines expressing MKKS were produced by G-418 (Geneticin; Invitrogen, Calrsbad, CA) selection of HEK293 cells transfected with pCAGGS-based expression vectors carrying the G-418 resistant gene.
Antibodies and Western Blot Analysis
Antibody against the carboxy terminus of human MKKS (MKC-1) was prepared by immunizing rabbits with the synthetic peptide CLDLSYVIEDKN. Rabbit antibody against full-length MKKS (MKF-1) was produced with recombinant His-tagged MKKS expressed in Escherichia coli. Rabbit polyclonal antibodies against FLAG tag (Sigma-Aldrich, St. Louis, MO), hemagglutinin (HA) tag (Y-11; Santa Cruz Biotechnology, Santa Cruz, CA), ubiquitin (Dako Denmark A/S, Glostrup, Denmark) and CHIP (PC711; Calbiochem, San Diego, CA), mouse monoclonal antibodies to FLAG tag (M2; Sigma-Aldrich), Myc tag (9E10; Santa Cruz Biotechnology), HSP70 (SPA810; Nventa Biopharmaceuticals, San Diego, CA), HSP90 (SPA835; Nventa Biopharmaceuticals), vimentin (V9; Sigma-Aldrich), and glyceraldehyde-3-phosphate dehydrogenase (6C5; HyTest, Turku, Finland), and rat monoclonal antibody to HSC70 (SPA815; Nventa Biopharmaceuticals) were purchased from the indicated sources. Western blotting was performed with alkaline phosphatase-conjugated anti-immunoglobulin (Ig)G, as described previously (Yokota et al., 1999), except that the HA tag was detected with horseradish peroxidase-conjugated anti-IgG and the ECL system (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom).
Fluorescence Recovery after Photobleaching (FRAP) Analysis
The green fluorescent protein (GFP)-MKKS expression vector was constructed using pEGFP-C1 (Clontech, Mountain View, CA). HEK293 cells (3.5-cm dish) were transfected with 0.6 μg of expression vector by using 10 μl of Effecten reagent (QIAGEN, Dusseldorf, Germany). Centrosomes (20 pixels) were bleached for 10 ms with a 405-nm diode (6-mW) laser set at 50% acoustical optical transmission filter (AOTF) transmission by using an FV1000 confocal microscope with a UPlanSApo 40×/0.9 numerical aperture (NA) objective (Olympus, Tokyo, Japan) at 37°C. For the recovery measurement, GFP was excited with a 488-nm Ar (30-mW) laser at 1% AOTF transmission. Signals were collected through a 505-nm long-path filter (64 ms/frame).
Fluorescence Loss in Photobleaching (FLIP) Analysis
After images were collected for five frames (3.93 s) by using a 488-nm Ar laser at 1% power, cytoplasmic region (10.2 μm2) of a GFP-MKKS–expressing cell was bleached for 10 iterations (6.94 s) by using 488-, 543-, and 633-nm laser sources set at 100% power by using an LSM 510 META confocal microscope with a C-Apochromat 40×/1.2 NA Corr. water immersion objective (Carl Zeiss, Jena, Germany) at 37°C. The scanning and bleaching cycles were repeated for 80 s. Fluorescence signals were collected using an HFT405/488/543/633 excitation dichroic mirror and an LP505 emission barrier filter. The pinhole size was corresponded to 3.99 Airy unit (284 μm in diameter). One frame was scanned at 512 × 512 pixels, and the zoom factor was 5×. Collected images were analyzed by AIM software release 4.2 (Carl Zeiss).
Photoconversion Analysis
Cells were transfected with a Dendra-MKKS expression vector constructed with the vector pDendra-2 (Evrogen, Moscow, Russia) encoding the green-to-red photoactivatable fluorescent protein of the octocoral Dendronephthya (Gurskaya et al., 2006). Preconversion green signals of Dendra were collected by excitation with a 488-nm Ar laser set at 1% power, and postconversion red signals were collected by excitation with a 543-nm He-Ne laser at 50% power by using a FV1000 laser scanning microscope with a UApo 40×/1.15 NA 340 water immersion objective (Olympus) at 37°C in a system controlled by Fluoview software version 1.6 (Olympus). Fluorescence signals were collected using a DM405/488/543 excitation dichroic mirror and an SDM560 emission dichroic mirror. Green signals were recorded through a BA505-525 band pass filter, and red signals were recorded through a BA560IF band pass filter. The pinhole size was 800 μm in diameter. Each frame of images was scanned at 256 × 256 pixels in 0.428 s, and the zoom factor was 15×. Before photoconversion, the position of centrosomes was determined by referring to the green fluorescence images. Dendra-MKKS in a centrosome-containing circular area (4.5 μm2) was photoconverted with a 405-nm diode laser set at 20% power for 1 ms through an ADM450 dichroic mirror, and images were continuously collected for 210 frames after conversion. Collected images were analyzed with ImageJ, version 1.37 (National Institute of Health, Bethesda, MD; http://rsb.info.nih.gov/ij/).
Immunofluorescence Microscopy
Cells were fixed in methanol, methanol/acetone (1:1) or 4% parformaldehyde on ice for 15 min and permeabilized with 0.2% Triton-X 100 in phosphate-buffered saline (PBS) at room temperature for 5 min. After washing and blocking, cells were incubated with primary antibodies and AlexaFluor 488- or 594-conjugated anti-IgG (Invitrogen). For double staining of FLAG-MKKS and HSC70, quantification of merged area in the centrosomal region was performed by the ImageJ program by using fluorescent intensities in the cytosol as background levels.
Metabolic Labeling and Immunoprecipitation
HEK293 cells expressing FLAG-MKKS were cultured in methionine (Met)- and cysteine (Cys)-free DMEM containing 10% dialyzed fetal bovine serum and 4.1 MBq/ml [35S]Met/[35S]Cys mixture and chased with excess unlabeled Met and Cys. Cells were lysed in lysis buffer (40 mM HEPES-KOH, pH 7.5, 100 mM NaCl, 5 mM EDTA, and 1% NP-40), and incubated with anti-FLAG M2 Affinity Gel (Sigma-Aldrich). After washing the gel with lysis buffer, proteins bound to the gel were eluted and separated by SDS-polyacrylamide gel electrophoresis (PAGE). Untagged MKKS was immunoprecipitated with the MKC-1 antibody and protein-G Sepharose.
Filter Trap Assay
Cells were lysed in PBS containing 0.5% Triton X-100, and the lysate was diluted in 1% SDS/PBS and boiled for 5 min. Immediately after cooling, samples were loaded onto a cellulose acetate membrane (0.2 μm) placed on a dot blotter (Bio-Rad, Hercules, CA). After blocking with 1% skim milk and 0.05% Tween 20 in PBS overnight, the membrane was incubated with the mouse monoclonal anti-FLAG antibody and alkaline-phosphatase conjugated anti-mouse IgG. Alkaline-phosphatase activity was detected by developing the membrane in nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate solution (Sigma-Aldrich).
Analysis of MKKS Polyubiquitination
Cells were cotransfected with HA-ubiquitin (Sato et al., 2006) and FLAG-MKKS expression vectors. Cell lysates were immunoprecipitated using the mouse anti-FLAG antibody, and polyubiquitination was analyzed by Western blotting by using the rabbit anti-HA antibody.
Overexpression and Knockdown of CHIP
For overexpression, cells were transfected with a vector expressing myc-tagged CHIP (Meacham et al., 2001). RNA interference (RNAi)-mediated knockdown of CHIP was performed as follows. Cells (3.5-cm dish) were transfected with 5 μl of 20 μM CHIP siRNA (Invitrogen oligo ID HSS145538) or nonspecific siRNA (12935-400; Invitrogen) by using 10 μl of Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions and cultured for 48 h. For cotransfection, cells were transfected with siRNA 6 h after transfection with the expression vectors.
Statistical Analysis
Significance of difference was analyzed by the Student's t test.
RESULTS
Rapid Degradation and Increased Insolubility of MKKS Mutants
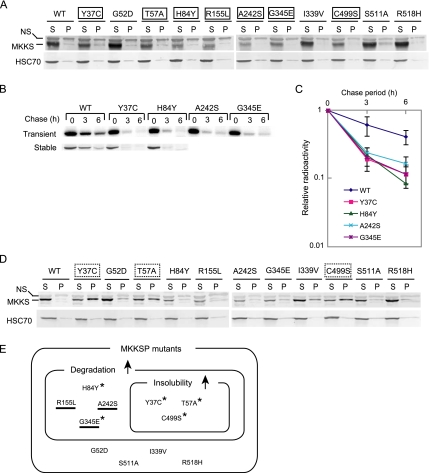
Many MKKS mutations are known to cause the diseases MKKS, BBS, or both. To examine whether mutant MKKS proteins are normally expressed in cultured cells, vectors expressing 11 typical disease-causing MKKS mutants fused to an amino-terminal FLAG tag were constructed. These vectors, or a control vector expressing FLAG-tagged wild-type MKKS, were transiently transfected into HEK293 cells. Soluble and insoluble fractions prepared by centrifugation of the cell lysates were analyzed by Western blotting. Under transfection conditions (0.3 μg of expression vector) that resulted in the recovery of FLAG-MKKS proteins in the soluble but not in the pellet fraction, seven MKKS mutants (Y37C, T57A, H84Y, R155L, A242S, G345E, and C499S) were expressed at clearly decreased levels relative to the wild-type protein (Figure 1A). As the reduced expression levels suggested that these mutants had undergone rapid degradation, four of them (Y37C, H84Y, A242S, and G345E) were examined for stability. Pulse-chase experiments followed by immunoprecipitation indicated that all four mutants underwent accelerated degradation relative to the wild-type protein (Figure 1B, top) and that the half-lives of these mutants were less than half of that of wild-type MKKS (Figure 1C). A similar enhancement of degradation was observed for the stable Y37C and H84Y transformants (Figure 1B, bottom). These results indicated that many MKKS mutants are rapidly degraded, suggesting that they are structurally unstable.
Figure 1.
Many disease-causing mutants of MKKS are rapidly degraded and prone to aggregation. (A) HEK293 cells were transiently transfected with 0.3 μg of FLAG-tagged wild-type or mutant MKKS expression vectors. Cell lysates were fractionated by centrifugation (15,000 × g; 15 min), and the resulting supernatant (S) and pellet (P) fractions were analyzed by Western blot analysis. Mutants clearly expressing lower levels than wild-type MKKS are boxed with a solid line. NS, nonspecific band. (B) FLAG-tagged MKKS proteins were transiently expressed in HEK293 cells as described in A (top). Alternatively, HEK293 cells stably expressing wild-type or mutant MKKS were produced (bottom). Cells were radiolabeled for 30 min and chased for 3 and 6 h, and cell lysates were analyzed by immunoprecipitation. (C) The radioactivity of the FLAG-MKKS bands (mean and SD) was determined from three transient expression experiments. (D) HEK293 cells were transiently transfected with 0.5 μg of FLAG-tagged wild-type or mutant MKKS expression vectors and analyzed as described in A. Mutants showing greatly enhanced insolubility (P/S ratio equal to or lager than 1.0) are boxed with a dotted line. (E) Classification of disease-causing MKKS mutants according to their enhanced degradation and insolubility characteristics. The upward arrows denote enhanced characteristics relative to the wild-type protein. Mutants failing to localize to the centrosome are underlined (Figure 3). Mutants exhibiting increased insolubility in the presence of MG-132 (Figure 4B) are indicated by asterisks.
Structurally unstable proteins often form insoluble aggregates when expressed at high levels. We examined the solubility of the 11 mutants after transfection with higher amounts of the expression vector (0.5 μg). Under these conditions, three mutants (Y37C, T57A, and C499S) formed insoluble fraction whereas the wild-type MKKS remained almost soluble (Figure 1D). These three mutants were among the seven mutants that were rapidly degraded under low expression conditions (see Figure 1E for summary), suggesting that aggregation occurs due to a particular type of structural instability.
In contrast, four mutants (G52D, I339V, S511A, and R518H) exhibited no significant differences relative to the wild-type protein with respect to their expression levels and tendency to occur in the insoluble fraction. These mutants probably have lost particular functions without losing the normal conformation.
The 11 MKKS mutants can be divided into three groups: 1) mutants exhibiting increases in both degradation and insolubility (T57A, C499S, and Y37C); 2) mutants exhibiting increases only in degradation (H84Y, A242S, R155L, and G345E); and 3) mutants unaffected in either process (G52D, S511A, I339V, and R518H). These observations indicate that the mutants in groups 1 and 2 are both unstable but differ with respect to their insolubility in vivo.
Rapid Shuttling of Wild-Type MKKS between the Centrosome and Cytosol and Mislocalization of Mutant Proteins
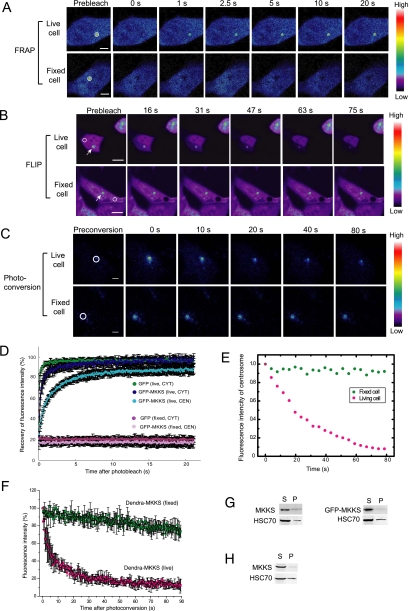
Many BBS proteins are known to localize to the centrosome/basal body, and MKKS was recently reported to localize to the centrosome (Kim et al., 2005). We also observed this MKKS distribution by immunostaining fixed cells (data not shown). It was recently reported that knockdown of MKKS in zebrafish results in delayed intracellular trafficking (Yen et al., 2006). We thus postulated that the highly concentrated MKKS in the centrosome may function by shuttling between the centrosome and cytosol. To analyze the centrosomal dynamics of MKKS, we performed live cell imaging (Lippincott-Schwartz et al., 2001) of MKKS tagged with fluorescent proteins by using three different techniques. We first examined the mobility of GFP-tagged MKKS at the centrosome by FRAP analysis. The intense fluorescence of GFP-MKKS at the centrosome was photobleached for 10 ms, and fluorescence recovery was recorded using a laser-scanning confocal microscope. The immediate recovery of strong fluorescence after photobleaching (Figure 2, A and D) indicates that MKKS moves rapidly from the bulk cytosol to the centrosome.
Figure 2.
Wild-type MKKS highly concentrated in the centrosome rapidly shuttles to and from the cytosol. (A) FRAP analysis of MKKS at the centrosome. GFP-MKKS at centrosomes, as indicated by circles, was bleached for 10 ms, and its subsequent movement from the cytosol was monitored. Bar, 5 μm. (B) FLIP experiment was performed by continuous bleaching of a small area (circles) in the cytosol. Bar, 10 μm. (C) Dendra-MKKS at the centrosome was photoconverted from green to red, and movement of the red color emission after the conversion was monitored. Bar, 5 μm. (D) Quantification of the fluorescence intensity of GFP-MKKS during the FRAP experiments (n = 20). CYT and CEN denote cytosol and centrosome, respectively. (E) Fluorescence intensity at centrosomes during FLIP analysis shown in B. (F) Red fluorescence intensity of Dendra-MKKS at the centrosome after photoconversion (n=3) (G and H) MKKS solubility test employing centrifugal fractionation. Transiently expressed untagged MKKS or GFP-MKKS (G) or untagged MKKS expressed in a stable cell line (H) were analyzed by centrifugal fractionation followed by Western blotting by using the MKF-1 anti-MKKS antibody. S, supernatant; P, pellet.
Next, to determine whether the centrosomal MKKS could move to the cytosol, we continuously bleached a small remote area in the cytosol in FLIP (fluorescence loss in photobleaching) experiments. The fluorescence of centrosomal GFP-MKKS gradually decreased during bleaching (Figure 2, B and E), indicating a movement of MKKS from the centrosome to the cytosol. Finally, to confirm the centrosome-to-cytosol movement of MKKS by a more direct method, we carried out photoconversion experiments at the centrosome. Photoconversion is a variation of the photoactivation method (Patterson and Lippincott-Schwartz, 2002), which allows the tracking of movement by fluorescently labeled molecules after pulse activation. MKKS tagged with Dendra, a green-to-red photoactivatable fluorescent protein (Gurskaya et al., 2006), was expressed in HEK293 cells. The green Dendra–MKKS protein concentrated at the centrosome was photoconverted to red by a pulse of laser beam. The red Dentra–MKKS signals rapidly disappeared from the centrosome after the conversion (Figure 2, C and F), showing that MKKS rapidly moves from the centrosome to the cytosol. We also confirmed that MKKS is highly soluble by centrifugal fractionation either in transiently transfected cells (Figure 2G) or in stable transformant (Figure 2H). These results clearly indicate that wild-type MKKS is highly mobile and rapidly shuttles between the centrosome and cytosol.
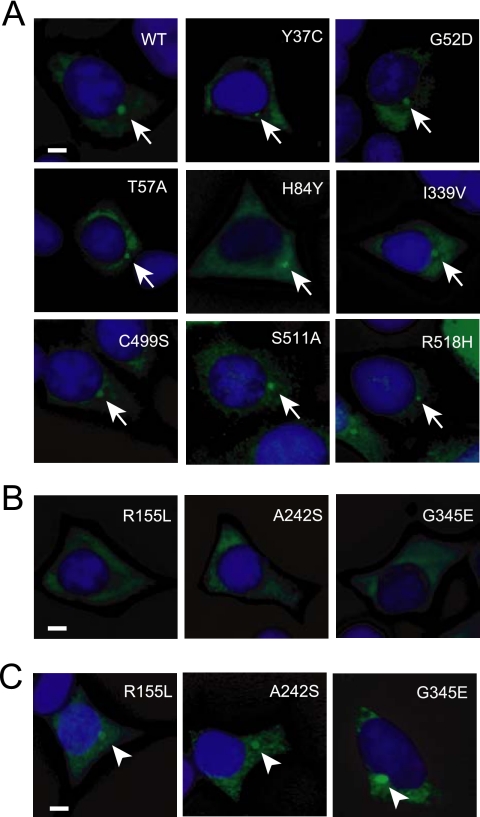
Kim et al. (2005) reported that four MKKS mutants (G52D, D285A, T325P, and G345E) tagged with GFP fail to localize to the centrosome in COS-7 cells. We determined the intracellular localization of the 11 mutants used for degradation/aggregation analysis (Figure 1) by immunostaining of FLAG-tagged MKKS mutants transiently expressed in HEK293 cells. Three rapidly degraded mutants (R155L, A242S, and G345E) did not localize to the centrosome (Figure 3). Although the localization of G345E is consistent with previous findings, the localization of G52D is not. This apparent discrepancy may be due to the differences in the tag or cell line used. Nevertheless, taken together with the results of live cell imaging of the wild-type MKKS, these observations suggest that at least three rapidly degraded MKKS mutants belonging to group 2 (stimulated for degradation but not for insolubility; Figure 1E) lack a centrosome-related dynamic function, which is probably due to their structural instability.
Figure 3.
Intracellular localization of MKKS mutants. (A and B) HEK293 cells were transiently transfected with FLAG-tagged MKKS expression vectors and stained with the anti-FLAG antibody. Counterstaining was performed with 4′,6-diamidino-2-phenylindole (DAPI). The mutants shown in B did not localize to the centrosome, whereas those in A did (indicated by arrows). (C) Cells were transfected with the same mutants as in B and treated with 10 μM of the proteasome inhibitor MG-132 for 6 h. Bar, 5 μm. Arrowheads indicate highly concentrated MKKS structures produced by proteasome inhibition.
Proteasome Inhibition Stimulates the Formation of Insoluble Structures by MKKS Mutants
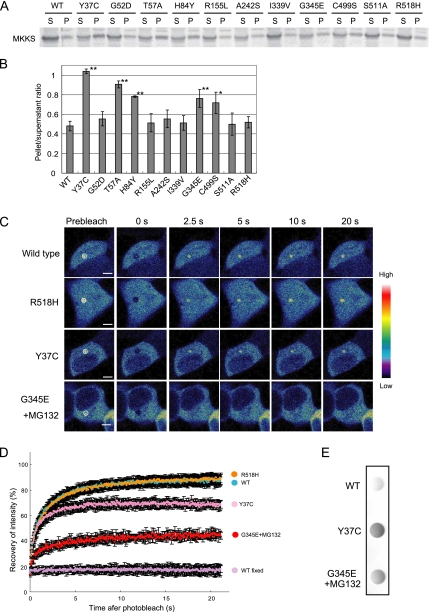
The ubiquitin–proteasome system is the major pathway that degrades misfolded proteins. To determine whether the MKKS mutants are processed by this system, we analyzed their expression levels in the presence of MG-132, a proteasome inhibitor. Although exposure to MG-132 frequently increased expression levels of MKKS mutants, this treatment resulted in the appearance of the MKKS mutants in the insoluble fraction (Figure 4A). Quantitation of these data indicated that the pellet/supernatant ratios of five mutants (Y37C, T57A, H84Y, C499S, and G345E) were significantly higher than that of wild-type MKKS (Figure 4B). Thus, these five mutants probably have a tendency to form insoluble structures even when they are expressed at low levels, but their insoluble forms are cleared by the ubiquitin-proteasome system (Figure 1E).
Figure 4.
Proteasome inhibition stimulates the formation of insoluble structures by several MKKS mutants. (A) HEK293 cells transfected with MKKS expression vectors (0.3 μg) were treated with 10 μM MG-132. Cell lysates were analyzed by centrifugal fractionation followed by Western blotting. S, supernatant; P, pellet. (B) Western blotting was performed as in A, and the band intensity was quantified (n = 3). Mutants exhibiting pellet/supernatant ratios significantly greater than the wild type are indicated by asterisks (*, p < 0.05; **, p < 0.01). (C) FRAP analysis of MKKS mutants. Cells were transfected with vectors (0.6 μg) expressing GFP-tagged MKKS mutants (Y37C, G345E, or R518H). G345E-expressing cells were treated with MG-132 to inhibit the rapid proteasome-dependent degradation. The centrosome region was bleached for 10 ms, and fluorescence recovery was recorded. (D) Quantification of the fluorescence intensity during FRAP analysis (n = 20). The data of GFP-tagged wild-type MKKS at the centrosome in living and fixed cells are taken from Figure 2D for comparison. (E) Cells transiently expressing FLAG-tagged wild-type or mutant MKKS were lysed in 0.5% TritonX-100/PBS and boiled in the presence of 1% SDS. Cell lysates were analyzed by the filter trap assay using anti-FLAG antibody.
One may speculate that the three mutants that failed to localize into the centrosome (R155L, A242S, and G345E; Figure 3B) would recover intrinsic localization if their expression levels were increased by inhibiting degradation. To test this possibility, we determined the localization of these mutants after MG-132 treatment. Cells exposed to the proteasome inhibitor exhibited a dense structure that contained the MKKS mutants at the centrosome (Figure 3C). However, FRAP analysis of the GFP-tagged G345E mutant indicated that this protein was almost immobilized at the centrosome in MG-132–treated cells (Figure 4, C and D), suggesting that G345E forms aggregates at the centrosome. Filter trap analysis of cell lysates after 1% SDS treatment indicated that the G345E mutant forms SDS-resistant large structures in the presence of MG-132 (Figure 4E).
Because FRAP analysis has been used to analyze the mobility of misfolded proteins (Nehls et al., 2000; Kim et al., 2002), we used this technique to assess the behavior of the Y37C mutant (increased insolubility; Figures 1, C and E, and 4, A and B) at the centrosome. Even in the absence of the proteasome inhibitor, the fluorescence recovery of this mutant was ∼70%, whereas the fluorescence recoveries of the R518 mutant (unaffected for degradation and insolubility) and the wild-type protein were both 90% (Figure 4, C and D). Thus, a significant portion of Y37C at the centrosome was immobilized and could not be replaced by cytosolic soluble protein. This is probably due to its tendency to aggregate, because the Y37C mutant forms SDS-resistant large structures even in the absence of the proteasome inhibitor (Figure 4E).
Together, these observations indicate that several MKKS mutants are rapidly degraded by a proteasome-dependent pathway as a result of structural instability. Among the mutants whose degradation was highly proteasome-dependent, G345E was rapidly degraded despite its potential to form insoluble structures. In contrast, Y37C formed insoluble structures more readily in living cells. These observations suggest that G345E degradation may be mediated by cytosolic proteins that function to prevent aggregation.
The G345E Mutant of MKKS Forms an Aggresome-like Structure at the Centrosome in a Microtubule-independent Manner after Proteasome Inhibition
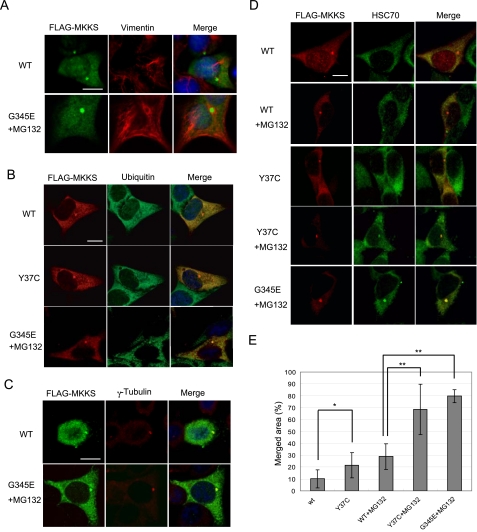
The insoluble and immobile characteristics of the centrosomal structure formed by MKKS mutants are reminiscent of the aggresome inclusion (Kopito, 2000). To determine whether the observed centrosomal structure is an aggresome, we performed immunofluorescence analysis of cells transiently expressing MKKS mutants by using antibodies against aggresome makers. Costaining of FLAG-tagged G345E with vimentin in MG-132–treated cells indicated that a significant number of cells form a cage-like vimentin structure surrounding G345E at the centrosome (Figure 5A). The centrosomal G345E structure was strongly stained by an anti-ubiquitin antibody (Figure 5B). Together with the fact that the G345E mutant forms SDS-resistant large structures in the presence of MG-132 (Figure 4E), these results strongly suggest that the G345E mutant forms an aggresome-like inclusion upon proteasome inhibition. However, disruption of microtubule organization by nocodazole treatment indicated that the microtubule network was not essential for the formation of the centrosomal structure at the centrosome (Figure 5C). Thus, this aggresome-like structure differs from typical aggresomes in that its formation is independent of the presence of microtubules.
Figure 5.
Immunofluorescence analysis of HEK293 cells expressing wild-type and mutant MKKS proteins. Cells were transfected with FLAG-tagged wild-type or mutant MKKS. G345E-expressing cells were treated with MG-132 to inhibit rapid degradation. (A–D) Cells were fixed and incubated with mouse (A and C) or rabbit (B and D) anti-FLAG antibodies along with rabbit antibodies against vimentin (A) or γ-tubulin (C), mouse antibody against ubiquitin (B), or rat antibody against HSC70. Cells were then incubated with AlexaFluor 488-conjugated anti-mouse IgG and AlexaFluor 594-conjugated anti-rabbit IgG (A and C), AlexaFluor 594-conjugated anti-rabbit IgG and AlexaFluor 488-conjugated anti-mouse IgG (B), or AlexaFluor 594-conjugated anti-rabbit IgG and AlexaFluor 488-conjugated anti-rat IgG (D). (C) Cells were treated with 20 μM nocodazole. Counterstaining was performed with DAPI except for D. Bar, 10 μm. (E) Double staining of FLAG-MKKS and HSC70 was performed as shown in D, and quantification of merged area in the centrosomal MKKS structure was carried out (n = 10). Asterisks indicate significance of difference (*, p < 0.02; **, p < 0.01).
CHIP-mediated Degradation of MKKS Mutants
Because structurally unstable proteins are known to be recognized by molecular chaperones, including HSP70/HSC70, we examined by immunostaining whether HSC70 colocalizes with G345E in the aggresome-like structure. Fluorescence signals from antibodies against FLAG and HSC70 were merged well in the centrosomal structure formed by FLAG-tagged G345E upon proteasome inhibition, whereas the same HSC70 antibody did not strongly recognize centrosomes that contain highly concentrated FLAG-tagged wild-type MKKS (Figure 5, D and E). The anti-HSC70 antibody also stained centrosomes harboring the Y37C mutant particularly when proteasomal activity was inhibited.
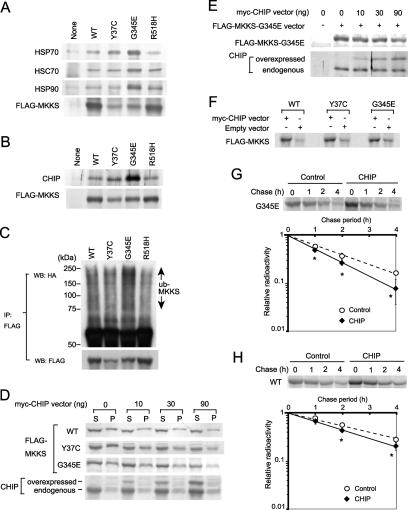
Molecular chaperones prevent protein aggregation, and CHIP is known to stimulate the degradation of misfolded proteins by ubiquitinating them in an HSP70/HSC70- or HSP90-dependent manner. Strong colocalization of HSC70 with the G345E mutant in the aggresome-like structure suggested that this mutant may be directly recognized by chaperones. Moreover, HSP70/HSC70 and HSP90 were detected as MKKS-binding proteins by mass spectroscopic analysis of immunoprecipitates (data not shown). We thus examined the association of endogenous HSP70, HSC70, and HSP90 with the G345E mutant by immunoprecipitation followed by Western blotting analysis, and we compared the results with those obtained using the wild-type protein and other mutants. Intriguingly, the rapidly degraded MKKS mutant G345E was much more strongly recognized by HSP70, HSC70, and HSP90 than were the wild-type protein or the other mutants, despite the fact that the expression level of G345E was much lower than that of the wild-type protein or R518H mutant (unaffected for degradation) (Figure 6A). The Y37C mutant exhibited enhanced binding of HSP70, but with an efficiency lower than that of the G345E mutant. We thus postulated that the degradation of MKKS mutants, particularly of the G345E mutant, may be mediated by these molecular chaperones and the chaperone-mediated ubiquitin ligase CHIP.
Figure 6.
Increased association of CHIP with unstable MKKS mutants and effect of CHIP overexpression on their degradation. (A and B) FLAG-tagged Y37C (unstable and highly insoluble), G345E (unstable and rapidly degraded), or R518H (normal for degradation and solubility) mutants or wild-type MKKS as a control were transiently expressed in HEK293 cells. Cell lysates were immunoprecipitated with the anti-FLAG antibody and analyzed by Western blotting by using antibodies against molecular chaperones (A) or CHIP (B). Note that the amounts of Y37C and G345E mutants recovered by immunoprecipitation are much lower than those of the R518H mutant and wild-type MKKS due to rapid degradation. (C) Enhanced polyubiquitination of unstable MKKS mutants. Cells were cotransfected with ubiquitin-HA and FLAG-MKKS, and cell lysates were immunoprecipitated with the anti-FLAG antibody. Recovered FLAG-MKKS was analyzed by Western blotting by using antibodies against HA or FLAG. (D) CHIP overexpression reduces the amount of MKKS in both soluble and insoluble fractions. To stimulate MKKS aggregation, transfection efficiency was enhanced by transfecting cells with Effecten instead of FuGENE 6. After cotransfection with FLAG-MKKS (200 ng) and myc-CHIP (0–90 ng), cells lysates were fractionated by centrifugation and analyzed by Western blotting by using antibodies against FLAG or CHIP. (E) Overexpressed and endogenous CHIP proteins were associated with the G345E mutant. Cells were transfected with FLAG-MKKS-G345E as described in D, and FLAG-MKKS-G345E was immunoprecipitated. FLAG-MKKS-G345E and coprecipitated CHIP were analyzed by Western blotting. (F) MKKS synthesis is stimulated by CHIP overexpression. Cells transfected with FLAG-tagged wild-type or mutant MKKS were pulse labeled for 30 min in the presence or absence of CHIP overexpression. Labeled FLAG-MKKS proteins were analyzed by immunoprecipitation with the anti-FLAG antibody. (G and H) Effect of CHIP overexpression on the degradation of the G345E mutant and wild-type MKKS. Cells transiently expressing FLAG-tagged G345E mutant (G) or wild-type MKKS (H) under CHIP overexpression conditions were analyzed by pulse-chase experiments followed by immunoprecipitation. Radioactivity of FLAG-tagged MKKS bands in pulse-chase experiments was determined and normalized against the value of chase at 0 h (n = 3). Asterisks indicate significance of difference (p < 0.05).
To test this possibility, we first determined whether endogenous CHIP associates with MKKS mutants. CHIP was much more efficiently associated with the G345E mutant than with the other mutants or wild-type MKKS (Figure 6B). The results of this analysis reflected the degree of association of HSP70 with MKKS proteins (Figure 6A). Thus, CHIP probably associates with MKKS via HSP70/HSC70 or HSP90, as has been shown for other substrate proteins (Connell et al., 2001; Meacham et al., 2001; Murata et al., 2001). Next, cells were cotransfected with HA-ubiquitin and FLAG-MKKS (WT, Y37C, G345E, or R518H) to test whether G345E is highly polyubiquitinated. Polyubiquitination of MKKS was assessed by immunoprecipitation of FLAG-MKKS followed by Western blotting with anti-HA antibody. Polyubiquitination signals detected at high molecular size regions were much stronger for G345E than for the other mutants, despite the lower expression level of this mutant compared with that of the wild-type protein or R518 mutant (Figure 6C). Although the level of Y37C polyubiquitination detected on Western blots was similar to the levels of the wild-type MKKS and the R518 mutant, the expression level of Y37C was much lower than that of either protein. Thus, the proportion of protein that is polyubiquitinated seems to be higher for the Y37C mutant than for the wild-type MKKS or R518 mutant. These results indicate that unstable MKKS mutants are highly polyubiquitinated in the following order: G345E > Y37C > wild type, R518H.
Overexpression of CHIP decreased the amount of Y37C and G345E in a dose-dependent manner (Figure 6, D and E), despite the general stimulation of MKKS synthesis by CHIP overexpression (Figure 6F). The increased protein synthesis following CHIP overexpression may be due to the enhanced association of the molecular chaperones including HSP70 as described previously (Kampinga et al., 2003). Increased HSP70 association during or immediately after translation may stabilize MKKS, but long-term posttranslational association probably causes enhanced degradation. In contrast, Cardozo et al. (2003) reported that CHIP overexpression reduced synthesis of the androgen receptor (Cardozo et al., 2003). Thus, effects of CHIP on translation may differ between proteins. Although overexpression of CHIP also decreased the level of wild-type MKKS, the effect was less pronounced compared with Y37C and G345E (Figure 6D). Because these results suggested that CHIP stimulates degradation of mutant MKKS more effectively than the wild type, we examined this possibility by performing pulse-chase experiments. Overexpression of CHIP stimulated degradation of the G345E mutant at a statistically significant level, although the effect was not very strong (Figure 6G). However, a weak stimulation of wild-type MKKS degradation by CHIP overexpression was also observed (Figure 6H). These results suggest that the degradation of unstable MKKS mutants is stimulated by CHIP. The degradation of wild-type MKKS may reflect the abnormally high level of CHIP produced by overexpression.
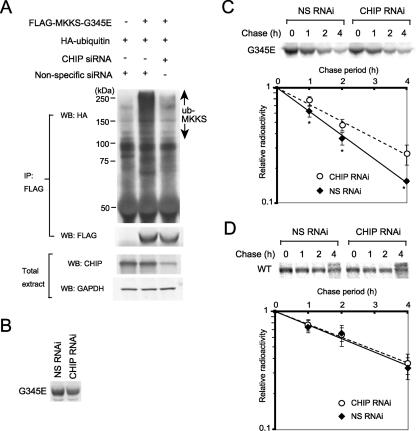
To determine whether endogenous CHIP affects polyubiquitination of the G345E mutant, CHIP was depleted by performing RNAi-mediated knockdown in HEK293 cells. HEK293 cells were cotransfected with FLAG-MKKS and HA-ubiquitin and then treated with a CHIP siRNA or a nonspecific siRNA as a control. FLAG-MKKS was immunoprecipitated, and polyubiquitination was analyzed by Western blotting with an anti-HA antibody. CHIP siRNA treatment moderately reduced the level of endogenous CHIP (Figure 7A, bottom), and inhibited polyubiquitination of FLAG-tagged G345E (Figure 7A, top). Pulse labeling followed by immunoprecipitation indicated that CHIP RNAi treatment weakly decreased the synthesis of G345E, supporting the notion that CHIP stimulates MKKS synthesis (Figure 6F). Pulse-chase experiments revealed that the degradation of G345E was moderately inhibited by modest knockdown of CHIP (Figure 7C), whereas that of wild-type MKKS was unaffected under these conditions (Figure 7D). These results clearly indicate that the polyubiquitination activity of endogenous CHIP plays a crucial role in the proteasome-dependent degradation of the unstable MKKS mutant G345E.
Figure 7.
Polyubiquitination and degradation of the G345E mutant are inhibited by RNAi-mediated knockdown of CHIP. (A) Strong inhibition of MKKS-G345E polyubiquitination by CHIP knockdown. After cotransfection with FLAG-tagged G345E and HA-tagged ubiquitin, cells were treated with CHIP siRNA or nonspecific siRNA as a control. FLAG-G345E was immunoprecipitated with the anti-FLAG antibody and analyzed by Western blotting by using antibodies against HA or FLAG. Expression levels of CHIP were analyzed by Western blotting of total cell extracts. (B) Decreased synthesis of MKKS-G345E by CHIP RNAi treatment. Transiently expressed FLAG-tagged G345E was pulse labeled with [35S]Met for 30 min. Cell lysates were immunoprecipitated with the anti-FLAG antibody and analyzed by SDS-PAGE. (C and D) Transiently expressed FLAG-tagged G345 mutant (C) and wild-type MKKS (D) were pulse labeled with [35S]Met for 30 min and chased for 1, 2, and 4 h in the presence of CHIP siRNA or nonspecific siRNA as a control. The radiolabeled FLAG-MKKS was immunoprecipitated and analyzed by SDS-PAGE. Radioactivity of FLAG-tagged MKKS bands in pulse-chase experiments was determined and normalized against the value of chase at 0 h (n = 5 in C and n = 4 in D). Asterisks indicate significance of difference (p < 0.02).
DISCUSSION
We found in this study that MKKS rapidly shuttles between the centrosome and cytosol by using live cell imaging techniques that included FRAP, FLIP, and photoconversion (Figure 2). The fully soluble nature of MKKS was supported by centrifugal fractionation, and it was independent of the presence or absence of different tags. These results are consistent with the notion that BBS family proteins, including MKKS plays a role in intracellular and/or intraflagellar motor-driven transport (Mykytyn and Sheffield, 2004; Badano et al., 2005; Beales, 2005; Blacque and Leroux, 2006). BBS4 and BBS8 localize to the centrosome/MTOC in mammalian cells, and BBS4 interacts with the p150glued subunit of the dynactin complex, which mediates association of cargoes with dynein motors (Ansley et al., 2003; Kim et al., 2004). Moreover, retrograde transport of melanosomes in zebrafish is slowed by knockdown of BBS2 or BBS4-8 (Yen et al., 2006). In C. elegans, mutations in BBS7 and BBS8 result in a delay in an intraflagellar transport driven by kinesin-II and OMS-3 kinesin motors (Ou et al., 2005). The BBSome containing BBS1, 2, 4, 5, and 7–9 was recently shown to cooperate with the Rab8 GTPase to promote ciliary membrane growth (Nachury et al., 2007), although this complex does not contain MKKS or the two other chaperonin-like BBS proteins. Thus, the rapid shuttling of MKKS at the centrosome is probably involved in intracellular transport, although its role in this transport may be different from that of the BBSome.
The phenotypes of the MKKS-knockout mouse (Fath et al., 2005) closely resemble those of mice disrupted for the BBS1, BBS2, or BBS4 genes (Kulaga et al., 2004; Mykytyn et al., 2004; Nishimura et al., 2004), and many of these phenotypes have been suggested to be caused by ciliary dysfunction. In the case of the BBS2-knockout mouse, the photoreceptor degeneration is preceded by mislocalization of rhodopsin, which indicates a defect in protein transport. Thus, rhodopsin is a candidate protein that would be transported with assistance of BBS proteins, including MKKS, although other target proteins also must exist. In addition to MKKS (Stone et al., 2000), BBS10 (Stoetzel et al., 2006), and BBS12 (Stoetzel et al., 2007) are now known to share weak but significant sequence similarity to the group II chaperonins, including CCT. These proteins might assist protein transport by performing chaperone-like activities, although the exact molecular mechanisms of MKKS-dependent transport remain to be investigated.
Experiments with 11 disease-causing MKKS mutants revealed that many of them are rapidly degraded and/or tend to form insoluble structures (Figure 1), indicating that these mutations reduce the levels of functional MKKS proteins. A recent study of MKKS-knockout mice indicated that they exhibit photoreceptor degeneration, obesity, and defects in olfaction, social dominance and sperm flagellum formation (Fath et al., 2005). Because these phenotypes closely resemble those associated with human BBS (Beales et al., 1999), the reduced levels of mutant MKKS proteins resulting from rapid degradation and increased insolubility are most likely a major cause of BBS symptoms.
The tendency to form insoluble structures by MKKS mutants when proteasome-dependent degradation is inhibited suggests that they have an abnormal conformation (Figures 3–5). Moreover, molecular chaperones were found to preferentially recognize these mutants. Because molecular chaperones recognize the exposed hydrophobic surfaces of unfolded and misfolded proteins, these mutants probably have exposed hydrophobic surfaces caused by their structural instability.
However, the degree of insolubility varied considerably between the different mutants. The G345E mutant is a representative of rapidly degraded mutants, whereas the Y37C mutant formed insoluble structures more readily in the cytosol. Although these two mutants were rapidly degraded at similar rates under conditions of low expression and both formed insoluble structures when the proteasome was inhibited, G345E was more strongly recognized by molecular chaperones HSP70/HSC70 and HSP90 than Y37C. The insoluble structures formed by the G345E mutant at the centrosome under proteasome inhibition conditions are likely to be aggresome-like inclusions because they were surrounded by a vimentin cage and highly ubiqutinated. Thus, the G345E protein may have more extensively exposed hydrophobic surfaces than the Y37C protein. Nevertheless, G345E was efficiently degraded without forming insoluble fractions in vivo. Together, these observations suggest that these chaperones maintain G345E in a soluble state in the cytosol. The chaperones seem to play an important role in the quality control of disease-causing proteins, although the detailed molecular mechanism for the preferential recognition of the G345E mutant by molecular chaperones remains to be investigated.
The Y37C mutant forms partially immobilized structures at the centrosome, but under the same conditions the G345E mutant does not. Y37C may be more toxic than G345E because it interferes with other centrosomal proteins. A heterozygote bearing the Y37C mutation and the 1215-1216delGG frame-shift mutation, which occurs in the middle of the coding region, showed MKKS symptoms (Stone et al., 2000) that were more severe than the BBS symptoms. In contrast, when homozygotic, the Y37C (Katsanis et al., 2000) or G345E (Slavotinek et al., 2002) mutation caused only BBS symptoms. Tendency of the Y37C mutant to aggregate when combined with the frame-shift mutant may result in more severe MKKS symptoms. This is unlikely to represent a loss-of-function phenotype because the MKKS-knockout mouse shows BBS phenotypes but not MKKS phenotypes (Fath et al., 2005). Sekijima et al. (2005) recently demonstrated that the thermodynamic and kinetic instability of transthyretin, a protein known to cause familial amyloidosis, affects the severity of the symptoms and that the most unstable mutants are less toxic than the moderately unstable mutants because the former undergo endoplasmic reticulum (ER)-associated protein degradation. In the MKKS mutants, G345E was more strongly recognized by cytosolic molecular chaperones and more easily degraded than Y37C. It is tempting to speculate that the cytosolic chaperone-dependent protein degradation system may affect the severity of the symptoms, as observed for transthyretin degradation in the ER, although this possibility remains to be investigated.
Detailed analysis of MKKS degradation demonstrated that the chaperone-assisted ubiquitin ligase CHIP (Connell et al., 2001; Meacham et al., 2001; Murata et al., 2001) plays a crucial role in the ubiquitin-proteasome dependent degradation of MKKS mutants. MKKS mutants are highly ubiquitinated on association with CHIP, and this probably occurs via a preferential recognition of the mutants by chaperones (Figures 6 and 7). Modest RNAi-mediated knockdown of CHIP inhibited ubiquitination of the G345E mutant and resulted in a modest delay in degradation, indicating that CHIP is required for the rapid clearance of the G345E mutant. It should be emphasized that the G345E mutant was strongly recognized by chaperones and CHIP and that it was heavily polyubiquitinated. It was recently shown by in vitro experiments that CHIP stimulates HSP70 activity to prevent huntingtin-Q53 aggregation and that CHIP can directly inhibit the aggregation of heat-denatured luciferase (Rosser et al., 2007). CHIP may maintain the MKKS G345E mutant in a degradation-competent soluble state via enhancement of HSP70 activity and the intrinsic chaperone-like activity of CHIP. The G345E mutant is potentially toxic to the cell because it forms insoluble fractions when proteasome activity is inhibited. However, this mutant was rapidly degraded by the CHIP-dependent system without forming insoluble structures. Thus, CHIP seems to contribute to the clearance of potentially toxic misfolded proteins. In contrast, the Y37C mutant was more resistant to the recognition by chaperones and polyubiquitination by CHIP than was the G345E mutant, despite its instability and insolubility, which were similar to those of G345E. The tendency that Y37C is inefficiently recognized by chaperones and CHIP may enhance its aggregation in vivo. These observations suggest that chaperone-dependent CHIP activity greatly modulates the aggregation and toxicity of disease-causing point mutations that result in structurally unstable proteins.
The CHIP-dependent degradation of misfolded protein has been well-studied in cystic fibrosis transmembrane conductance regulator (CFTR) Δ508 mutant proteins (Tateishi et al., 2004; Younger et al., 2004, 2006; Qian et al., 2006), which make insoluble aggregates in the ER (Meacham et al., 2001). However, the role of CHIP in cystic fibrosis seems to differ from its role in BBS. CFTR Δ508 localization and function are known to be restored by CHIP inhibition; thus, CHIP is considered to be a contributory factor to this disease. In contrast, inhibition of CHIP in cells expressing MKKS mutants resulted in insoluble aggregates in the cytosol, suggesting that CHIP protects cells from the toxicity of MKKS mutant aggregation.
In conclusion, these observations indicate that MKKS function, which may act during a centrosome-associated protein transport, is lost in MKKS mutants because of their rapid degradation and increased insolubility. CHIP plays an essential role in the rapid degradation of MKKS mutants and this role is assisted by chaperone-dependent recognition of protein misfolding. CHIP-dependent clearance of misfolded proteins seems to greatly contribute to the cellular homeostasis of the protein folding environment.
ACKNOWLEDGMENTS
We thank Mana Kadota for technical assistance and Dr. Keiji Tanaka (Tokyo Metropolitan Institute of Medical Science, Japan) for providing the HA-ubiquitin expression vector. We are grateful to Drs. Takao Yoshida, Takuo Yasunaga, and Tamotsu Yoshimori for helpful discussion. S.H., A.K., and Y.O. were supported by fellowships from the Japan Society for Promotion of Science. H.K. and K.N. were supported by grant-in-aid for scientific research and grant-in-aid for creative scientific research programs in Japan, respectively.
Abbreviations used:
- BBS
Bardet–Biedl syndrome
- CCT
chaperonin containing t-complex polypeptide 1
- CHIP
carboxy terminus of HSC70 interacting protein
- ER
endoplasmic reticulum
- FRAP
fluorescence recover after photobleaching
- FLIP
fluorescence loss in photobleaching
- HSP60
60-kDa heat-shock protein.
- MKKS
McKusick–Kaufman syndrome.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-07-0631) on December 19, 2007.
REFERENCES
- Ansley S. J., et al. Basal body dysfunction is a likely cause of pleiotropic Bardet-Biedl syndrome. Nature. 2003;425:628–633. doi: 10.1038/nature02030. [DOI] [PubMed] [Google Scholar]
- Badano J. L., Ansley S. J., Leitch C. C., Lewis R. A., Lupski J. R., Katsanis N. Identification of a novel Bardet-Biedl syndrome protein, BBS7, that shares structural features with BBS1 and BBS2. Am. J. Hum. Genet. 2003;72:650–658. doi: 10.1086/368204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badano J. L., Leitch C. C., Ansley S. J., May-Simera H., Lawson S., Lewis R. A., Beales P. L., Dietz H. C., Fisher S., Katsanis N. Dissection of epistasis in oligogenic Bardet-Biedl syndrome. Nature. 2006;439:326–330. doi: 10.1038/nature04370. [DOI] [PubMed] [Google Scholar]
- Badano J. L., Teslovich T. M., Katsanis N. The centrosome in human genetic disease. Nat. Rev. Genet. 2005;6:194–205. doi: 10.1038/nrg1557. [DOI] [PubMed] [Google Scholar]
- Barral J. M., Broadley S. A., Schaffar G., Hartl F. U. Roles of molecular chaperones in protein misfolding diseases. Semin. Cell Dev. Biol. 2004;15:17–29. doi: 10.1016/j.semcdb.2003.12.010. [DOI] [PubMed] [Google Scholar]
- Beales P. L. Lifting the lid on Pandora's box: the Bardet-Biedl syndrome. Curr. Opin. Genet Dev. 2005;15:315–323. doi: 10.1016/j.gde.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Beales P. L., Elcioglu N., Woolf A. S., Parker D., Flinter F. A. New criteria for improved diagnosis of Bardet-Biedl syndrome: results of a population survey. J. Med. Genet. 1999;36:437–446. [PMC free article] [PubMed] [Google Scholar]
- Beales P. L., et al. Genetic and mutational analyses of a large multiethnic Bardet-Biedl cohort reveal a minor involvement of BBS6 and delineate the critical intervals of other loci. Am J. Hum. Genet. 2001;68:606–616. doi: 10.1086/318794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrends C., et al. Chaperonin TRiC Promotes the Assembly of polyQ Expansion Proteins into Nontoxic Oligomers. Mol Cell. 2006;23:887–897. doi: 10.1016/j.molcel.2006.08.017. [DOI] [PubMed] [Google Scholar]
- Bennett E. J., Bence N. F., Jayakumar R., Kopito R. R. Global impairment of the ubiquitin-proteasome system by nuclear or cytoplasmic protein aggregates precedes inclusion body formation. Mol Cell. 2005;17:351–365. doi: 10.1016/j.molcel.2004.12.021. [DOI] [PubMed] [Google Scholar]
- Blacque O. E., Leroux M. R. Bardet-Biedl syndrome: an emerging pathomechanism of intracellular transport. Cell Mol. Life Sci. 2006;63:2145–2161. doi: 10.1007/s00018-006-6180-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacque O. E., et al. Loss of C. elegans BBS-7 and BBS-8 protein function results in cilia defects and compromised intraflagellar transport. Genes Dev. 2004;18:1630–1642. doi: 10.1101/gad.1194004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo C. P., Michaud C., Ost M. C., Fliss A. E., Yang E., Patterson C., Hall S. J., Caplan A. J. C-terminal Hsp-interacting protein slows androgen receptor synthesis and reduces its rate of degradation. Arch. Biochem. Biophys. 2003;410:134–140. doi: 10.1016/s0003-9861(02)00680-x. [DOI] [PubMed] [Google Scholar]
- Chiang A. P., et al. Homozygosity mapping with SNP arrays identifies TRIM32, an E3 ubiquitin ligase, as a Bardet-Biedl syndrome gene (BBS11) Proc. Natl. Acad. Sci. USA. 2006;103:6287–6292. doi: 10.1073/pnas.0600158103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang A. P., et al. Comparative genomic analysis identifies an ADP-ribosylation factor-like gene as the cause of Bardet-Biedl syndrome (BBS3) Am J. Hum. Genet. 2004;75:475–484. doi: 10.1086/423903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A., Brundin P. The ubiquitin proteasome system in neurodegenerative diseases: sometimes the chicken, sometimes the egg. Neuron. 2003;40:427–446. doi: 10.1016/s0896-6273(03)00606-8. [DOI] [PubMed] [Google Scholar]
- Connell P., Ballinger C. A., Jiang J., Wu Y., Thompson L. J., Hohfeld J., Patterson C. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat. Cell Biol. 2001;3:93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- Farr G. W., Scharl E. C., Schumacher R. J., Sondek S., Horwich A. L. Chaperonin-mediated folding in the eukaryotic cytosol proceeds through rounds of release of native and nonnative forms. Cell. 1997;89:927–937. doi: 10.1016/s0092-8674(00)80278-0. [DOI] [PubMed] [Google Scholar]
- Fath M. A., et al. Mkks-null mice have a phenotype resembling Bardet-Biedl syndrome. Hum. Mol. Genet. 2005;14:1109–1118. doi: 10.1093/hmg/ddi123. [DOI] [PubMed] [Google Scholar]
- Gurskaya N. G., Verkhusha V. V., Shcheglov A. S., Staroverov D. B., Chepurnykh T. V., Fradkov A. F., Lukyanov S., Lukyanov K. A. Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat. Biotechnol. 2006;24:461–465. doi: 10.1038/nbt1191. [DOI] [PubMed] [Google Scholar]
- Holmberg C. I., Staniszewski K. E., Mensah K. N., Matouschek A., Morimoto R. I. Inefficient degradation of truncated polyglutamine proteins by the proteasome. EMBO J. 2004;23:4307–4318. doi: 10.1038/sj.emboj.7600426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwich A. L., Fenton W. A., Chapman E., Farr G. W. Two families of chaperonin: physiology and mechanism. Annu. Rev. Cell Dev. Biol. 2007;23:115–145. doi: 10.1146/annurev.cellbio.23.090506.123555. [DOI] [PubMed] [Google Scholar]
- Kampinga H. H., Kanon B., Salomons F. A., Kabakov A. E., Patterson C. Overexpression of the cochaperone CHIP enhances Hsp70-dependent folding activity in mammalian cells. Mol. Cell. Biol. 2003;23:4948–4958. doi: 10.1128/MCB.23.14.4948-4958.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsanis N., Beales P. L., Woods M. O., Lewis R. A., Green J. S., Parfrey P. S., Ansley S. J., Davidson W. S., Lupski J. R. Mutations in MKKS cause obesity, retinal dystrophy and renal malformations associated with Bardet-Biedl syndrome. Nat. Genet. 2000;26:67–70. doi: 10.1038/79201. [DOI] [PubMed] [Google Scholar]
- Kim J. C., et al. The Bardet-Biedl protein BBS4 targets cargo to the pericentriolar region and is required for microtubule anchoring and cell cycle progression. Nat. Genet. 2004;36:462–470. doi: 10.1038/ng1352. [DOI] [PubMed] [Google Scholar]
- Kim J. C., et al. MKKS/BBS6, a divergent chaperonin-like protein linked to the obesity disorder Bardet-Biedl syndrome, is a novel centrosomal component required for cytokinesis. J. Cell Sci. 2005;118:1007–1020. doi: 10.1242/jcs.01676. [DOI] [PubMed] [Google Scholar]
- Kim S., Nollen E. A., Kitagawa K., Bindokas V. P., Morimoto R. I. Polyglutamine protein aggregates are dynamic. Nat. Cell Biol. 2002;4:826–831. doi: 10.1038/ncb863. [DOI] [PubMed] [Google Scholar]
- Kitamura A., Kubota H., Pack C. G., Matsumoto G., Hirayama S., Takahashi Y., Kimura H., Kinjo M., Morimoto R. I., Nagata K. Cytosolic chaperonin prevents polyglutamine toxicity with altering the aggregation state. Nat. Cell Biol. 2006;8:1163–1170. doi: 10.1038/ncb1478. [DOI] [PubMed] [Google Scholar]
- Kopito R. R. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- Kubota H. Function and regulation of cytosolic molecular chaperone CCT. Vitam. Horm. 2002;65:313–331. doi: 10.1016/s0083-6729(02)65069-1. [DOI] [PubMed] [Google Scholar]
- Kubota H., Hynes G., Carne A., Ashworth A., Willison K. Identification of six Tcp-1-related genes encoding divergent subunits of the TCP-1-containing chaperonin. Curr. Biol. 1994;4:89–99. doi: 10.1016/s0960-9822(94)00024-2. [DOI] [PubMed] [Google Scholar]
- Kubota S., Kubota H., Nagata K. Cytosolic chaperonin protects folding intermediates of Gbeta from aggregation by recognizing hydrophobic beta-strands. Proc. Natl. Acad. Sci. USA. 2006;103:8360–8365. doi: 10.1073/pnas.0600195103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulaga H. M., Leitch C. C., Eichers E. R., Badano J. L., Lesemann A., Hoskins B. E., Lupski J. R., Beales P. L., Reed R. R., Katsanis N. Loss of BBS proteins causes anosmia in humans and defects in olfactory cilia structure and function in the mouse. Nat. Genet. 2004;36:994–998. doi: 10.1038/ng1418. [DOI] [PubMed] [Google Scholar]
- Li J. B., et al. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 2004;117:541–552. doi: 10.1016/s0092-8674(04)00450-7. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J., Snapp E., Kenworthy A. Studying protein dynamics in living cells. Nat. Rev. Mol. Cell Biol. 2001;2:444–456. doi: 10.1038/35073068. [DOI] [PubMed] [Google Scholar]
- Llorca O., McCormack E. A., Hynes G., Grantham J., Cordell J., Carrascosa J. L., Willison K. R., Fernandez J. J., Valpuesta J. M. Eukaryotic type II chaperonin CCT interacts with actin through specific subunits. Nature. 1999;402:693–696. doi: 10.1038/45294. [DOI] [PubMed] [Google Scholar]
- McKusick V. A., Bauer R. L., Koop C. E., Scott R. B. Hydrometrocolpos as a simply inherited malformation. Jama. 1964;189:813–816. doi: 10.1001/jama.1964.03070110015003. [DOI] [PubMed] [Google Scholar]
- Meacham G. C., Patterson C., Zhang W., Younger J. M., Cyr D. M. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat. Cell Biol. 2001;3:100–105. doi: 10.1038/35050509. [DOI] [PubMed] [Google Scholar]
- Meyer A. S., Gillespie J. R., Walther D., Millet I. S., Doniach S., Frydman J. Closing the folding chamber of the eukaryotic chaperonin requires the transition state of ATP hydrolysis. Cell. 2003;113:369–381. doi: 10.1016/s0092-8674(03)00307-6. [DOI] [PubMed] [Google Scholar]
- Murata S., Minami Y., Minami M., Chiba T., Tanaka K. CHIP is a chaperone-dependent E3 ligase that ubiquitylates unfolded protein. EMBO Rep. 2001;2:1133–1138. doi: 10.1093/embo-reports/kve246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mykytyn K., Mullins R. F., Andrews M., Chiang A. P., Swiderski R. E., Yang B., Braun T., Casavant T., Stone E. M., Sheffield V. C. Bardet-Biedl syndrome type 4 (BBS4)–null mice implicate Bbs4 in flagella formation but not global cilia assembly. Proc. Natl. Acad. Sci. USA. 2004;101:8664–8669. doi: 10.1073/pnas.0402354101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mykytyn K., Sheffield V. C. Establishing a connection between cilia and Bardet-Biedl Syndrome. Trends Mol. Med. 2004;10:106–109. doi: 10.1016/j.molmed.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Nachury M. V., et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- Nehls S., Snapp E. L., Cole N. B., Zaal K. J., Kenworthy A. K., Roberts T. H., Ellenberg J., Presley J. F., Siggia E., Lippincott-Schwartz J. Dynamics and retention of misfolded proteins in native ER membranes. Nat. Cell Biol. 2000;2:288–295. doi: 10.1038/35010558. [DOI] [PubMed] [Google Scholar]
- Nishimura D. Y., et al. Bbs2-null mice have neurosensory deficits, a defect in social dominance, and retinopathy associated with mislocalization of rhodopsin. Proc. Natl. Acad. Sci. USA. 2004;101:16588–16593. doi: 10.1073/pnas.0405496101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura D. Y., et al. Comparative genomics and gene expression analysis identifies BBS9, a new Bardet-Biedl syndrome gene. Am. J. Hum. Genet. 2005;77:1021–1033. doi: 10.1086/498323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H., Yamamura K., Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Nollen E. A., Morimoto R. I. Chaperoning signaling pathways: molecular chaperones as stress-sensing ‘heat shock’ proteins. J. Cell Sci. 2002;115:2809–2816. doi: 10.1242/jcs.115.14.2809. [DOI] [PubMed] [Google Scholar]
- Ou G., Blacque O. E., Snow J. J., Leroux M. R., Scholey J. M. Functional coordination of intraflagellar transport motors. Nature. 2005;436:583–587. doi: 10.1038/nature03818. [DOI] [PubMed] [Google Scholar]
- Patterson G. H., Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- Pickart C. M. Back to the future with ubiquitin. Cell. 2004;116:181–190. doi: 10.1016/s0092-8674(03)01074-2. [DOI] [PubMed] [Google Scholar]
- Qian S. B., McDonough H., Boellmann F., Cyr D. M., Patterson C. CHIP-mediated stress recovery by sequential ubiquitination of substrates and Hsp70. Nature. 2006;440:551–555. doi: 10.1038/nature04600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosser M. F., Washburn E., Muchowski P. J., Patterson C., Cyr D. M. Chaperone functions of the E3 ubiquitin ligase CHIP. J. Biol. Chem. 2007;282:22267–22277. doi: 10.1074/jbc.M700513200. [DOI] [PubMed] [Google Scholar]
- Sato S., Chiba T., Sakata E., Kato K., Mizuno Y., Hattori N., Tanaka K. 14–3-3eta is a novel regulator of parkin ubiquitin ligase. EMBO J. 2006;25:211–221. doi: 10.1038/sj.emboj.7600774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawano A., Miyawaki A. Directed evolution of green fluorescent protein by a new versatile PCR strategy for site-directed and semi-random mutagenesis. Nucleic Acids Res. 2000;28:E78. doi: 10.1093/nar/28.16.e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffar G., Breuer P., Boteva R., Behrends C., Tzvetkov N., Strippel N., Sakahira H., Siegers K., Hayer-Hartl M., Hartl F. U. Cellular toxicity of polyglutamine expansion proteins: mechanism of transcription factor deactivation. Mol. Cell. 2004;15:95–105. doi: 10.1016/j.molcel.2004.06.029. [DOI] [PubMed] [Google Scholar]
- Sekijima Y., Wiseman R. L., Matteson J., Hammarstrom P., Miller S. R., Sawkar A. R., Balch W. E., Kelly J. W. The biological and chemical basis for tissue-selective amyloid disease. Cell. 2005;121:73–85. doi: 10.1016/j.cell.2005.01.018. [DOI] [PubMed] [Google Scholar]
- Slavotinek A. M., et al. Mutation analysis of the MKKS gene in McKusick-Kaufman syndrome and selected Bardet-Biedl syndrome patients. Hum. Genet. 2002;110:561–567. doi: 10.1007/s00439-002-0733-3. [DOI] [PubMed] [Google Scholar]
- Slavotinek A. M., Stone E. M., Mykytyn K., Heckenlively J. R., Green J. S., Heon E., Musarella M. A., Parfrey P. S., Sheffield V. C., Biesecker L. G. Mutations in MKKS cause Bardet-Biedl syndrome. Nat. Genet. 2000;26:15–16. doi: 10.1038/79116. [DOI] [PubMed] [Google Scholar]
- Spiess C., Meyer A. S., Reissmann S., Frydman J. Mechanism of the eukaryotic chaperonin: protein folding in the chamber of secrets. Trends Cell Biol. 2004;14:598–604. doi: 10.1016/j.tcb.2004.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoetzel C., et al. BBS10 encodes a vertebrate-specific chaperonin-like protein and is a major BBS locus. Nat. Genet. 2006;38:521–524. doi: 10.1038/ng1771. [DOI] [PubMed] [Google Scholar]
- Stoetzel C., et al. Identification of a novel BBS gene (BBS12) highlights the major role of a vertebrate-specific branch of chaperonin-related proteins in Bardet-Biedl syndrome. Am. J. Hum. Genet. 2007;80:1–11. doi: 10.1086/510256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone D. L., Slavotinek A., Bouffard G. G., Banerjee-Basu S., Baxevanis A. D., Barr M., Biesecker L. G. Mutation of a gene encoding a putative chaperonin causes McKusick-Kaufman syndrome. Nat. Genet. 2000;25:79–82. doi: 10.1038/75637. [DOI] [PubMed] [Google Scholar]
- Tam S., Geller R., Spiess C., Frydman J. The chaperonin TRiC controls polyglutamine aggregation and toxicity through subunit-specific interactions. Nat. Cell Biol. 2006;8:1155–1162. doi: 10.1038/ncb1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateishi Y., Kawabe Y., Chiba T., Murata S., Ichikawa K., Murayama A., Tanaka K., Baba T., Kato S., Yanagisawa J. Ligand-dependent switching of ubiquitin-proteasome pathways for estrogen receptor. EMBO J. 2004;23:4813–4823. doi: 10.1038/sj.emboj.7600472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G., Vainberg I. E., Tap W. D., Lewis S. A., Cowan N. J. Specificity in chaperonin-mediated protein folding. Nature. 1995;375:250–253. doi: 10.1038/375250a0. [DOI] [PubMed] [Google Scholar]
- Yen H. J., Tayeh M. K., Mullins R. F., Stone E. M., Sheffield V. C., Slusarski D. C. Bardet-Biedl syndrome genes are important in retrograde intracellular trafficking and Kupffer's vesicle cilia function. Hum. Mol. Genet. 2006;15:667–677. doi: 10.1093/hmg/ddi468. [DOI] [PubMed] [Google Scholar]
- Yokota S., Yanagi H., Yura T., Kubota H. Cytosolic chaperonin is up-regulated during cell growth. Preferential expression and binding to tubulin at G(1)/S transition through early S phase. J. Biol. Chem. 1999;274:37070–37078. doi: 10.1074/jbc.274.52.37070. [DOI] [PubMed] [Google Scholar]
- Young J. C., Agashe V. R., Siegers K., Hartl F. U. Pathways of chaperone-mediated protein folding in the cytosol. Nat. Rev. Mol. Cell Biol. 2004;5:781–791. doi: 10.1038/nrm1492. [DOI] [PubMed] [Google Scholar]
- Younger J. M., Chen L., Ren H. Y., Rosser M. F., Turnbull E. L., Fan C. Y., Patterson C., Cyr D. M. Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell. 2006;126:571–582. doi: 10.1016/j.cell.2006.06.041. [DOI] [PubMed] [Google Scholar]
- Younger J. M., Ren H. Y., Chen L., Fan C. Y., Fields A., Patterson C., Cyr D. M. A foldable CFTR{Delta}F508 biogenic intermediate accumulates upon inhibition of the Hsc70-CHIP E3 ubiquitin ligase. J. Cell Biol. 2004;167:1075–1085. doi: 10.1083/jcb.200410065. [DOI] [PMC free article] [PubMed] [Google Scholar]