Abstract
Mitosis in animals starts with the disassembly of the nuclear pore complexes and the breakdown of the nuclear envelope. In contrast to many fungi, the corn smut fungus Ustilago maydis also removes the nuclear envelope. Here, we report on the dynamic behavior of the nucleoporins Nup214, Pom152, Nup133, and Nup107 in this “open” fungal mitosis. In prophase, the nuclear pore complexes disassembled and Nup214 and Pom152 dispersed in the cytoplasm and in the endoplasmic reticulum, respectively. Nup107 and Nup133 initially spread throughout the cytoplasm, but in metaphase and early anaphase occurred on the chromosomes. In anaphase, the Nup107-subcomplex redistributed to the edge of the chromosome masses, where the new envelope was reconstituted. Subsequently, Nup214 and Pom152 are recruited to the nuclear pores and protein import starts. Recruitment of nucleoporins and protein import reached a steady state in G2 phase. Formation of the nuclear envelope and assembly of nuclear pores occurred in the absence of microtubules or F-actin, but not if both were disrupted. Thus, the basic principles of nuclear pore complex dynamics seem to be conserved in organisms displaying open mitosis.
INTRODUCTION
In interphase, the nuclear envelope surrounds the chromosomes and separates the nucleoplasm from the cytoplasm. Communication between the chromosomes and the cytoplasm is mediated by nuclear pore complexes (NPCs), large protein assemblies that provide gates for controlled exchange of mRNA and proteins (Rout et al., 2000). Although the number of proteins in assembled NPCs from yeast and animal cells might be different, the basal architecture and function of nuclear pores is evolutionarily conserved (Mans et al., 2004; Devos et al., 2006). However, the importance of several nucleoporins in yeast and animals differs in a few respects. For example, components of the Nup107-160 complex, e.g., nucleoporins Nup107 and Nup133, play important roles in NPC formation after mitosis and they are essential proteins in animal cells (Harel et al., 2003; Walther et al., 2003). In contrast, the Nup107 homologue Nup84p is not essential in the yeast Saccharomyces cerevisiae (Pemberton et al., 1995; Siniossoglou et al., 1996) and in the filamentous fungus Aspergillus nidulans (Osmani et al., 2006).
In animal cells, the nuclear envelope breaks down at the onset of mitosis (Margalit et al., 2005). The removal of the envelope is accompanied by the disassembly of the nucleoporins, which either disperse in the endoplasmic reticulum (ER; Ellenberg et al., 1997; Yang et al., 1997) or are released into the cytoplasm (Rabut et al., 2004). A subcomplex of nucleoporins that includes Nup107 and Nup133 is recruited to the chromosomes and the kinetochores in metaphase (Belgareh et al., 2001; Loiodice et al., 2004). The role of these nucleoporins at the kinetochores is not fully understood, but it seems to be involved in spindle assembly (Orjalo et al., 2006; Zuccolo et al., 2007). Later in mitosis, the Nup107-160 complex leaves the chromosomes and it is thought to participate in NPC reassembly (Walther et al., 2003; Margalit et al., 2005).
In contrast to animals, in the fungi S. cerevisiae and Schizosaccharomyces pombe the mitotic spindle assembles inside the intact nuclear envelope (De Souza et al., 2004; Sazer, 2005). During this “closed” mitosis, the NPCs remain largely assembled and mediate import of factors needed for mitotic progression (Makhnevych et al., 2003). In the filamentous fungus A. nidulans, the nuclear envelope also persists, but the NPCs partially disassemble (De Souza et al., 2004). However, the scaffold component Nup107 remains part of the incomplete NPC (Osmani et al., 2006).
Our laboratory recently reported that a dynein-based mechanism removes the mitotic nuclear envelope in the basidiomycete fungus Ustilago maydis (Straube et al., 2005). The nuclear envelope ruptures at the tip and remains in the mother cell, whereas the chromosomes leave the old envelope and migrate into the daughter cell, where the spindle forms. Thus, similar to animal cells, this fungus undergoes an “open” mitosis. Here, we investigated the dynamics of several NPC components during this process. We found that NPCs disassemble at the beginning of mitosis and that Nup107-160 components cycle between the nuclear envelope and the chromosomes. Our data show that the basic principles of nucleoporin dynamics are conserved between U. maydis and animal cells and provide insight into the evolution of open and closed forms of mitosis.
MATERIALS AND METHODS
Sequence Analysis
U. maydis nucleoporin homologues were identified in the PubMed (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi) and MIPS (http://mips.gsf.de/genre/proj/ustilago/) public databases by using BLAST with the respective homologues from S. cerevisiae and Homo sapiens (National Center for Biotechnology Information database). U. maydis protein sequences were compared with the protein sequences from S. cerevisiae and H. sapiens by using Lalign (http://www.ch.embnet.org/software/LALIGN_form.html). Protein domains were analyzed by SMART (http://smart.embl-heidelberg.de/).
Strains and Plasmids
Constructs used to visualize the endoplasmic reticulum (ER), nuclear import, α-tubulin, and histone 4 were described previously (Steinberg et al., 2001; Wedlich-Söldner et al., 2002; Straube et al., 2003, 2005). Analogously, a plasmid carrying monomeric red fluorescent protein (RFP) was fused N-terminally to calreticulin and C-terminally to the HDEL signal to visualize the ER. Nucleoporin fusion proteins were constructed by attaching green fluorescent protein (GFP), yellow fluorescent protein (YFP), or monomeric RFP, to the C terminus of the respective protein to GFP by homologous recombination. Correct integration was confirmed by Southern blot analysis. The inducible crg-promoter was introduced immediately upstream of nup107 in place of the presumed wild-type promoter. Correct integration was confirmed by Southern blot analysis according to standard protocols.
Western Blot Analysis
Protein extracts were obtained from Ustilago cells grown to OD600 <0.6 in their respective medium as described in Straube et al. (2001). Proteins were separated in 8% polyacrylamide gels and transferred to nitrocellulose membranes for 1 h at 400 mA in a wet blot chamber. Anti-GFP antibody (Roche Diagnostics, Mannheim, Germany) and horseradish peroxidase (HRP)-coupled anti-mouse antibody (Promega, Madison, WI) were used to address Nup107-GFP levels. Antibody detection followed standard procedures.
Growth Conditions
Strains were grown overnight at 28°C in CM liquid medium (Holliday, 1974) supplemented with 1% glucose (CM-G). Solid medium contained 2% (wt/vol) bacto-agar. Strains with nup107 under the control of the crg-promoter were propagated on CM agar plates containing 1% arabinose. These strains were incubated at 28°C for at least 16 h in CM-G. Control cultures were grown in CM containing 1% arabinose (CM-A).
Spore Analysis
To test whether nup107 is an essential gene, a diploid strain was created in which one copy of the gene was replaced with the nourseothricin-resistance cassette. Maize (Zea mays) plants were infected with an overnight culture supplemented with 0.5% Tween 20 before injection following a standard protocol (Fuchs et al., 2006). Plants were grown for 25 d at 28°C before harvesting tumors. Spores from these tumors were isolated as described in Fuchs et al., 2006. Spores were germinated on CM-G agar plates at 22°C. Fifty-seven spores were selected from these plates and placed on CM-G agar plates containing 150 μg ml−1 nourseothricin. Thirty-six colonies that grew in the presence of the antibiotic were subjected to open reading frame (ORF)-specific polymerase chain reaction (PCR) and Southern blot analysis. All colonies showed at least either PCR product or a wild-type band in the Southern blot analysis.
Staining Procedure
For 4,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich, St. Louis, MO) staining of nuclei, cells expressing nucleoporin-GFP fusion proteins and ER markers were fixed by incubation with 0.5% formaldehyde (Polysciences, Warrington, PA) for 10 min (Nup107-GFP and Pom152-GFP) or 2 min, respectively (Nup214-GFP). Cells were mounted on poly-l-lysine coverslips, incubated for 3 min, and washed in phosphate-buffered saline (PBS). Samples were then stained with 0.5 μg ml−1 DAPI for 15 min at 37°C, and washed several times in PBS before analysis.
Microscopy, Image Processing, and Quantitative Analysis
Cells were placed on a thin layer of 2% agarose, covered with a coverslip, and immediately observed using a Zeiss Axioplan II microscope (Carl Zeiss, Oberkochen, Germany). Epifluorescence was observed using filter sets for fluorescein isothiocyanate (FITC) (BP470/20, FT510, and BP515-565; Carl Zeiss), DsRed (BP565/30, FT585, and BP620/60; Carl Zeiss), YFP (BP500/20, FT515, and BP535/30; Carl Zeiss), and cyan fluorescent protein (CFP) (BP436, FT455, and BP480-500; Carl Zeiss) or for GFP (HC470/40, T495LP, and HC525/50; AHF, Tübingen, Germany) and Texas-Red (HC562/40, BS 593, and HC624/40; AHF). Frames were taken with a cooled coupled device camera (CoolSNAP HQ, Photometrics, Tucson, AZ) controlled by MetaMorph software (Molecular Devices, Sunnyvale, CA). Fluorescence measurements, line-scan analyses, and image processing, including adjustment of brightness, contrast, and γ-values, were carried out with MetaMorph (Molecular Devices) and Photoshop (Adobe Systems, San Jose, CA). For colocalization studies of nucleoporins, cells were fixed with 0.4% formaldehyde and 0.1% glutaraldehyde (Polysciences).
To obtain Z-stacks, cells were fixed with 0.5% formaldehyde (Polysciences) and imaged every 0.5 μm. Z-streams of nuclei were processed by 100 iterations of three-dimensional (3-D) deconvolution; maximum projections, and 3-D reconstructions of the resulting images were created using AutoQuantX software (AutoQuant Imaging, Troy, NY). To determine the amount of nucleoporin-GFP fusion proteins, the integrated intensity of fluorescence signal in the nuclear envelope was measured and the cytoplasmic background fluorescence was subtracted. The same method was used for measuring NLS-3xRFP and NPC reassembly in telophase. The fluorescence signal in the cytoplasm during mitosis was determined by average intensity measurements. The Nup107-GFP signal on the DNA in metaphase was measured against the fluorescence intensity of the cytoplasmic background. Statistical analysis by two-tailed t test at α = 0.05 used Prism software (GraphPad Software, San Diego, CA). All values are given as means ± SD. Prism software was also used to generate regression lines.
Inhibitor Studies
Benomyl-induced depolymerization of microtubules was monitored by placing 1 μl of logarithmically growing cells on a 2% agarose cushion containing 30 μM benomyl (Sigma Chemie, Taufkirchen, Germany) as described previously (Fink and Steinberg, 2006). Analogously, depolymerization of the actin cytoskeleton was achieved by adding 10 μM latrunculin A (kindly provided by Dr. P. Crews, University of California, Santa Cruz, Santa Cruz, CA) to the agarose cushion. Cells in anaphase were identified under the microscope by the cloud-like appearance of Nup107-GFP/Nup133-GFP in the daughter cell or completed nuclear envelope breakdown in the case of strains expressing Nup214-GFP or Pom152-GFP, and the first image pair taken. The second image pair was obtained 7 min after exposure of cells to the inhibitor. A third image pair was taken at 20 min of inhibitor treatment to observe effects of inhibitor treatment on NPC reassembly and functionality.
RESULTS
Nucleoporins in U. maydis Exhibit Homology to Those in Budding Yeast and Human Cells
In the fungus U. maydis, the nuclear envelope remains in the mother cell during mitosis, whereas chromosomes migrate into the daughter bud, where the spindle forms within the cytoplasm (Straube et al., 2005; Supplemental Movie 1). As a first step toward an understanding of the behavior of NPCs in this fungal open mitosis, we set out to identify nucleoporins by screening the published U. maydis genome with sequences of known NPC proteins from H. sapiens (Cronshaw et al., 2002) and S. cerevisiae (Rout et al., 2000). Twenty putative nucleoporins with either significant sequence similarity to both organisms (BLAST value <1.0e−05) or high similarity to either yeast or human (BLAST value <1.0e−20; Table 2) were found. One protein, UM06099, exhibited only weak sequence similarity to its human counterpart Nup188 (BLAST value <1.0e−04) and was therefore excluded from the table. Two putative nucleoporins (Ndc1, UM06416; Nup88, UM00333; all annotations according to MIPS http://mips.gsf.de/genre/proj/ustilago/) were only found when nucleoporins from A. nidulans or S. pombe were compared (UM06416, BLAST value <1.0e−04; UM00333, BLAST value <1.0e−14; not included in Table 2).
Table 2.
Comparison of nucleoporins in U. maydis, S. cerevisiae, and H. sapiens
| U. maydis | S. cerevisiae | H. sapiens | Protein domaina |
|---|---|---|---|
| um04509 | Nsp1p (25.9) | Nup62 (28.4) | C-terminal Nsp1_C domain |
| um05489 | Nup1p (21.4) | Nup153 (21.3) | |
| um02688 | Nup2p (18.2) | Nup50 (14.3) | C-terminal Ran binding domain |
| um02245 | Sec13p (45.3) | Sec13R (45.7) | WD repeats |
| um01308 | Nup42p (26.5) | NupL2 (21.9) | N-terminal zinc finger |
| um01418 | Nup49p (23.8) | NupL1 (20.4) | Coiled coil domain |
| um11701 | Nup57p (23.4) | Nup54 (17.6) | |
| um04795 | Nup84p (21.2) | Nup107 (23.6) | Nup84_Nup100 domain |
| um04624 | Nup85p (16.9) | Nup85 (22.6) | N-terminal Nup85 domain |
| um03813 | Nic96p (27.3) | Nup93 (27.3) | NIC domain |
| um00639 | Nup120p (16.5) | Nup160 (19.7) | |
| um02855 | Nup133p (15.9) | Nup133 (18.5) | C-terminal Nup133 domain |
| um05158 | Nup100p (18.3) | Nup98-96 (25.5) | Central nucleoporin 2 domain |
| Nup116p (18.8) | |||
| N/C-Nup145 (19.0) | |||
| um03853 | Nup157p (20.2) | Nup155 (24.2) | Nup170 domain |
| Nup170p (20.9) | |||
| um01089 | Nup159p (16.2) | Nup214 (20.2) | |
| um02524 | Nup192p (15.9) | Nup205 (21.5) | |
| um01075 | Mlp1 (18.9) | TPR (18.5) | C-terminal TPR_Mlp1_2 domain |
| Mlp2 (19.6) | |||
| um03950 | Yrb2p (24.7) | RanBP3 (21.3) | C-terminal Ran binding domain |
| um03762 | Gle2p (34.7) | RAE1 (37.3) | WD repeats |
| um03963 | Pom152p (22.5) | 3 N-terminal TM domains |
Sequences of full-length U. maydis proteins were compared with sequences of S. cerevisiae and H. sapiens in NCBI databases. Percentage of identity of full-length U. maydis proteins to their respective homologues is given in parentheses.
a Protein domains were predicted by the SMART server.
As expected, we found that many putative nucleoporins found in U. maydis are similar to their homologues in S. cerevisiae (Nup2, Nup42, Nup49, Nup57, and Yrb2; Table 2), and one putative nucleoporin was only represented in yeast (Pom152; Table 2), but not in the human proteome. However, numerous putative U. maydis proteins showed unexpected high sequence similarity with their human homologues (e.g., Nup107 shares 24% identity with human Nup107, but only 21% with Nup84p from S. cerevisiae; Table 2). Most of these nucleoporins, including Nup107, Nup133, Nup160, Nup96, Nup85, and Sec13, are expected to be part of the Nup107-160 complex (Siniossoglou et al., 2000; Lutzmann et al., 2002; Berke et al., 2004).
To investigate NPC dynamics in the open mitosis of U. maydis, we chose to observe Nup107 (UM04795, Table 1), the cytoplasm-facing Nup214 homologue (UM01089), and the transmembrane domain-anchored Pom152-like protein (UM03963). These predicted proteins share significant sequence similarity and a comparable domain structure with their homologues in both human and budding yeast (Figure 1A), indicating that they are part of the NPC. However, it is important to note that no Pom152 homologue exists in vertebrates, but the Pom121 protein (depicted in Figure 1) may execute a similar function (Mans et al., 2004).
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or phenotype | Reference |
|---|---|---|
| FB1ERY_H4C | a1b1/pERYFP/pH4CFP | Straube et al. (2005) |
| FB2N107G_ER | a2b2 nup107-gfp, bleR/pERRFP | This study |
| FB2P152G_ER | a2b2 pom152-gfp, bleR/pERRFP | This study |
| FB2N214G_ER | a2b2 nup214-gfp, hygR/pERRFP | This study |
| FB2N133G_ER | a2b2 nup133-gfp, natR/pERRFP | This study |
| FB2N2G_ER | a2b2 nup2-gfp, hygR/pERRFP | This study |
| FB2N107R-N214G | a2b2 nup107-mrfp, bleRnup214-gfp, hygR | This study |
| FB2N107G-P152R | a2b2 nup107-gfp, bleRpom152-rfp, hygR | This study |
| FB2P152G_214R | a2b2 nup214-mrfp, hygRpom152-gfp, bleR | This study |
| FB2 | a2b2 | Banuett and Herskowitz (1989) |
| FBD11 | a1b1 a2b2 | Banuett and Herskowitz (1989) |
| FBD11ΔN107 | a1b1 a2b2 Δnup107:: natR | This study |
| FB2N107Y_EC | a2b2 nup107-yfp, bleR/pERCFP | This study |
| FB2N107Y_H4C | a2b2 nup107-yfp, bleR/pH4CFP | This study |
| FB2N107R_Mis12G | a2b2 nup107-mrfp, bleR/pMis12-3xgfp, natR | This study |
| FB2N107R_Nup133G | a2b2 nup107-mrfp, bleRnup133-gfp, natR | This study |
| FB2N107R_H4G | a2b2 nup107-mrfp, bleR/pH4GFP | This study |
| FB2N214R_H4G | a2b2 nup214-mrfp, hygR/pH4GFP_C | This study |
| FB2YT1-H4C | a2b2/pYFPTub1/pH4CFP | Straube et al. (2005) |
| FB2N107Y_CT | a2b2 nup107-yfp, bleR/pCFPTub1 | This study |
| FB2N107G_RT | a2b2 nup107-gfp, bleR/pN_RFPTub1 | This study |
| FB2nRFP_ERG | a2b2/pC_NLS3xRFP/pN_ERGFP | Straube et al. (2005) |
| FB2P152G_nR | a2b2/pC_NLS3xRFP pom152-gfp, hygR | This study |
| FB2N107G_nR | a2b2 nup107-gfp, bleR/pH_NLS3xRFP | This study |
| FB2N214G_nR | a2b2/pC_NLS3x RFP nup214-gfp, hygR | This study |
| FB2Fim2G | a2b2 fim2-gfp, bleR | This study |
| FB2rN107G | a2b2 Pcrg-nup107-gfp, bleR hygR | This study |
| FB2rN107_N214G_nR | a2b2 Pcrg-nup107, bleR, nup214-gfp, hygR/pC_NLS3xRFP | This study |
| pERYFP | Potef-cals-yfp-HDEL, natR | Straube et al. (2005) |
| pH4CFP | Potef-his4-cfp, hygR | Straube et al. (2005) |
| pERRFP | Potef-cals-mrfp-HDEL, cbxR | This study |
| pERCFP | Potef-cals-cfp-HDEL, cbxR | Adamikova et al. (2004) |
| pC_NLS3 × RFP | Potef-gal4s-mrfp-mrfp-mrfp, cbxR | Straube et al. (2005) |
| pH_NLS3 × RFP | Potef-gal4s-mrfp-mrfp-mrfp, hygR | Straube et al. (2005) |
| pH4GFP | Potef-his4-gfp, hygR | Straube et al. (2005) |
| pH4GFP_C | Potef-his4-gfp, cbxR | This study |
| pH4CFP | Potef-his4-cfp, hygR | Straube et al. (2005) |
| pH4CFP_C | Potef-his4-cfp, cbxR | This study |
| pYFPTub1 | Potef-yfp-tub1, cbxR | Straube et al. (2005) |
| pCFPTub1 | Potef-cfp-tub1, hygR | Wedlich-Soldner et al. (2002) |
| pN_RFPTub1 | Potef-2xmrfp-tub1, natR | This study |
| pERRFP_H | Potef-cals-mrfp-HDEL, hygR | This study |
| pERGFP | Potef-cals-gfp-HDEL, cbxR | Wedlich-Soldner et al. (2002) |
| pN_ERGFP | Potef-cals-gfp-HDEL, natR | Straube et al. (2005) |
| pN107Y | nup107-yfp, bleR | This study |
| pN107G | nup107-gfp, bleR | This study |
| pN107R | nup107-mrfp, bleR | This study |
| pP152G_B | pom152-gfp, bleR | This study |
| pP152G_H | pom152-gfp, hygR | This study |
| pP152R_B | pom152-mrfp, bleR | This study |
| pP152R_H | pom152-mrfp, hygR | This study |
| pN214G | nup214-gfp, hygR | This study |
| pN214R | nup214-mrfp, hygR | This study |
| pN133G | nup133-gfp, natR | This study |
| pN2G | nup2-gfp, hygR | This study |
| pMis12G | mis12-3xgfp, natR | This study |
| pDnup107 | nup107::natR | This study |
| Pcrgnup107 | Pcrg-nup107, bleR | This study |
a, b, mating type loci; Δ, deletion; P, promoter; ::, homologous replacement; -, fusion; hygR, hygromycine resistance; bleR, phleomycine resistance; natR, nourseothricin resistance; cbxR, carboxin resistance; /, ectopically integrated; otef, constitutive promoter; crg, conditional arabinose-induced promoter; gfp, green fluorescent protein; cfp, cyan fluorescent protein; yfp, yellow-shifted fluorescent protein; mrfp, monomeric red fluorescent protein; tub1, α -tubulin; his4, histone 4; nup107, pom152, nup214, nup133, nup2 nucleoporins; mis12 kinetochore marker; cals, signal sequence of calreticulin; HDEL, ER retention signal.
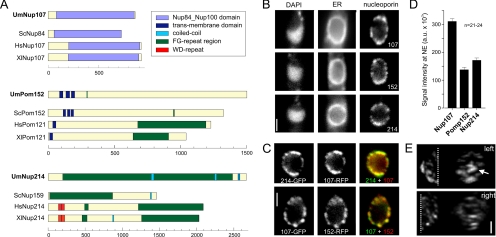
Figure 1.
Nucleoporins in U. maydis. (A) U. maydis homologues of nucleoporins were identified by BLAST search, and domain models were predicted by the SMART server. Note that Pom121 is no homologue of Pom152, but it may execute similar functions. Um, Ustilago maydis; Sc, S. cerevisiae; Hs, Homo sapiens; and Xl, Xenopus laevis. (B) GFP-fusion proteins of U. maydis Nup107, Nup214, and Pom152 localize to the nuclear envelope, which is visualized by monomeric RFP targeted to the ER. DNA is stained with DAPI. Cells were briefly fixed with 0.5% formaldehyde before observation. Bar, 1.5 μm. (C) Nup107, Nup214, and Pom152 proteins were either fused to GFP or monomeric RFP. Pom152 and Nup214 colocalize with Nup107, indicating that they are part of the same structures. Cells were fixed with 0.4% formaldehyde and 0.1% glutaraldehyde before observation. Bar, 1.5 μm. (D) Stoichiometry of nucleoporins. The signal intensities of Nup107-GFP, Pom152-GFP, and Nup214-GFP in the nuclear envelope were quantified. The nuclear envelope was identified by the presence ER-RFP (strains FB2N107_ER, FB2P152_ER, and FB2N214_ER). All values are mean value ± SD. (E) Distribution of NPCs, labeled with Nup107-GFP, in the nuclear envelope. Cells were fixed with 0.5% formaldehyde; images were taken in z-axis at 0.5-μm intervals. Image stacks were deconvolved, and the individual halves of the nucleus are presented as maximum projections in xy- or as yz-projection. Arrow points to an accumulation of Nup107-signals (yz-projection), which is found on one side of the nucleus. Bar, 1 μm. Also see Supplemental Material.
Homologues of Nup107, Nup214, and Pom152 Colocalize at the Nuclear Envelope
To test whether our chosen proteins are indeed nucleoporins, we fused a single GFP to the C terminus of the endogenous Nup107, Nup214, and Pom152 proteins and coexpressed these with a monomeric RFP targeted to the ER by a C-terminal retention signal (Wedlich-Söldner et al., 2002; strains FB2N107G_ER, FB2N214G_ER, FB2P152G_ER). All GFP-fusion proteins localized to the nuclear envelope, which surrounded the interphase DNA stained with DAPI (Figure 1B). Coexpression of Nup107-RFP and Nup214-GFP (strain FB2N107R-N214G) and Nup107-GFP and Pom152-RFP (strain FB2N107G-P152R) confirmed that the fusion proteins colocalize (Figure 1C). A quantitative analysis of the integrated signal intensity within the nuclear envelope in a central focal plane indicated a Nup107:Pom152:Nup214 stoichiometry of 2:1:1 (Figure 1D; strains FB2N107G_ER, FB2N214G_ER, and FB2P152G_ER). We also noted that all nucleoporins distributed unevenly within the nuclear envelope, and they had a tendency to form large aggregates, which became most obvious in deconvolved Z-stacks (Figure 1E, arrow; and Supplemental Movie 2). Thus, our data suggest that the analyzed nucleoporins accumulate in the same NPC cluster.
Nucleoporins Disperse in Mitosis
Mitosis in U. maydis begins with the elongation of the nucleus. After it reaches into the daughter cell, chromosomes start to condense and migrate within the closed nuclear envelope into this nuclear extension (O'Donell and McLaughlin, 1984a; Straube et al., 2005). At this stage Nup107 and Pom152 colocalize (Supplemental Movie 3), and the same was found for Nup107 and Nup214 (data not shown), which suggested that NPC structure is largely intact in the prophase nucleus.
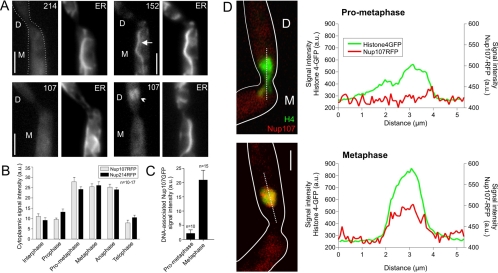
At the end of prophase, the chromosomes leave the old envelope, which collapses and falls back into the mother cell. The peripheral component Nup214 dispersed evenly in the cytoplasm, where it remained throughout mitosis (Figure 2, A and B; strains FB2N214G_ER and FB2N214R_H4G; and Supplemental Movie 4). The transmembrane domain-anchored Pom152-GFP stayed associated with ER membranes and diffusely labeled the collapsed envelope (Figure 2A, arrowhead; strain FB2P152G_ER; and Supplemental movie 5). When chromosomes left the old envelope in prophase, Nup107 dispersed evenly in the cytoplasm (Figure 2, A and B; strains FB2N107G_ER and FB2N107R_H4G; see Supplemental Movie 6). However, in metaphase, a fraction of Nup107-RFP appears at the condensed chromosomes (Figure 2, C and D; strain FB2N107R_H4G), whereas most Nup107-RFP remained cytoplasmic (Figure 2B).
Figure 2.
Localization of nucleoporins in mitosis. (A) In late prometaphase, the nuclear envelope falls back into the mother cell. Nup214-GFP leaves the envelope (214), whereas Pom152-GFP (152) disperses in the ER, with the most prominent signal in the old nuclear envelope (arrow). Nup107-GFP (107) first distributes into the cytoplasm, but it rapidly reappears in the daughter cell (arrowhead). The outline of the cell is indicated by a profile in the first image. D, daughter cell; M, mother cell. Bar, 3 μm. (B) Average signal intensities of Nup107-RFP and Nup214-RFP within the cytoplasm. Mitotic stages were identified by coexpression of histone 4-GFP (strains FB2N107R_H4G and FB2N214R_H4G). Values are mean ± SD. Sample size is indicated. (C) Average signal intensity of Nup107-GFP on the DNA over background cytoplasmic fluorescence in prometaphase and metaphase. Values are mean ± SD. Sample size is indicated. (D) Line-scan analyses of prometaphase and metaphase cells demonstrate that Nup107 associates with DNA after chromosome migration is completed and cells enter metaphase. The outline of the cells is indicated by a profile. D, daughter cell; and M, mother cell. Bar, 2 μm.
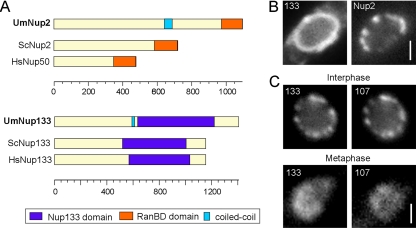
These results suggested that the NPC disassembles in prophase. To gain further evidence for this assumption, we tagged the putative homologues of Nup2 and Nup133 (UM02688 and UM02855), which again share similar domain structures with their counterparts in human cells and budding yeast (Figure 3A and Table 2). The C-terminal fusion proteins also localized to the nuclear envelope in interphase (Figure 3, B and C; strains FB2N2G_ER and FB2N107R_Nup133G). Both nucleoporins were released from the envelope at the onset of mitosis (Supplemental Movie 7), again confirming the idea that the nuclear pore disassembles when the chromosomes move into the daughter cell. However, similar to Nup107 the Nup133-like nucleoporin occurred at the chromosomes in metaphase and anaphase (Figure 3C, and Supplemental Movie 8). In both yeast and animal cells, Nup133 and Nup107 are part of the Nup107-160 subcomplex (Siniossoglou et al., 2000; Vasu et al., 2001; Lutzmann et al., 2002); therefore, we consider it likely that a similar Nup107/Nup133-containing protein complex is recruited to the chromosomes in U. maydis.
Figure 3.
Nup107 subcomplex components colocalize in U. maydis metaphase. (A) The U. maydis homologues of Nup133 and Nup2 were identified by BLAST search, and the domain models were constructed according to SMART predictions (Pfam Nup133, BLAST value = 1.0 e−06; and RanBD, BLAST value = 2.9 e−09). Um, U. maydis; Sc, S. cerevisiae; Hs, H. sapiens. (B) Nup2-GFP localizes to the nuclear envelope, identified by ER-RFP, in a punctuate pattern like the other nucleoporin fusion proteins investigated. Bar, 1 μm. (C) The U. maydis homologue of Nup133 colocalizes with nuclear pore component Nup107 in interphase at the rim of the nucleus and in mitosis (metaphase) on the DNA in the bud. Bar, 1 μm.
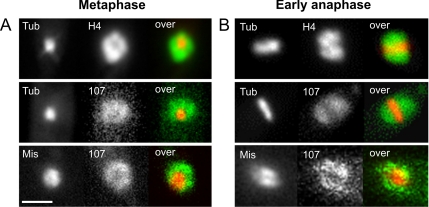
In animal cells, it has been described that the Nup107-160 subcomplex associates with the kinetochores (Loiodice et al., 2004). However, in U. maydis Nup107-GFP diffusely localized on the entire chromosome mass that surrounds the spindle in metaphase and early anaphase (Figure 4, A and B; chromosomes were labeled by histone4-CFP [H4], microtubules were visualized by YFP-α-tubulin [Tub]; strains FB2YT1-H4C and FB2N107Y_CT). This raised doubt about a recruitment of Nup107-160 complex to the fungal kinetochores. Therefore, we generated a strain that contained Nup107-RFP and a fusion protein of the U. maydis homologue of kinetochore protein Mis12 (UM04180; 14% identity to human Mis12 and 21% to Mtw1 from S. cerevisiae), fused to 3xGFP (strain FB2N107R_Mis12G; Figure 4, A and B). Indeed, the kinetochore marker only partially overlaps with Nup107. Thus, Nup107 localizes to the chromosomal DNA, rather than specifically to kinetochores.
Figure 4.
Nup107 in U. maydis does not colocalize with kinetochores in mitosis. (A) In metaphase, a short spindle, lableded by YFP-αtubulin (Tub), is formed in the daughter cell that is surrounded by the condensed chromosomes, labeled by histone 4-CFP (H4). Nup107-YFP (107) also encompassed the metaphase spindle. Nup107-RFP (107) does not seem to colocalize with the kinetochore protein Mis12 (Mis), labeled with 3xGFP. Bar, 2 μm. (B) In anaphase A, the spindle (Tub) slowly elongates, and the chromosomes (H4) align along the spindle. Nup107-YFP (107) shows a similar distribution, indicating that the nucleoporin remains at the DNA during early anaphase. Nup107-RFP again does not colocalize with Mis12-3xGFP (Mis).
Nup107 Occurs at the Edge of the Separating DNA in Anaphase B
In anaphase A, a short spindle forms; the spindle rapidly elongates in anaphase B, thereby segregating the chromosomes between the mother and the daughter cell (Fink et al., 2006). The new envelopes are reestablished from the distal rim of the separating chromosome masses (Straube et al., 2005). Interestingly, spindle elongation seemed to coincide with the recruitment of existing ER to the condensed chromosomes (Figure 5A, arrow; DNA is stained with histone 4 CFP and the ER with YFP-HDEL; strain FB1ERY_H4C; and Supplemental Movies 9 and 10). While the spindle elongated, increasing amounts of Nup107-RFP accumulated at the rim of the dividing DNA (Figure 5B; strain FB2N107R_H4G), whereas minor traces were still found on the separating chromosomes (Figure 5C, arrow). First, patches of Nup107 occurred in late anaphase (Figure 5B). In contrast, Nup214 and Pom152 levels above background in the cytoplasm/endoplasmic reticulum were only detectable after nuclear envelopes (NEs) had closed and began to expand with a circumference of >4 μm (Figure 5D; strains FB2P152G_ER and FB2N214G_ER). Thus, our experiments suggest that Nup107 is the first of the markers investigated that returns to the nuclear envelope. A similar order of addition has been found in vertebrate cells (Harel et al., 2003; Walther et al., 2003). However, we cannot exclude that undetectable traces of other nucleoporins arrive with Nup107 at the nuclear envelope.
Figure 5.
Nup107 rearrangement in anaphase. (A) As mitosis progresses through anaphase, the outer rims of the DNA masses (labeled by histone 4-CFP; H4) seem to collect ER membranes (marked by YFP with an ER retention signal) that are later incoporated into the newly forming nuclei (arrow). The outline of the cell is indicated by a profile in the first image. D, daughter cell; and M, mother cell. Time is given in minutes. Bar, 3 μm. Also see Supplemental Material. (B) Colocalization of Nup107-RFP (107) and histone 4-GFP-labeled chromosomes (H4) shows that the nucleoporin leaves the DNA and occurs at the outward edge of the separating chromosome masses during spindle elongation. In late anaphase B, Nup107 concentrates in small areas. Bar, 3 μm. (C) Line-scan analysis of an anaphase B spindle demonstrates that traces of Nup107-RFP are still found on the chromosomes (arrow), whereas most of the protein concentrates at the poles of the separating DNA. Bar, 3 μm. (D) In anaphase B, the new envelope begins to form at the outward rim of the chromosomes. Although Nup107 becomes incorporated into the newly forming NE, the other nucleoporins (Nup214, Pom152) are not yet concentrated at the new NE. Bar, 1.5 μm. Also see Supplemental Material.
In human cells and Xenopus laevis egg extracts depletion of Nup107-160 leads to failure to incorporate NPCs into nuclear envelopes (Harel et al., 2003; Walther et al., 2003). To test whether Nup107 is essential in U. maydis, we used a genetic analysis using a viable diploid strain that contained only one wild-type allele of nup107. Analysis of the offspring after plant infection revealed that of 57 germinated strains none carried the deletion. This result strongly suggests that nup107 is essential in U. maydis. To confirm this finding, we generated a conditional promoter mutant of nup107 by using the crg-promoter, which is repressed in the presence of glucose (Bottin et al., 1996). Although low levels of Nup107-GFP persisted in these mutants under restrictive conditions, cells showed clustering of NPCs and morphological defects (Supplemental Figure 1; strain FB2rN107G).
Ordered Reassembly of NPCs in Telophase
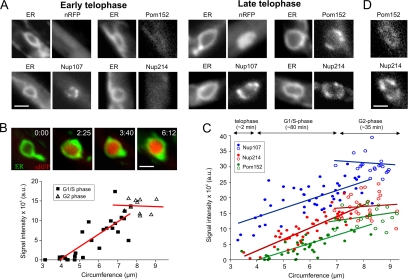
In anaphase, membranes are recruited to form the new envelopes. Nup107 occurred at the poles of the dividing chromosome masses. At this stage, Nup214 was still undetectable and Pom152 only diffusely labeled the envelopes (Figure 6A; strains FB2nRFP_ERG, FB2N107G_ER, FB2N214G_ER, and FB2P152G_ER; and Supplemental Movies 11 and 12). Subsequently, Nup214-RFP appeared slightly before Pom152-GFP levels increased above background ER fluorescence (Figure 6, C and D; strain FB2P152G_214R; and Supplemental Movie 13). All nucleoporins continuously accumulated until they reached a maximum in G2 phase (Figure 6C; strains FB2N107G_nR, FB2N214G_nR, and FB2P152G_nR; Supplemental Movie 14). Protein import, monitored by the appearance of the nlsRFP reporter protein, was not found until the diffusely distributed Pom152 started to accumulate in NPCs (Figure 6A, nlsRFP; and Supplemental Movie 14), which did not occur before NE closure and the onset of NE expansion. Import continued while the nuclei enlarged during chromosomes decondensation (Figure 6B; strain FB2nRFP_ERG) and a steady-state level was reached in cells with growing buds, which are considered to be in G2 phase (Snetselaar and McCann, 1997; Figure 6B). Together, these data suggest that nuclear pores assemble in a stepwise manner, with Nup107 being the first of the nucleoporins investigated at the new forming envelope. However, nuclear import begins in telophase, when the envelope closes (O'Donell and McLaughlin, 1984b) and all nucleporins started to accumulate in the NPCs.
Figure 6.
Ordered reassembly of NPCs in telophase. (A) The new envelope closes in telophase (O'Donell and McLaughlin, 1984b). At this stage, Nup107 concentrates in the NE, whereas only traces of Nup214 and Pom152 are found. Note that low levels of Pom152 are present all over the ER. The nuclear marker NLS-3xRFP (nRFP) does not yet occur in the lumen of the small envelopes, indicating that protein import has not yet started. Bar, 1.5 μm. (B) As the chromosomes decondense, NLS-3xRFP occurs in the interior of the expanding nucleus, indicating that active protein import takes place. Nuclear import proceeds until cells begin to grow and form apical buds (integrated intensity; triangles), which indicates the beginning of G2 phase (Snetselaar and McCann, 1997; open symbols). Image series was taken from a single cell; the graph is based on the analysis of numerous nuclei whose circumference at a central plane was taken as an indication of their size and degree of chromosome decondensation. Elapsed time is given in minutes:seconds. Bar, 1.5 μm. (C) Quantitative analysis of the signal intensity of Nup107-GFP, Pom152-GFP, and Nup214-GFP (strains strains FB2N107G_ER, FB2N214G_ER, and FB2P152G_ER) in relation to the size of the nucleus, indicated by its circumference. An estimate of the timing and correlation to the cell cycle is presented above the graph. The amount of all nucleoporins continuously increases until cells begin to bud and reach G2 phase (open circles). (D) Colocalization of Pom152-GFP and Nup214-RFP in nuclei with a circumference of ∼4 μm shows that Pom152-GFP gives only a weak punctuate localization as indication of incorporation into NPCs, whereas the signal of Nup214-RFP is more pronounced (strain FB2P152G_214R), suggesting that Pom152 accumulates slightly before Nup214. Bar, 1 μm.
NPC Reassembly Proceeds Independently of Spindle Elongation
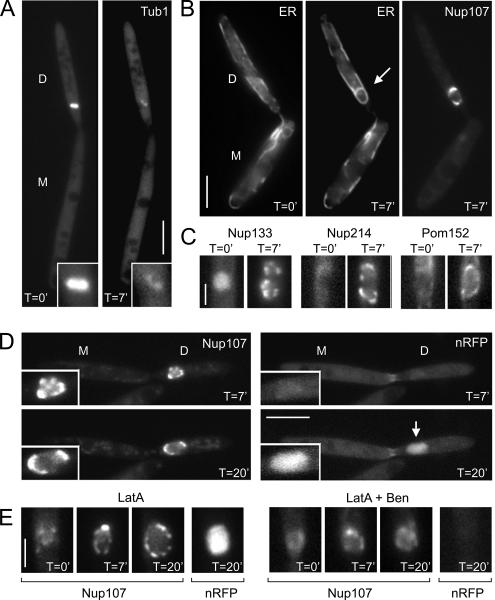
We also set out to gain further insight in the mechanism of nuclear envelope reassembly in living U. maydis cells. The formation of the new envelope seemed to involve recruitment of existing ER tubules by the elongating anaphase spindle (Figure 5A and Supplemental Movies 9 and 10), which is expected to involve microtubules. To investigate whether nuclear envelope formation depends on spindle elongation, we treated mitotic cells with benomyl, an inhibitor that disrupts fungal microtubules within 5 min (Fink and Steinberg, 2006). Unfortunately, we have no means to synchronize U. maydis; therefore, we selected large-budded cells from logarithmically growing cultures that were characterized by the presence of Nup107-GFP on chromosomes flanking a short spindle in the daughter cell (Figure 4B; also see Materials and Methods). These cells have entered anaphase (Fink and Steinberg, 2006); therefore, we expected that the benomyl treatment should not activate mitotic spindle assembly checkpoints that would halt all processes associated with cell cycle progression. After 7 min on a benomyl-containing cushion (see Materials and Methods for details), most of the RFP-tubulin fluorescence was cytoplasmic (Figure 7A). This confirmed previous data showing that this method leads to disassembly of the mitotic spindle microtubules in U. maydis (Fink and Steinberg, 2006). Consequently, chromosome segregation was abolished, and the DNA remained in the daughter cell (data not shown). Despite the absence of the microtubules and an elongating anaphase B spindle, a nuclear envelope was formed within 7 min (Figure 7B). This envelope contained nucleoporins (Figure 7C) and imported the nlsRFP reporter protein (Figure 7D), indicating that these new envelopes contained functional NPCs. Next, we tested for a role of F-actin in NE formation. We previously demonstrated that the inhibitor latrunculin A rapidly depolymerizes F-actin in U. maydis (Fuchs et al., 2005). In many fungi, phalloidin does not bind to F-actin (Heath et al., 2003), and this includes U. maydis (unpublished data). Therefore, we placed control cells with fimbrin-GFP labeled actin patches (strain FB2Fim2G; Castillo-Lluva et al., 2007) on agarose pads containing 10 μM latrunculin. This treatment led to disassembly of the fimbrin-GFP–stained actin patches within 5 min (data not shown), but it had no obvious effect on nuclear envelope or pore formation (Figure 7E, LatA). However, when cells were simultaneously treated with both 10 μM latrunculin A and 30 μM benomyl, Nup107 did not accumulate into dots, suggesting that functional pores did not assemble. Consequently, we observed no import of nRFP (Figure 7E, LatA + Ben). These results suggest that microtubules and F-actin cooperate in reformation of the NE, but neither component of the cytoskeleton is essential on its own.
Figure 7.
Role of the cytoskeleton in nuclear envelope reassembly. (A) Cells that express 2xRFP-Tub1 (Tub1) were placed on agarose cushions containing 30 μM benomyl. After 7 min (T = 7′), microtubules of the spindle are mainly disrupted (inset). D, daughter cell; and M, mother cell. Bar, 5 μm. (B) Anaphase cells are characterized by a cloud-like Nup107-GFP signal in the daughter cell that is divided by a cleft representing the spindle (see Figure 4). When placed on benomyl cushions (T = 0′), no ER membranes, marked by ER-RFP (ER), were surrounding the chromosomes. However, after 7-min treatment with benomyl (T = 7′), ER membranes surround the undivided chromosomes. These membranes are also decorated with a distinct Nup107-GFP signals, suggesting that they are newly formed NEs. Note that nuclear division and migration stopped in the absence of microtubules. Bar, 5 μm. (C) All nucleoporins tested reappear after 7 min of treatment with benomyl (T = 7′; strains FB2N133G_ER, FB2N214G_ER, and FB2P152G_ER). Bar, 2 μm. (D) In the absence of microtubules, chromosomes still decondese, and the Nup107-GFP containing nuclei enlarge. In agreement with the appearance of nucleoporins, protein import occurs, which is indicated by increasing levels of the NLS-3xRFP reporter protein (nRFP; T = 20′), suggesting that functional pores are formed. Time is given in minutes. Bar, 5 μm. (E) Treatment with 10 μM of the F-actin inhibitor latrunculin A did not have an obvious effect on NPC formation (Nup107) and protein import (nRFP; left, LatA), whereas simultaneous treatment with latrunculin and benomyl (right, LatA + Ben) abolished the formation of Nup107-labeled NPCs and nuclear import, suggesting that microtubules and F-actin cooperate in NE formation in U. maydis. Time is given in minutes. Bar, 2 μm.
DISCUSSION
A fundamental difference between mitosis in animals and fungi is the behavior of the nuclear envelope and its nuclear pores (Figure 8). In animal cells, the nuclear envelope breaks down in prophase, and it is removed by a microtubule-based mechanism that involves the motor protein dynein (Beaudouin et al., 2002; Salina et al., 2002). Simultaneously, the NPCs disassemble and transmembrane domain-anchored compounds disperse into the ER, whereas peripheral nucleoporins are released into the cytoplasm or are recruited to the chromosomes and kinetochores (Terasaki et al., 2001; Loiodice et al., 2004). Subsequently, the nucleoporins of the Nup107-160 complex return from the chromosomes to the newly forming envelope in anaphase, where they participate in the assembly of the NPC (Belgareh et al., 2001). In contrast, in fungi the mitotic spindle is formed within the intact nuclear envelope (Sazer, 2005). In such closed mitosis in S. cerevisiae, the NPCs remain intact (Hetzer et al., 2005), whereas in A. nidulans, NPCs partially disassemble (De Souza et al., 2004). However, central compounds such as the Nup107-160 complex still remain in the envelope (Osmani et al., 2006), suggesting that the recruitment of the Nup107-160 complex to the chromosomes might be linked to the mechanism of the open mitosis.
Figure 8.
Nuclear envelope behavior and NPC disassembly/reassembly in U. maydis. The interphase nucleus resides in the mother cell, and NPCs mediate communication between the cytoplasm and the interior of the nucleus. At the onset of mitosis, the spindle pole bodies become active, and astral microtubules pull them into the daughter cell. Chromosomes and some of the NPCs concentrate near the tip of this extension, whereas other NPCs already start to disassemble. In prometaphase, NPCs completely disassemble, and Nup107/Nup133 subcomplex and Nup214 disperse in the cytoplasm, whereas the integral Pom152 remains in the ER and the old envelope. In metaphase, the Nup107/Nup133 containing subcomplex partially returns to the DNA, where it also localizes during anaphase A. In late anaphase, the separating chromosomes begin to form new envelopes, in which Nup107 becomes concentrated. Nuclear envelopes close in telophase, and protein import starts after Nup214 and Pom152 are recruited. NPC assembly continues until G2 phase, when cells start to form a bud.
In this study, we set out to investigate the behavior of nucleoporins in the open mitosis of U. maydis, which was reported recently (Straube et al., 2005). In agreement with our expectations, most of the 23 nucleoporins that we identified in the U. maydis genome were found to have close homologues in S. cerevisiae. However, some nuclear pore proteins in U. maydis showed up to 5–6% higher identity with their human counterpart. Among these are Nup107 and Nup133, which are components of the Nup107-160 complex in human cells. We describe here that both proteins leave the nuclear envelope at the onset of mitosis, but they return at the chromosomes in metaphase and are the first that reappear at the rim of the segregating chromosomes in anaphase. This dynamic rearrangement is in agreement with the behavior of their orthologues in human cells (Belgareh et al., 2001; Loiodice et al., 2004). However, in contrast to human Nup107/133, the fungal counterparts do not seem to colocalize with Mis12, which is an important kinetochore component in humans and fission yeast (Goshima et al., 1999; Amor et al., 2004). The biological role of U. maydis Nup107/133 at the meta- and anaphase chromosomes in not known. In later stages in mitosis, Nup107 occurs first at the newly formed envelopes, and nup107 null mutants are not viable. This behavior is reminiscent of animal cells, in which Nup107 participates in the reformation of the NPCs (Boehmer et al., 2003; Harel et al., 2003; Walther et al., 2003). However, Nup107 is also essential in fission yeast, which might be related to defects in its function in transporting poly(A)+ RNA across the nuclear envelope (Bai et al., 2004). Consequently, the observed lethality of mutations in nup107 in U. maydis could also be due to a malfunction of the NPCs in interphase.
Open or Closed Mitosis: Which Came First?
Our studies on the mechanism of open mitosis in U. maydis suggest that this organism combines features of the ascomycete fungi, such as S. cerevisiae and A. nidulans, and animal cells. As a fungus U. maydis shares much sequence similarity with its fungal cousins, which is best illustrated by the presence of Pom152 that is absent from animals. In contrast, U. maydis removes the nuclear envelope (Straube et al., 2005), a process that is not found in the other ascomycete model fungi. The data presented in this study place U. maydis mitosis even more similar to animals. We demonstrate that the NPCs disassemble in prophase and that the Nup107-160 complex is recruited to the DNA, albeit not to the kinetochores. In view of these results, we consider it likely that the open mitosis resembles an evolutionarily original mode of chromosome inheritance. This is supported by ultrastructural data in zygomycete fungi. This ancient group of fungi is considered to be the ancestors of ascomycetes, including S. cerevisiae and A. nidulans, and of basidiomycetes, such as U. maydis (Fitzpatrick et al., 2006). Ultrastructural data indicate that zygomycete fungi also undergo an open mitosis, whereas this has not been observed in ascomycetes (Heath, 1980). Thus, it is most likely that the removal of the envelope is an ancient process, which was abandoned in ascomycete fungi. It is presently not known, why U. maydis undergoes an open mitosis, but further studies on this organism promise to give new insight into the biological meaning of this fascinating process.
Supplementary Material
ACKNOWLEDGMENTS
We thank Daniela Aßmann for technical support and Gero Fink for helpful discussions. Dr. Karen Brune is acknowledged for improving the manuscript. U.T. is supported by a grant of the Studienstiftung des Deutschen Volkes.
Abbreviations used:
- NPC
nuclear pore complex.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-02-0130) on January 2, 2008.
REFERENCES
- Amor D. J., Kalitsis P., Sumer H., Choo K. H. Building the centromere: from foundation proteins to 3D organization. Trends Cell Biol. 2004;14:359–368. doi: 10.1016/j.tcb.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Bai S. W., Rouquette J., Umeda M., Faigle W., Loew D., Sazer S., Doye V. The fission yeast Nup107-120 complex functionally interacts with the small GTPase Ran/Spi1 and is required for mRNA export, nuclear pore distribution, and proper cell division. Mol. Cell. Biol. 2004;24:6379–6392. doi: 10.1128/MCB.24.14.6379-6392.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudouin J., Gerlich D., Daigle N., Eils R., Ellenberg J. Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell. 2002;108:83–96. doi: 10.1016/s0092-8674(01)00627-4. [DOI] [PubMed] [Google Scholar]
- Belgareh N., et al. An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J. Cell Biol. 2001;154:1147–1160. doi: 10.1083/jcb.200101081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berke I. C., Boehmer T., Blobel G., Schwartz T. U. Structural and functional analysis of Nup133 domains reveals modular building blocks of the nuclear pore complex. J. Cell Biol. 2004;167:591–597. doi: 10.1083/jcb.200408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehmer T., Enninga J., Dales S., Blobel G., Zhong H. Depletion of a single nucleoporin, Nup107, prevents the assembly of a subset of nucleoporins into the nuclear pore complex. Proc. Natl. Acad. Sci. USA. 2003;100:981–985. doi: 10.1073/pnas.252749899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottin A., Kamper J., Kahmann R. Isolation of a carbon source-regulated gene from Ustilago maydis. Mol. Gen. Genet. 1996;253:342–352. doi: 10.1007/pl00008601. [DOI] [PubMed] [Google Scholar]
- Castillo-Lluva S., Alvarez-Tabarés I., Weber I., Steinberg G., Pérez-Martín. J. Sustained cell polarity and virulence in the phytopathogenic fungus Ustilago maydis depends on an essential cyclin-dependent kinase from the Cdk5/Pho85 family. J. Cell Sci. 2007;120:1584–1595. doi: 10.1242/jcs.005314. [DOI] [PubMed] [Google Scholar]
- Cronshaw J. M., Krutchinsky A. N., Zhang W., Chait B. T., Matunis M. J. Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza C. P., Osmani A. H., Hashmi S. B., Osmani S. A. Partial nuclear pore complex disassembly during closed mitosis in Aspergillus nidulans. Curr. Biol. 2004;14:1973–1984. doi: 10.1016/j.cub.2004.10.050. [DOI] [PubMed] [Google Scholar]
- Devos D., Dokudovskaya S., Williams R., Alber F., Eswar N., Chait B. T., Rout M. P., Sali A. Simple fold composition and modular architecture of the nuclear pore complex. Proc. Natl. Acad. Sci. USA. 2006;103:2172–2177. doi: 10.1073/pnas.0506345103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenberg J., Siggia E. D., Moreira J. E., Smith C. L., Presley J. F., Worman H. J., Lippincott-Schwartz J. Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J. Cell Biol. 1997;138:1193–1206. doi: 10.1083/jcb.138.6.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink G., Schuchardt I., Colombelli J., Stelzer E., Steinberg G. Dynein-mediated pulling forces drive rapid mitotic spindle elongation in Ustilago maydis. EMBO J. 2006;25:4897–4908. doi: 10.1038/sj.emboj.7601354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink G., Steinberg G. Dynein-dependent motility of microtubules and nucleation sites supports polarization of the tubulin array in the fungus Ustilago maydis. Mol. Biol. Cell. 2006;17:3242–3253. doi: 10.1091/mbc.E05-12-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick D. A., Logue M. E., Stajich J. E., Butler G. A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol. Biol. 2006;6:99. doi: 10.1186/1471-2148-6-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs U., Hause G., Schuchardt I., Steinberg G. Endocytosis is essential for pathogenic development in the corn smut fungus Ustilago maydis. Plant Cell. 2006;18:2066–2081. doi: 10.1105/tpc.105.039388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs U., Manns I., Steinberg G. Microtubules are dispensable for the initial pathogenic development but required for long-distance hyphal growth in the corn smut fungus Ustilago maydis. Mol. Biol. Cell. 2005;16:2746–2758. doi: 10.1091/mbc.E05-03-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Saitoh S., Yanagida M. Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes Dev. 1999;13:1664–1677. doi: 10.1101/gad.13.13.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel A., Orjalo A. V., Vincent T., Lachish-Zalait A., Vasu S., Shah S., Zimmerman E., Elbaum M., Forbes D. J. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol. Cell. 2003;11:853–864. doi: 10.1016/s1097-2765(03)00116-3. [DOI] [PubMed] [Google Scholar]
- Heath I. B. Variant mitosis in lower eukaryotes: indicators of the evolution of mitosis? Int. Rev. Cytol. 1980;64:1–80. doi: 10.1016/s0074-7696(08)60235-1. [DOI] [PubMed] [Google Scholar]
- Heath I. B., Bonham M., Akram A., Gupta G. D. The interrelationships of actin and hyphal tip growth in the ascomycete Geotrichum candidum. Fungal Genet. Biol. 2003;38:85–97. doi: 10.1016/s1087-1845(02)00511-x. [DOI] [PubMed] [Google Scholar]
- Hetzer M., Walther T. C., Mattaj I. W. Pushing the envelope: structure, function, and dynamics of the nuclear periphery. Annu. Rev. Cell Dev. Biol. 2005;21:347–380. doi: 10.1146/annurev.cellbio.21.090704.151152. [DOI] [PubMed] [Google Scholar]
- Holliday R. Molecular aspects of genetic exchange and gene conversion. Genetics. 1974;78:273–287. doi: 10.1093/genetics/78.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiodice I., Alves A., Rabut G., Van Overbeek M., Ellenberg J., Sibarita J. B., Doye V. The entire Nup107–160 complex, including three new members, is targeted as one entity to kinetochores in mitosis. Mol. Biol. Cell. 2004;15:3333–3344. doi: 10.1091/mbc.E03-12-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutzmann M., Kunze R., Buerer A., Aebi U., Hurt E. Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins. EMBO J. 2002;21:387–397. doi: 10.1093/emboj/21.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhnevych T., Lusk C. P., Anderson A. M., Aitchison J. D., Wozniak R. W. Cell cycle regulated transport controlled by alterations in the nuclear pore complex. Cell. 2003;115:813–823. doi: 10.1016/s0092-8674(03)00986-3. [DOI] [PubMed] [Google Scholar]
- Mans B. J., Anantharaman V., Aravind L., Koonin E. V. Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle. 2004;3:1612–1637. doi: 10.4161/cc.3.12.1316. [DOI] [PubMed] [Google Scholar]
- Margalit A., Vlcek S., Gruenbaum Y., Foisner R. Breaking and making of the nuclear envelope. J. Cell. Biochem. 2005;95:454–465. doi: 10.1002/jcb.20433. [DOI] [PubMed] [Google Scholar]
- O'Donell K. L., McLaughlin D. J. Postmeiotic mitosis, basidiospore development, and septation in Ustilago maydis. Mycologia. 1984a;76:486–502. [Google Scholar]
- O'Donell K. L., McLaughlin D. J. Ultrastructure of meiosis in Ustilago maydis. Mycologia. 1984b;76:468–485. [Google Scholar]
- Orjalo A. V., Arnaoutov A., Shen Z., Boyarchuk Y., Zeitlin S. G., Fontoura B., Briggs S., Dasso M., Forbes D. J. The Nup107-160 nucleoporin complex is required for correct bipolar spindle assembly. Mol. Biol. Cell. 2006;17:3806–3818. doi: 10.1091/mbc.E05-11-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani A. H., Davies J., Liu H. L., Osmani S. A. Systematic deletion and mitotic localization of the nuclear pore complex proteins of Aspergillus nidulans. Mol. Biol. Cell. 2006 doi: 10.1091/mbc.E06-07-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton L. F., Rout M. P., Blobel G. Disruption of the nucleoporin gene NUP133 results in clustering of nuclear pore complexes. Proc. Natl. Acad. Sci. USA. 1995;92:1187–1191. doi: 10.1073/pnas.92.4.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabut G., Lenart P., Ellenberg J. Dynamics of nuclear pore complex organization through the cell cycle. Curr. Opin. Cell Biol. 2004;16:314–321. doi: 10.1016/j.ceb.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Rout M. P., Aitchison J. D., Suprapto A., Hjertaas K., Zhao Y., Chait B. T. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salina D., Bodoor K., Eckley D. M., Schroer T. A., Rattner J. B., Burke B. Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell. 2002;108:97–107. doi: 10.1016/s0092-8674(01)00628-6. [DOI] [PubMed] [Google Scholar]
- Sazer S. Nuclear envelope: nuclear pore complexity. Curr. Biol. 2005;15:R23–R26. doi: 10.1016/j.cub.2004.12.015. [DOI] [PubMed] [Google Scholar]
- Siniossoglou S., Lutzmann M., Santos-Rosa H., Leonard K., Mueller S., Aebi U., Hurt E. Structure and assembly of the Nup84p complex. J. Cell Biol. 2000;149:41–54. doi: 10.1083/jcb.149.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S., Wimmer C., Rieger M., Doye V., Tekotte H., Weise C., Emig S., Segref A., Hurt E. C. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell. 1996;84:265–275. doi: 10.1016/s0092-8674(00)80981-2. [DOI] [PubMed] [Google Scholar]
- Snetselaar K. M., McCann M. P. Using microdensitometry to correlate cell morphology with the nuclear cycle in Ustilago maydis. Mycologia. 1997;89:689–697. [Google Scholar]
- Steinberg G., Wedlich-Söldner R., Brill M., Schulz I. Microtubules in the fungal pathogen Ustilago maydis are highly dynamic and determine cell polarity. J. Cell Sci. 2001;114:609–622. doi: 10.1242/jcs.114.3.609. [DOI] [PubMed] [Google Scholar]
- Straube A., Brill M., Oakley B. R., Horio T., Steinberg G. Microtubule organization requires cell cycle-dependent nucleation at dispersed cytoplasmic sites: polar and perinuclear microtubule organizing centers in the plant pathogen Ustilago maydis. Mol. Biol. Cell. 2003;14:642–657. doi: 10.1091/mbc.E02-08-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straube A., Enard W., Berner A., Wedlich-Söldner R., Kahmann R., Steinberg G. A split motor domain in a cytoplasmic dynein. EMBO J. 2001;20:5091–5100. doi: 10.1093/emboj/20.18.5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straube A., Weber I., Steinberg G. A novel mechanism of nuclear envelope break-down in a fungus: nuclear migration strips off the envelope. EMBO J. 2005;24:1674–1685. doi: 10.1038/sj.emboj.7600644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasaki M., Campagnola P., Rolls M. M., Stein P. A., Ellenberg J., Hinkle B., Slepchenko B. A new model for nuclear envelope breakdown. Mol. Biol. Cell. 2001;12:503–510. doi: 10.1091/mbc.12.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasu S., Shah S., Orjalo A., Park M., Fischer W. H., Forbes D. J. Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export. J. Cell Biol. 2001;155:339–354. doi: 10.1083/jcb.200108007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther T. C., et al. The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell. 2003;113:195–206. doi: 10.1016/s0092-8674(03)00235-6. [DOI] [PubMed] [Google Scholar]
- Wedlich-Söldner R., Schulz I., Straube A., Steinberg G. Dynein supports motility of endoplasmic reticulum in the fungus Ustilago maydis. Mol. Biol. Cell. 2002;13:965–977. doi: 10.1091/mbc.01-10-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Guan T., Gerace L. Integral membrane proteins of the nuclear envelope are dispersed throughout the endoplasmic reticulum during mitosis. J. Cell Biol. 1997;137:1199–1210. doi: 10.1083/jcb.137.6.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccolo M., et al. The human Nup107-160 nuclear pore subcomplex contributes to proper kinetochore functions. EMBO J. 2007;26:1853–1864. doi: 10.1038/sj.emboj.7601642. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.