Abstract
Antibody single-chain Fv fragment (scFv) molecules that are specific for fluorescein have been engineered with a C-terminal cysteine for a directed immobilization on a flat gold surface. Individual scFv molecules can be identified by atomic force microscopy. For selected molecules the antigen binding forces are then determined by using a tip modified with covalently immobilized antigen. An scFv mutant of 12% lower free energy for ligand binding exhibits a statistically significant 20% lower binding force. This strategy of covalent immobilization and measuring well separated single molecules allows the characterization of ligand binding forces in molecular repertoires at the single molecule level and will provide a deeper insight into biorecognition processes.
Atomic force microscopy (AFM) has been a versatile tool for imaging the surface structure of individual molecules and of molecular assemblies of a wide variety of biological specimens (1–5). Furthermore, molecular binding forces between a small number of various ligand and receptor molecules have been measured by AFM (6–15). In these force spectroscopy studies, one interacting partner is immobilized on the AFM cantilever and the other on a surface, and the interaction is probed with force–distance curves. The rupture force, the maximum force at the moment of detachment, which is obtained upon withdrawing the cantilever from the surface, is taken as a measure of the interaction between ligand and receptor.
In this paper we report on combining AFM imaging and force spectroscopy at the level of individually selected and addressed molecules. We used recombinant antibody single-chain Fv (scFv) fragments as a versatile model system to study unbinding forces of single molecules. Antibody scFv fragments, in which the variable domain of the heavy chain (VH) is fused by means of a (Gly4Ser)3 linker to the variable domain of the light chain VL, are the minimal size antibody molecules that still comprise the complete antigen binding site (16). Unlike whole antibodies, they do not contain additional domains whose unfolding under force may give rise to structural changes (17, 18) that might influence the unbinding event. scFv fragments are particularly interesting models, because they can be generated against all conceivable antigenic targets, and mutants with various binding properties can be engineered.
The scFv proteins were immobilized in a directed orientation on an ultraflat gold surface, their position was detected with AFM, and then the binding force of a spatially well isolated molecule was measured by using a tip endowed with immobilized antigen. To achieve correct determinations of the interaction force between scFv fragments and the cognate antigen fluorescein, their immobilization was carefully designed such that no detachment from the surface occurred within the time of the experiment. Furthermore, by sufficiently spacing the individual molecules on the surface, interactions could be restricted to single protein molecules. Thus, individual molecular binding forces could be obtained directly, and not only by interpreting multiple maxima arising from molecules interacting with several partners.
METHODS
Tip Modification.
Pilot experiments were first carried out with pieces of oxidized silicon wafers to determine the optimal density of the antigen fluorescein, which was assessed by binding tests using radioiodinated antibody fragments. The modification of the AFM cantilever was then carried out as follows: Cantilevers (Si3N4-Microlever, Park Scientific Instruments, Sunnyvale, CA; k ≈ 0.03 N/m) were first activated by dipping for 10 s in concentrated nitric acid and silanized in a solution of 2% aminopropyltriethoxysilane (Sigma) in dry toluene for 2 h. After washing with toluene, the cantilevers were incubated with 1 mg/ml fluorescein-poly(ethylene glycol)-OCH2CH2CO2-N-hydroxysuccinimide (Fluor-NHS5000; Shearwater Polymers, Huntsville, AL) in 50 mM sodium phosphate buffer, pH 8.5, overnight at 4°C. The cantilevers were washed with phosphate buffer and used for AFM imaging and force–distance experiments. Modified tips were stable for at least 2 weeks if stored in the refrigerator. The cantilever spring constants were determined by three independent methods (19–21), which agreed within 15%.
Preparation of scFv Molecules.
The fluorescein binding wild-type scFv antibody FITC-E2 (22, 23) and the His(H58)Ala mutant, each in the orientation VH-(Gly4Ser)3-VL, were cloned in the secretion vector pAK400 (23, 24). At the C terminus of VL an EcoRI–HindIII cassette encoding the sequence SGAEFPKPSTP2GS2G2APH6G4C was introduced. The proteins were expressed in Escherichia coli SB536 (25) and purified from the periplasm as described previously (23). Briefly, a Ni2+-NTA column (Qiagen) was followed by Sepharose-SP (Pharmacia). No dithiothreitol was added to the column buffers, but the protein was stored in 20 mM Hepes/150 mM NaCl/2 mM EDTA/5 mM dithiothreitol, pH 6.8, at 4°C. Final yields were about 200 μg of protein per liter of bacterial culture. For immobilization to gold, the protein stock solution was diluted to 1 μg/ml with 50 mM phosphate buffer, pH 7.4, directly before use.
Surface Preparation and Characterization.
Freshly prepared flat gold surfaces (26) were incubated for 2 h in 50 mM mercaptoethanesulfonate, dissolved in water/ethanol (1:1, vol/vol). After drying under a stream of nitrogen, a drop of 1 μg/ml protein in 50 mM phosphate buffer, pH 7.4, was added. After 20 min the surfaces were rinsed with buffer and used directly or after storage in the phosphate buffer at 4°C up to 2 weeks.
To check whether the number of observed spots in AFM corresponds to single molecules, the surface density was determined by labeling experiments. The scFv fragments, with and without the C-terminal cysteine, were labeled with the 125I-Bolton–Hunter reagent, and the surface-bound radioactivity was measured by electronic autoradiography as described in ref. 27. The number of molecules counted from an AFM image agrees within a factor of 2 with this radiotracer method; dimerization of scFv proteins has not been observed in solution. It is worth noting that protein molecules appear enlarged in AFM due to convolution effects (28).
AFM Measurements.
Force–distance measurements were performed by a commercial AFM (Topometrix Explorer, Santa Clara, CA) using modified tips in three sequential steps: (i) scanning of a 1-μm range with 2 μm/s in contact mode, applying a weak force (<500 pN); (ii) selection of one to four isolated protein molecules for parallel measurements of about 50 force–distance curves at velocity of 1 μm/s and complete retraction of the cantilever after each curve; and (iii) rescanning to check the lateral xy-drift. For control experiments the samples were incubated and blocked with 0.1 mM fluorescein for 1 h.
RESULTS AND DISCUSSION
To immobilize the scFv proteins in a directed orientation on an ultraflat gold surface, they were engineered to carry a single free thiol group at the C terminus of a spacer. We have used a fluorescein-binding scFv fragment, FITC-E2 (25), whose VL domain was extended with the sequence SGAEFPKPSTP2GS2G2APH6G4C (Fig. 1), as a model system. This system displays a high affinity (22, 23) and allows convenient determination of binding kinetics and affinity by fluorescence titration.
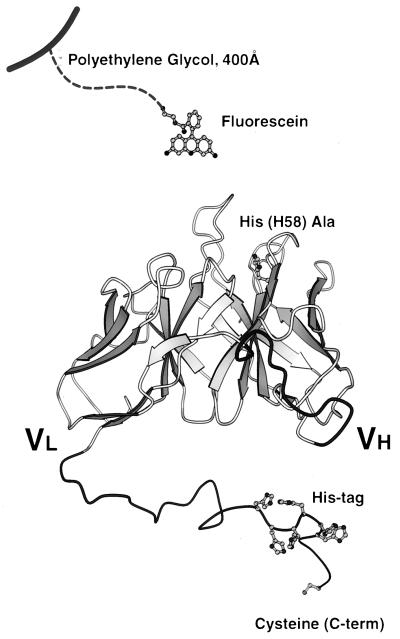
Figure 1.
Model of the single-chain anti-fluorescein fragment. In the mutant His(H58)Ala, a His residue in the heavy chain is replaced by Ala. The C-terminal tail, consisting of an IgG3 upper hinge, a Gly-Ser spacer, a His6 tag, Gly4, and a single C-terminal Cys, is shown (for sequence see text).
The antigen fluorescein was covalently immobilized on a Si3N4-AFM-tip with aminosilane and an extended hydrophilic poly(ethylene glycol) spacer (Fig. 2). This spacer is crucial to minimize nonspecific interactions and to provide enough reach for the antigen to enter the binding pocket (13). The scFv fragment was directly immobilized on the gold surface by forming a strong, quasicovalent bond between the thiol group of the engineered cysteine at the C terminus and gold. To prevent the denaturation of the scFv fragment on the metal surface, the gold surface was modified with negatively charged mercaptoethanesulfonate (27). Because the gold surface is very flat, individual molecules can be easily discerned by AFM. scFv fragments missing the C-terminal cysteine used for immobilization could not be visualized. This observation confirms that the molecules with the C-terminal cysteine do not simply stick in a random orientation on protected gold.
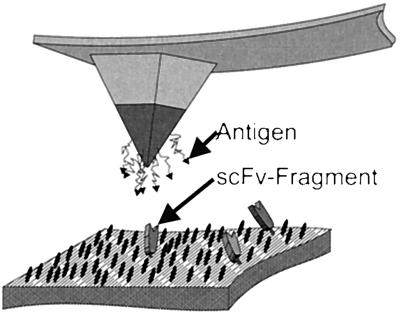
Figure 2.
Scheme of the immobilization concept. The antigen fluorescein is covalently bound by means of a poly(ethylene glycol) linker to the silanized silicon nitride AFM tip. Single-chain Fv′-fragments (scFv) of an anti-fluorescein antibody are attached directly to a gold surface by an engineered C-terminal Cys residue. The black ovoids on the surface represent the protection layer of negatively charged thiol compound needed to avoid denaturation of proteins on gold.
The equilibrium binding constants of the wild-type and 11 mutants have been measured by fluorescence titration (23). The dissociation constant Kd was found to be 0.75 nM for the wild-type and 8.94 nM for the His(H58)Ala mutant, converting to Gibbs free energies ΔG of −50.8 kJ/mol and −44.8 kJ/mol, respectively, at standard states.
To determine unbinding forces, surfaces with immobilized wild-type scFv or its His(H58)Ala mutant (Fig. 1) have first been imaged by using the antigen-functionalized tip. Isolated protein molecules were selected and multiple force–distance cycles were recorded. Before and after each cycle the position of the tested molecule was checked by rescanning (see Fig. 4 A and B Insets). More than 250 binding–unbinding cycles can be performed with the same protein molecule without observing any loss of binding activity. A typical unbinding curve is displayed in Fig. 3A. The detectable minimum force is given by thermal noise: Fmin = (kbkleverT)1/2 ≈ 12 pN. The unbinding force values were always the same, independent of the distance from the surface at which the antigens detached. This can be explained by the interaction of the binding site with different antigen molecules, located at different places on the pyramidal tip.
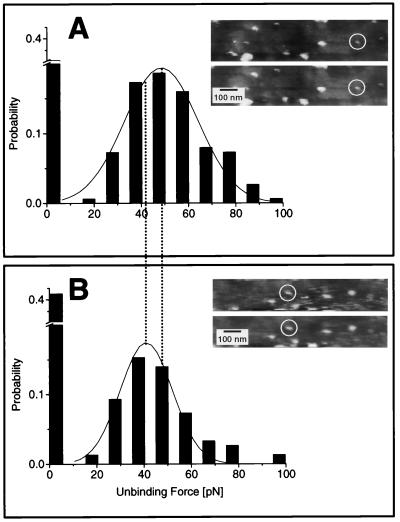
Figure 4.
Histograms of the unbinding forces of a wild-type (A) and a mutant (B) scFv. Each histogram comprises 3 × 50 force–distance curves of the same molecule. The interval width in the histograms is 10 pN and the unbinding forces Fu were determined by a Gaussian fit. All data shown were obtained with the identical cantilever. (Insets) The images next to the histograms show the selected binding protein before and after the measurement.
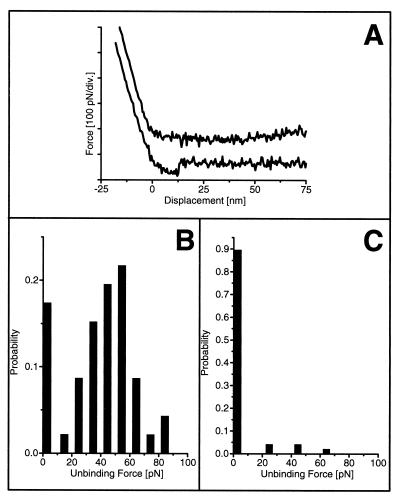
Figure 3.
(A) Typical force–distance curves before (lower curve) and after (upper curve) blocking of a wild-type scFv. Only the retracting traces are shown. (B and C) Histograms of unbinding forces of an scFv wild-type molecule before (B) and after (C) blocking with free antigen.
An important control is to verify the absence of binding events at positions on the surface where no protein molecules are present. Furthermore, blocking of binding sites is achieved by the addition of free fluorescein to the immobilized fragments prior to the measurement. When the interaction of the scFv molecule with the antigen on the cantilever was measured in the presence of free fluorescein, binding forces >15 pN completely disappeared (Fig. 3C). When the antigen solution was exchanged again by buffer before measuring, the binding probability of unbinding forces >15 pN was restored to about 50% of all events, and the force distribution was the same as before blocking (data not shown). These observations give us confidence that the molecular recognition of the antigen–antibody interaction was measured and not an unspecific binding process.
The statistical distribution of 150 measured force values is represented in histograms for the wild type (Fig. 4A) and the mutant (Fig. 4B), respectively. Each histogram shows a single maximum, which is a decisive advantage for the determination of mean values of unbinding forces by using mathematical fitting procedures (15). The lack of multiple force peaks in the histograms confirms that single molecules are measured by AFM imaging, as anticipated. Thus, a reliable mean value for the unbinding force can be determined by fitting a Gaussian distribution to the single peak, a process that is very difficult for histograms with multiple maxima. To ensure that the mean values are independent of the histogram arrangement, different classes of variable width were compared, and no significant differences in the maxima were found.
The power of this technique is demonstrated in a clear discrimination of mutant scFvs from wild type: The ratio of unbinding forces was calculated to be 0.80 ± 0.08, at a statistically significant difference of P < 0.001 in the Student t test. To assess the variation between individual molecules and the experimental reproducibility, four profiles from two mutant molecules, obtained in a first and a second distance–force experiment, are shown (Fig. 5). Wild-type and mutant molecules differ in their affinity constant by roughly one order of magnitude, which corresponds to a ratio of 0.88 in Gibbs free energies (Table 1). This ratio is in the same range as the ratio of the binding forces. Determining the relationship of forces to thermodynamic quantities (8, 11), however, will require further studies. To exclude any error from cantilever calibration, the binding experiments of the wild-type and mutant fragments have been carried out with the identical antigen-functionalized cantilever.
Figure 5.
(A and B) Histograms of unbinding force values of (50 force–distance cycles) from two individual mutant scFv molecules. (C and D). Histograms showing the values of the same proteins measured in a second experiment.
Table 1.
Unbinding force values (Fu), dissociation constants (Kd), and Gibbs free energies of binding (ΔG) of wild-type and mutant His(H58)Ala
| Cys-scFv-fragment | Fu, pN | Kd, nM | ΔG, kJ/mol |
|---|---|---|---|
| Wild type | 50 ± 4 | 0.75 | −50.8 |
| Mutant His(H58)Ala | 40 ± 3 | 8.94 | −44.8 |
Fu values with standard deviations are calculated from the means of five histograms.
We have been able to limit the interaction of functionalized AFM tips and surfaces to single, spatially well separated ligand–receptor complexes, and we showed that this ability is essential to obtain the discriminatory power necessary to study closely related binding proteins. To confirm this finding, control experiments with the same tip, but high coating densities of scFv fragments, were performed. There was a higher proportion of the stronger unbinding forces, tailing up to 300 pN, which can be interpreted as multiple interactions (data not shown). When histograms from wild type and mutant are compared, a shift of the mean value was found as observed in the single-molecule experiments described above. It was found in independent experiments in solution (23) that only 29% of mutant scFv molecules are functional, compared with 86% of the wild-type protein. This difference in activities influences the number of multiple binding events, the profile of the histogram, and most important, the position of the maximum, a problem that does not exist in the single-molecule experiments. Furthermore, a quantitative evaluation at the high density of immobilization was impossible because of the overlap of putative force maxima, even when a higher number of data points was available.
A stable immobilization of the binding partners turns out to be crucial for maintaining functional antibody proteins as well as for preventing a detachment of the whole complex when the tip is retracted. In systems where antibody molecules have been stably immobilized on a surface, values of 60 ± 10 pN for biotin/anti-biotin (12) and 49 ± 10 pN for ferritin/anti-ferritin (15) have been found. The values determined in this study, 50 ± 4 pN for the wild type and 40 ± 3 pN for the mutant His(H58)Ala, are in the same range as those estimated in previous studies where no individual molecules could be observed.
The high relative precision achieved by the strategy of spatially separating individual molecules as described here will permit us now to study a wider range of scFv mutants and unrelated scFv molecules. One of the fundamental questions in ligand binding, which may now be experimentally addressed, is the existence of a quantitative relationship between the unbinding force measured by AFM and thermodynamic parameters, such as enthalpy or free energy of ligand binding or the kinetic parameters of ligand dissociation. The precise understanding of such a relationship will be pivotal for the potential use of AFM in ligand screening.
Previously, in a model system with avidin and biotin and biotin analogs (8), correlations have been suggested to exist between binding forces and enthalpy. When streptavidin mutants were used, the rupture force was proposed to directly probe the enthalpic activation barrier to biotin dissociation (11). The rupture mechanism of this complex has also been simulated on a molecular level: five major unbinding steps have been proposed and the contribution of water molecules has been emphasized (29). Nevertheless, further precise measurements with different systems will be required to test and further develop the correlation of unbinding forces with structural, thermodynamic, and kinetic parameters.
We consider such measurements to be crucial for deeper insight into macromolecular interactions, a key point in ligand and receptor design. This technique will allow us in the future to investigate the binding properties of molecules from large libraries at the single-molecule level. Furthermore, such measurements pave the way for the use of functionalized tips in investigating biological specimens.
Acknowledgments
We are grateful to J.-J. Hefti and D. Bächle for preparation of materials; to J. Fritz for help with cantilever calibration and writing the force analysis software; to A. Honegger for help with Fig. 1; to C. Padeste, L. Scandella, and H.-J. Güntherodt for inspiring discussions; and to W. Gutmannsbauer for technical support. This work was supported by the Swiss National Research Program 36 Nanoscience, Grant 4036–43973.
ABBREVIATIONS
- AFM
atomic force microscopy
- scFv
single-chain Fv
- VH and VL
variable domains of heavy and light immunoglobulin chains
References
- 1.Parpura V, Haydon P G, Henderson E. J Cell Sci. 1993;104:427–432. doi: 10.1242/jcs.104.2.427. [DOI] [PubMed] [Google Scholar]
- 2.Hansma H G, Sinsheimer R L, Groppe J, Bruice T C, Elings V, Gurley G, Bezanilla M, Mastrangelo I A, Hough P V C, Hansma P K. Scanning. 1993;15:296–299. doi: 10.1002/sca.4950150509. [DOI] [PubMed] [Google Scholar]
- 3.Haydon P G, Henderson E, Stanley E. Neuron. 1994;13:1275–1280. doi: 10.1016/0896-6273(94)90414-6. [DOI] [PubMed] [Google Scholar]
- 4.Hansma H G, Hoh J. Ann Rev Biophys Biomol Struct. 1994;23:115–139. doi: 10.1146/annurev.bb.23.060194.000555. [DOI] [PubMed] [Google Scholar]
- 5.Gaub, H. & Engel, A., eds. (1997) J. Struct. Biol. 119 (2), 83–237. [DOI] [PubMed]
- 6.Lee G, Kidwell D A, Colton R J. Langmuir. 1994;10:354–357. [Google Scholar]
- 7.Florin E-L, Moy V T, Gaub H E. Science. 1994;264:415–417. doi: 10.1126/science.8153628. [DOI] [PubMed] [Google Scholar]
- 8.Moy V T, Florin E-L, Gaub H. Science. 1994;266:257–259. doi: 10.1126/science.7939660. [DOI] [PubMed] [Google Scholar]
- 9.Florin E L, Rief M, Lehmann H, Ludwig M, Dornmair C, Moy V T, Gaub H E. Biosens Bioelectron. 1995;10:895–901. [Google Scholar]
- 10.Dammer U, Popescu O, Wagner P, Anselmetti D, Güntherodt H-J, Misevic G N. Science. 1995;267:1173–1175. doi: 10.1126/science.7855599. [DOI] [PubMed] [Google Scholar]
- 11.Chilkoti A, Boland T, Ratner B D, Stayton P S. Biophys J. 1995;69:2125–2130. doi: 10.1016/S0006-3495(95)80083-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dammer U, Hegner M, Anselmetti D, Wagner P, Dreier M, Huber W, Güntherodt H-J. Biophys J. 1996;70:2437–2441. doi: 10.1016/S0006-3495(96)79814-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hinterdorfer P, Baumgartner W, Gruber H J, Schilcher K, Schindler H. Proc Natl Acad Sci USA. 1996;93:3477–3481. doi: 10.1073/pnas.93.8.3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fritz J, Anselmetti D, Jarchow J, Fernandez-Busquets X. J Struct Biol. 1997;119:165–171. doi: 10.1006/jsbi.1997.3887. [DOI] [PubMed] [Google Scholar]
- 15.Allen S, Chen X, Davies J, Davies M C, Dawkes A C, Edwards J C, Roberts C J, Sefton J, Tendler S J B, Williams P M. Biochemistry. 1997;36:7457–7463. doi: 10.1021/bi962531z. [DOI] [PubMed] [Google Scholar]
- 16.Plückthun A. Immunol Rev. 1992;130:151–188. doi: 10.1111/j.1600-065x.1992.tb01525.x. [DOI] [PubMed] [Google Scholar]
- 17.Rief M, Gautel M, Oesterhelt F, Fernandez J M, Gaub H E. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 18.Rief M, Oesterhelt F, Heymann B, Gaub H E. Science. 1997;275:1295–1297. doi: 10.1126/science.275.5304.1295. [DOI] [PubMed] [Google Scholar]
- 19.Gibson C T, Watson G S, Myhra V. Nanotechnology. 1996;65:259–263. [Google Scholar]
- 20.Neumeister J M, Ducker W A. Rev Sci Instrum. 1994;64:2527–2531. [Google Scholar]
- 21.Hutter J L, Bechhoefer J. Rev Sci Instrum. 1993;7:1868–1873. [Google Scholar]
- 22.Vaughan T J, Williams A J, Pritchard K, Osbourn J K, Pope A R, Earnshaw J C, McCafferty J, Hodits R A, Wilton J, Johnson K S. Nat Biotechnol. 1996;14:309–314. doi: 10.1038/nbt0396-309. [DOI] [PubMed] [Google Scholar]
- 23.Pedrazzi G, Schwesinger F, Honegger A, Krebber D, Plückthun A. FEBS Lett. 1997;415:289–293. doi: 10.1016/s0014-5793(97)01143-5. [DOI] [PubMed] [Google Scholar]
- 24.Krebber A, Bornhauser S, Burmester J, Honegger A, Willuda J, Bosshard H R, Plückthun A. J Immunol Methods. 1997;201:35–55. doi: 10.1016/s0022-1759(96)00208-6. [DOI] [PubMed] [Google Scholar]
- 25.Bass S, Gu Q, Christen A. J Bacteriol. 1996;178:1154–1161. doi: 10.1128/jb.178.4.1154-1161.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wagner P, Kernen P, Hegner M, Ungewickell E, Semenza G. FEBS Lett. 1994;356:267–271. doi: 10.1016/0014-5793(94)01296-2. [DOI] [PubMed] [Google Scholar]
- 27.Tiefenauer L X, Kossek S, Padeste C, Thiébaud P. Biosens Bioelectron. 1997;12:213–223. doi: 10.1016/s0956-5663(97)85339-0. [DOI] [PubMed] [Google Scholar]
- 28.Kossek S, Padeste C, Tiefenauer L. Biosens Bioelectron. 1998;13:31–43. doi: 10.1016/s0956-5663(97)85339-0. [DOI] [PubMed] [Google Scholar]
- 29.Grubmüller H, Heymann B, Tavan P. Science. 1996;271:997–999. doi: 10.1126/science.271.5251.997. [DOI] [PubMed] [Google Scholar]